ABSTRACT
Middle East respiratory syndrome coronavirus (MERS‐CoV) is a single‐stranded RNA virus that causes severe respiratory disease in humans with a high fatality rate. Binding of the receptor binding domain (RBD) of the spike (S) glycoprotein to dipeptidyl peptidase 4 is the critical step in MERS‐CoV infection of a host cell. No vaccines or clinically applicable treatments are currently available for MERS‐CoV. Therefore, rapid diagnosis is important for improving patient outcomes through prompt treatment and protection against viral outbreaks. In this study, the aim was to establish two ELISA systems for detecting antigens and antibodies against MERS‐CoV. Using a recombinant full‐length S protein, an indirect ELISA was developed and found to detect MERS‐CoV‐specific antibodies in animal sera and sera of patient with MERS. Moreover, MAbs were induced with the recombinant S protein and RBD and used for sandwich ELISA to detect the MERS‐CoV S protein. Neither ELISA system exhibited significant intra‐assay or inter‐assay variation, indicating good reproducibility. Moreover, the inter‐day precision and sensitivity were adequate for use as a diagnostic kit. Thus, these ELISAs can be used clinically to diagnose MERS‐CoV.
Keywords: ELISA, Middle East respiratory syndrome coronavirus, receptor binding domain, spike protein
List of Abbreviations
- cELISA
competitive ELISA
- CV
coefficient of variation
- IPTG
isopropyl β‐d‐1‐thiogalactopyranoside
- LLD
lower limit of detection
- MERS
Middle East respiratory syndrome
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- PRNT
plaque reduction neutralization
- RBD
receptor binding domain
- RID
RNA interaction domain
- S protein
spike protein
Middle East respiratory syndrome coronavirus is a single‐stranded positive RNA virus belonging to the genus Betacoronavirus within the family Coronaviridae. MERS‐CoV causes severe respiratory disease and organ failure in humans, leading to death. In 2012, the first patient with MERS was identified in Saudi Arabia 1. Since then, MERS in humans has been reported in 24 countries 2, and has had high fatality rates 3. MERS‐CoV has a significant impact on public health in endemic areas. For example, there was a large outbreak in Korea in 2015. Since the first patient with MERS was identified in Korea on 20 May 2015, 186 of 16,752 suspected cases have been confirmed and 38 deaths reported. In this outbreak, the spread of MERS‐CoV was extremely rapid, patients being reported from May to June, 2015 4. The many reasons for the unexpectedly fast spread of MERS‐CoV in Korea in 2015 included late diagnoses, failure to quarantine “super spreaders”, familial care‐giving and visiting, non‐disclosure by patients, poor communication by the Korean government, inadequate hospital infection management and “doctor shopping” 4.
There are currently no approved therapeutic agents or vaccines against MERS‐CoV in clinical use. The S glycoprotein is known to be a major target for neutralizing antibodies in viral infections 5. The RBD region of the MERS‐CoV S protein binds to dipeptidyl peptidase 4 on the surfaces of host cells, enabling infection of the cell 6, 7. MERS‐CoV vaccines under development are therefore mostly based on the S protein and specifically the RBD 8, 9. A DNA vaccine expressing the full‐length S sequence is the only product that has advanced to human trials 10. Thus, rapid diagnosis of the virus remains critical in controlling spread of the disease and providing prompt treatment.
Currently, RT‐RCR is a common method for diagnosing MERS‐CoV 11, major target genes being the envelope, open reading frame 1A/1B and nucleocapsid genes 12. RT‐PCR is highly sensitive and specific; however, it is time‐consuming and does not provide viral titers. Serological tests for detecting antibodies against MERS‐CoV, such as ELISAs, microneutralization, immunofluorescence and PRNT assays, are also widely used to diagnose MERS‐CoV infection 13, 14, 15. However, microneutralization, immunofluorescence and PRNT assays are based on cell culture and therefore require Biosafety Level 3 containment.
In this study, we aimed to establish specific antibodies and antigens to develop ELISAs for MERS‐CoV diagnosis. We prepared two recombinant proteins, one containing the RBD alone and one containing the full‐length S protein, and mAbs against MERS‐CoV antigens. We used the S protein for MERS‐CoV‐specific antibody detection ELISA and mAbs for MERS‐CoV antigen detection ELISA. Finally, we validated the precision, sensitivity and reproducibility of both ELISAs.
MATERIALS AND METHODS
Ethical statement
Sera from patients with MERS were used in this study with the ethical approval of the Institutional Review Boards of Chungnam National University Hospital (IRB no. 2015‐08‐029) and Seoul National University Hospital (IRB no. 1509‐103‐705). This study was performed in accordance with the ethical standards laid down in the 1964 declaration of Helsinki and all subsequent revisions. All patients provided written, informed consent to participate.
Expression and purification of S proteins in a baculovirus system
The MERS‐CoV S protein was prepared with a Bac‐to‐Bac Baculovirus Expression System (Invitrogen, Carlsbad, CA, USA). The MERS‐CoV S gene (GenBank Accession No. KF186567) was cloned into the pFastBac1 vector and transferred to a Bacmid vector by Tn7 transposition. Baculovirus was obtained by transfection of the vector into Sf9 cells derived from the insect Spodoptera frugiperda. Prepared baculoviruses were used to infect Sf9 cells at an MOI of 0.01–0.8 and harvested after 3 days. Cell pellets were separated from the supernatant via centrifugation at 5000 g for 20 min and lysed with lysis buffer. Cell membranes containing MERS‐CoV proteins were obtained by differential centrifugation and ultracentrifugation. Soluble membrane extracts were purified by anion exchange chromatography and glucose affinity chromatography. Proteins were concentrated by ultrafiltration and analyzed by SDS‐PAGE and Coomassie blue staining.
Expression and purification of RBD in Escherichia coli
The pGE‐mRID vector was used for expression of the RBD 4. This expression vector is composed of a RID of ∼70 aa from the N‐terminal of the mouse LyRS protein (nonspecific transfer RNA interaction domain). The RBD fragment (367–606 aa) was prepared from the S gene of MERS‐CoV by PCR and inserted between the KpnI and SalI sites of the vector. The plasmid was transformed into E. coli strain BL2 Star (DE3) pLysS, which was then cultured overnight in 50 mL LB medium containing 40 μg/mL ampicillin and 40 μg/mL chloramphenicol at 37°C. Then, 50 mL culture broth was used to inoculate 500 mL LB medium and incubated at 37°C until an OD600 of 0.5–0.6 had been reached. Protein expression was induced by adding of 1 mM IPTG at 16°C. Cells were centrifuged at 3000 g for 10 min at an OD600 of 1.4–1.6. Cell lysates were obtained after sonication and centrifuged at 12,000 g for 10 min to separate soluble fractions from pellet fractions.
Proteins were purified by using a HisTrap HP column (GE Healthcare, Marlborough, MA, USA). After equilibration of the column with wash buffer, proteins were obtained with a linear gradient of imidazole in elution buffer, concentrated by Centriprep (Millipore, Billerica, MA, USA) and analyzed by SDS‐PAGE and Coomassie blue staining. Purified samples were quantified using a known concentration of BSA (Amresco, Solon, OH, USA) as a control.
Production of polyclonal antibodies against MERS‐CoV S protein
S protein (50 μg) was used to immunize 8‐week‐old Wistar rats (n = 5) from Daehan Bio Link (Chungbuk, Korea) twice at 3‐week intervals. Three weeks after the second immunization, the rats were killed humanely in accordance with an official protocol and polyclonal antibodies in their sera harvested by a previously reported method 16. All animal experiments were performed in accordance with the relevant ethical guidelines and regulations established by the Korean Association for Laboratory Animals 17. All rats were housed under specific‐pathogen‐free conditions with a standard 12 hr light/dark cycle and maintained according to protocols approved by the Institutional Animal Care and Use Committee of Chonbuk National University.
Production of mAbs against MERS‐CoV S and RBD
Mouse MAbs against MERS‐CoV S and RBD proteins were produced by ATGen (Seoul, Korea) using a previously reported protocol 18. In brief, 50 μg antigen was injected s.c. into BALB/c mice and boosted with 50 μg antigen after a 2‐week interval. Splenocytes were obtained 3 days after the last immunization and fused to FO myeloma cells to generate hybridoma cells. The culture supernatants of the hybridoma cells were screened for the presence of antigen‐specific antibodies by ELISA using MERS‐CoV S or RBD proteins as a coating antigen. Hybridoma cells that specifically reacted with antigens were selected and subcloned by the limiting dilution method. All MAbs from cell culture supernatants were purified with a Protein G column (GE Healthcare) according to the manufacturer's instructions.
ELISA for detection of MERS‐CoV‐specific antibodies
An indirect ELISA method was developed to measure MERS‐CoV‐specific antibodies. ELISA plates were coated overnight at 4°C with 0.25 μg/well of MERS‐CoV S protein. For the human sera, the plates were washed with PBS‐T (0.05% Tween 20 in PBS) three times and incubated for 2 hr at 37°C with blocking solution (1% BSA in PBS). After four washes, the serum samples were diluted with 1% BSA in PBS and added to the wells. The plates were then incubated for 2 hr at 37°C. After three washes, HRP‐conjugated goat anti‐human immunoglobulin G was diluted 1:10,000 in 1% BSA containing PBS and added to the plates, followed by incubation for 2 hr at 37°C. The plates were then washed five times and 3,3′,5,5′‐tetramethylbenzidine solution added. After 5 min, 1 N H2SO4 was added to stop the reaction and the OD450 measured with a microplate reader. Except for the use of a secondary antibody (1:10,000 dilution; Invitrogen) that specifically binds the IgG Fc of rat or rabbit after incubation with the animal serum sample, the same procedures were followed for the rat or rabbit sera.
ELISA for detection of MERS‐CoV antigen (S protein)
A sandwich ELISA method was established to quantify MERS‐CoV S protein. To determine detector and capture antibodies, MAbs against both RBD and S protein were evaluated with pairwise tests. The detector antibody was biotinylated by dialysis for 1 hr at 4°C three times with 10 ng/mL biotin solution. ELISA plates were coated overnight at 4°C with 0.25 μg/well of capture antibody. Plates were then washed with PBS‐T three times and incubated for 2 hr at 37°C with blocking solution (1% BSA in PBS). After four washes, the antigen samples were diluted with 1% BSA in PBS and added to the wells. The plates were then incubated for 2 hr at 37°C. After washing, biotinylated detector antibody was added to the plates, followed by incubation for 1 hr at 37°C. After three washes, avidin‐HRP was diluted 1:5000 in 1% BSA containing PBS and added to the plates, followed by incubation for 30 min at 37°C. The plates were then washed five times and 3,3′,5,5′‐tetramethylbenzidine solution added. After 5 min, 1 N H2SO4 was added to stop the reaction and the OD450 measured with a microplate reader.
Electron microscopy
Purified S protein was loaded onto a formvar‐coated grid for 5 min. The grid was then negatively stained with 2% uranyl acetate and dried by aspiration. S protein was observed under a Tecnai G2 Spirit transmission electron microscope (FEI, Hillsboro, OR, USA) operating at 120 kV, images being digitally obtained with a CCD camera at 1 k × 1 k resolution.
RESULTS
Production of MERS‐CoV S protein in insect cells
The full‐length MERS S protein was expressed in a recombinant baculovirus system, harvested from infected insect cells and purified by chromatography. Purified MERS‐CoV S protein was analyzed by SDS‐PAGE, resulting in identification of a major band with a molecular weight of 180 kDa (Fig. 1a). Under an electron microscope, particles 20–30 nm in diameter were observed (Fig. 1b), this size being similar to that previously reported 19.
Figure 1.
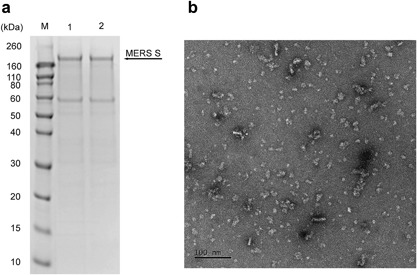
Characterization of MERS‐CoV S protein expressed in a baculovirus system. (a) Purified S protein was loaded onto SDS‐PAGE in duplicate lanes and stained with Coomassie blue. (b) Purified S protein was negatively stained with uranyl and examined at 67,000× magnification under an electron microscope.
Production of MERS‐CoV RBD in E. coli
The RBD was expressed after fusion with mRID in E. coli to improve protein solubility and folding, as previously reported 20, 21. The mRID‐RBD protein (molecular weight 48 kDa) was expressed in pellet fractions after IPTG induction. (Fig. 2a) The amount of protein sample was calculated based on the band intensity of the BSA control (Fig. 2b).
Figure 2.
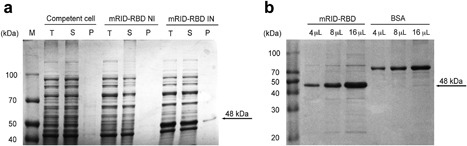
Characterization of MERS‐CoV RBD expressed in E. coli. (a) Mouse RID‐RBD protein was expressed in the soluble fraction of IPTG‐induced E. coli. IN, IPTG induction; M, protein marker; NI, no IPTG induction; P, cell pellets; S, supernatant; T, total lysate. (b) Purified mRID‐RBD and BSA were loaded onto SDS‐polyacrylamide gels in each lane.
Specific detection of polyclonal antibodies against MERS‐CoV S protein
Heat‐inactivated MERS‐CoV‐immunized rabbit polyclonal antibody was purchased from JoongKyeom (Ansan, Korea). This polyclonal antibody was used to determine whether the MERS‐CoV S protein produced in this study specifically binds to MERS‐CoV‐immunized serum. In addition, a MERS‐CoV S protein‐immunized rat polyclonal antibody that was produced in this study was tested. The MERS‐CoV S protein was used as the coating antigen for an indirect ELISA to detect MERS‐CoV‐specific antibodies in animal sera. The S protein was coated at concentrations of 100, 50, 25, 12.5, 6.25, 3.13 and 1.56 ng/well. All animal sera, both heat‐inactivated MERS‐CoV‐immunized rabbit sera and S protein‐immunized rat sera, were also serially diluted twice from 1:200. The S protein bound specifically to the diluted serum (Fig. 3), indicating that the recombinant S protein produced in this study could be used to develop a diagnostic system for MERS‐CoV.
Figure 3.
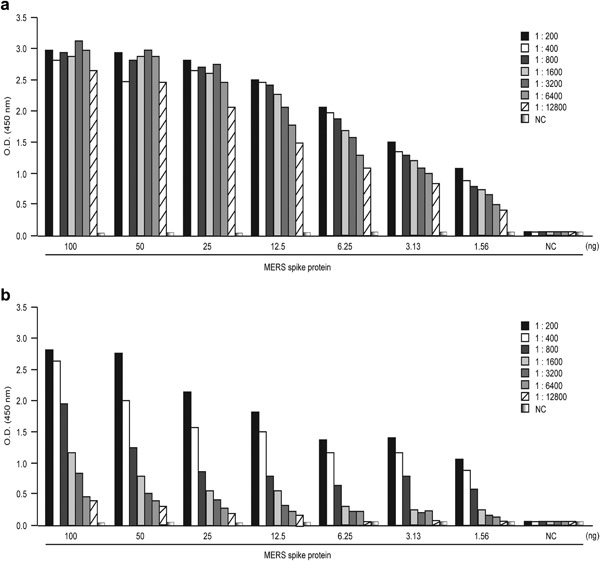
Detection of MERS‐CoV antibodies in animal sera with S protein. Purified S protein was used to coat the plate and ELISA performed to identify antibodies against MERS‐CoV in rat and rabbit sera. (a) Sera of rats immunized with S protein were serially diluted and tested. (b) Sera of rabbits immunized with heat‐inactivated MERS‐CoV were analyzed under the same conditions. NC, negative control.
Specific detection of antibodies in MERS‐CoV patient serum using MERS S protein
As mentioned above, recombinant S protein specifically binds to both MERS‐CoV‐derived and S protein‐derived antibodies in rat and rabbit sera. Next, whether the S protein can be used to diagnose MERS‐CoV infection in patient sera was determined. For this analysis, whether the recombinant S protein is able to specifically bind to sera of patients with MERS was investigated by two independent hospitals. Patients were diagnosed as having MERS by real‐time RT‐PCR targeting upE and orf1a, this being performed at the Korean Center for Disease Control laboratory 22. The hospitals used their own protocols for the dilution of patient sera, one of them diluting patient sera at a ratio of 1:32 twice (n = 6) (Fig. 4a), whereas the other diluted patient sera at a ratio of 1:100 twice (n = 5) (Fig. 4b). All steps in the rest of the ELISA protocol, such as incubation time, were as detailed in the Materials and Methods, 100 ng of S protein being used to coat each well, and normal human serum being used as a negative control.
Figure 4.
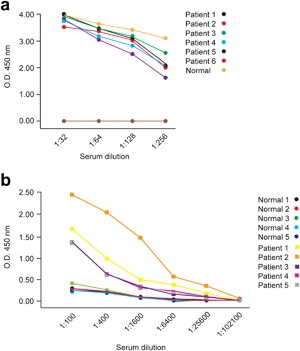
Detection with S protein of MERS‐CoV antibodies in sera of patients with MERS. MERS‐CoV‐specific antibodies in sera of patients with MERS from two hospitals were measured by ELISA using S proteins as coating antigens. (a) Patient sera from one hospital were serially diluted, and antibody titers against MERS‐CoV S protein measured. (b) Patient sera from the other hospital were analyzed under the same conditions except for use of different dilutions. [Color figure can be viewed at http://wileyonlinelibrary.com]
Titers of antibody against MERS‐CoV S protein in sera of patients with MERS were measured by indirect ELISA at different dilutions (Fig. 4). ELISA titers were significantly higher than those of the negative control, even at a serum dilution of 1:256, in all patient sera from one hospital (Fig. 4a). However, only four of the five sera examined in the other hospital exhibited higher ELISA titers than five negative controls at a dilution of 1:100 (Fig. 4b). It is possible that the fifth of these patients had an extremely low MERS‐CoV antibody titer. Thus, at patient serum dilutions of 1:200 and 1:300, the ELISA successfully distinguished sera of patients with MERS from healthy human serum, indicating that the recombinant MERS S protein produced in this study specifically binds MERS‐CoV‐derived antibodies in human samples and that the developed assay can be used clinically to diagnose MERS‐CoV infection.
Development and validation of sandwich ELISA to detect MERS‐CoV S protein
To develop a sandwich ELISA for detecting MERS‐CoV S protein, MAbs that specifically bind MERS‐CoV S protein were first produced and characterized. ATGen developed 12 MAbs against the S protein and 29 MAbs against the RBD. To select capture and detector antibodies, pairwise tests were performed with each MAb in an ELISA using 2.5 μg/mL detector and 1 μg/mL capture antibody (Fig. 5a). As shown in Figure 5b, c, two combinations were able to detect 100 ng of S protein: #16 S MAb as the capture antibody and #89 RBD MAb as the detector, and #9 RBD MAb as the capture antibody and #16D4 S MAb as the detector. However, only the #9/#16D4 combination, and not the #16/#89 combination, specifically bound to a serial dilution of S protein (Fig. 5d). Thus, the combination of the #16D4 MAb (induced by the S protein in the baculovirus system) as the detector antibody and the #9 MAb (induced by the RBD in the E. coli system) as the capture antibody was determined to be the optimal pairing for development of the sandwich ELISA.
Figure 5.
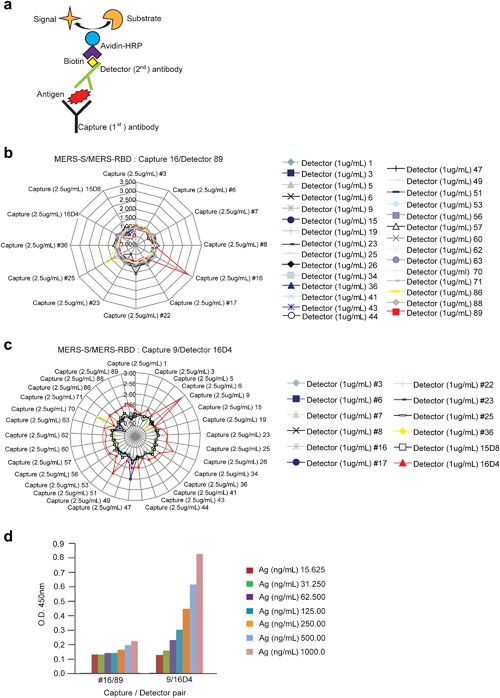
Determination of detector and capture antibodies for MERS‐CoV ELISA. (a) Schematic representation of sandwich ELISA method for the pairwise MAb tests. (b) Distribution of values using #16 MAb as the capture antibody and #89 MAb as the detector with 100 ng of S protein as the antigen (Ag). Results are shown as OD450. (c) Distribution of values using #9 MAb as the capture antibody and #16D4 MAb as the detector. ELISA conditions were the same as above. (d) Two combinations (#16/#89 and #9/#16D4) were tested to identify the optimal MAb pair. ELISA was performed using serially diluted S protein. [Color figure can be viewed at http://wileyonlinelibrary.com]
To assess the sandwich ELISA developed in this study, amounts of intra‐assay variation (within‐run precision) and inter‐assay variation (between‐run precision) were determined to confirm good linearity and reproducibility. Both intra‐ and inter‐assay R2 values were > 0.9, and both coefficients of variation (CV%) were < 10% (Fig. 6a, b). Inter‐day precision tests to assess differences between tests run at two time points (7 days apart) showed comparable R2 values (0.9316 vs. 0.9178) and CV% values < 10% (Fig. 6c). Theoretically, the LLD of the sandwich ELISA is 5.89 ng/mL of MERS‐CoV S protein (Fig. 6d).
Figure 6.
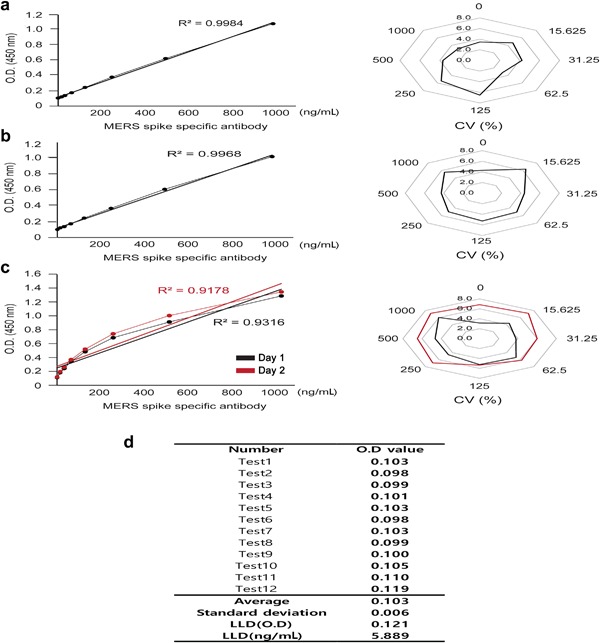
Validation of MERS‐CoV antigen ELISA. MERS‐CoV antigen ELISA was tested with various S protein dilutions to confirm linearity and reproducibility. MAbs #9 and #16D4 were used as capture and detector antibodies, respectively. (a) Intra‐assay test results showing variations between wells (n = 12). (b) Inter‐assay test results showing variations between plates (n = 12). (c) Inter‐day precision test results showing variations between assays conducted at different time points. Black: Day 0, red: Day 7. (d) The LLD was determined by adding 3 × SD of the blank to the average of the blank (n = 12). OD values were used in the standard curve formula to calculate the minimum detectable concentration. [Color figure can be viewed at http://wileyonlinelibrary.com]
Development and validation of indirect ELISA for detecting MERS‐CoV S‐induced antibodies
As mentioned above, an indirect ELISA to detect MERS‐CoV‐induced antibodies in animal and human patient sera was developed by coating wells with 100 ng of MERS S protein. Twelve mAbs derived from the S protein were tested against S protein to select standard antibodies. Among these, #36 MAb showed the best performance and serially diluted #36 MAb exhibited binding activity with 100 ng of S protein (data not shown). Therefore, #36 MAb was used to validate the indirect ELISA.
Intra‐assay and inter‐assay variations showed that both R2 values (0.9197 and 0.9279, respectively) were > 0.9, indicating good linearity. Moreover, both CV% values were < 10%, indicating good reproducibility (Fig. 7a, b). Tests for inter‐day precision (the difference between two time points) showed similar R2 and CV% values, indicating acceptable reproducibility with a 1‐week interval (Fig. 7c). As to sensitivity, the LLD was 0.04 ng/mL of MERS‐CoV‐specific antibody (Fig. 7d).
Figure 7.
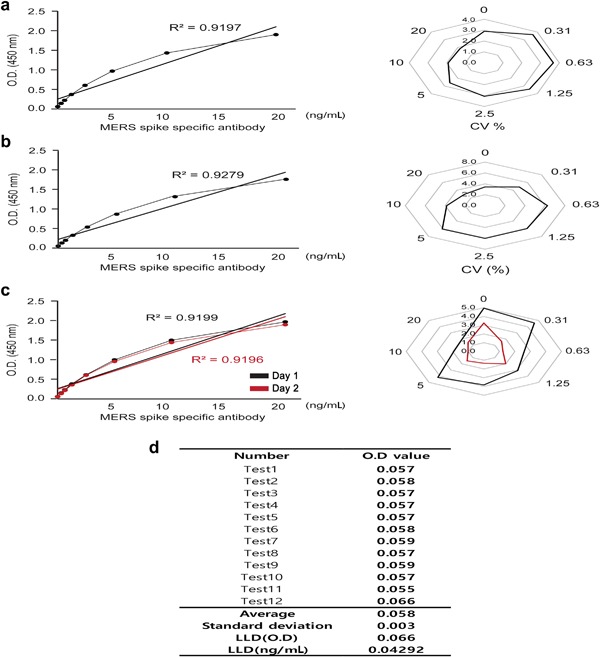
Validation of MERS‐CoV antibody ELISA. MERS‐CoV antibody ELISA was tested with various MERS‐CoV‐specific MAb dilutions to confirm linearity and reproducibility. S protein was used as a coating antigen. (a) Intra‐assay test results showing variations between wells (n = 12). (b) Inter‐assay test results showing variations between plates (n = 12). (c) Inter‐day precision test results showing variations between assays conducted at different time points. Black: Day 0, red: Day 7. (d) Lower limit of detection (LLD) was calculated by adding 3 × the standard deviation of the blank to the average of the blank (n = 12). OD values were used in the standard curve formula to calculate the minimum detectable concentration. [Color figure can be viewed at http://wileyonlinelibrary.com]
DISCUSSION
Although MERS causes acute respiratory disease and represents a considerable threat, there are currently no approved vaccines or drugs for this condition 23. Fast and accurate diagnosis is therefore the most promising means of controlling MERS outbreaks. Traditionally, PRNT and/or microneutralization tests have been used as the gold standards for detecting MERS‐CoV‐specific antibodies. However, these tests are labor‐intensive and time‐consuming and require high‐level biosafety facilities because they involve handling of live viruses 14, 24, 25. Therefore, more convenient methods are required. Recently, a cELISA in which a labeled MAb competes with test serum antibodies for target epitopes was developed and compared with the PRNT assay; Pearson's correlation coefficients were reportedly 0.71–0.76 depending on the cELISA serum dilution 26. Although this correlation is positive, there were considerable variations at the same PRNT titers 26.
In the present study, we developed two ELISA systems: an indirect ELISA for detecting MERS‐CoV‐induced antibodies and a sandwich ELISA for detecting the MERS‐CoV S protein. We expressed the MERS‐CoV S protein and RBD in recombinant baculovirus and E. coli systems, respectively. The indirect ELISA involved coating with recombinant full‐length S protein, which specifically bound to antibodies in MERS patient sera, showing good linearity and reproducibility, as well as high precision and sensitivity. Therefore, the indirect ELISA developed in this study can be used clinically to detect MERS‐specific antibodies in patient sera. In addition, we used serial dilutions of S protein‐immunized rat sera to compare indirect ELISA titers with PRNT titers; human sera were not used because insufficient volumes of patient sera were available. However, we did not find a positive correlation between the two titers because of differences in the sensitivities of the two assays; specifically, a dilution of MERS‐CoV antibody of over 1280‐fold still showed a high titer in the indirect ELISA, whereas it was not detectable in the PRNT assay (data not shown). It should be noted that it is also possible that there were no neutralizing antibodies in the rat serum samples. Therefore, as stated in a previous report 26, further studies are necessary to develop a cELISA using MERS‐CoV S protein‐derived MAbs in order to determine the correlation between the cELISA and PRNT, the latter currently being used clinically to diagnose MERS‐CoV infection.
In this study, we established a sandwich ELISA to detect the MERS‐CoV S protein using MAbs induced by the S protein and RBD. We validated the assay with recombinant S protein and demonstrated good reproducibility, precision and sensitivity. Therefore, although we did not test whether the recombinant spike protein‐induced mAb detects authentic MERS‐CoV, this sandwich ELISA can be used clinically to detect the presence of MERS‐CoV in biological samples from patients suspected of having MERS and in various contaminated materials. Recent studies have shown that qRT‐PCR can also accurately detect the MERS‐CoV genome in a variety of biological samples, even when there are extremely small amounts of the virus. Moreover, the biological hazard levels required for the devices and facilities capable of performing sandwich ELISA and real‐time PCR are similar. Thus, sandwich ELISA offers no significant advantage over real‐time PCR in a clinical setting. However, the sandwich ELISA developed in this study can directly detect and quantify MERS‐CoV S protein. Given that most MERS‐CoV vaccine candidates target the S protein because of its promise as a vaccine target 27, 28, 29, the sandwich ELISA could be used for quality‐control testing, such as measuring the antigen content of vaccine candidates, rather than detecting MERS‐CoV in clinical settings.
In conclusion, we have developed indirect and sandwich ELISAs that can detect MERS‐CoV‐induced antibody and S protein, respectively. Both ELISAs have adequate reproducibility, precision and sensitivity for clinical use. However, further intensive testing of these assays with sera from more patients is required.
DISCLOSURE
The authors declare that they have no conflicting interests associated with this study.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Food and Drug Safety in 2016–2017 (16172MFDS268), the Ministry of Health and Welfare, Korea (HI15C2955) and Basic Science Research Program through the National Research Foundation funded by the Ministry of Science, Information and Communication Technology and Future Planning (NRF‐2015M3A9B5030157).
Kunse Lee and Hae Li Ko contributed equally to this work.
REFERENCES
- 1. Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367: 1814–20. [DOI] [PubMed] [Google Scholar]
- 2. Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R., Pebody R.G., Green H.K., Boddington N.L., Gopal R., Price N., Newsholme W., Drosten C., Fouchier R.A., Zambon M. (2012) Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill 17: 20290. [PubMed] [Google Scholar]
- 3. Van Den Brand J.M., Smits S.L., Haagmans B.L. (2015) Pathogenesis of Middle East respiratory syndrome coronavirus. J Pathol 235: 175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim K.H., Tandi T.E., Choi J.W., Moon J.M., Kim M.S. (2017) Middle East respiratory syndrome coronavirus (MERS‐CoV) outbreak in South Korea, 2015: Epidemiology, characteristics and public health implications. J Hosp Infect 95: 207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer‐Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., Von Hahn T., Simmons G., Hofmann H., Pohlmann S. (2013) The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 87: 5502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y., Rajashankar K.R., Yang Y., Agnihothram S.S., Liu C., Lin Y.L., Baric R.S., Li F. (2013) Crystal structure of the receptor‐binding domain from newly emerged Middle East respiratory syndrome coronavirus. J Virol 87: 10777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. (2013) Molecular basis of binding between novel human coronavirus MERS‐CoV and its receptor CD26. Nature 500: 227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang N., Jiang S., Du L. (2014) Current advancements and potential strategies in the development of MERS‐CoV vaccines. Expert Rev Vaccines 13: 761–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., Lees C.R., Zhou T., Yassine H.M., Kanekiyo M., Yang Z.Y., Chen X., Becker M.M., Freeman M., Vogel L., Johnson J.C., Olinger G., Todd J.P., Bagci U., Solomon J., Mollura D.J., Hensley L., Jahrling P., Denison M.R., Rao S.S., Subbarao K., Kwong P.D., Mascola J.R., Kong W.P., Graham B.S. (2015) Evaluation of candidate vaccine approaches for MERS‐CoV. Nat Commun 6: 7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cockrell A.S., Baric R.S. (2016) An effective DNA vaccine platform for Middle East respiratory syndrome coronavirus. Ann Transl Med 4: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Al Johani S., Hajeer A.H. (2016) MERS‐CoV diagnosis: An update. J Infect Public Health 9: 216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corman V.M., Muller M.A., Costabel U., Timm J., Binger T., Meyer B., Kreher P., Lattwein E., Eschbach‐Bludau M., Nitsche A., Bleicker T., Landt O., Schweiger B., Drexler J.F., Osterhaus A.D., Haagmans B.L., Dittmer U., Bonin F., Wolff T., Drosten C. (2012) Assays for laboratory confirmation of novel human coronavirus (hCoV‐EMC) infections. Euro Surveill 17: 20334. [DOI] [PubMed] [Google Scholar]
- 13. Meyer B., Drosten C., Muller M.A. (2014) Serological assays for emerging coronaviruses: Challenges and pitfalls. Virus Res 194: 175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perera R.A., Wang P., Gomaa M.R., El‐Shesheny R., Kandeil A., Bagato O., Siu L.Y., Shehata M.M., Kayed A.S., Moatasim Y., Li M., Poon L.L., Guan Y., Webby R.J., Ali M.A., Peiris J.S., Kayali G. (2013) Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill 18: 20574. [DOI] [PubMed] [Google Scholar]
- 15. Spanakis N., Tsiodras S., Haagmans B.L., Raj V.S., Pontikis K., Koutsoukou A., Koulouris N.G., Osterhaus A.D., Koopmans M.P., Tsakris A. (2014) Virological and serological analysis of a recent Middle East respiratory syndrome coronavirus infection case on a triple combination antiviral regimen. Int J Antimicrob Agents 44: 528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cooper H.M., Paterson Y. (2009) Production of polyclonal antisera. Curr Protoc Neurosci Chapter 5: Unit 5.5. [DOI] [PubMed] [Google Scholar]
- 17. Cho A., Seok S.H. (2013) Ethical guidelines for use of experimental animals in biomedical research. J Bacterial Virol 43: 18–26. [Google Scholar]
- 18. He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. (2005) Receptor‐binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation‐dependent epitopes that induce highly potent neutralizing antibodies. J Immunol 174: 4908–15. [DOI] [PubMed] [Google Scholar]
- 19. Coleman C.M., Liu Y.V., Mu H., Taylor J.K., Massare M., Flyer D.C., Smith G.E., Frieman M.B. (2014) Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine 32: 3169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi S.I., Han K.S., Kim C.W., Ryu K.S., Kim B.H., Kim K.H., Kim S.I., Kang T.H., Shin H.C., Lim K.H., Kim H.K., Hyun J.M., Seong B.L. (2008) Protein solubility and folding enhancement by interaction with RNA. PLoS ONE 3: e2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang S.W., Jang Y.H., Kwon S.B., Lee Y.J., Chae W., Byun Y.H., Kim P., Park C., Lee Y.J., Kim C.K., Kim Y.S., Choi S.I., Seong B.L. (2018) Harnessing an RNA‐mediated chaperone for the assembly of influenza hemagglutinin in an immunologically relevant conformation. FASEB J 32: 2658–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Min C.K., Cheon S., Ha N.Y., Sohn K.M., Kim Y., Aigerim A., Shin H.M., Choi J.Y., Inn K.S., Kim J.H., Moon J.Y., Choi M.S., Cho N.H., Kim Y.S. (2016) Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep 6: 25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chafekar A., Fielding B.C. (2018) MERS‐CoV: Understanding the latest human coronavirus threat. Viruses 10: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hemida M.G., Perera R.A., Al Jassim R.A., Kayali G., Siu L.Y., Wang P., Chu K.W., Perlman S., Ali M.A., Alnaeem A., Guan Y., Poon L.L., Saif L., Peiris M. (2014) Seroepidemiology of Middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Euro Surveill 19: 20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reusken C.B., Ababneh M., Raj V.S., Meyer B., Eljarah A., Abutarbush S., Godeke G.J., Bestebroer T.M., Zutt I., Muller M.A., Bosch B.J., Rottier P.J., Osterhaus A.D., Drosten C., Haagmans B.L., Koopmans M.P. (2013) Middle East Respiratory Syndrome coronavirus (MERS‐CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro Surveill 18: 20662. [DOI] [PubMed] [Google Scholar]
- 26. Fukushi S., Fukuma A., Kurosu T., Watanabe S., Shimojima M., Shirato K., Iwata‐Yoshikawa N., Nagata N., Ohnishi K., Ato M., Melaku S.K., Sentsui H., Saijo M. (2018) Characterization of novel monoclonal antibodies against the MERS‐coronavirus spike protein and their application in species‐independent antibody detection by competitive ELISA. J Virol Methods 251: 22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiaming L., Yanfeng Y., Yao D., Yawei H., Linlin B., Baoying H., Jinghua Y., Gao G.F., Chuan Q., Wenjie T. (2017) The recombinant N‐terminal domain of spike proteins is a potential vaccine against Middle East respiratory syndrome coronavirus (MERS‐CoV) infection. Vaccine 35: 10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muthumani K., Falzarano D., Reuschel E.L., Tingey C., Flingai S., Villarreal D.O., Wise M., Patel A., Izmirly A., Aljuaid A., Seliga A.M., Soule G., Morrow M., Kraynyak K.A., Khan A.S., Scott D.P., Feldmann F., Lacasse R., Meade‐White K., Okumura A., Ugen K.E., Sardesai N.Y., Kim J.J., Kobinger G., Feldmann H., Weiner D.B. (2015) A synthetic consensus anti‐spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med 7: 301ra132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coleman C.M., Venkataraman T., Liu Y.V., Glenn G.M., Smith G.E., Flyer D.C., Frieman M.B. (2017) MERS‐CoV spike nanoparticles protect mice from MERS‐CoV infection. Vaccine 35: 1586–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


