Abstract
Biosensors are analytical devices which find extensive applications in fields such as the food industry, defense sector, environmental monitoring, and in clinical diagnosis. Similarly, intrinsically conducting polymers (ICPs) and their composites have lured immense interest in bio‐sensing due to their various attributes like compatibility with biological molecules, efficient electron transfer upon biochemical reactions, loading of bio‐reagent, and immobilization of biomolecules. Further, they are proficient in sensing diverse biological species and compounds like glucose (detection limit ≈0.18 nm), DNA (≈10 pm), cholesterol (≈1 µm), aptamer (≈0.8 pm), and also cancer cells (≈5 pm mL−1) making them a potential candidate for biological sensing functions. ICPs and their composites have been extensively exploited by researchers in the field of biosensors owing to these peculiarities; however, no consolidated literature on the usage of conducting polymer composites for biosensing functions is available. This review extensively elucidates on ICP composites and doped conjugated polymers for biosensing functions of copious biological species. In addition, a brief overview is provided on various forms of biosensors, their sensing mechanisms, and various methods of immobilizing biological species along with the life cycle assessment of biosensors for various biosensing applications, and their cost analysis.
Keywords: biological molecules, biosensors, conductive polymer composites, conductive polymers, immobilization, nanomaterials
Conductive polymer composites are widely employed in fabricating biosensors with different reinforcements such as gold nanoparticles, carbon nanotubes, graphene, and also hybrid nanocomposites. Thus, working principles, sensing mechanisms, classification, and immobilization techniques of biosensors are discussed in detail. Life cycle assessment of biosensors is also discussed in the final section.

1. Introduction
Human life has been surrounded by the use of sensors for the simplicity of living. Over the last few decades, progression and investigation in the arena of sensors have augmented in an exponential manner in terms of published literature and capital spent on it.1, 2 Various sensing devices based on temperature sensor3, 4, 5 (thermometer), gas sensor6, 7, 8 (vehicle's emission control system), motion sensor9, 10, 11 (home security lights), photosensor12, 13, 14 (for detection of infrared and ultraviolet light), and tactile sensor15, 16, 17 (touchscreen devices) have been developed and comprehensively explored by the scientific community. However, an ingenious concept to combine the recognition characteristics of a biological macromolecule to an electronic device brings about the occurrence of the biosensor. The rapid advancements in the field of material science and innovation coupled with environmental concerns have brought about new opportunities for the advancement of biosensors.
Biosensors are analytical sensing devices that integrate i) a bio‐receptor, which is an immobilized biomolecule (e.g., enzyme, ss‐DNA, antibody) that recognizes the analyte (e.g., antigen, complementary DNA) and ii) a transducer (e.g., conductive polymers, self‐assembled monolayers, metal nanoparticles, etc.) that produces a biochemical signal upon reaction of biological entity with the analyte which can be fetched by a detector. A diagrammatic representation of a biosensor is shown in Figure 1 , where various components of the biosensor are being depicted. As stated by the new market research, the biosensors market by 2022 is estimated to value USD 27.06 billion and is developing at an annual growth rate of ≈9% between 2017 and 2022.18 This market growth is motivated by the need for quantification and exploitation of the biological species in the field of environmental monitoring,19 clinical analysis for diagnosis of various diseases,20, 21 food quality evaluation,22 drug development23 as well as in defense sector (for the detection of biowarfare agents and nerve gases).24 Biosensors promise to render an economical and robust alternative to some conventional analytical chromatographic strategies for analyzing the chemical species in some complex matrices, thanks to their ability to distinguish the target analyte from interfering and inert biological species without the necessity for separation and subsequent identification of all constituents of the analyte. Usually, biosensors are supposed to be independent of some physical parameters like temperature, pH, pressure, etc. to acquire precise results and the data obtained must be efficiently reproducible and reliable. However, as a result of their highly integrated compact structure, the biosensing devices are at a juncture of distinct areas of knowledge. Biotechnology deals with one of the key components of biosensor, that is, biomolecule, which provides with specificity for the test. The field of chemical physics is linked with the transducing component that converts the biochemical recognition signal to electrical one. Additionally, microelectronic circuits and micro‐fabricated structures are associated with small volumes of samples in microliters required to carry out the process at a small scale. And finally, the field of nanotechnology has a significant role in the advancement of biosensor owing to their nanostructure property such as high electrical conductivity, large surface area, and so on. Thus, the field of biosensor is truly a multidisciplinary one.
Figure 1.

Diagrammatic representation of a biosensor.
The term “biosensor” was coined by Clark and associates in 1962 where they developed an enzyme‐based glucose sensing device and they quantified the concentration of glucose using glucose oxidase immobilized on the oxygen electrode via a semi‐permeable membrane.25 Since then, due to their adaptability, biosensors have been urbanized for the recognition of some complex biological species like virus,26 pathogens,27 insulin,28 neurotransmitters,29 and hormones.30 In recent times, menacing diseases caused by viruses, for instance, human immunodeficiency virus (HIV), severe acute respiratory syndrome coronavirus (SARS‐CoV), Nipah, avian influenza, Hendra, etc. have affected human life significantly, thus biosensor can be a beneficial tool for their detection due to their on‐site, real‐time monitoring and early detection capability.31, 32, 33
A biosensor performance evaluation is primarily executed on the basis of the limit of detection, reproducibility, sensitivity, selectivity, linear dynamic range, and other peculiarities.24 Also, other parameters like portability, response time, accessibility, storage, and operational stability need to be considered. Numerous materials such as semiconductor,34 magnetic nanoparticle,35 metal oxide,36 carbon‐based nanomaterial,37 and intrinsically conducting polymer29 (ICP) have been developed for biosensing applications. Among the abovementioned material, ICP is much‐admired for biosensors due to its paramount characteristics such as biocompatibility,38 inimitable electronic structure39 (for facile transfer of electrons), tunable electrical conductivity40 (via doping), environmental stability,41 facile corrosion‐free synthesis,42 and sensitivity to small perturbations.43 Because of their efficient electron transfer capability, formed upon biochemical reactions, they have been comprehensively utilized as transducers in biosensor that forms an intermediary layer between the biological molecule and electronics that are used for signal readout. A number of investigations on ICPs have been carried out in the field of biosensors where a typical example is in aptamer sensing. Gao et al. exploited an enzymatically catalyzed‐polyaniline, where they found the limit of detection of 1 fM for DNA sensing.44 Another researcher, Komaravo et al., synthesized polypyrrole‐based biosensor for the recognition of Variola major virus and achieved 16 pM detection limit.45 Similarly, Krishnamoorthy et al. electrochemically synthesized poly(3,4‐ethylene dioxythiophene) based biosensor and developed a label‐free DNA sensor with a detection limit of 8 × 10−8 g mL−1.46 The scope of diverse application of biosensors based on conducting polymer is illustrated in Figure 2 .
Figure 2.

Scope of biosensing application based on conducting polymers.
Multifunctional composites of ICP have also been extensively explored as it combines the attributes of conjugated polymer and the reinforcement material giving rise to a synergetic effect, by demonstrating improved properties, which can be utilized for complex biomolecule sensing.47, 48 Moreover, composites of ICP can arrest agglomeration of nanoparticles due to dispersion in conductive organic matrix and also prevent restacking due to steric hindrance and electrostatic interactions.49 Simultaneously, ICP composites enhance the electron transfer rates during biochemical reactions thereby proving as an excellent transducer. Various materials, for instance, gold nanoparticle, carbon nanotube, graphene, graphene oxide, metal oxide, etc. have been explored as a reinforcement material, thanks to their unique chemical, electrical, mechanical, and optical properties.50 Nanomaterial integrated into biosensing device bids the possibility of realizing economical, easy‐to‐use, and portable device due to the proficiency of miniaturization of the material as well as transduction system.2 Alternatively, nanoparticles can prove as an ideal remedy to various contradictory issues in optimizing immobilized biomolecules such as high effectual loading, minimal diffusion limitations, and maximal surface area per unit of mass.51
In this context, extensive literature analysis reveals that no consolidated literature is as such available on doped‐ICP and ICP‐composite materials for biosensing application to the best of our perception. Thus, we have discussed in detail on the significance of ICPs and their composites based on gold nanoparticles, carbon nanotubes, graphene and their derivatives, and hybrid composites along with doped‐ICP for biosensing of copious biomolecules. Simultaneously, we have described the sensing mechanisms of different biosensors based on enzymes, DNAs, and antibodies and their interaction with their organic counterpart that leads to signal generation. Further, we have briefly discussed the importance of ICP and their composites in various methodologies for immobilization of biomolecules on the electrode surface which is mandatory to increase the performance of the biosensor. Finally, we conclude the review by discussing the life cycle assessment of different biosensing devices being developed in the world.
2. Conducting Polymers
ICP is a class of organic polymer that exhibits metallic characteristics such as electronic, electrical, magnetic, and optical while retaining the attributes of traditional organic polymers such as facile synthesis, low cost, and corrosion resistance.52, 53, 54, 55 They can be either insulator or semi‐conductor in undoped or neutral form that can be transformed into doped form by redox reaction forming delocalized charge carriers. In general, ICPs possess alternating single (σ) and double (π) bonds in the main chain and these π‐conjugated systems impart the ICPs their inherent electrochemical, optical, and electrical or electronic properties. The benefit that ICP possesses over several organic polymers is tunable chemical structure, which can be modified to alter the conductivity and solubility of these polymers. For example, adopting poly(3‐hexylthiophene) as a functional group can enhance the processibility and solubility of some insoluble polymers.56 Among the copious ICPs, aromatic‐based conductive polymers have attracted great attention of researchers because it offers high conductivity, good chemical and thermal stability.57
As in saturated polymer, where all the four valence electrons of carbon are covalently bonded, the electronic configuration of conjugated ICP is totally different. The chemical bonding, here, leads to one unpaired π‐electron per atom of carbon. Also, this π‐bonding leads to delocalization of electron by lateral overlapping of p‐orbitals alongside the backbone of the ICP, providing the “highway” for charge mobility. This unique electronic structure of ICP contributes to low ionization potential, high electron affinity, and low energy optical transition which in turn leads to their electrical conductivity.58 However, conjugation alone is not sufficient enough to contribute to conductivity of such polymers. Thus, doping is a process where the conductivity is augmented up to several orders of magnitude by introducing different doping agents that are either partial oxidizing (p‐doping) or partial reducing agents (n‐doping).59 With the help of such process, charged defects such as soliton, polaron (radical ions), and bipolaron can be introduced into the ICP which are then available as charge carriers.
2.1. Conducting Polymer Composites
To date, biosensors modified with the aid of a solitary material were not commercialized owing to their surface poisoning because of adsorbed intermediates, low sensitivity, poor selectivity, and intervention from additional species. Also, though ICPs provide copious benefits, the progress in increasing the properties has not been commensurate with those of their carbon‐based, metallic, and metal oxide counterparts.60 Thus, forming composites of ICP with different materials avoid these difficulties and find attractive features in the field of biosensing.61 ICPs have also been hybridized and modified with other heterogeneous components to overcome their limitations such as long‐term stability,62 solubility,63 and processing.24 These components not only provide augmented mechanical and thermal stability, but also distinct functionalities that depend on chemical nature, crystallinity, size, and structure of the reinforcement.38 Strategic coupling of the ICP with other counterparts can result in many epitome properties providing opportunities in myriad applications ranging from storage device to sensors and biosensors.64, 65, 66 Successful incorporation of this particle in polymer framework also provides an enhanced rate of electron transfer at the altered surface/electrolyte interface.67
In the field of nanoscience and nanotechnology, incorporation of nanoparticles into ICP matrix has attracted considerable attention of scientists as well as researchers.68 The arena of ICP‐based nanocomposite has expanded enormously because of their potential characteristics such as superior electrocatalytic activity, augmented electrical conductivity, and decent chemical stability in aqueous solution.69 Interest in integrating metal nanoparticle was motivated by their application in biosensors due to their nano‐range size, unique physical, electronic, and chemical properties (different from their bulk material), flexibility to build amended and novel sensing device.70 Incorporation of carbon‐based material (such as graphene [GN] and carbon nanotube [CNT]) leads to interaction of CNT and GN with ICP, which is an important attribute that regulates the efficiency and working of a carbon‐based ICP biosensor.71 Conjugation along with the highly hydrophobic surface of CNT and GN allows them to interact with the ICPs via hydrophobic and electrostatic interactions. Hence, a synthetic customized approach for developing a novel type of biosensor using this combination will give an entirely different carbon‐based ICP composite with a new set of fascinating properties.72, 73, 74 Selectivity of a biosensor largely depends upon the recognition element (e.g., antibody) and also on the type of host matrix and the interaction between them. Thus, integration of nanoparticle is convenient for attaining adequate stability and sensitivity since nanoparticle acts as a redox mediator of biological molecule and ICP acts as a selective adsorbate for these molecules.75, 76, 77
3. Biosensors
A specific definition of biosensor is given by IUPAC as a self‐contained integral device capable of issuing specific quantitative/semi‐quantitative analytical information with the aid of a bio‐recognition element that is retained by a transduction element via direct spatial contact.78 As mentioned earlier, a biosensor consists of a bio‐receptor that is capable of determining specific target analyte with the aid of a biological molecule, and a transducer for the conversion of the signal into readable output. The reaction amid analyte and the biomolecule causes chemical alterations such as change in mass or pH, release of heat, formation of different chemical or flowing of electrons that can be transformed into an electrical output by transducer. The aim of a biosensor is generation of a digital signal that is relative to the concentration of the peculiar analyte. The type of signal that is being transduced by ICP depends on the type of ICP, doped state of ICP, and the binding molecule used. For example, small molecule can diffuse into the polymeric chains that can give rise to chemical reaction with ICP, thereby varying its state of doping.
3.1. Working Principle of Biosensor versus Natural Sensor
In the initial stage, the bio‐recognition molecule is immobilized on the transducer surface that forms a specific complex with the target analyte. In context to its binding partner, the immobilized biomolecule often undergoes conformational change which can be immediately detected and transduced with the aid of a transducer. The effects of interaction among the immobilized biomolecule and target analyte are quantified and recognized by the use of different transducers and electronic section of the biosensor. The transduction of the biochemical signal can be carried out in the form of electrochemical, optical, thermometric, and piezoelectric transducer combined with bioreceptors.79 The idea of incorporating the receptor into the biosensor system has actually originated from Mother Nature since it has developed some exceptional sensing capabilities with the aid of the evolution of biological species almost billions of years ago. The most typical example of natural sensor is human beings having five senses such as touch, smell, taste, hearing, and sight using the ability of skin, nose, tongue, ear, and eyes, respectively. Thus, inspired by nature, researchers have been trying to “mimic” the natural counterparts to produce devices, termed as “Biomimetic,”80, 81, 82 for example, an analog of eardrum of mammalians to produce flexible diaphragms have been utilized in microphone and pressure sensors.83 Some of the examples on the natural sensor and their working principle compared to biosensors are depicted in Figure 3 .
Figure 3.
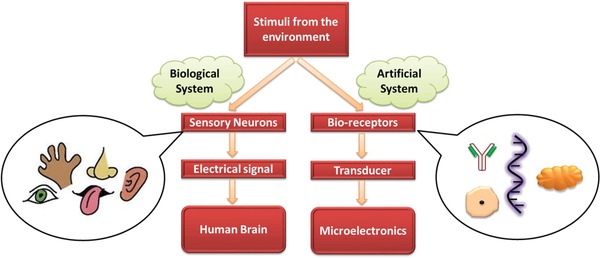
A schematic comparison between artificial and biological system.
3.2. Classification of Biosensor
Biosensors can be classified based on the type of bio‐recognition element but keeping in context the diversity of structural biological molecule, it is difficult to classify the newer biosensor strictly on structural basis.2 Thus based on the mode in which the physiochemical signal is transmitted and sensed, biosensors are categorized into subsequent types: electrochemical, piezoelectric, calorimetric, and optical transducers which have been discussed in section 3.2.1, 3.2.2, 3.2.3, and 3.2.4, resp.
3.2.1. Electrochemical Biosensor
Electrochemical biosensor is a constitutional concept that is based on an electrochemical transducer coupled with a matrix‐bound biological molecule. Enzymes are, most often, being used as bio‐recognition elements on account of their biocatalytic activity and specific binding aptitudes. Apart from them, other elements such as antigens, micro‐organisms, and antibody fragments are also being used.84, 85, 86, 87 A schematic of the principle of an electrochemical biosensor is depicted in Figure 4 . The underlying principle of this type of biosensor is that the electrochemical reaction produces or consumes electrons causing changes in the electrical characteristics of the analyte that can be determined and used as a parameter for measuring it.78 This change in electrical properties can be correlated with the amount of the biological molecules or the rate at which it is produced or consumed.88
Figure 4.

Principle of recognition in an electrochemical biosensor.
The electrochemical reaction being monitored would cause generation of a detectable current (amperometric), a detectable charge/potential accumulation (potentiometric), or alter the conductivity (conductometric) of the analyte medium between the electrodes. A review by Moon et al. reveals the use of electrochemical biosensor based on conductive polymer for recognizing neurotransmitters that control the physiological and behavioral function in the peripheral and central nervous system.29 Gui et al. excellently assessed the use of electrochemical biosensor based on molecularly imprinted polymers (MIP) that showed superior capacity for detecting selective biomolecules.89 A new level in clinical diagnosis is also being attained in the detection of cancer‐causing cells with the aid of electrochemical biosensor. Recently, Saeed et al. synthesized a label‐free ultrasensitive Polypyrrole‐3‐carboxylic acid/reduced graphene oxide based electrochemical biosensor for recognizing BRCA1 gene with a detection limit of around 3 fM.90 On the basis of the nature of electrochemical changes occuring during a bio‐recognition event, they fall into different categories like amperometry, conductometry, and potentiometry which will be discussed hereby. A comparison of all the three types of electrochemical biosensor is shown in Table 1 .
Table 1.
Comparison of the distinct types of biosensors based on electrochemical measurements
| Type of electrochemical biosensor | Transduction mechanism | Measured property | Ref. |
|---|---|---|---|
| Amperometric | Electron transfer reaction | Electric current | 91 |
| Potentiometric | Charge density accumulation | Potential difference | 92 |
| Conductometric | Ionic strength alteration | Conductance | 93 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Amperometric Biosensor: Amperometry is a prevalent approach that is being used in biosensing based on ICPs thanks to their fast response and simplicity of measurement. Amperometric biosensor relies on determining the current produced by electrochemical redox reaction of electro‐active species, as a function of time, with a fixed applied potential on the electrode surface, most commonly an ICP. The applied potential aids in efficient electron transfer reaction and the resulting current determined is directly proportional to the electro‐active product's concentration over a linear applied potential range.91 They actually hold leading position among other biosensor systems, thus the majority of work in biosensors has been done in the field of amperometric biosensor based on various electrochemically deposited ICP.
The limitation of this type of biosensor is the generation of false current reading because of the electro‐active interference from the sample matrix. However, these effects can be eliminated with the aid of selective membranes that control the charge of compounds having access to the electrode.94 Continuous signal that is provided by the amperometric biosensor can be utilized for steady in vivo surveillance of patients. Hence, such measurements are highly craved for real‐time in intensive care unit, at the time of surgeries, or for managing diabetes as they offer an alternative of premature warning of an abrupt deterioration.
A tangible application of amperometric immunosensor is for the measurement of β‐HCG level in advanced pregnancy testing.95 In another work, German et al. established a glucose‐based amperometric sensor by utilizing gold nanoparticle along with electron transfer mediator and different immobilized glucose oxidase that were sourced from Penicillium funiculosum, Penicillium adametzii, or Aspergillus niger that were cross‐linked with glutaraldehyde.96 They observed a linear concentration range and the glucose oxidase from Penicillium funiculosum exhibited a limit of detection of 0.024 mmol L−1. Introduction of the ICP layer augmented the linear detection range on the biosensor 0.1 to 25 mmol L−1. Another researcher grafted Prussian Blue onto the composite of MWCNT/Poly (4‐vinyl pyridine) for their application in amperometric biosensor. The biosensor unveiled rapid response to the detection of hydrogen peroxide with a limit of detection 25 nM.97 Also, Lupu et al. proposed PEDOT and tyrosinase based amperometric biosensor for the detection of dopamine and catechol.98
Potentiometric Biosensor: Potentiometry is considered to be one of the oldest analytical methods that enable the estimation of physicochemical quantities like pH, dissociation constant, activity of coefficients, and solubility products. Potentiometric biosensor has the following potential characteristics as mentioned below99:
Inherent selectivity
Analyte is not consumed
Simple instrumentation
Logarithmic response
However, it is rarely used for detection in a biosensor with immobilized enzymes on ICP layer, though certain advantages of this type of biosensor over amperometric biosensor for polypyrrole based glucose oxidase has been demonstrated.92
Potentiometric biosensor relies on the utilization of ion‐selective electrode (ISE) to obtain analytical information. ISEs are working electrode constructed of permselective ion‐conductive membrane on the surface. They find the majority of their application in the field of clinical chemistry in physiological fluids for determining biological electrolytes.88 Potentiometric biosensor works on the principle of accumulation of charge density (during the recognition process) that results in the formation of potential at that electrode while drawing negligible current. In such a sensor, the bioreceptor transforms the recognition process to a potential signal providing an analytical signal. The development of charge can be related to the concentration that is governed by Nernst relation.
| (1) |
where E = Actual cell potential at zero current, E 0 = Standard cell potential, T = Absolute temperature (in K), R = Universal gas constant, n = number of charges, F = Faraday constant, Q = Ratio of ion concentration at the anode to ion concentration at the cathode, + and − are for cations and anions, respectively.
The biological function of a living cell and an enzyme can be modulated based on the electrode potential adjustment by implementing the use of polypyrrole molecular interface. Often additionally obtained discernment of electrochemical interfaces by employing ICPs as matrices for enzymes permits the use of such biosensors for scrutinizing natural samples, for example, flow injection perseverance of lactate in whole blood.92 Apart from the advantage of decent polymer layer formation, this method yields some bottlenecks, associated with a considerable electrochemical activity of ICPs because of the similarity in these materials toward redox equilibria and ion‐exchange process. It was reported in the year 1969 where an enzyme‐based potentiometric biosensor was established for sensing urea.88 Karyakin et al. investigated the use of economical and highly sensitive potentiometric pH transducer based on processible polyaniline for glucose detection where they acquired a maximum response value of ≈80 mV.100
Conductometric Biosensor: On the course of biochemical reaction, charges are produced that causes changes in the overall conductance or resistance of the analyte. Thus, conductometric biosensor provides analytical information by measuring the conductivity of analyte between two electrodes that varies with the concentration of ionic species. Conductometric biosensors, in most cases, are strongly associated with enzymes where the conductivity (as a result of ionic strength) of the analyte alters owing to the enzymatic reaction between two electrodes hence, they can be utilized to examine the enzymatic reaction that yield deviations in the concentration of charged species.93 But the applicability of this enzyme‐based conductometric biosensor is limited because of the mutable ionic background of diverse samples and also the necessity for evaluating minor variation of conductivity in media of different ionic strength.101, 102
Though conductometric biosensor has not been implemented extensively, they have found their successful application in the field of foodborne pathogen detection where Zarini et al. synthesized a rapid, versatile, specific, and highly sensitive polyaniline based conductometric biosensor where they found a lower detection limit of 7.9 × 101 colony forming units (CFU) per milliliter for Escherichia coli O157: H7 and Salmonella.103
3.2.2. Piezoelectric Biosensor
Sauerbrey, in the year 1959, depicted that a change in the resonance frequency of a thickness‐shear‐mode resonator can be directly related to mass placed on it.104 This was considered as the inaugurating idea for the construction of new generation piezoelectric mass‐sensitive appliance. Also, in the past few decades, immobilization of biomolecule onto the functionalized surface turned out to be the paramount application of this biosensor. They also allow for label‐free recognition of biomolecules however, they are considered a lot more than a mere mass biosensor because the response of biosensor is also swayed on the surface charge, viscoelastic attributes of biomolecule, surface roughness, and interfacial phenomena. These new insights have opened up the use of this biosensor in the adhesion of liposomes, proteins, and cells onto the surface thus allowing for determination in morphological changes in cells.105
Measurement in the piezoelectric biosensor is based on measuring variation in resonance frequency of an ICP‐coated piezoelectric crystal that relies on the minute mass changes on the crystal surface. The basic principle of this biosensor is the transmission of an electrical signal through an ICP‐coated piezoelectric crystal that causes vibration (as shown in Figure 5 ) at a certain resonant frequency which relies on its mass, size, shape, and chemical structure.2 The mass change, as a result of biomolecule adsorption on the electrode surface, is used for measuring the specific biological activity. These are brought about by the interaction between the analyte and immobilized biomolecule on the crystal surface. The vibration frequency of the oscillating crystal usually decreases when the analyte binds to bioreceptor on the surface. Such biosensors normally operate by propagation of acoustoelectric waves, either through the bulk or along the crystal surface.106
Figure 5.

A diagrammatic illustration of a piezoelectric biosensor.
Based on this, piezoelectric biosensors are of two types: surface acoustic wave and bulk wave. However, they have not received much attention and are compared inferior to the electrochemical biosensor.88 Such type of biosensor has been employed for the detection of hydrogen sulfide, carbon monoxide, ammonia, and also caffeine.107 A modern approach for the preparation of molecularly imprinted polypyrrole in combination with piezoelectric quartz crystal based caffeine sensor was prepared with a detection limit of 0.024 µm.108 A general decorum for immobilizing the human immunoglobulin G on polyaniline was utilized for the development of the piezoelectric immunosensor that was able of distinguishing the concentration of target analyte within 500 ng mL−1 and 25 µg mL−1.109
3.2.3. Calorimetric Biosensor
Even though they provide with weak sensitivity and unspecific heating, calorimetric biosensor has drawn considerable attention for the determination of biomolecule. This type of biosensor is developed for studying the enzyme reaction in reference to enzymatic conversion because most of the biochemical reaction have exothermic characteristics. In this context, the total heat absorbed or evolved during the biochemical reaction is proportional to the enthalpy and the amount of products formed upon reaction.110
| (2) |
| (3) |
where Q is total heat, np is total moles of product, ΔH is enthalpy change, and Cp is specific heat of solvent included system.
The variation in temperature is measured by thermistor is inversely proportional to Cp and directly to ΔH.
| (4) |
Here, augmented sensitivities and limit of detections in organic solvents are obtained provided the enthalpy change remains unaltered.111
So, calorimetric biosensor is based on the measurement of enthalpy changes (total heat produced) that occurs due to biochemical reaction that can be quantified and related to the amount of analyte, which is measured by thermistor as depicted in Figure 6 . Some major benefits of this type of biosensor are stability, possibility of miniaturization, and increased sensitivity.112 Additionally, they can be miniaturized effortlessly and incorporated with the microfluidics for augmenting the sensitivity. Most of the biochemical reactions are exothermic and have an enthalpy of ≈80 kJ mole−1.113
Figure 6.

Schematic representation of a calorimetric biosensor.
In real sample determination of glucose, interference is often encountered from an electro‐active substance in the blood. However, calorimetric biosensors are insensitive to electro‐active and optical interference that is present in clinical samples. Recently, Xie et al. combined the intrinsic advantages of pyrroloquinoline quinone glucose dehydrogenase (PQQGDH) and calorimetric biosensor that resulted in a 0.009 to 100 mm of linear range for glucose.114
3.2.4. Photometric Biosensor
Photometric biosensors are constructed on the basis of measuring the variation in optical properties of analyte owing to the biochemical reaction that occurs during the biological recognition event. Different optical properties such as absorption, fluorescence, bioluminescence, chemiluminescence, internal reflection, surface plasmon resonance can be exploited for monitoring the bio‐recognition event in biosensors.115 For example, a chemiluminescence‐based device measures the changes in the frequency of emission of light upon biochemical reaction which is caused by the origination of excited states that lasts for a very short duration.91 The biochemical reaction continuously alters the optical properties of the analyte causing light emission which is continuously monitored with the aid of optical fiber or by optical waveguide device.106, 116 In this type of biosensor, there is resilience in the measurement modes, for example, surface reflectance, evanescent wave, and embryonic technologies as surface plasmon resonance (SPR). Also, there is a drive toward the incorporation of some optical components (waveguides, detectors, source, and sampling region) onto the planar platform.117, 118
Figure 7 demonstrates the working of an optical/photometric biosensor. Recently, a novel doped polyaniline‐dioctyl sodium sulfosuccinate film coated on fiber bragg grating was utilized as an optical fiber‐based sensor for the detection of chloroform. This biosensor exhibited good recyclability of up to ten cycles with a swift response period of 7 s and a detection limit of 9.22 ppm.119 Another researcher, Angelica et al., utilized a copolymer of aniline and ortho‐phenylenediamine for the determination of the degree of freshness of tilapia (Orechromis niloticus) using colorimetric method. The optical sensor was sensitive to alkaline vapors such as volatile amines that generate during spoilage of fish and the sensor displayed changes in color as a function of pH.120
Figure 7.

Working of a photometric biosensor.
SPR is the most widely used optical biosensor which uses surface plasmon waves (SPW) for detecting changes upon interaction of target analyte and bio‐recognition element.121 Upon exposure of SPR to any alterations, it induces a change in refractive index upon biochemical reaction that is used to measure and observe the reaction. This change brings about variation in propagation constant of SPW that can be measured to produce a reading. The important feature of SPR is the label‐free recognition without the aid of fluorescence and radioactivity making it extremely appealing for real‐time monitoring. To date, SPR is being widely used in drug discovery, health science research, fundamental biological studies, clinical and environmental diagnosis.118
3.3. Sensing Mechanism of Different Analyte‐Based Biosensors
Based on the type of biomolecule utilized for immobilization, the mechanisms of enzymatic biosensor, immunosensor, and genosensor are being discussed further.
3.3.1. Enzymatic Biosensor
Enzymes, generally, are orbicular proteins consisting of chains of amino acid producing a 3D structure that acts as a catalyst which accelerates the biochemical reaction.122, 123 In the enzymatic reaction, the initial stage of reaction in which a molecule reacts with the active site of enzyme is known as a substrate. Upon biochemical reaction, the substrate is converted by the enzymes to form another distinct molecule known as products. The enzyme electrodes are formed by immobilizing a thin layer of enzyme onto the surface of the working electrode. Enzyme‐based biosensor operates particularly in a four‐step procedure (Figure 8 ):
The substrate in the analyte is diffused to the immobilized enzyme on the electrode from the bulk of solution
Reaction on the surface of the electrode occurs between the active site of enzyme and substrate
Upon enzymatic reaction, formation of product occurs which is then transported to the transducer
Measurement of the product on the ICP coated electrode surface with the aid of a transducer124
Figure 8.
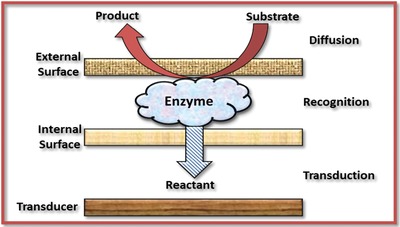
Configuration of an enzymatic biosensor.
3.3.2. Immunosensors
Immunosensor or antibody‐antigen biosensor is a compact analytical device that utilizes the immunochemical reaction of antibody (or antigen) as an immobilized element and antigen (or antibody) as the analyte, especially of body fluids, and also in disparate medias such as in groundwater for trinitrotoluene (TNT) quantification via TNT‐antibody complex formation.125
Antibodies (weighing 150 kDa), also known as immunoglobulin, are “Y” shaped complex protein molecule composed of two heavy chains and two light chains (Figure 9 a).126 An antibody, which is precise to a particular antigen, fits in a highly specific way to that antigen. This peculiar characteristic of antibodies is vital for their utilization in the immunosensors where specific antigen fits into the antibody binding site.88 The binding interaction between the antibody‐antigen (Figure 9b) is high and thus it is possible to recognize the analyte even in the existence of interfering species. Thus, biosensor utilizing antibodies as a bio‐receptor are being used extensively as it provides high specificity, versatility, and strong affinity (but of covalent nature) toward the target antigens. However, this high affinity can be a bottleneck as the antigen cannot be easily detached from antibody as a result of the formation of highly stable antibody‐antigen complex upon measurement being done, thus many immunosensors are considered to be for single‐use.127
Figure 9.
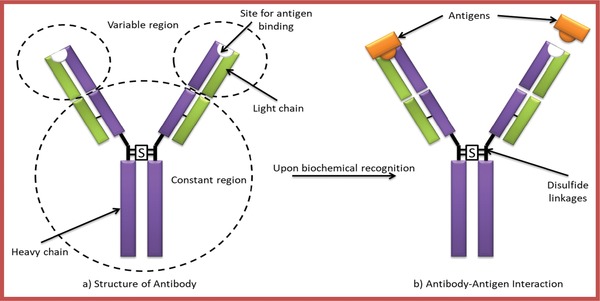
a) Structure of an antibody and b) Antibody–antigen interaction.
3.3.3. Genosensor/Nucleic Acid Sensor
In recent years, advancement in the technology of a genosensor has enticed great interest as a result of its importance in early infection diagnosis (and genetic diseases) and also in sequencing DNA.128 A genosensor is a biosensor capable of detecting individual nucleic acids comprising of a genome (or DNA) molecule. The nucleic acid immobilization on the ICP surface results in the formation of biosensor and the recognition of the conformational change in nucleic acid can be accomplished with the aid of different transduction principles.127
Basic sensing mechanism of DNA biosensor is based on the formation of high‐affinity binding among two single‐stranded DNA (ssDNA) chains resulting in the formation of double‐stranded DNA (dsDNA) helix which can be employed in DNA biosensing by utilizing ssDNA as a recognition element. The working principle of DNA biosensors is based on the determination of the complementary DNA by immobilization of ssDNA probe that creates hydrogen bonding among the two nucleic acids by the formation of adenine‐thymine and guanidine‐cytosine pairs (Figure 10 ). This exposure of the probe to target results in hybridization of both nucleic acid that causes biochemical reaction to occur, thus allowing the transducer to detect the signal.
Figure 10.

Schematic representation of double helix DNA along with base pairing.
3.4. Immobilization Techniques of Biomolecule Using Conductive Polymers
As the biomolecule, attached to the electrode surface, is being held exterior to their natural environmental conditions, it may decrease its biological activity, thus it is essential to preserve its integrity. Hence, immobilization of the biomolecule on to the solid interface is the central step for the advancement of a competent biosensor with appropriate performances such as acquiring high reproducibility, high sensitivity, high reliability, short response time, and good operational stability.129 Actually, the utilization of an immobilized biomolecule is preferred because it offers robustness, prolonged availability, ease of handling, and resistance to environmental variations.130 The activity of a biosensor relies on the materials characteristics (surface area, biocompatibility, electron transfer efficiency, porosity) and methods used for immobilization but eventually, the immobilized biomolecules must retain their structure, sensitivity, and biological activity and simultaneously, it should not decay or desorb over the use of biosensor.131, 132
However, some methods of immobilization may decrease the sensitivity of biosensor caused due to various issues such as conformational change and denaturation of biomolecule, lower efficacy in the presence of insoluble substrates, and mass transfer limitations.133 A better sensitivity can be obtained by appropriate orientation of enzymes (as shown in Figure 11 ) on the surface of the transducer that exposes the active site in the solution phase.134
Figure 11.

Schematic representation of the orientation of immobilized biomolecule on the surface.
Copious materials such as magnetic hydrogels, sol‐gels, resins, clays have been utilized for the immobilization of biomolecule but most of the mentioned materials have various limitations such as poor biocompatibility, difficult synthesis, poor controllability, and poor response properties.132, 135, 136, 137, 138 Thus, there is a substantial need for the development of biosensor electrodes that has improved compatibility with biomolecules and quick response time. ICPs provide an organized molecular structure on electrode surface that permits their utilization for immobilizing the active biomolecules because they function as a 3D matrix that preserves the activity of biomolecule for a prolonged period. This characteristic of ICP, along with their viability as a membrane, has furnished with new opportunities for exploring a biosensor.139 Simultaneously, they render an excellent platform for immobilizing biomolecule since they are known to offer durability, porosity, selectivity, excellent electron transfer rate, biocompatibility, and better signal transduction.140, 141 Integrating enzymes into the organic ICP matrix prevents the biomolecule from leaching out of the matrix while retaining the accessibility of catalytic active sites as a result of the permeability of films to analyte.142
In vivo biomedical application requires biocompatibility of the material that is being utilized for a specific purpose.143 Thus, immobilized biomolecule alongside with organic nature of ICP renders them biocompatible in neutral aqueous solution, which makes the possibility of in vivo application for continuous metabolites and drug monitoring in biological fluids.144, 145 Despite the copious benefits provided by ICP, they have stability and property issues that are affected by ambient conditions such as oxygen, redox reactant, acidic and basic media.146 Moreover, the physical and electronic characteristics of ICP are also a matter of concern for certain application. For example, in a bioelectrochemical application, it is essential to retain the conductivity of ICP at a pH value of above 4.147, 148 Also, it is obligatory to ponder the consequence of integration of counter ion as a dopant on pH stability of ICP during synthesis.149
Recently, researchers have demonstrated the benefit of various nanomaterial for the immobilization of biomolecule that aids in enhancing the stability and performance of immobilized biomolecule.150, 151, 152 Similar to ICP, which have ascertained to show good sensing characteristics, nanomaterial has also offered attractive feature providing with good electronic and surface area characteristics for further advancement of the biosensor. The physicochemical property of nanomaterial renders an ideal remedy to some conflicting disputes employed for the optimization of enzymes such as maximum surface area for effective enzyme loading, minimum diffusion limitation, along with the polymeric characteristics such as porosity, stability, and good conductivity.51 Thus, integration of nanomaterial into ICP matrix can generate a powerful ICP‐composite that offers numerous benefits such as tunable physical and chemical properties along with structural and functional flexibility which aids in immobilization of biomolecule.132
For the sake of higher stability and sensitivity of biosensor, intense research efforts have been executed for the advancement of successful approaches for immobilization. Various procedures, to date, have been developed for immobilizing biomolecules, viz., covalent attachment, physical adsorption, cross‐linking, and entrapment as shown in Figure 12 .
Figure 12.

Depiction of different approaches for immobilizing biomolecule and the reaction mechanism in a biomolecule (here, enzyme).
Physical adsorption represents one of the simplest methods for the immobilization of biomolecule on ICP matrix that effectively maintains the enzymatic activity under moderate experimental conditions. Here, the adsorption of biomolecule takes place as a result of the electrostatic interaction between ICP and biomolecule since they exhibit distinct surface charge property in accordance with their functional groups. Also, other non‐covalent interactions such as hydrophobic, van der Walls forces, hydrogen bonds also contribute to the adsorption process.133 This technique is dispensable of any functionalization of monomer/polymer or biomolecule and is noninvasive for the activity of enzyme. Though this process induces little enzyme inactivation, the relative weak electrostatic forces comprised in adsorption causes the biomolecule to leach out from the polymer matrix during a long experimental run. As the immobilization is limited to only one mono‐layer of ICP, very less amount of biomolecules are being incorporated. Also, there might be nonspecific adsorption of other biomolecular species or substance during this technique.
Covalent anchoring of biomolecule on ICP is considered to be one of the widely utilized chemical immobilization techniques where the biomolecule is covalently linked to ICP via the functional groups on the surface generating a robust immobilized biomolecule devoid of the problem of leaching. This type of biosensor usually exhibits magnificent stability during long experimental measurements because of covalent attachment on ICP surface.153, 154 The covalent bonding between ICP and biomolecule utilizes the chemistry of N‐hydroxysuccinimide (NHS)/1‐ethyl‐3‐(3‐dimethylaminopropyl) carbodiimide (EDC) for coupling carboxylic acid (—COOH) with an amine group (—NH2) on ICP and biomolecule, respectively.155 Recently, Sarac et al. utilized (—COOH) modified poly(m‐anthranilic acid) and (—NH2) modified DNA probe for covalent immobilization on poly(m‐anthranilic acid) that evokes reaction between (—COOH) and (—NH2) group forming amide linkages.156 They manifested good electrochemical activity in a broad range of pH, good processibility, and mechanical properties. Covalent immobilization may eliminate or either decrease some common difficulties such as diffusion, instability, or inactivation of biomolecule that occurs when the biomolecule is immobilized on the biosensor surface.157
Immobilization of biomolecule by cross‐linking on ICP matrix with bi‐functional or multi‐functional agents such as glutaraldehyde, hexamethylene diamine, glyoxal, toluene diisocyanates is another approach for the development of biosensor.158 This coupling agents generally cross‐links with phenolic groups of tyrosine, amino groups lysine, sulfhydryl groups of cysteine, or imidazole groups of histidine for enzyme binding.133, 157 Though this method offers short response time and strong chemical binding among biomolecules, there are many bottlenecks associated with this technique: i) it results in multilayer formation of biomolecules, resulting in decrease in activity; ii) it causes loss of biomolecules due to uncontrollable reaction; iii) it may also cause deterioration in activity as a result of distortion of active biomolecule conformation and chemical variation of active site during cross‐linking; iv) poor stability because of the biomolecule exposed directly to the solution; v) the layer of biomolecule is not rigid; vi) large diffusional barriers causing delay in interactions.159, 160 Toppare et al. synthesized a novel poly(6‐(4,7‐di(thiophen‐2‐yl)‐2H‐benzo[d][1,2,3]triazol‐2‐yl)hexan‐1‐amine) functional conductive polymer matrix for immobilization of choline oxidase with the aid of glutaraldehyde as a cross‐linking and bi‐functional agent. They achieved an overwhelmingly robust linkage among the polymer matrix and biomolecule with the aid of glutaraldehyde. Also, they optimized the amount of cross‐linker of up to 2.5% that is required for achieving ideal cross‐linking.161
Apart from the conventional procedures of immobilization, the coupling of biomolecule in electropolymerized films has sought significant importance due to its nonmanual procedure and reproducibility. Electrochemical entrapment is a one‐step method where an appropriate potential is applied to the transducer submerged in an aqueous solution of electropolymerizable monomer and buffer containing biomolecule forming polymer layers integrated with homogenously distributed biomolecules/bio probes.162 Near the vicinity of transducer surface, the biomolecules are incorporated into the growing polymer network by physical means. This simple one‐step immobilization technique does not affect the biomolecule activity since there is no chemical reaction involved between the biomolecule and in situ polymerized ICP. This process offers numerous benefits like reproducibility and potentiality of amalgamation of co‐enzymes or some another enzyme concurrently by directly embodying them into the solution.163 This well‐controlled technique for biomolecule immobilization is greatly significant in the construction of multi‐enzyme and micro‐sensor. Over conventional procedures, electrochemical entrapment offers numerous advantages such as one‐step facile fabrication, accurate control over film thickness, and greater reproducibility, however drawbacks that this procedure provides are possibility of blemishing the biomolecule because of the potential applied while polymerization, poor target approachability due to biomolecule incorporation amid bulk ICP film and the process is suitable only for water‐soluble monomer.164, 165 The most commonly used ICP is polypyrrole. Recently, Ganjali et al. constructed a novel and selective biosensor based on electropolymerized polypyrrole for the entrapment of cytochrome C aptamer on a screen printed electrode. It also exhibited a linear response over a concentration range of 10 pm to 1 nm and limit of detection 5 pm.166
4. Intrinsically Conductive Polymer Composites for Biosensing Application
For the past few years, profound research effort has been made in biosensor's field in the quest for exceptional design strategy of biosensors proficient to dispense superior analytical characteristics with regard to reliability, sensitivity, facile fabrication, selectivity, and cost‐effectiveness. These days, it is customary that the implementation of the biosensor will rely considerably on the impact levied by immobilization on biomolecules. In this context, exploiting the nanomaterials for the fabrication of biosensors constitutes for exciting approaches.
Composites of ICP utilize ICP as a matrix and a secondary component that can be an organic or inorganic material that includes gold, silver, carbon nanotubes, graphene, graphene oxide, etc. The main objective of developing composite material is to perceive novel diverse characteristics which are usually not observed in their singular counterparts.
4.1. Gold Based ICP Composites
Definite chemical and physical characteristics of gold nanoparticles (AuNPs) make them splendid candidates for the construction of biological sensors. Few of the common attributes of utilizing AuNPs for the fabrication of biosensor are mentioned:
Facile, straightforward, and green synthesis method167
Unique optoelectronic and electrochemical properties168
Exceptional biocompatibility with high surface‐to‐volume ratio169
Tunable properties of AuNPs by varying size, shape, and ambient microenvironment170
Good electrodeposition171 offering biosensing platform for multi‐functionalization with a broad range of biomolecules for selective determination of biological targets.
Due to each of the characteristics, AuNP has sought tremendous interest of scientists to develop unusual sensing strategies with augmented selectivity and sensitivity toward target analyte. With high surface energy and surface to volume ratio, AuNPs provide persistent immobilization of great amount of biomolecule preserving their biological activity along with direct and fast electron transfer during biochemical reaction between the biomolecule and electrode due to their unique conductive property thus acting as “electron wires.”172, 173, 174
Because of the suitable characteristics of ICP, viz., film thickness, charge transport, and permselective properties, electropolymerization is one of the most widely accepted protocols for manufacturing suitable immobilization matrix. Though biomolecule immobilization onto various ICPs matrix has acquired considerable attention as a result of these inimitable attributes, the primary constraint faced is a large number of monomer and biomolecule essential for immobilization. Thus, AuNPs in ICP matrix provides with increased surface area providing a high amount of area for biomolecule immobilization as shown in Figure 13 . Simultaneously, it also provides with superior electrochemical activity and avoids biomolecule leakage while permitting rapid diffusion of substrates and products utilizing less amount of biomolecule.
Figure 13.

High surface area of AuNP causing augmented biomolecule loading.
Au‐ICP nanocomposites have been used for determining numerous biological species, however, practical application of Au‐ICP composite was far from straightforward mainly due to poor solubility of ICPs and distinct optical properties of ICP and AuNPS. Thus, Hoonacker et al. synthesized a composite of encapsulated gold nanoparticles in water‐soluble poly(aniline‐2‐carboxylic acid) via electrostatic adsorption. They proposed the as‐synthesized composite for their use in biosensing because the individual counter‐parts revealed matching optical characteristics that permitted charge transfer to occur, upon light absorption, between them exhibiting a significant enhancement in photonic reactivity. The composite showed enhanced photonic reactivity toward the environmental stimuli such as redox, pH, or change in refractive index.175 Apart from this, the fact that this composite remained stable in aqueous solution allowed for their development in the field of biological science. One of the fascinating applications of Au‐ICP based nanocomposite in DNA sensing is for the detection of the gender of arowana fish (or dragon fish) on‐site which is necessary for optimal production of this cultured fish. So, Marugan et al. prepared a kappa‐carrageenan‐AuNP‐polypyrrole based DNA biosensor where they covalently anchored the ssDNA probe of the dragon fish on the composite surface.176 Since polypyrrole has fragile properties,177 kappa‐carrageenan was used for developing a mechanically strong ICP composite for biosensing. Genomic DNA samples collected from the scales of fish were subjected to DNA extraction and the resulting biosensor revealed wide response range, good stability, and low detection limit of 5 × 10−18 m. Mikolaj et al. demonstrated a smooth film of polypyrrole with non‐aggregated AuNPs for the detection of DNA. The modification of electrode with this composite layer allowed for the deposition of a higher number of DNA probes (up to two orders of magnitude) in comparison with the traditional thiol self‐assembled monolayers.178 De et al. fabricated a platform for biosensing of three different biomolecules viz., glucose, DNA, and protein by immobilizing glucose oxidase, single‐stranded DNA, and Lamin A antibody, respectively by using polyaniline nanowire decorated with gold nanoparticles. Method of detection and immobilization for each system was different and the biosensor displayed excellent specificity, higher sensitivity, greater stability, and low detection limit which proved the Au‐polyaniline composite suitable as general biosensor platform.179 Recently, Denkbas et al. produced a disposable, stable, and cost‐effective biosensor of self‐assembled peptide nanotube, AuNP, and polyaniline on pencil graphite electrode for recognizing prostate specific antigen (PSA). They found the detection limit to be 0.68 ng mL−1.180 The practicality of the biosensor was also shown by successfully applying the modified biosensor to blood serum samples for detecting PSA. Omidfar et al. fabricated an ultrasensitive sensing platform for detecting E. coli utilizing surface modified screen‐printed carbon electrode by first polyaniline film and then AuNP. In the next step, avidin, followed by biotinylated DNA probe was immobilized covalently on the modified electrode by avidin–biotin interaction.181 The described electrochemical assay was found to detect 4*106 to 4 CFU of E. coli and concluded that this geneosensor had potential for accurate and rapid diagnosis of E. coli imposed infections. Au‐ICP composites have also been widely used in medical diagnosis for the recognition of destructive cancer cells. Shim et al. designed a novel, facile, and biocompatible aptamer‐based nano‐biosensor for the detection of non‐small‐cell lung cancer (NSCLC) cells. The authors covalently immobilized MUC1 aptamer probe on the ICP composite synthesized by self‐assembly of 4‐([2,2′:5′,2″‐terthiophen]‐3′‐yl) benzoic acid layer on AuNPs and obtained a detection limit as low as 8 cell mL−1.182 In another research, they established an immunosensor for the recognition of hypoxia‐inducible factor1 alpha (HIF1α) tumor by covalently immobilizing anti‐HIF1α by using the same functionalized ICP with AuNPs and observed a limit of detection 5.35 ± 0.02 pm mL−1.183 Another researcher, Wang et al. proposed an electrochemical nanobiosensor for early determination of breast cancer by direct recognition of microRNA‐21 in clinical samples without extraction or amplification of RNA. They reported a label‐free self‐assembled polypyrrole‐gold nanocomposite biosensor with toluidine blue as a signal amplifier with a lower limit of detection of 78 am.184 Self‐assembly of polypyrrole coated gold nanoparticles formed a superlattice that exhibited the close‐packed type, thereby generating maximum current. Brain‐derived neurotrophic factor (or BDNF) is a neurotrophin that is being involved in major psychiatric and neurological disorders.185 So recently, Chandra et al. synthesized a micro‐fluidic immunosensor composed of gold nanoparticle, functionalized polyterthiophene based ICP, and an anti‐BDNF probe immobilized on composite layer for detecting BDNF from the extracellular matrix of neuronal cells.186 The immunosensor showed a dynamic linear range with limit of detection 1.5 ± 0.012 pg mL−1.
Gold based ICP composites have also been utilized in the biological fluids for the simultaneous detection of co‐existing compounds. Folic acid (water‐soluble vitamin) and dopamine (a neurotransmitter) play a vital part in human metabolism and central nervous system. Stephen et al. fabricated a poly(o‐methoxyaniline)‐Au based nanocomposites by in situ chemical oxidative polymerization for novel simultaneous determination of folic acid and dopamine. The sensor was stable, precise, and sensitive to dopamine and folic acid in pharmaceutical and human urine samples and the limit of detection was obtained 0.062 and 0.090 µm, respectively.187 Baoxian et al. investigated on simultaneous detection of uric acid (2 × 10−7 mol L−1 detection limit) and epinephrine (8*10−8 mol L−1 detection limit) in human blood serum by depositing AuNP/polyaniline film prepared by Langmuir–Blodgett technology that showed high stability, high sensitivity, and wide linear range.188
ICP‐based hydrogels have been acquiring the interest of scientists189, 190, 191 as a matrix for biosensor owing to its 3D network structure that can accumulate both, the nanoparticle as well as the biomolecule and short charge diffusion pathway. This hydrogels of ICP not only maintain the unique characteristics of ICP but simultaneously retain the nanomaterials attributes such as a 3D perpetual conducting network and large surface area that enhances the benefits of ICP hydrogels for designing novel biosensor. So, Zhangfang et al. utilized the gains of ICP hydrogels and synthesized a network composite of polypyrrole hydrogel and AuNPs for the construction of an amperometric label‐free immunosensor by utilizing carcinoembryonic antigen (CEA) as a model analyte (Figure 14 ).192 The resulting biosensor exhibited a good selectivity, broad linear range, and ultralow limit of detection of 0.16 fg mL−1.
Figure 14.

Illustration of an immunoassay protocol.
A consolidated data of various biosensors based on gold particles ICP composites are presented in Table 2 .
Table 2.
Characteristics of several gold‐based ICP composite biosensors
| ICP | Nanomaterial | Target analyte | Linear range | Detection limit | Ref. |
|---|---|---|---|---|---|
| Poly (o‐methoxyaniline) | AuNP |
Dopamine folic acid |
10.0–300.0 µm 0.5–900.0 µm |
0.062 µm 0.090 µm |
187 |
| Polyaniline | AuNP‐peptide nanotube |
Prostate specific antigen |
1–100 ng mL−1 | 0.68 ng mL−1 | 180 |
| Polyaniline | AuNP | E. coli | — | 4*106 to 4 CFU | 181 |
| 4‐([2,2′:5′,2″‐terthiophen]‐3′‐yl) benzoic acid | AuNP | Non‐small‐cell lung cancer cells | 15 to 1 × 106 cells mL−1 | 8 cells mL−1 | 182 |
| [2,2:5,2‐terthiophene‐3‐(p‐benzoic acid)] | AuNP | Hypoxia inducible factor 1 alpha (HIF1α) | 25–350 Pm mL−1 | 5.35 ± 0.02 pm mL−1 | 183 |
| Polypyrrole | AuNP+TB | MicroRNA‐21 | 100 am–1 nm | 78 am | 184 |
| Functionalized Polyterthiophene | AuNP | Brain‐derived neutrophic factor | 4.0 to 600.0 pg mL−1 | 1.5 ± 0.012 pg mL−1 | 186 |
| Polyaniline nanowire | AuNP |
Glucose ssDNA strand Lamin A protein |
1–20 mm | 1 µm | 179 |
| Polypyrrole | Kappa‐carrageen‐AuNP | DNA of fish | 5 × 10−18 to 5 × 10−12 m | 5 × 10−18 m | 176 |
| Polypyrrole | AuNP | DNA | 2 × 10−13 to 2 × 10−6 m | 8.4 × 10−13 m | 178 |
| Polyaniline | AuNP |
Uric acid epinephrine |
4.0 × 10−7 to 6 × 10−5 mol L−1 4.0 × 10−7 to 1 × 10−5 mol L−1 |
2 × 10−7 mol L−1 8 × 10−8 mol L−1 |
188 |
| Polypyrrole Hydrogel | AuNP | Carcinoembryonic antigen | 1 fg mL−1 to 200 ng mL−1 | 0.16 fg mL−1 | 192 |
| Polypyrrole | Luminol functional‐AuNP | Carcinoembryonic antigen | 0.01 pg mL−1 to 10 ng mL−1 | 3 fg mL−1 | 193 |
Abbreviations: Gold nanoparticle (AuNP), deoxyribonucleic acid (DNA), single‐stranded DNA (ssDNA), Escherichia coli (E. coli), toluidine blue (TB).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
4.2. Carbon Nanotube Based ICP Composites
Ever since their discovery,194 CNT has become a vast research activity globally considering their exceptional mechanical, electrochemical, and electrical attributes along with ultra‐high surface area. CNTs basically are of two forms: single‐walled (SWCNT) and multi‐walled (MWCNT) that consists of two or more cylindrical concentric shells of graphite sheets. One of the important attributes is the helicity of honeycomb of carbon in respect to tube axes.194 Also, it is well prominent that the characteristics of CNTs are sensitive to be influenced by their exposure to biomolecules that has led to their exploration as a sensing element for biological sensor.195, 196, 197 Considering their exceptional surface‐to‐volume ratio, wide electrochemical window, efficient electron transfer, and large specific surface area, they have been renowned as an ideal material for the fabrication of biosensor since it is conceivable to attain ultra‐fast detection of biomolecule at very low concentrations.198, 199 Hence, CNT‐based biosensors are one of the best contenders for ultra‐sensitive sensing methodology. Though CNTs provide with copious benefits, some limitations such as poor processibility hinder their application because of the randomly lying CNTs on the electrode surface that assist in easy peel‐off thus depressing the reproducibility of biosensor.200 Thus many methodologies such as multi‐functionalization and composite formation with ICPs have been implemented to make CNTs processable and minimize the agglomeration. It has also been reported that wrapping CNTs with conjugated polymers render them processable. Briefly, CNTs exhibit π‐conjugated structure along with a highly hydrophobic surface that allows their interaction with the aromatic ICPs via π–π and hydrophobic interactions that form new structures known as composites that can be utilized for biosensing.201, 202
One of the common fields of CNT‐based biosensor is in glucose level monitoring for diagnosing diabetes203 where Kang et al. utilized chitosan‐coupled CNT/polyaniline composite for the detection of glucose. Polyaniline layer was prepared by oxidative graft polymerization on which chitosan coupled CNT were covalently anchored with ICP layer with the aid of glutaraldehyde as a bi‐functional linker. Pendant hydroxyl groups of chitosan were used for covalently immobilizing glucose oxidase by using a bifunctional linker. The electrode was able to provide biocompatible environment due to the 3D network structure of the composite electrode that enhanced the biocatalytic activity of the enzyme.204 Basu et al. developed a novel bi‐enzymatic (horseradish peroxidase and glucose oxidase) glucose biosensor by utilizing polypyrrole and carboxy modified multi‐walled CNT to form composite where they observed superior performance with respect to sensitivity, longer shelf‐life, and linearity compared to single enzyme biosensor. Thus, the enhanced performances of the biosensor are attributed to the presence of horseradish peroxidase along with glucose oxidase that enhances overall biochemical reaction.205 They also observed that the sensitivity of the bi‐enzymatic electrode is three times as good as mono‐enzymatic. A comparison of mono and bi‐enzymatic type biosensor is shown in Table 3 .205
Table 3.
Comparative performance evaluation of mono and bi‐enzymatic biosensors
| Sr. No. | Characteristics | GOx/MWCNT/PPy | GOx‐HRP/MWCNT/PPy |
|---|---|---|---|
| 1 | Detection limit | 0.3 mm | 0.1 mm |
| 2 | Linear range | 1–6 mm | 1–10 mm |
| 3 | Sensitivity | 4.4 µA mm −1 | 13.8 µA mm −1 |
| 4 | Response time | 15 s | 10 s |
| 5 | Shelf life | 2.5 weeks | 5 weeks |
| 6 | K m | 0.52 mA mm −1 | 0.42 mA mm −1 |
Adapted with permission.205 Copyright 2012, John Wiley and Sons.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Lihua et al. utilized poly(o‐aminophenol)‐CNT composite based biosensor by immobilizing glucose oxidase. The enzyme was immobilized by copolymerization of glucose oxidase, o‐aminophenol, and CNT at the surface of the electrode in weakly acidic medium. They equated the two biosensors with and without CNT where they found that CNT based electrode exhibited twice higher sensitivity, 1.5 times response current, and twice lower limit of detection along with fast amperometric response, good anti‐interferent ability, good stability, and excellent reproducibility.206
Hydrogen peroxide (H2O2) is the product that is formed in the course of the enzymatic reaction during biosensing so, Xia et al. initially studied the biosensing of hydrogen peroxide by utilizing polyvinylpyridine/CNT/Prussian blue‐based composite based on synergy of the electrode that showed a good detection limit of 25 nm.97 They extended this strategy for the determination of glucose by recognizing the hydrogen peroxide formed during the enzymatic reaction between glucose and immobilized glucose oxidase. Another researcher utilized cytochrome c‐immobilized on polyaniline/carboxylated MWCNT for detecting hydrogen peroxide at trace level.207 The modified electrode was sensitive to H2O2 and showed a good limit of detection of 0.2 µmol L−1.
Usually, kidney dysfunction is caused due to increased concentration of urea in blood and decrease in the level of urine. Thus, urea determination in blood samples is of great interest by the medical field. So, Hassan et al. developed a unique technique for continuous and direct monitoring of urea in blood samples where they utilized poly (o‐toluidine)/CNT nanocomposite by immobilizing urease enzyme on the composite film.208 The working electrode was modified by electropolymerization and in situ preparation where the latter displayed good electrochemical response with a detection limit of 0.03 mm for urea in blood samples. Shim et al. developed an amperometric l‐lactate biosensor because it is necessary to sustain the concentration of l‐lactate in human blood which otherwise may cause clinical disorders like hypoxia and acute heart disease.209, 210 They fabricated the biosensor using functionalized carboxylic acid‐based polythiophene and multi‐walled CNT on which lactate dehydrogenase and nicotinamide adenine dinucleotide (NAD) were immobilized subsequently. Immobilization occurred among the acid group of composite and amine group of enzyme by covalent bond formation.211 The resulting biosensor provided enormous active sites and displayed good response range of 5–90 µm and detection limit around 1 µm and was demonstrated successfully in milk and human serum samples. Another researcher synthesized the same biosensor using poly(3‐methylthiophene)/MWCNT based composite on which lactate dehydrogenase was immobilized. Under optimized conditions, the resulting biosensor exhibited over 1 × 10−6 m to 5 × 10−4 m concentration range with 5.6 × 10−7 m detection limit.212
Estimation of cholesterol or triglyceride in blood is extremely important since its excess concentration may cause hyperlipidemia.213 So, a nanocomposite consisting of polyaniline and single‐walled CNT was fabricated by Malhotra et al. by co‐immobilizing lipase and glycerol dehydrogenase and their application in triglyceride sensing was explored. A good storage stability of 13 weeks was observed with a good response time of 12 s.214 In their other work, they fabricated nanocomposite electrode for cholesterol biosensing comprised of polypyrrole and carboxy functionalized MWCNT. Cholesterol esterase and oxidase were immobilized on the composite surface for the recognition of total cholesterol since most of the research work comprises of detecting 30% free cholesterol.215 The proposed biosensor exhibited short response time (9 s) and 0.04 mm L−1 of limit of detection.
In another work, polypyrrole and carboxylic‐based MWCNT based biosensor were synthesized for the determination of DNA. Such COOH modification of CNT is necessary for providing a linkage between the amino group of ssDNA and (—COOH) group of MWCNT that assists in covalent immobilization. Due to the synergetic attributes, both the selectivity and sensitivity were improved and the complementary DNA sequence of 5.0 × 10−12 mol L−1 was detected by the proposed biosensor.200 Gonorrhea is another disease that is transmitted sexually and is also identified as a co‐factor in HIV transmission thus it is an important purpose for timely and accurate detection of gonorrhea.216 Thus, Malhotra et al. used polyaniline/CNT composite for the detection of Neisseria Gonnorhoeae (NG) by immobilizing its bio probe using glutaraldehyde as a cross‐linker. This bioelectrode exhibited good response time of 60 s and 75 days stability under refrigerated conditions with low limit of detection 1.2 × 10−17 m and displayed negative response to non‐NG species and also to gram‐negative bacteria.217 Shim et al. established an amperometric immunosensor for detecting Immunoglobulin G (IgG) by covalent immobilization of anti‐IgG on MWCNT‐embedded carboxylic acid‐based polyterthiophene composite. They applied another hydrazine‐labeled secondary antibody conjugate for the purpose of reducing H2O2. The proposed immunosensor presented a linear range (0.1 to 10 ng mL−1) with low limit of detection (0.084 ± 0.004 ng mL−1) and was applied for evaluation in rabbit serum sample.218
Characteristics of various biosensors based on carbon nanotube ICP are outlined in Table 4 .
Table 4.
Characteristics of several carbon nanotube‐based ICP composite biosensors
| ICP | Nanomaterial | Target analyte | Linear range | Detection limit | Ref. |
|---|---|---|---|---|---|
| POT | CNT | Urea | 0.1–11 mm | 0.03 mm | 208 |
| Poly(o‐aminophenol) | CNT | Glucose | Upto 5 mm | 0.01 mm | 206 |
| Polyaniline | Chitosan‐CNT | Glucose | 1–20 mm | 0.1 mm | 204 |
| Polypyrrole | cMWCNT | Glucose | 1–10 mm | 0.1 mm | 205 |
| Poly‐5,2′‐5′,2′′‐terthiophene‐3′‐carboxylic acid | MWCNT | Glucose | 5–90 µm | 1 µm | 211 |
| Polyaniline | SWCNT | Triglyceride | 50 to 400 mg dL−1 | ‐ | 214 |
| PVP | MWCNT | Glucose | 10–700 µm | 2 µm | 97 |
| Polyaniline | cMWCNT | H2O2 | 2 to 600 µmol L−1 | 0.2 µmol L−1 | 207 |
| Poly (3‐methylthiophene) | MWCNT | Lactate | 1 × 10−6 to 5 × 10−4 m | 5.6 × 10−7 m | 212 |
| Polyaniline | CNT | Nisseria Gonnorhoeae | 1 × 10−6 to 5 × 10−17 m | 1.2 × 10−17 m | 217 |
| Polypyrrole | cMWCNT | Cholesterol | 4 × 10−4 to 6.5 × 10−3 m L−1 | 0.04 mm L−1 | 215 |
| poly‐5,2′:5′′,2′′‐terthiophene‐3′‐carboxylic acid | MWCNT | IgG | 0.1 to 10 ng mL−1 | 0.084 ± 0.004 ng mL−1 | 218 |
| Polyaniline | MWCNT | Pesticide | 10 to 50 nmol L−1 | 5 nmol l−1 | 219 |
| Polypyrrole | cMWCNT | DNA | — | 5.0 × 10−12 mol L−1 | 200 |
Abbreviations: Carbon nanotubes (CNTs), single‐walled CNT (SWCNT), multiwalled CNT (MWCNT), carboxy functionalized MWCNT (cMWCNT), immunoglobulin G (IgG), poly(o‐toluidine) (POT), polyvinylpyridine (PVP).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
4.3. Graphene and Their Derivatives Based ICP Composites
Graphene (GR), a 2D honeycomb lattice with an atomic thickness of sp2 bonded carbon atom, is a monolayer of graphite that has sought considerable attention as a result of its optical, mechanical, electrical, thermal, electrochemical, and sensing properties.220, 221 GR structure caters with multitudinous attributes including excellent electrical and thermal conductivity, high mechanical strength, large specific surface area, tunable band gap, high electron mobility, and room‐temperature hall effect.221, 222, 223 Principally, the structure of GR and performance benefits such as high charge mobility and surface‐to‐volume ratio can be elucidated into immensely sensitive sensing application.224 The conjugated π‐electrons in GR sheets are delocalized that provides ballistic charge transport throughout the sheet with trifling optical absorption and based on these exceptional attributes, they have sought application in composites, aerospace, solar cells, supercapacitors, sensors, and biosensors.225, 226, 227 Unlike the tubular structure of CNTs, GR is a 2D sheet with open structure thus both the sides of GR sheets can be employed for supporting biomolecules.228 However, it has been found that the 2D graphene layers, owing to their intrinsic van der Walls interactions, results in restacking to form graphite or cause irreversible agglomeration. Thus, this drawback can be overcome by the addition of other macromolecule or by the use of polymers.229, 230, 231, 232 Unique attributes of ICP and GR in coalition have attracted the interest of researchers in recent times as a result of the creation of new composite material for its application in next‐generation biosensor devices. This GR‐based ICP composite provides with biomolecule loading platform and amplify or harness the adsorption, redox reaction, catalytic reaction, and transport behavior of the system. ICPs act as conducting conduits that interact with GR and analytes and enhance sensitivity, selectivity, response time, lower detection limit, and ultimately give rise to extraordinary biosensors as the grafted ICP with GR alters the in‐plane characteristics of composite materials.233, 234, 235 As GR‐sheets incorporated ICP matrix have poor adhesion between them, numerous approaches have been developed to boost the adhesion characteristics of GR by modifying their structure. Also, there exists chemical inertness of GR that makes it challenging to attain multi‐functional and altered hybrid nanomaterials. Thus chemically modified GR such as graphene oxide (GO) and reduced graphene oxide (rGO) have been developed for multi‐functionalization. For instance, covalent immobilization of the biomolecule, defective plane, and rich edge reaction sites contribute to the recognition of target analyte.236, 237 Various composites of GR and chemically modified GR and their applications in various biosensors are shown in Figure 15 .
Figure 15.

Illustration of development of graphene‐based ICP composite biosensors.
Ho et al. prepared a novel composite containing reduced GO/PEDOT nanotubes for electrochemical biosensing of hydrazine and hydrogen peroxide where they observed noticeable improvement in electrochemical activity compared to pristine PEDOT and rGO electrode. The proposed biosensor manifested good selectivity and sensitivity with limit of detection of 2.2 µm for hydrazine and 3.2 µm for hydrogen peroxide.238 Another researcher, Wei et al. depicted large‐scale one‐step electrochemical synthesis of layered GR/polyaniline composite for the recognition of hydrogen peroxide by the reduction process. Horseradish peroxidase was immobilized on this prepared composite on the electrode that showed a good limit of detection 0.8 × 10−7 m.239 Pilan et al. presented an efficient path for the advancement of a biosensing platform based on sulfonated GR/polypyrrole nanocomposite for the detection of glucose. The nanocomposite surface was modified with subsequent carboxyphenyl groups for immobilizing glucose oxidase. The proposed biosensor enabled good selectivity toward glucose in the existence of the interferents (paracetamol, uric acid, ascorbic acid, cysteine) because of the permselective characteristics of grafted carboxyphenyl and overoxidized polypyrrole layer, which eliminated the need of additional membranes that simplified the biosensor fabrication.240 Hanongbua et al. constructed a MIP by co‐electropolymerization of o‐phenylenediamine and aminophenyl boronic acid. This MIP was constructed on a GR‐PEDOT:PSS composite modified electrode for determining salbutamol. The aim of the use of composite, here, was to boost the current response to improve the sensitivity of the current which showed exceptional detection limit of 100 pm with a wide linear range of 1 nm–1.2 µm.241 The biosensor displayed good reusability of at least ten repetitive measurements without any substantial loss of electrochemical response and it revealed good recovery results for tangible sample analysis (real swine meat) without the need of pretreatment steps.
Levofloxacin is an antibiotic that belongs to the quinolone group antibiotic used for the treatment of various bacterial infections. However, rampant frequent consumption of levofloxacin leads to diseases such as tendon injury, heart disease, and muscle wasting.242 So, Feng et al. cultivated a sensitive and selective levofloxacin sensor with the aid of novel poly(sulfanilic acid)‐rGO composite film. Based on the synergetic effect, the proposed biosensor exhibited excellent selectivity and electrocatalytic activity, broad linear range, and low limit of detection (0.12 µm).243 The constructed sensor also showed good reproducibility and reliability and was successfully applied in real urine samples of a human for the detection of levofloxacin. Shasheng et al. validated a DNA biosensor based on oxidized GR and polyaniline nanowires by immobilizing the complementary DNA sequence. The resulting composite exhibited a good electrochemical behavior toward this complementary DNA with limit of detection as low as 3.25 × 10−13 mol L−1 and good storage stability.244 Nasrabadi et al. anticipated a label‐free and enzyme‐free electrochemical immunosensor constructed by poly p‐phenylene diamine (PPD) and GR nanocomposite for the recognition of neuron‐specific enolase biomarker in human serum.245 The proposed non‐enzymatic biosensor not only increased the stability, but also the sensitivity with a broad linear range and limit of detection as low as 0.3 ng mL−1 for biomarker detection.
Dopamine is a neurotransmitter that causes neurological disorders if there are abnormalities present in the level of dopamine.246 The significant problem faced while its determination is the interference caused by ascorbic and uric acid that co‐exist in biological samples. Thus, Jadranka et al. fabricated a highly selective PEDOT modified laser scribed graphene biosensor for the recognition of dopamine in the occurrence of these interfering compounds. This electrode produced by direct laser scribing on sheets of polyimide followed by electrodeposition of PEDOT showed 3D porous morphology with excellent electrochemical characteristics that exhibited a limit of detection 0.33 µm.247 Luo et al. fabricated PEDOT/GO nanocomposite for selectively determining dopamine free from common interference such as uric and ascorbic acid.246 Electrodeposition of nanocomposite on the electrode surface was performed, followed by electrochemical reduction to procure reduced nanocomposite‐modified electrode that displayed excellent electrocatalytic activity toward dopamine oxidation.
Pereira et al. validated a facile electrochemical technique for recognizing uric acid in human urine by immobilizing uricase on the polypyrrole/uricase/GR composite. The suggested biosensor presented good reproducibility, stability, and considerable values of its wide linear range (0.002 to 0.024 µmol L−1) along with low limit of detection (0.541 nmol L−1).248 Weilu et al. reported the usage of MIP/reduced GO composite for the concurrent recognition of tyrosine and uric acid. The MIP layer was electropolymerized by using 2‐amino‐5‐mercapto‐1,3,4‐thiadiazole monomer on the electrode modified by rGO and by immobilizing the dual template of uric acid and tyrosine. They explained the sensing mechanism of biosensor by first the recognition of target molecule and thereafter catalyzation of the oxidation reaction on the composite of MIP/rGO by evaluating the electrochemical behavior of tyrosine and uric acid.249 Under optimal conditions, the resulting nanocomposite exhibited enhanced electron transfer rate, more recognition binding sites, low limit of detection for tyrosine (0.046 µm) and uric acid (0.0032 µm), and wide linear range.
l‐lysine is one of the eight crucial amino acids required by the human body, however, they are the most endangered amino acid that can be damaged easily. Thus upon storage of food for a longer duration, it is essential to determine the nutritional value of food. So, Esma et al. synthesized an amperometric biosensor for determining l‐lysine by immobilizing l‐lysine‐α‐oxidase (LyOx) on GR/poly (vinyl ferrocene). The resulting biosensor exhibited long term stability due to the suitable microenvironment provided by the biosensor that helps in preserving the bioactivity of LyOx.250 The presented biosensor showed good analytical properties such as good sensitivity, low limit of detection (2.3 × 10−7 m), and short response time (<5 s). Wen et al. constructed an amperometric organophosphate pesticide biosensor by immobilizing acetylcholinesterase (AChE) on GR/polyaniline composite.251 The obtained composite showed a large specific area, high conductivity, increase in surface loading of AChE, and have perfect encapsulated and layered structures with detection limit of 20 ng L−1.
Characteristics of various biosensors based on graphene and their derivative based‐ICP are presented in Table 5 .
Table 5.
Attributes of several graphene and their derivative‐based ICP composite biosensor
| ICP | Nanomaterial | Target analyte | Linear range | Detection limit | Ref. |
|---|---|---|---|---|---|
| PEDOT:PSS | GR+MIP | Salbutamol | 1 nm–1.2 µm | 100 pm | 241 |
| Poly p‐phenylene diamine | GR | Neuron‐specific enolase biomarker | 1.0–1000 ng mL−1 | 0.3 ng mL−1 | 245 |
| PEDOT | Laser scribbed GR | Dopamine | 1–150 µm | 0.33 µm | 247 |
| Polypyrrole | Sulfonated GR | Glucose | 0.02–12 mm | 0.02 mm | 240 |
| Polypyrrole | GR | Uric acid | 0.002 to 0.024 µmol L−1 | 0.541nmol L−1 | 248 |
| Poly 2‐amino‐5‐mercapto‐1,3,4‐thiadiazole | rGO |
Tyrosine uric acid |
0.1–400 μm 0.01–100 μm |
0.046– 0.0032 µm |
249 |
| PEDOT | rGO | Dopamine | 0.1–175 µm | 39 nm | 246 |
| Poly(sulfanilic acid) | rGO | Levofloxacin |
2.0–30.0 and 30.0–500.0 µm |
0.12 µm | 243 |
| Polyaniline | GR | Organophosphate pesticide | ‐ | 20 ng L−1 | 251 |
| PEDOT | GO |
Hydrazine H2O2 |
0.1–1.9 mm 0.2–3.2 mm |
2.2 µm 3.2 µm |
238 |
| Poly (vinylferrocene) | GR | l‐lysine | 9.9 × 10−7 – 3.1 × 10−4 m | 2.3 × 10−7 m | 250 |
| Polyaniline | GR | H2O2 | 1.0 × 10−6 to 1.6 × 10−4 m | 0.8 × 10−7 m | 239 |
| Polyaniline Nanowires | GO | DNA |
2.12 × 10−6 to 2.12 × 10−12 mol L−1 |
3.25 × 10−13 mol L−1 | 244 |
| Poly(ionic liquids) functionalized Polypyrrole | GO | Dopamine | 4–18 μm | 73.3 nm | 252 |
| Poly (o‐anisidine) | GR | NADH | – | 1.3 μm | 253 |
| Polyvinyl pyrrolidone/Polyaniline | GR | Cholesterol | 50 μm–10 mm | 1 μm | 254 |
Abbreviation: Graphene (GR), GR oxide (GO), reduced GR oxide (rGO), molecularly imprinted polymer (MIP), poly(3,4‐ethylene dioxythiophene) (PEDOT), PEDOT:polystyrene sulfonate (PEDOT:PSS), dihydronicotinamide adenine dinucleotide (NADH).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
4.4. Hybrid ICP Composites
Since the nanomaterials discussed are having one or the other bottlenecks either during biosensors fabrication or at the time of sensing, it is necessary to fabricate hybrid ICP composites using more than one nanomaterial. So, different combinations of most of the existing nanomaterials have been tried, of which most of the amalgamations are with CNTs and GR‐based materials. Sheikhha et al. described the synthesis of a stable enzyme‐free poly (catechol)/GR sheets/biosynthesized AuNP for the recognition of acute lymphoblastic leukemia (ALL) with linear range of measurement of 100 µm to 10 pm and ultralow detection limit of 1 pm.255 Graphene in combination with ICP created a surface with superior electrical conductivity that resulted in a selective and sensitive biosensor and AuNP provided high affinity toward DNA for immobilization. Mucin 1 is a type of protein biomarker that aids in the detection of breast cancer thus Prabhakar et al. produced an aptasensor using PEDOT/GO/AuNP composite for the detection of mucin 1. The aptasensor displayed good reusability (eight times), stability (14 days), and a low limit of detection (0.031 fm) and it was applied in spiked human serum samples showing 85–93% recovery.256 Aflatoxins B1 (AfB1) is another mycotoxin that are considered to be hepatocarcinogenic so Raju et al. developed a biosensing platform of PEDOT/GO composite decorated with AuNPs for electrochemical detection of AfB1.257 The reason for the decoration of AuNP on GO was to augment the sensitivity of the electrode as GO is known to exhibit low sensitivity as a result of its poor conductivity.258 Also, AuNP aids in better immobilization thereby providing a benign microenvironment that retains their activity and prevents them from leaching. The resulting PEDOT/GO/AuNP composite‐based biosensor exhibited excellent sensitivity within linear range of 0.5–20 ng mL−1. Another group of researchers synthesized immunosensor for the detection of same mycotoxins using 2,5‐di‐(2‐thienyl)‐1‐pyrrole‐1‐(p‐benzoic acid)/GO/AuNP/ionic liquid composite.259 A native microenvironment is produced for the antibody because of the integration of ionic liquid that resulted in higher stability, sensitivity, reproducibility, and low detection limit of 1 fm and dynamic range of 3.2 fm–0.32 pm.
Quantitative determination of glucose is of great importance because of their increasing demand in diabetes diagnosis, food industry, and biotechnology. So Luo et al. reported a two‐step electrochemical strategy for synthesizing PEDOT/RGO/nickel nanoparticles for the construction of non‐enzymatic glucose biosensor. Nickel‐based materials have emerged as an electrode active material for non‐enzymatic glucose biosensor since they offer environmental benignity, low cost, and excellent electrocatalytic activity.260 The glucose sensor, under optimum conditions, exhibited linear range and low limit of detection 0.8 µm. Fisher et al. presented a scalable nanostructured biosensor on the basis of multilayered graphene petal nanosheets (MGPNs)/platinum nanoparticle (PtNPs)/PEDOT composite and glucose oxidase as a biorecognition element. The sensor platform was created with the combination of 0D NPs with 2D support arrayed in third dimension with outstanding characteristics.261 The work enabled the robust design of biosensor that exhibited exceptional performance with enhanced sensitivity toward glucose with 0.3 µm limit of detection. The performance of the glucose biosensor was exemplary with persistent glucose sensitivity after 5 weeks usage and the performance was benchmarked against the different collection of glucose biosensor. For enzymatic glucose detection, Tang et al. fabricated a glucose sensor based on layer‐by‐layer assemblage of polyaniline/MWCNT/AuNP composite film on which glucose oxidase was immobilized. The resulting biosensor retained its 92% of initial activity after a period of 30 days and showed low detection limit of 0.19 µm.262 Recently, traditional metal oxides, copper oxide, have drawn huge attention because it offers many benefits to biosensor as good stability, facile synthesis, and remarkable redox process at various potential ranges under certain reaction conditions.263 Alias et al. synthesized nanocomposite of copper oxide/RGO/polypyrrole for their application in glucose detection where the ensuing biosensor revealed low limit of detection at 0.03 µm.264
Tanver et al. synthesized sensitive amperometric biosensor for selective monitoring of K+ induced dopamine liberated from dopaminergic cells based on immobilization of ethylenediaminetetraacetic acid (EDTA) on poly(1,5‐diaminonaphthalene) layer comprised of GO/AuNP. They optimized the experimental variables and found limit of detection 5 nm and the resulting biosensor was applied for the surveillance of released dopamine from dopaminergic cells.265 Decha et al. fabricated a one‐pot facile and green approach for the synthesis of PEDOT:PSS/AuNPs/graphene nanocomposites for selective determination of uric acid and dopamine in the co‐occurrence of ascorbic acid.236 The composite was prepared by initially electro‐exfoliating graphene in polystyrene sulfonate (PSS) followed by in situ formation of AuNP and PEDOT. The as‐synthesized dispersion of PEDOT:PSS‐AuNP‐graphene was used for the fabrication of biosensor on the electrode surface by drop casting and the resulting biosensor unveiled linear response of (1 nm–300 mm) and (10–1 mm) with low limit of detection 100 pm and 10 mm of dopamine and uric acid, respectively and was tested in real pharmaceutical and human serum samples. Baohe et al. determined simultaneous detection of dopamine, serotonin, and tryptophan by electrodeposition synthesis of novel PEDOT/CNT/nickel oxide composite.237 The composite was found to be 1D co‐axial structured nanotube of CNT and nickel oxide/PEDOT composite that exhibited high electrocatalytic activity and a linear response was obtained with low detection limit of 0.026, 0.063, and 0.210 µm for dopamine, serotonin, and tryptophan, respectively, which was then utilized for simultaneous detection in serum samples.
Zhang et al. established a unique strategy for constructing a sensitive levofloxacin (LEV) sensor by incorporation of AuNP‐GR on MIP polypyrrole where they found the detection limit of 0.53 µmol L−1 and good linear range.266 In their study, GR‐AuNP served as an electrooxidation promoter for LEV on electrode whereas MIP polypyrrole acted as a recognition element. Paracetamol is another analgesic and antipyretic drug whose overdose can cause acute kidney and liver failure.267 Thus, Luo et al. synthesized an Au/GR nanoparticles with core‐shell structure having high stability and catalytic activity doped into PEDOT to construct a nanocomposite by electrodeposition method. The modified electrode exhibited gratifying sensitivity toward paracetamol over wide range of concentrations with detection limit of as low as 41 nm and was capable of recognizing paracetamol in real samples along with good accuracy.268 Orlando et al. carried out simultaneous detection of levofloxacin and paracetamol detection by utilizing silver nanoparticles, carbon black, and PEDOT:PSS as an efficient voltammetric biosensor. Carbon black was utilized because of its cheaper cost compared to graphene along with its catalytic and electrocatalytic properties that ameliorate the sensor performance.269 The subsequent biosensor exhibited good reproducibility, stability with good detection limit of 1.2 × 10−8 and 1.4 × 10−8 mol L−1 for paracetamol and levofloxacin, respectively.
Characteristics of various biosensors based on hybrid ICP are summarized in Table 6 .
Table 6.
Characteristics of several hybrid ICP composite biosensors
| ICP | Nanomaterial 1 | Nanomaterial 2 | Target analyte | Range | Limit | Ref. |
|---|---|---|---|---|---|---|
| Poly(cathechol) | GR sheets | AuNP | Acute lymphoblastic leukemia | 100.0 µm–10.0 pm | 1 pm | 255 |
| PEDOT:PSS | GR | AuNP |
Dopamine uric acid |
(1 nm–300 mm) (10 mm–1 mm) |
100 pm 10 mm |
236 |
| PEDOT | GO | AuNP | Aflatoxins B1 | 0.5 ng mL−1 to 20 ng mL−1 | 0.109 ng mL−1 | 257 |
| 2,5‐di‐(2‐thienyl)‐1‐pyrrole‐1‐(p‐benzoic acid) | GO | AuNP | Aflatoxins B1 | 3.2 fm–0.32 pm | 1fm | 259 |
| PEDOT | GO | AuNP | Mucin 1 | 3.13 am–31.25 nm | 0.031 fm | 256 |
| PEDOT | RGO | NiNP | Glucose | 1.0 µm–5.1 mm | 0.8 µm | 260 |
| Polyaniline | MWCNT | AuNP | Glucose | 0.0625–1.19 mm | 0.19 mm | 262 |
| Polypyrrole | rGO | CuO | Glucose | 0.1–100 mm | 0.03 µm | 264 |
| Poly(1,5‐diaminonaphthalne) | GO | AuNP | Dopamine | 10 nm–1 mm | 5 nm | 265 |
| PEDOT | CNT | NiO |
Dopamine Serotonin Tryptophan |
0.03–20 µm 0.3–35 µm 1–41 µm |
0.026 µm 0.063 µm 0.210 µm |
237 |
| MIP Polypyrrole | GR | AuNP | Levofloxacin | 1.0 to 100 µmol L−1 | 0.53 µmol L−1 | 266 |
| PEDOT | GR | AuNP | Paracetamol | 0.15 µm–5.88 mm | 41 nm | 268 |
| PEDOT | MGPN | PtNP | Glucose | 0.01–50 mm | 0.3 µm | 261 |
| PEDOT | Carbon black | AgNP |
Paracetamol levoflaxocin |
6.2 × 10−7 to 7.1 × 10−6 mol L−1 6.7 × 10−7 to 1.2 × 10−5 mol L−1 |
1.2 × 10−8 mol L−1 1.4 × 10−8 mol L−1 |
269 |
Abbreviations: Gold nanoparticles (AuNP), nickel nanoparticle (NiNP), silver nanoparticle (AgNP), platinum nanoparticle (PtNP), nickel oxide (NiO), copper oxide (CuO), multilayered GR petal nanosheets (MGPN), graphene (GR), carbon nanotubes (CNT), multiwalled CNT (MWCNT), GR oxide (GO), reduced GO (rGO), molecularly imprinted polymer (MIP).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
4.5. Doped ICP Based Biosensors
Doping of ICPs with acids is usually preferred since it augments the electrical conductivity and also they act as a functional material that possesses functional groups that tend to bond with multifunctional biomolecules for the construction of biosensor.270 Kaneto et al. developed a urea biosensor by covalent immobilization of urease on the poly(N‐3‐aminopropyl pyrrole‐co‐pyrrole) doped with para‐toluene sulfonate (PTS). The PTS doped ICP had NH2 group that covalently bonded with the urease enzyme on the surface via carbodiimide anchoring reaction. Apart from having NH2 functional group, the ICP exhibited porous morphology as a result of large PTS dopant that resulted in high enzyme loading that leads to good performance of enzyme electrode with regard to dynamic range, long life stability, and short response time.271 Luo et al. developed a 3D macroporous structure poly(sodium 4‐styrene sulfonate) (PSS) doped polyaniline for the recognition of alpha‐fetoprotein. The sensitive and selective biosensor exhibited excellent conductivity, high surface area, and many functional groups that aided in excellent immobilization of antibodies with low detection limit of 3.7 fg mL−1. Polyaniline was doped with PSS since the lone polyaniline composite displays redox activities in acidic condition that depresses the sensitivity of most of the biosensing system.272 Another acid dopant, camphor sulfonic acid, was used for doping polyaniline by Mathiyarasu et al. for their use as potentiometric glucose biosensor by covalently immobilizing glucose oxidase. The resulting ICP displayed uniform and smooth micro‐porous structure that resulted in a homogenous composite with consistent enzyme loading offering a better conductive platform for electron transfer. The modified ICP electrode displayed better selectivity, admirable sensitivity, and good linear concentration range of 1–50 mm.273 Another researcher, Wernet et al., synthesized flexible polypyrrole doped with sulfated poly(β‐hydroxy ethers) for its determination. The resulting biosensor displayed higher H2O2 oxidation potential at lower applied current with linear range of up to 100 mm.274 Marisa et al. developed a bi‐enzymatic biosensor based on PAAMPSA doped polyaniline for the recognition of gluconic acid by co‐immobilizing 6‐phospo‐d‐gluconate dehydrogenase and gluconate kinase (6PGDH and GK, respectively). Gluconic acid acts as a biomarker for quantitative determination of grape infection by Botrytis cinerea and the consequent biosensor showed high sensitivity, linear concentration range, low limit of detection (5 µm), and good storage stability.275 Nicholas et al. utilized poly(2‐acrylamide‐2‐methyl propane sulfonic acid) as a dopant for doping polyaniline (PANI‐PAAMPSA) for the recognition studies of bovine serum albumin and lysozyme as the target protein. They also performed experiments for assessing the response of ICP to proteins at physiological pH for measuring the conductivity alterations of PANI‐PAAMPSA with respect to varying pH for testing the range of pH over which the doping mechanism is active.276 Additionally, the incorporation of PAAMPSA as an acid dopant resulted in water‐soluble ICP with high conductivity. Noemi et al. fabricated an aptasensor for label‐free recognition of kanamycin A and ampicillin by utilizing tosylate doped PEDOT. The consequent biosensor was highly selective and sensitive to the analyzed target with a dynamic concentration range of 10 nm–1 mm for kanamycin A and 100 pm–1 µm for ampicillin. Real samples of ampicillin‐spiked low‐fat milk were successfully analyzed to assess the functionality of biosensor with real samples.277 Erslan et al. utilized polyvinylsulfonate (PVS) doped PEDOT film for the determination of acetylcholine by using amperometric biosensor. Immobilization was carried out by cross‐linking of acetylcholinesterase‐choline oxidase with bovin serum albumin and glutaraldehyde. PEDOT was doped with PVS since it increases conductivity, improves environmental stability, maintains charge neutrality during reduction reaction, and augments the polymerization rate when such large dopants are used. As a result of these attributes, the ICP showed broad linear range and low limit of detection of 5.0 × 10−9 m with response time of 200 s.278
Characteristics of various biosensors based on doped ICP are summarized in Table 7 .
Table 7.
Characteristics of several Doped ICP composite biosensors
| ICP | Dopant | Target Analyte | Range | Limit | Ref. |
|---|---|---|---|---|---|
| Poly (N‐3‐amino propyl pyrrole‐co‐pyrrole) | Para‐toluene sulfonate | Urea | 0.16–5.02 mm | 0.020 mm | 271 |
| Polyaniline | Camphor sulfonic acid | Glucose | 1–50 mm | – | 273 |
| Polyaniline | Poly (sodium 4‐styrene sulfonate) | Alpha‐fetoprotien | 0.01–1000 pg mL‐1 | 3.7 fg mL | 272 |
| Polypyrrole | Sulfated poly(β‐hydroxy ethers) | Glucose | 0–100 mm | – | 274 |
| PEDOT | Tosylate | Ampicillin Kanamycin A |
100 pM to 1 µm 10 nM to 1 mm |
– | 277 |
| Polyaniline | Poly(2‐acrylamido‐2‐methyl‐1‐propane sulfonic acid) | Gluconic acid | 4 × 10‐3 to 1 mm | 5 µm | 275 |
| PEDOT | Poly vinylsulfonate | Acetylcholine | 0.01–1 µm | 5.0 × 10‐9 m | 278 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
5. Life Cycle Assessment of a Biosensor Device
As the biosensors have found their applications in diverse fields from environmental to biodefense, it is difficult to obtain open literature on the cost estimation for their development.
In one instance, Schmidt proposed a multi‐analyte biosensor for Environmental Protection Agency (EPA) for recognition of phenolic compounds. The aim was to curtail the analysis time from 19 days to 15 min and annual cost by more than USD 20 million in the United States. The investigator emphasized on the major bottleneck in the development of biosensor that is discrepancy of the market for environmental measurements hence no one analyte is enough to justify the advancement cost of a specific biosensor. Thus Schmidt proposed that the required biosensor that is capable of determining dozens of analytes can be developed in 6–7 months with ≈USD 70 000.279
In another instance, Rescue Service Agency (Sweden) sought an “artificial dog nose” for localizing minefields and free other zones of land. The biosensor application was intended to deliver a prototype that would cost roughly USD 100 000 in August1999. The field testing was planned in Sweden's military field and they also considered its actual utilization in Angola.280
The University of Mississippi proposed the detection of steroidogenic activity (endocrine disrupting compounds) in environmental chemicals in Basin of Mississippi River with an estimated time period of 1–2 years with USD 250 000–300 000 for the construction of biosensor.281, 282
Rogers et al. indicated the development of new methods and instruments for a promising in situ monitoring of toxic chemicals. They compared two distinct detection systems (biosensor and immunoassay kit) for the surveillance of a groundwater pump‐and‐treat system. The projections were acquired from initial investment cost versus cost for each sample wherein the estimated cost for biosensor was USD 8 and that of immunoassay kit from USD 50 to 75 per assay. The cost of the biosensor was slightly offset from start‐up cost for the kits, which was substantially cheaper.283
One of the major markets of the biosensor is glucose monitoring in human urine and blood samples. This market is unique and large enough to stimulate stand‐alone business with sales in excess of USD 100 million per year (1996).284 The cost of this product ranges from cents (for paper strips) to almost USD 1 (for disposable electrodes) utilized in commercially available electrochemical biosensors.
As the last example, the advancement of a biosensor that mimics, closely, the biological sensory function is discussed.285 Initially, no costs were presented but to convert the concept of ion‐channel switch biosensor to practical device,286 it took around 10 years and 60 scientists and engineers. The change in ion flux through the membrane is detected as a change in conductance of membrane. The benefit this sensor provides is direct functional test of the interaction between an artificial cell membrane and the potential drug. Recently, researchers from Stanford University have developed a wearable biosensor that determines the level of Cortisol (a hormone linked to stress) from the sweat of human being that can help to maintain physical fitness and optimum health status.287
One of the major hindrances that the biosensor market has come across is the inadequacy of mass market available, barring the exceptional case of glucose market thus, from the viewpoint of profit, it causes severe resistance for investors to invest in biosensor technology. It was indicated that the development cost for biosensor, as a result of its complex fabrication route, lies in the range of USD 20–30 million within a time limit of 7–10 years and hence, small industries are bound to hamper from arriving in the market as a result of huge investment amount along with the losses suffered in the early phase.288 Some of the limitations of this may include the lack of reliability to manufacture a competitive product, complex fabrication route, and mercantile advancement of biosensor technology required for the production of an enormous amount of devices.
6. Conclusion
Biosensors are analytical sensing devices that transmit the biochemical signal occurring upon interaction between the analyte and the bio‐receptor that have sought their application in diverse fields from environmental monitoring to clinical diagnosis, food quality evaluation to drug development, and also in the defense sector. Since the discovery of glucose biosensor by Clark, research in this field has augmented and never looked back. Of the various materials developed for the construction of biosensor, intrinsically conductive polymers have acquired a considerable interest of scientists and industrialists, thanks to their paramount characteristics such as inimitable electronic structure, biocompatibility, and tunable electrical conductivity. Though they provide with copious attributes, ICPs have stability and property issues in different ambient conditions such as oxygen media, acidic or basic media. Thus, materials such as AuNP, GR and their derivatives, CNTs, and hybrid materials, due to their exceptional attributes, have been amalgamated with ICP to improve the performance characteristics such as rapid response, good selectivity, increased sensitivity, long‐term stability, and lower detection limit. Thus, we have meticulously consolidated the existing literature on various composites of ICPs with different materials along with doped conductive polymer based biosensor. Simultaneously, diverse forms of biosensors and the importance of the ICP and their composites for immobilizing the biomolecule have also been highlighted. ICPs along with nanomaterials provide a 3D matrix that preserves the activity of biomolecule and renders excellent platform for immobilization since they offer excellent durability, porosity, selectivity, excellent electron transfer rate, biocompatibility, and better signal transduction. As described previously, various ICP‐based composites have been reconnoitered to fabricate unique biosensors that utilize ICP composite as transducer elements, however, the opportunity for extra development is still provided. The clever amalgamation of diverse nanostructured materials with best ICPs will pave the way for utilizing novel ICP composites for manufacturing enhanced sensing platform with superior performance. The major interferences that the biosensor market came across were the inadequacy of availability of mass market thus, from the viewpoint of profit, it causes severe resistance for investors to invest in biosensor technology. Complex fabrication route and mercantile advancement of biosensor technology required for manufacturing many devices can be considered as the limitation for mass production.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
The authors would like to thank Dr. C. P. Ramanarayanan, Vice‐Chancellor, DIAT (DU), Pune, for constant encouragement and support. The authors would also like to acknowledge Prakash Gore, Swaroop Gharde, Jay Korde, and Rushikesh Ambekar for technical discussions and support. The authors are thankful to anonymous reviewers for improving the quality of the manuscript by their valuable suggestions and comments.
Biographies
Deepak G. Prajapati is presently pursuing his Master's degree in metallurgical and materials engineering from the Defense Institute of Advanced Technology, Pune. He received his Bachelor's degree in plastic and polymer engineering from Maharashtra Institute of Technology, Aurangabad during 2014–2017. He has completed his diploma in plastics engineering from Shri Bhagubhai Mafatlal Polytechnic, Mumbai, during 2010–2014. His research interests include conductive polymers, composites, phase changing materials, biosensors for food, diagnosis, and ecological applications.

Balasubramanian Kandasubramanian is professor and head of department of Metallurgical and Materials Engineering, DIAT(DU), Ministry of Defense, India, and has been highly acclaimed for his contribution toward polymer processing and fabrication for various applications including smart textiles, hydrophobic coatings, ablative materials, fire retardant fabrics, waste‐water treatments, etc. for defense applications. He was actively involved in the development of various research laboratories including Nano Texturing, Advanced Characterization (FESEM, HRTEM, SAXS), Additive Manufacturing, etc. The research activity of his research group can be classified under five categories: water management; smart textiles; biomimicking of polymers; structural composites; and advanced coatings for defense applications.

Prajapati D. G., Kandasubramanian B., Progress in the Development of Intrinsically Conducting Polymer Composites as Biosensors. Macromol. Chem. Phys. 2019, 220, 1800561 10.1002/macp.201800561
References
- 1. Sadana A., Sadana N., in Handbook of Biosensors and Biochips, (Eds: Marks R. S., Cullen D. C., Karube I., Lowe C. R., Weetall H. H.), John Wiley & Sons, Chichester, UK: 2008. [Google Scholar]
- 2. Li S., Singh J., Li H., Banerjee I. A., in Biosensor Nanomaterials, (Eds: Li S., Singh J., Li H., Banerjee I. A.), Wiley‐VCH, Weinheim, Germany: 2011. [Google Scholar]
- 3. Liu X. Y., Cheng F., Liu Y., Li W. G., Chen Y., Pan H., Liu H. J., J. Mater. Chem. 2010, 20, 278. [Google Scholar]
- 4. Davaji B., Cho H. D., Malakoutian M., Lee J. K., Panin G., Kang T. W., Lee C. H., Sci. Rep. 2017, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu C., Chortos A., Wang Y., Pfattner R., Lei T., Hinckley A. C., Pochorovski I., Yan X., To J. W.‐F., Oh J. Y., Tok J. B.‐H., Bao Z., Murmann B., Nat. Electron. 2018, 1, 183. [Google Scholar]
- 6. Wales D. J., Grand J., Ting V. P., Burke R. D., Edler K. J., Bowen C. R., Mintova S., Burrows A. D., Chem. Soc. Rev. 2015, 44, 4290. [DOI] [PubMed] [Google Scholar]
- 7. Thakur S., Patil P., RSC Adv. 2016, 6, 45768. [Google Scholar]
- 8. Chen G., Paronyan T. M., Pigos E. M., Harutyunyan A. R., Sci. Rep. 2012, 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li C., Cui Y. L., Tian G. L., Shu Y., Wang X. F., Tian H., Yang Y., Wei F., Ren T. L., Sci. Rep. 2015, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu H., Lu Y. F., Xiang J. X., Zhang M. K., Zhao Y. J., Xie Z. Y., Gu Z. Z., Nanoscale 2018, 10, 2090. [DOI] [PubMed] [Google Scholar]
- 11. Yamada T., Hayamizu Y., Yamamoto Y., Yomogida Y., Izadi‐Najafabadi A., Futaba D. N., Hata K., Nat. Nanotechnol. 2011, 6, 296. [DOI] [PubMed] [Google Scholar]
- 12. Yu G., Cao Y., Wang J., McElvain J., Heeger A. J., Synth. Met. 1999, 102, 904. [Google Scholar]
- 13. Lavery L. L., Whiting G. L., Arias A. C., Org. Electron. Phys., Mater. Appl. 2011, 12, 682. [Google Scholar]
- 14. Manouras T., Vamvakaki M., Polym. Chem. 2017, 8, 74. [Google Scholar]
- 15. Engel J., Chen J., Fan Z., Liu C., Sens. Actuators, A 2005, 117, 50. [Google Scholar]
- 16. Xu M., Qi J., Li F., Liao X., Liu S., Zhang Y., RSC Adv. 2017, 7, 30506. [Google Scholar]
- 17. Chun S., Hwang I., Son W., Chang J. H., Park W., Nanoscale 2018, 10, 10545. [DOI] [PubMed] [Google Scholar]
- 18. Rohan , Biosensors Market worth 27.06 Billion USD by 2022, https://www.marketsandmarkets.com/PressReleases/biosensors.asp (accessed: July 2018).
- 19. Upadhyay L. S. B., Verma N., in Environmental Microbial Biotechnology, Vol. 45 (Eds: Sukla L. B., Pradhan N., Panda S., Mishra B. K.), Springer, Cham, Switzerland: 2015, p. 77. [Google Scholar]
- 20. Cennamo N., Varriale A., Pennacchio A., Staiano M., Massarotti D., Zeni L., DAuria S., Sens. Actuators, B 2013, 176, 1008. [Google Scholar]
- 21. Mat Zaid M. H., Abdullah J., Yusof N. A., Sulaiman Y., Wasoh H., Md Noh M. F., Issa R., Sens. Actuators, B 2017, 241, 1024. [Google Scholar]
- 22. Mercante L. A., Scagion V. P., Migliorini F. L., Mattoso L. H. C., Correa D. S., TrAC Trends Anal. Chem. 2017, 91, 91. [Google Scholar]
- 23. Yu D., Blankert B., Viré J. C., Kauffmann J. M., Anal. Lett. 2005, 38, 1687. [Google Scholar]
- 24. Kaur G., Adhikari R., Cass P., Bown M., Gunatillake P., RSC Adv. 2015, 5, 37553. [Google Scholar]
- 25. Clark L. C., Lyons C., Ann. N. Y. Acad. Sci. 1962, 102, 29. [DOI] [PubMed] [Google Scholar]
- 26. Amano Y., Cheng Q., Anal. Bioanal. Chem. 2005, 381, 156. [DOI] [PubMed] [Google Scholar]
- 27. Caygill R. L., Blair G. E., Millner P. A., Anal. Chim. Acta 2010, 681, 8. [DOI] [PubMed] [Google Scholar]
- 28. Frasconi M., Tortolini C., Botrè F., Mazzei F., Anal. Chem. 2010, 82, 7335. [DOI] [PubMed] [Google Scholar]
- 29. Moon J. M., Thapliyal N., Hussain K. K., Goyal R. N., Shim Y. B., Biosens. Bioelectron. 2018, 102, 540. [DOI] [PubMed] [Google Scholar]
- 30. Kausaite‐Minkstimiene A., Ramanaviciene A., Ramanavicius A., Analyst 2009, 134, 2051. [DOI] [PubMed] [Google Scholar]
- 31. Pejcic B., De Marco R., Parkinson G., Analyst 2006, 131, 1079. [DOI] [PubMed] [Google Scholar]
- 32. Neethirajan S., Ahmed S. R., Chand R., Buozis J., Nagy É., Nanotheranostics 2017, 1, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang J., Cai X., Rivas G., Shiraishi H., Farias P. A. M., Dontha N., Anal. Chem. 1996, 68, 2629. [DOI] [PubMed] [Google Scholar]
- 34. Hideshima S., Kobayashi M., Wada T., Kuroiwa S., Nakanishi T., Sawamura N., Asahi T., Osaka T., Chem. Commun. 2014, 50, 3476. [DOI] [PubMed] [Google Scholar]
- 35. Braham Y., Barhoumi H., Maaref A., Anal. Methods 2013, 5, 4898. [Google Scholar]
- 36. Mondal K., Sharma A., RSC Adv. 2016, 6, 94595. [Google Scholar]
- 37. Wang Z., Dai Z., Nanoscale 2015, 7, 6420. [DOI] [PubMed] [Google Scholar]
- 38. Kai D., Prabhakaran M. P., Jin G., Ramakrishna S., J. Mater. Chem. B 2013, 1, 2305. [DOI] [PubMed] [Google Scholar]
- 39. Pron A., Rannou P., Prog. Polym. Sci. 2002, 27, 135. [Google Scholar]
- 40. Bhandari S., Singha N. K., Khastgir D., J. Appl. Polym. Sci. 2013, 129, 1264. [Google Scholar]
- 41. Jen K. Y., Miller G. G., Elsenbaumer R. L., J. Chem. Soc., Chem. Commun. 1986, 5, 1346. [Google Scholar]
- 42. Bhandari S., Khastgir D., Int. J. Polym. Mater. Polym. Biomater. 2016, 65, 543. [Google Scholar]
- 43. Rahman M. A., Kumar P., Park D.‐S., Shim Y.‐B., Sensors 2008, 8, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao Z., Rafea S., Lim L. H., Adv. Mater. 2007, 19, 602. [Google Scholar]
- 45. Komarova E., Aldissi M., Bogomolova A., Biosens. Bioelectron. 2005, 21, 182. [DOI] [PubMed] [Google Scholar]
- 46. Krishnamoorthy K., Gokhale R. S., Contractor A. Q., Kumar A., Chem. Commun. 2004, 820. [DOI] [PubMed] [Google Scholar]
- 47. Le Goff A., Holzinger M., Cosnier S., Analyst 2011, 136, 1279. [DOI] [PubMed] [Google Scholar]
- 48. de Oliveira Farias E. A., Nogueira S. S., de Oliveira Farias A. M., de Oliveira M. S., de Fátima Cardoso Soares M., da Cunha H. N., dos Santos Junior J. R., da Silva D. A., Eaton P., Eiras C., J. Mater. Sci. 2017, 52, 13365. [Google Scholar]
- 49. Naveen M. H., Gurudatt N. G., Shim Y. B., Appl. Mater. Today 2017, 9, 419. [Google Scholar]
- 50. Serrano‐Cinca C., Fuertes‐Callén Y., Mar‐Molinero C., in Decision Support Systems, Vol. 38, John Wiley & Sons, Hoboken, NJ: 2005, p. 557. [Google Scholar]
- 51. Ramanavicˇius A., Kaušaite A., Ramanavicˇiene A., Sens. Actuators, B Chem. 2005, 532, 111. [Google Scholar]
- 52. Min G., Synth. Met. 1999, 102, 1163. [Google Scholar]
- 53. Garcia‐Alvarez J., Curr. Org. Chem. 2008, 12, 1199. [Google Scholar]
- 54. Yamamoto T., Fukumoto H., Koizumi T., J. Inorg. Organomet. Polym. Mater. 2009, 19, 3. [Google Scholar]
- 55. Patil A. O., Ikenoue Y., Wudl F., Heeger A. J., J. Am. Chem. Soc. 1987, 109, 1858. [Google Scholar]
- 56. Kim J., Lee J., You J., Park M.‐S., Al Hossain M. S., Yamauchi Y., Kim J. H., Mater. Horiz. 2016, 3, 517. [Google Scholar]
- 57. Schopf G., Koßmehl G., Polythiophenes—Electrically Conductive Polymers, Advances in Polymer Science, Vol. 129, Springer, Berlin: 1997. [Google Scholar]
- 58. Bredas J. L., Street G. B., Acc. Chem. Res. 1985, 18, 309. [Google Scholar]
- 59. Heeger A. J., Curr. Appl. Phys. 2001, 1, 247. [Google Scholar]
- 60. Le T.‐H., Kim Y., Yoon H., Polymers. 2017, 9, 150. [Google Scholar]
- 61. Prakash S., Chakrabarty T., Singh A. K., Shahi V. K., Biosens. Bioelectron. 2013, 41, 43. [DOI] [PubMed] [Google Scholar]
- 62. Billingham N. C., Calvert P. D., Foot P. J. S., Mohammad F., Polym. Degrad. Stab. 1987, 19, 323. [Google Scholar]
- 63. Li Y., Organic Optoelectronic Materials, Lecture Notes in Chemistry, Vol. 91 (Ed: Li Y.), Springer, Cham, Switzerland: 2015. [Google Scholar]
- 64. Hansora D. P., Shimpi N. G., Mishra S., RSC Adv. 2015, 5, 107716. [Google Scholar]
- 65. Herrmann S., Ritchie C., Streb C., Dalton Trans. 2015, 44, 7092. [DOI] [PubMed] [Google Scholar]
- 66. Cheung W., Chiu P. L., Parajuli R. R., Ma Y., Ali S. R., He H., J. Mater. Chem. 2009, 19, 6465. [Google Scholar]
- 67. Ballarin B., Masiero S., Seeber R., Tonelli D., J. Electroanal. Chem. 1998, 449, 173. [Google Scholar]
- 68. Mishra A. K., Mishra S. B., Tiwari A., in Biosensor Nanomaterials, Wiley‐VCH, Weinheim, Germany: 2011, p. 255. [Google Scholar]
- 69. Gangopadhyay R., De A., Chem. Mater. 2000, 12, 608. [Google Scholar]
- 70. Guo S., Dong S., J. Mater. Chem. 2011, 21, 16704. [Google Scholar]
- 71. Salavagione H. J., Díez‐Pascual A. M., Lázaro E., Vera S., Gómez‐Fatou M. A., J. Mater. Chem. A 2014, 2, 14289. [Google Scholar]
- 72. Jia H., Su X., Hou G., Ma F., Bi S., Liu Z., Integr. Ferr. 2013, 145, 130. [Google Scholar]
- 73. Wang B., Zheng J., He Y., Sheng Q., Sens. Actuators, B 2013, 186, 417. [Google Scholar]
- 74. Ostrovsky S., Hahnewald S., Kiran R., Mistrik P., Hessler R., Tscherter A., Senn P., Kang J., Kim J., Roccio M., Lellouche J. P., RSC Adv. 2016, 6, 41714. [Google Scholar]
- 75. Gopalan A. I., Lee K. P., Ragupathy D., Lee S. H., Lee J. W., Biomaterials 2009, 30, 5999. [DOI] [PubMed] [Google Scholar]
- 76. Prakash S., Shahi V. K., Anal. Methods 2011, 3, 1331. [Google Scholar]
- 77. Ragupathy D., Gopalan A. I., Lee K. P., Electrochem. Commun. 2009, 11, 397. [Google Scholar]
- 78. Thévenot D. R., Toth K., Durst R. A., Wilson G. S., Anal. Lett. 2001, 34, 635. [Google Scholar]
- 79. Willner I., Katz E., Bioelectronics: From Theory to Applications, Wiley‐VCH, Weinheim, Germany: 2005. [Google Scholar]
- 80. Sahoo B. N., Kandasubramanian B., RSC Adv. 2014, 4, 22053. [Google Scholar]
- 81. Mishra N., Kandasubramanian B., Polym. Plast. Technol. Eng. 2017, 57, 1. [Google Scholar]
- 82. Yadav R., Naebe M., Wang X., Kandasubramanian B., RSC Adv. 2016, 6, 115145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bogue R., Sens. Rev. 2009, 29, 107. [Google Scholar]
- 84. Wu H., Lee C. J., Wang H., Hu Y., Young M., Han Y., Xu F. J., Cong H., Cheng G., Chem. Sci. 2018, 9, 2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bilgi M., Ayranci E., J. Electroanal. Chem. 2018, 823, 588. [Google Scholar]
- 86. David M., Barsan M. M., Brett C. M. A., Florescu M., Sens. Actuators, B 2018, 255, 3227. [Google Scholar]
- 87. Liu Y., Turner A. P. F., Zhao M., Mak W. C., Biosens. Bioelectron. 2018, 100, 374. [DOI] [PubMed] [Google Scholar]
- 88. Perumal V., Hashim U., J. Appl. Biomed. 2014, 12, 1. [Google Scholar]
- 89. Gui R., Jin H., Guo H., Wang Z., Biosens. Bioelectron. 2018, 100, 56. [DOI] [PubMed] [Google Scholar]
- 90. Shahrokhian S., Salimian R., Sens. Actuators, B 2018, 266, 160. [Google Scholar]
- 91. Taylor R., Schultz J. S., Handbook of Chemical and Biological Sensors, Institute of Physics, Bristol, Pennsylvania: 1996. [Google Scholar]
- 92. Gerard M., Chaubey A., Malhotra B. D., Biosens. Bioelectron. 2002, 17, 345. [DOI] [PubMed] [Google Scholar]
- 93. D'Orazio P., Clin. Chim. Acta 2003, 334, 41. [DOI] [PubMed] [Google Scholar]
- 94. Rogers K. R., Mascini M., Field Anal. Chem. Technol. 1998, 2, 317. [Google Scholar]
- 95. Santandreu M., Alegret S., Fàbregas E., Anal. Chim. Acta 1999, 396, 181. [Google Scholar]
- 96. German N., Kausaite‐Minkstimiene A., Ramanavicius A., Semashko T., Mikhailova R., Ramanaviciene A., Electrochim. Acta 2015, 169, 326. [Google Scholar]
- 97. Li J., Qiu J. D., Xu J. J., Chen H. Y., Xia X. H., Adv. Funct. Mater. 2007, 17, 1574. [Google Scholar]
- 98. Lupu S., Lete C., Balaure P., Caval D., Mihailciuc C., Lakard B., Hihn J.‐Y., Campo F., Sensors 2013, 13, 6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hare D. O., in Body Sensor Networks (Ed: Yang G.‐Z.), Springer, London: 2014. [Google Scholar]
- 100. Karyakin A. A., Lukachova L. V., Karyakina E. E., Orlov A. V., Karpachova G. P., Anal. Commun. 1999, 36, 153. [DOI] [PubMed] [Google Scholar]
- 101. Grieshaber D., MacKenzie R., Vörös J., Reimhult E., Sensors 2008, 8, 1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Thévenot D. R., Toth K., Durst R. A., Wilson G. S., Biosens. Bioelectron. 2001, 16, 121. [DOI] [PubMed] [Google Scholar]
- 103. Muhammad‐Tahir Z., Alocilja E. C., Biosens. Bioelectron. 2003, 18, 813. [DOI] [PubMed] [Google Scholar]
- 104. Sauerbrey G., Z. Physik 1959, 155, 206. [Google Scholar]
- 105. Janshoff A., Galla H. J., Steinem C., Angew. Chem. 2000, 39, 4004. [DOI] [PubMed] [Google Scholar]
- 106. Sethi R. S., Biosens. Bioelectron. 1994, 9, 243. [Google Scholar]
- 107. Borole D. D., Kapadi U. R., Mahulikar P. P., Hundiwale D. G., Des. Monomers Polym. 2006, 9, 1. [Google Scholar]
- 108. Ebarvia B. S., Cabanilla S., Sevilla F., Talanta 2005, 66, 145. [DOI] [PubMed] [Google Scholar]
- 109. Sai V. V. R., Mahajan S., Contractor A. Q., Mukherji S., Anal. Chem. 2006, 78, 8368. [DOI] [PubMed] [Google Scholar]
- 110. Ramanathan K., Danielsson B., Biosens. Bioelectron. 2001, 16, 417. [DOI] [PubMed] [Google Scholar]
- 111. Zhou Y., Chiu C. W., Liang H., Sensors 2012, 12, 15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ahmad L. M., Towe B., Wolf A., Mertens F., Lerchner J., Sens. Actuators, B. 2010, 145, 239. [Google Scholar]
- 113. Ripka P., Tipek A., Modern Sensors Handbook, John Wiley & Sons, London, UK: 2010. [Google Scholar]
- 114. Xie W., Bülow L., Xie B., J. Therm. Anal. Calorim. 2018, 134, 1921. [Google Scholar]
- 115. Damborsky P., Vitel J., Katrlik J., Essays Biochem. 2016, 60, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kuswandi B., Andres R., Narayanaswamy R., Analyst 2001, 126, 1469. [DOI] [PubMed] [Google Scholar]
- 117. Wan D., Chen H. L., Lai Y. T., Yu C. C., Lin K. F., Adv. Funct. Mater. 2010, 20, 1742. [Google Scholar]
- 118. Gao S., Koshizaki N., Tokuhisa H., Koyama E., Sasaki T., Kim J. K., Ryu J., Kim D. S., Shimizu Y., Adv. Funct. Mater. 2010, 20, 78. [Google Scholar]
- 119. Mohamad Ahad I. Z., Wadi Harun S., Gan S. N., Phang S. W., Sens. Actuators, B 2018, 261, 97. [Google Scholar]
- 120. Domínguez‐Aragón A., Olmedo‐Martínez J. A., Zaragoza‐Contreras E. A., Sens. Actuators, B 2018, 259, 170. [Google Scholar]
- 121. Barnes W. L., Dereux A., Ebbesen T. W., Nature 2003, 424, 824. [DOI] [PubMed] [Google Scholar]
- 122. Rodrigues R. C., Ortiz C., Berenguer‐Murcia Á., Torres R., Fernández‐Lafuente R., Chem. Soc. Rev. 2013, 42, 6290. [DOI] [PubMed] [Google Scholar]
- 123. Itoh T., Shibuya Y., Yamaguchi A., Hoshikawa Y., Tanaike O., Tsunoda T., Hanaoka T. A., Hamakawa S., Mizukami F., Hayashi A., Kyotani T., Stucky G. D., J. Mater. Chem. A 2017, 5, 20244. [Google Scholar]
- 124. Baˇnicaˇ F. G., Chemical Sensors and Biosensors: Fundamentals and Applications, John Wiley & Sons, Chichester, UK: 2012. [Google Scholar]
- 125. Bromage E. S., Vadas G. G., Harvey E., Unger M. A., Kaattari S. L., Environ. Sci. Technol. 2007, 41, 7067. [DOI] [PubMed] [Google Scholar]
- 126. Winter G., Milstein C., Nature 1991, 349, 293. [DOI] [PubMed] [Google Scholar]
- 127. Serrano‐Cinca C., Fuertes‐Callén Y., Mar‐Molinero C., in Measuring DEA Efficiency in Internet Companies, Vol. 38 (Eds: Mitchell R., Gu J.‐D.), John Wiley & Sons, Chichester, UK: 2005. [Google Scholar]
- 128. Marazuela M. D., Moreno‐Bondi M. C., Anal. Bioanal. Chem. 2002, 372, 664. [DOI] [PubMed] [Google Scholar]
- 129. Sassolas A., Blum L. J., Leca‐Bouvier B. D., Biotechnol. Adv. 2012, 30, 489. [DOI] [PubMed] [Google Scholar]
- 130. Datta S., Christena L. R., Rajaram Y. R. S., 3 Biotech 2013, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Teles F. R. R., Fonseca L. P., Mater. Sci. Eng., C 2008, 28, 1530. [Google Scholar]
- 132. Singh S., Jain D. V. S., Singla M. L., Anal. Methods 2013, 5, 1024. [Google Scholar]
- 133. Dwevedi A., Enzyme Immobilization: Advances in Industry, Agriculture, Medicine, and the Environment, Springer, Berlin: 2016. [Google Scholar]
- 134. Brena B., González‐Pombo P., Batista‐Viera F., in Methods in Molecular Biology, Vol. 1051, Wiley‐VCH, Weinheim, Germany: 2013, p. 15. [DOI] [PubMed] [Google Scholar]
- 135. Chawla S., Rawal R., Shabnam, R. C. Kuhad , Pundir C. S., Anal. Methods 2011, 3, 709. [DOI] [PubMed] [Google Scholar]
- 136. Li Y., Huang G., Zhang X., Li B., Chen Y., Lu T., Lu T. J., Xu F., Adv. Funct. Mater. 2013, 23, 660. [Google Scholar]
- 137. H. Zejli , Hidalgo‐Hidalgo de Cisneros J. L., Naranjo‐Rodriguez I., Liu B., Temsamani K. R., Marty J. L., Anal. Chim. Acta 2008, 612, 198. [DOI] [PubMed] [Google Scholar]
- 138. Fan Q., Shan D., Xue H., He Y., Cosnier S., Biosens. Bioelectron. 2007, 22, 816. [DOI] [PubMed] [Google Scholar]
- 139. Santhanam K. S. V., Pure Appl. Chem. 1998, 70, 1259. [Google Scholar]
- 140. Selampinar F., Akbulut U., Özden M. Y., Toppare L., Biomaterials 1997, 18, 1163. [DOI] [PubMed] [Google Scholar]
- 141. Malhotra B. D., Chaubey A., Singh S. P., Anal. Chim. Acta 2006, 578, 59. [DOI] [PubMed] [Google Scholar]
- 142. Ho P. K. H., Kim J.‐S., Burroughes J. H., Becker H., Li S. F. Y., Brown T. M., Cacialli F., Friend R. H., Nature 2000, 404, 481. [DOI] [PubMed] [Google Scholar]
- 143. Korde J. M., Kandasubramanian B., Biomater. Sci. 2018, 6, 1691. [DOI] [PubMed] [Google Scholar]
- 144. Harwood G. W. J., Pouton C. W., Adv. Drug Delivery Rev. 1996, 18, 163. [Google Scholar]
- 145. Green R. A., Baek S., Lovell N. H., Poole‐Warren L. A., Nanostructured Conductive Polymers, John Wiley & Sons, Chichester, UK: 2010. [Google Scholar]
- 146. Xia L., Wei Z., Wan M., J. Colloid Interface Sci. 2010, 341, 1. [DOI] [PubMed] [Google Scholar]
- 147. Michalska A., Anal. Bioanal. Chem. 2005, 384, 391. [DOI] [PubMed] [Google Scholar]
- 148. Bobacka J., Electroanalysis 2006, 18, 7. [Google Scholar]
- 149. Brett C. M. A., Thiemann C., J. Electroanal. Chem. 2002, 215, 538. [Google Scholar]
- 150. Cui R., Liu C., Shen J., Gao D., Zhu J. J., Chen H. Y., Adv. Funct. Mater. 2008, 18, 2197. [Google Scholar]
- 151. Qian L., Yang X., Adv. Funct. Mater. 2007, 17, 1353. [Google Scholar]
- 152. Walcarius A., Minteer S. D., Wang J., Lin Y., Merkoçi A., J. Mater. Chem. B 2013, 1, 4878. [DOI] [PubMed] [Google Scholar]
- 153. Madakbas¸ S., Danis¸ Ö., Demir S., Kahraman M. V., Starch 2013, 65, 146. [Google Scholar]
- 154. Rahman M. A., Kothalam A., Choe E. S., Won M. S., Shim Y. B., Anal. Chem. 2012, 84, 6654. [DOI] [PubMed] [Google Scholar]
- 155. Xia N., Xing Y., Wang G., Feng Q., Chen Q., Feng H., Sun X., Liu L., Int. J. Electrochem. Sci. 2013, 8, 2459. [Google Scholar]
- 156. Guler Gokce Z., Akalın P., Kok F. N., Sarac A. S., Sens. Actuators, B 2018, 254, 719. [Google Scholar]
- 157. Monošík R., Stred'anský M., Šturdík E., Acta Chim. Slovaca 2012, 5, 109. [Google Scholar]
- 158. Bezerra C. S., De Farias Lemos C. M. G., De Sousa M., Gonçalves L. R. B., J. Appl. Polym. Sci. 2015, 132, 42125. [Google Scholar]
- 159. Ji T. H., BBA—Rev. Biomembr. 1979, 559, 39. [Google Scholar]
- 160. Collings A. F., Caruso F., Rep. Prog. Phys. 1997, 60, 1397. [Google Scholar]
- 161. Kanik F. E., Rende E., Timur S., Toppare L., J. Mater. Chem. 2012, 22, 22517. [Google Scholar]
- 162. Cosnier S., Biosens. Bioelectron. 1999, 14, 443. [DOI] [PubMed] [Google Scholar]
- 163. Vidal J. C., Garcia‐Ruiz E., Castillo J. R., Microchim. Acta 2003, 143, 93. [Google Scholar]
- 164. Cosnier S., Handbook of Biosensors and Biochips, John Wiley & Sons, Chichester, UK: 2008. [Google Scholar]
- 165. Cosnier S., Anal. Bioanal. Chem. 2003, 377, 507. [DOI] [PubMed] [Google Scholar]
- 166. Shafaat A., Faridbod F., Ganjali M. R., New J. Chem. 2018, 42, 6034. [Google Scholar]
- 167. Balasubramanian K., Yadav R., Prajith P., Int. J. Plast. Technol. 2015, 19, 363. [Google Scholar]
- 168. Gonte R. R., Balasubramanian K., J. Hazard. Mater. 2012, 447, 217. [DOI] [PubMed] [Google Scholar]
- 169. Balasubramanian R. K., Colloids Surf. A 2014, 455, 174. [Google Scholar]
- 170. Saha K., Agasti S. S., Kim C., Li X., Rotello V. M., Chem. Rev. 2012, 112, 2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Yadav R., Balasubramanian K., RSC Adv. 2015, 5, 24990. [Google Scholar]
- 172. Li Y., Schluesener H. J., Xu S., Gold Bull. 2010, 43, 29. [Google Scholar]
- 173. Liu S., Leech D., Ju H., Anal. Lett. 2003, 36, 1. [Google Scholar]
- 174. Tahalyani J., Rahangdale K. K., Aepuru R., Kandasubramanian B., Datar S., RSC Adv. 2016, 6, 36588. [Google Scholar]
- 175. Englebienne P., Van Hoonacker A., J. Colloid Interface Sci. 2005, 292, 445. [DOI] [PubMed] [Google Scholar]
- 176. Esmaeili C., Heng L. Y., Chiang C. P., Rashid Z. A., Safitri E., Malon Marugan R. S. P., Sens. Actuators, B 2017, 242, 616. [Google Scholar]
- 177. Zeng J., Huang Z., Yin G., Qin J., Chen X., Gu J., Colloids Surf., B 2013, 110, 450. [DOI] [PubMed] [Google Scholar]
- 178. Nowicka A. M., Fau M., Rapecki T., Donten M., Electrochim. Acta 2014, 140, 65. [Google Scholar]
- 179. Chowdhury A. D., Gangopadhyay R., De A., Sens. Actuators, B 2014, 190, 348. [Google Scholar]
- 180. Vural T., Yaman Y. T., Ozturk S., Abaci S., Denkbas E. B., J. Colloid Interface Sci. 2018, 510, 318. [DOI] [PubMed] [Google Scholar]
- 181. Shoaie N., Forouzandeh M., Omidfar K., Microchim. Acta 2018, 185, 1. [DOI] [PubMed] [Google Scholar]
- 182. Mir T. A., Yoon J. H., Gurudatt N. G., Won M. S., Shim Y. B., Biosens. Bioelectron. 2015, 74, 594. [DOI] [PubMed] [Google Scholar]
- 183. Hussain K. K., Gurudatt N. G., Mir T. A., Shim Y. B., Biosens. Bioelectron. 2016, 83, 312. [DOI] [PubMed] [Google Scholar]
- 184. Tian L., Qian K., Qi J., Liu Q., Yao C., Song W., Wang Y., Biosens. Bioelectron. 2018, 99, 564. [DOI] [PubMed] [Google Scholar]
- 185. Suzuki G., Tokuno S., Nibuya M., Ishida T., Yamamoto T., Mukai Y., Mitani K., Tsumatori G., Scott D., Shimizu K., PLoS One 2014, 9, e89455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Akhtar M. H., Hussain K. K., Gurudatt N. G., Chandra P., Shim Y. B., Biosens. Bioelectron. 2018, 116, 108. [DOI] [PubMed] [Google Scholar]
- 187. Sangamithirai D., Munusamy S., Narayanan V., Stephen A., Mater. Sci. Eng., C 2018, 91, 512. [DOI] [PubMed] [Google Scholar]
- 188. Zou L., Li Y., Cao S., Ye B., Talanta 2013, 117, 333. [DOI] [PubMed] [Google Scholar]
- 189. Zhao Y., Liu B., Pan L., Yu G., Energy Environ. Sci. 2013, 6, 2856. [Google Scholar]
- 190. Wang K., Zhang X., Li C., Zhang H., Sun X., Xu N., Ma Y., J. Mater. Chem. A 2014, 2, 19726. [Google Scholar]
- 191. Tang Z., Gao L., Wu Y., Su T., Wu Q., Liu X., Li W., Wang Q., J. Mater. Chem. B 2013, 1, 5393. [DOI] [PubMed] [Google Scholar]
- 192. Rong Q., Han H., Feng F., Ma Z., Sci. Rep. 2015, 5, 11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Zhu W., Wang Q., Ma H., Lv X., Wu D., Sun X., Du B., Wei Q., Sci. Rep. 2016, 6, 24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Iijima S., Nature 1991, 354, 56. [Google Scholar]
- 195. Kong J., Franklin N. R., Zhou C., Chapline M. G., Peng S., Cho K., Dai H., Science 2000, 287, 622. [DOI] [PubMed] [Google Scholar]
- 196. Yun Y. H., Dong Z., Shanov V., Heineman W. R., Halsall H. B., Bhattacharya A., Conforti L., Narayan R. K., Ball W. S., Schulz M. J., Nano Today 2007, 2, 30. [Google Scholar]
- 197. Odom T. W., Huang J. L., Kim P., Lieber C. M., Nature 1998, 391, 62. [Google Scholar]
- 198. Balasubramanian K., Burghard M., Anal. Bioanal. Chem. 2006, 385, 452. [DOI] [PubMed] [Google Scholar]
- 199. Yang N., Chen X., Ren T., Zhang P., Yang D., Sens. Actuators, B 2015, 207, 690. [Google Scholar]
- 200. Xu Y., Ye X., Yang L., He P., Fang Y., Electroanalysis 2006, 18, 1471. [Google Scholar]
- 201. Umasankar Y., Chen S. M., Anal. Lett. 2008, 41, 210. [Google Scholar]
- 202. Li Q., Zhang J., Yan H., He M., Liu Z., Carbon 2004, 42, 287. [Google Scholar]
- 203. Wu D., Hu D., Chen H., Shi G., Fetahu I. S., Wu F., Rabidou K., Fang R., Tan L., Xu S., Liu H., Argueta C., Zhang L., Mao F., Yan G., Chen J., Dong Z., Lv R., Xu Y., Wang M., Ye Y., Zhang S., Duquette D., Geng S., Yin C., Lian C. G., Murphy G. F., Adler G. K., Garg R., Lynch L., et al., Nature 2018, 559, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Wan D., Yuan S., Li G. L., Neoh K. G., Kang E. T., ACS Appl. Mater. Interfaces 2010, 2, 3083. [DOI] [PubMed] [Google Scholar]
- 205. Singh K., Singh B. P., Chauhan R., Basu T., J. Appl. Polym. Sci. 2012, 125, E235. [Google Scholar]
- 206. Pan D., Chen J., Yao S., Tao W., Nie L., Anal. Sci. 2005, 21, 367. [DOI] [PubMed] [Google Scholar]
- 207. Aghamiri Z. S., Mohsennia M., Rafiee‐Pour H. A., Thin Solid Films 2018, 660, 484. [Google Scholar]
- 208. Hassan R. Y. A., Kamel A. M., Hashem M. S., Hassan H. N. A., Abd El‐Ghaffar M. A., J. Solid State Electrochem. 2018, 22, 1817. [Google Scholar]
- 209. Artiss J. D., Karcher R. E., Cavanagh K. T., Collins S. L., Peterson V. J., Varma S., Zak B., Am. J. Clin. Pathol. 2000, 114, 139. [DOI] [PubMed] [Google Scholar]
- 210. Ichai C., Leverve X., Orban J. C., Acute Heart Failure, Springer, London: 2008. [Google Scholar]
- 211. Rahman M. M., Shiddiky M. J. A., Rahman M. A., Shim Y. B., Anal. Biochem. 2009, 384, 159. [DOI] [PubMed] [Google Scholar]
- 212. Agüí L., Eguílaz M., Peña‐Farfal C., Yáñez‐Sedeño P., Pingarrón J. M., Electroanalysis 2009, 21, 386. [Google Scholar]
- 213. Ericsson S., Eriksson M., Angelin B., Eur. J. Clin. Invest. 1990, 20, 261. [DOI] [PubMed] [Google Scholar]
- 214. Dhand C., Solanki P. R., Datta M., Malhotra B. D., Electroanalysis 2010, 22, 2683. [Google Scholar]
- 215. Singh K., Solanki P. R., Basu T., Malhotra B. D., Polym. Adv. Technol. 2012, 23, 1084. [Google Scholar]
- 216. Goire N., Speers D. J., Lahra M. M., Whiley D. M., Human Emerging and Re‐emerging Infections, John Wiley & Sons, Hoboken, NJ: 2015. [Google Scholar]
- 217. Singh R., Dhand C., Sumana G., Verma R., Sood S., Gupta R. K., Malhotra B. D., J. Mol. Recognit. 2010, 23, 472. [DOI] [PubMed] [Google Scholar]
- 218. Zhu Y., Koh W. C. A., Shim Y. B., Electroanalysis 2010, 22, 2908. [Google Scholar]
- 219. Cesarino I., Moraes F. C., Machado S. A. S., Electroanalysis 2011, 23, 2586. [Google Scholar]
- 220. Bhalara P. D., Balasubramanian K., Banerjee B. S., Mater. Focus 2015, 4, 154. [Google Scholar]
- 221. Yadav R., Subhash A., Chemmenchery N., Kandasubramanian B., Ind. Eng. Chem. Res. 2018, 57, 9333. [Google Scholar]
- 222. Sobolewski P., Piwowarczyk M., El Fray M., Macromol. Biosci. 2016, 16, 944. [DOI] [PubMed] [Google Scholar]
- 223. Joshi A., Bajaj A., Singh R., Alegaonkar P. S., Balasubramanian K., Datar S., Nanotechnology 2013, 24, 455705. [DOI] [PubMed] [Google Scholar]
- 224. Pumera M., Ambrosi A., Bonanni A., Chng E. L. K., Poh H. L., TrAC Trends Anal. Chem. 2010, 29, 954. [Google Scholar]
- 225. Parveen S., in Fundamentals of Conjugated Polymer Blends, Copolymers and Composites: Synthesis, Properties and Applications (Ed: Saini P.), Scrivener Publishing, Beverly, MA: 2015. [Google Scholar]
- 226. Tirumali M., Balasubramanian K., Kumaraswamy A., Mater. Focus 2017, 6, 691. [Google Scholar]
- 227. Balasubramanian K., IOP Conf. Ser.: Mater. Sci. Eng. 2012, 40, 012022. [Google Scholar]
- 228. Qiu J. D., Huang J., Liang R. P., Sens. Actuators, B 2011, 160, 287. [Google Scholar]
- 229. McAllister M. J., Li J. L., Adamson D. H., Schniepp H. C., Abdala A. A., Liu J., Herrera‐Alonso M., Milius D. L., Car R., Prud'homme R. K., Aksay I. A., Chem. Mater. 2007, 19, 4396. [Google Scholar]
- 230. Stankovich S., Dikin D. A., Piner R. D., Kohlhaas K. A., Kleinhammes A., Jia Y., Wu Y., Nguyen S. B. T., Ruoff R. S., Carbon 2007, 45, 1558. [Google Scholar]
- 231. Stankovich S., Piner R. D., Chen X., Wu N., Nguyen S. T., Ruoff R. S., J. Mater. Chem. 2006, 16, 155. [Google Scholar]
- 232. Li D., Müller M. B., Gilje S., Kaner R. B., Wallace G. G., Nat. Nanotechnol. 2008, 3, 101. [DOI] [PubMed] [Google Scholar]
- 233. Dey R. S., in Graphene‐Based Polymer Nanocomposites in Electronics (Eds: Sadasivuni K. K., Ponnamma D., Kim J., Thomas S.), Springer, Cham, Switzerland: 2015, p. 277. [Google Scholar]
- 234. Hangarter C. M., Chartuprayoon N., Hernández S. C., Choa Y., Myung N. V., Nano Today 2013, 8, 39. [Google Scholar]
- 235. Zheng Z., Du Y., Feng Q., Wang Z., Wang C., J. Mol. Catal. A Chem. 2012, 80, 353. [Google Scholar]
- 236. Pananon P., Sriprachuabwong C., Wisitsoraat A., Chuysinuan P., Tuantranont A., Saparpakorn P., Dechtrirat D., RSC Adv. 2018, 8, 12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Sun D., Li H., Li M., Li C., Dai H., Sun D., Yang B., Sens. Actuators, B 2018, 259, 433. [Google Scholar]
- 238. Huang T. Y., Kung C. W., Wang J. Y., Lee M. H., Chen L. C., Chu C. W., Ho K. C., Electrochim. Acta 2015, 172, 61. [Google Scholar]
- 239. Feng X. M., Li R. M., Ma Y. W., Chen R. F., Shi N. E., Fan Q. L., Huang W., Adv. Funct. Mater. 2011, 21, 2989. [Google Scholar]
- 240. Ott C., Raicopol M. D., Andronescu C., Vasile E., Hanganu A., Pruna A., Pilan L., Synth. Met. 2018, 235, 20. [Google Scholar]
- 241. Dechtrirat D., Sookcharoenpinyo B., Prajongtat P., Sriprachuabwong C., Sanguankiat A., Tuantranont A., Hannongbua S., RSC Adv. 2018, 8, 206. [Google Scholar]
- 242. Famularo G., Pizzicannella M., Gasbarrone L., J. Am. Geriatr.Soc. 2014, 62, 2018. [DOI] [PubMed] [Google Scholar]
- 243. Han L., Zhao Y., Chang C., Li F., J. Electroanal. Chem. 2018, 817, 141. [Google Scholar]
- 244. Bo Y., Yang H., Hu Y., Yao T., Huang S., Electrochim. Acta 2011, 56, 2676. [Google Scholar]
- 245. Amani J., Maleki M., Khoshroo A., Sobhani‐Nasab A., Rahimi‐Nasrabadi M., Anal. Biochem. 2018, 548, 53. [DOI] [PubMed] [Google Scholar]
- 246. Wang W., Xu G., Cui X. T., Sheng G., Luo X., Biosens. Bioelectron. 2014, 58, 153. [DOI] [PubMed] [Google Scholar]
- 247. Xu G., Jarjes Z. A., Desprez V., Kilmartin P. A., Travas‐Sejdic J., Biosens. Bioelectron. 2018, 107, 184. [DOI] [PubMed] [Google Scholar]
- 248. de Fátima Giarola J., Mano V., Pereira A. C., Electroanalysis 2018, 30, 119. [Google Scholar]
- 249. Zheng W., Zhao M., Liu W., Yu S., Niu L., Li G., Li H., Liu W., J. Electroanal. Chem. 2018, 813, 75. [Google Scholar]
- 250. Kaçar C., Erden P. E., Kılıç E., Electroanalysis 2017, 29, 2114. [Google Scholar]
- 251. Li Y., Zhang Y., Han G., Xiao Y., Li M., Zhou W., Chin. J. Chem. 2016, 34, 82. [Google Scholar]
- 252. Mao H., Liang J., Zhang H., Pei Q., Liu D., Wu S., Zhang Y., Song X. M., Biosens. Bioelectron. 2015, 70, 289. [DOI] [PubMed] [Google Scholar]
- 253. Sangamithirai D., Narayanan V., Muthuraaman B., Stephen A., Mater. Sci. Eng., C 2015, 55, 579. [DOI] [PubMed] [Google Scholar]
- 254. Ruecha N., Rangkupan R., Rodthongkum N., Chailapakul O., Biosens. Bioelectron. 2014, 52, 13. [DOI] [PubMed] [Google Scholar]
- 255. Mazloum‐Ardakani M., Barazesh B., Khoshroo A., Moshtaghiun M., Sheikhha M. H., Bioelectrochemistry 2018, 121, 38. [DOI] [PubMed] [Google Scholar]
- 256. Gupta P., Bharti A., Kaur N., Singh S., Prabhakar N., J. Electroanal. Chem. 2018, 813, 102. [Google Scholar]
- 257. Sharma A., Kumar A., Khan R., Synth. Met. 2018, 235, 136. [Google Scholar]
- 258. Zhu Y., Murali S., Cai W., Li X., Suk J. W., Potts J. R., Ruoff R. S., Adv. Mater. 2010, 22, 3906. [DOI] [PubMed] [Google Scholar]
- 259. Linting Z., Ruiyi L., Zaijun L., Qianfang X., Yinjun F., Junkang L., Sens. Actuators, B 2012, 174, 359. [Google Scholar]
- 260. Hui N., Wang S., Xie H., Xu S., Niu S., Luo X., Sens. Actuators, B 2015, 221, 606. [Google Scholar]
- 261. Claussen J. C., Kumar A., Jaroch D. B., Khawaja M. H., Hibbard A. B., Porterfield D. M., Fisher T. S., Adv. Funct. Mater. 2012, 22, 3399. [Google Scholar]
- 262. Zeng X., Zhang Y., Du X., Li Y., Tang W., New J. Chem. 2018, 42, 11944. [Google Scholar]
- 263. Lin M., Cho M., Choe W. S., Yoo J. B., Lee Y., Biosens. Bioelectron. 2010, 26, 940. [DOI] [PubMed] [Google Scholar]
- 264. Moozarm Nia P., Meng W. P., Lorestani F., Mahmoudian M. R., Alias Y., Sens. Actuators, B 2015, 209, 100. [Google Scholar]
- 265. Mir T. A., Akhtar M. H., Gurudatt N. G., Kim J. I., Choi C. S., Shim Y. B., Biosens. Bioelectron. 2015, 68, 421. [DOI] [PubMed] [Google Scholar]
- 266. Wang F., Zhu L., Zhang J., Sens. Actuators, B 2014, 192, 642. [Google Scholar]
- 267. Clark R., Borirakchanyavat V., Davidson A. R., Williams R., Thompson R. P. H., Widdop B., Goulding R., Lancet 1973, 301, 66. [DOI] [PubMed] [Google Scholar]
- 268. Li M., Wang W., Chen Z., Song Z., Luo X., Sens. Actuators, B 2018, 260, 778. [Google Scholar]
- 269. Wong A., Santos A. M., Fatibello‐Filho O., Sens. Actuators, B 2018, 255, 2264. [Google Scholar]
- 270. Luo X., Weaver C. L., Tan S., Cui X. T., J. Mater. Chem. B 2013, 1, 1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Rajesh, Bisht V., Takashima W., Kaneto K., Biomaterials 2005, 26, 3683. [DOI] [PubMed] [Google Scholar]
- 272. Liu S., Ma Y., Cui M., Luo X., Sens. Actuators, B Chem 2018, 255, 2568. [Google Scholar]
- 273. Sowmiya T., Ananthi A., Anandhakumar S., Mathiyarasu J., Anal. Methods 2012, 4, 1838. [Google Scholar]
- 274. Khan G. F., J. Electrochem. Soc. 1996, 143, 3336. [Google Scholar]
- 275. Albanese D., Malvano F., Sannini A., Pilloton R., Di Matteo M., Sensors 2014, 14, 11097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Bayer C. L., Konuk A. A., Peppas N. A., Biomed. Microdevices 2010, 12, 435. [DOI] [PubMed] [Google Scholar]
- 277. Daprà J., Lauridsen L. H., Nielsen A. T., Rozlosnik N., Biosens. Bioelectron. 2013, 43, 315. [DOI] [PubMed] [Google Scholar]
- 278. Aynaci E., Yas¸ar A., Arslan F., Sens. Actuators, B Chem 2014, 202, 1028. [Google Scholar]
- 279. Rubio F. M., Development of Antibodies for the Detection of the Toxin Anatoxin by Immunoassay, https://cfpub.epa.gov/ncer_abstracts/index.cfm/fuseaction/display.abstractDetail/abstract/7932/report/F (accessed: August 2018).
- 280. Barnard H., Development Cost of A Biosensor, https://www.barnardhealth.us/fractal-analysis/development-cost-of-a-biosensor.html (accessed: August 2018).
- 281. Wang G., Ma P., Zhang Q., Lewis J., Lacey M., Furukawa Y., O'Reilly S. E., Meaux S., McLachlan J., Zhang S., J. Environ. Monit. 2012, 14, 1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Sadana A., Engineering Biosensors: Kinetics and Design Applications (Google eBook), Academic Press; 2001. [Google Scholar]
- 283. Rogers K. R., Gerlach C. L., Environ. Sci. Technol. 1996, 30, 486A. [DOI] [PubMed] [Google Scholar]
- 284. Weetall H. H., Biosens. Bioelectron. 1996, 11, i. [DOI] [PubMed] [Google Scholar]
- 285. Downard A., Anal. Chem. 1998, 70, 774A. [Google Scholar]
- 286. Cornell B. A., Braach‐Maksvytis V. L. B., King L. G., Osman P. D. J., Raguse B., Wieczorek L., Pace R. J., Nature 1997, 387, 580. [DOI] [PubMed] [Google Scholar]
- 287. Parlak O., Keene S. T., Marais A., Curto V. F., Salleo A., Sci. Adv. 2018, 4, eaar2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Sadana A., Introduction BT—Fractal Binding and Dissociation Kinetics for Different Biosensor Applications, Elsevier, Amsterdam: 2005. [Google Scholar]
- 289. Yuan W., Liu A., Huang L., Li C., Shi G., Adv. Mater. 2013, 25, 766. [DOI] [PubMed] [Google Scholar]
- 290. Pillai C. K. S., Paul W., Sharma C. P., Prog. Polym. Sci. 2009, 34, 641. [Google Scholar]


