Abstract
The objective of this study was to investigate the use of chloroquine (CLQ) as an antiviral agent against dengue. Chloroquine, an amine acidotropic drug known to affect intracellular exocytic pathways by increasing endosomal pH, was used in the in vitro treatment of U937 cells infected with dengue virus type 2 (DENV‐2). Viral replication was assessed by quantification of virus produced through detection of copy numbers of DENV‐2 RNA, plaque assay and indirect immunofluorescence. qRT‐PCR and plaque assays were used to quantify the DENV‐2 load in infected U937 cells after CLQ treatment. It was found that a dose of 50 μg/mL of CLQ was not toxic to the cells and resulted in significantly less virus production in infected U937 cells than occurred in untreated cells. In the present work, CLQ was effective against DENV‐2 replication in U937 cells, and also caused a statistically significant reduction in expression of proinflammatory cytokines. The present study indicates that CLQ may be used to reduce viral yield in U937 cells.
Keywords: chloroquine, DENV‐2, plaque assay, quantitative reverse transcription polymerase chain reaction
List of Abbreviations
- cDNA
complementary DNA
- CLQ
chloroquine
- DENV‐2
dengue virus type 2
- DEPC
diethylpyrocarbonate
- RNA
ribonucleic acid
Dengue fever, a benign syndrome caused by dengue virus, is transmitted by arthropods (Aedes aegypti and Aedes albopictus) and characterized by a biphasic fever, myalgia or arthralgia, rash, leukopenia and lymphadenopathy. There are four antigenically distinct members of the dengue subgroup of the Flavivirus genus, named types 1–4 1. Each year it is estimated that around 50–100 million dengue cases occur worldwide, including about 500,000 cases of hemorrhagic dengue fever, the most severe manifestation of the disease 2. This form of the disease reportedly occurs in 5–10% of secondary infections, with a lethality rate of 10% 3.
Dengue viruses contain three structural proteins: capsid, which involves the virus genome, membrane and envelope, which are embedded in a lipid bilayer of the viral envelope 4, 5. The dengue viruses are single positive‐stranded RNA filaments of about 11 kb length. The genome codifies an open reading frame of nearly 10,200 nucleotides, which is processed by proteases of viral and cellular origin into three structural proteins codified by the 5′ region of the genome and seven non‐structural proteins (NS1‐NS5) codified by the remaining genomic region, in the order 5′ C‐prM‐E‐NS1‐NS2A‐NS2B‐NS3‐NS4A‐NS4B‐NS5 6, 7. Dengue viruses enter target cells via receptor‐mediated endocytosis through direct interaction of viral E glycoprotein with host cell receptors, including DC‐specific ICAM‐3‐grabbing non‐integrin 8, 9 and a mannose receptor 10, followed by acidic pH‐dependent fusion with the membranes of the endocytic vesicles 11.
Chloroquine, a 9‐aminoquinolin, was synthesized for the first time in Germany by Bayer (then Barmen, now Leverkusen, Germany) in 1934. By preventing the low pH‐induced fusion of viral envelope and cell endosome membranes, CLQ is potentially able to block the entry of certain viruses into the cytosol 12. The present study examined the effect of CLQ on DENV‐2 replication in U937 (mammalian) cells in order to define whether or not CLQ could be used to reduce viral yield.
MATERIALS AND METHODS
Ethics statement
This study was carried out in strict accordance with the Brazilian Government's ethical and animal experiments regulations. The procedures for using animals in this research project were in accordance with the ethical principles established by the Brazilian College of Animal Experimentation and the experimental protocol was approved by the Committee on the Ethics of Animal Experiments of the University of Sao Paulo (CETEA/USP, Permit Protocol Number 080/2006).
Cell cultures
Vero cells (an African green monkey kidney cell line) were grown in Leibovitz‐15 (L‐15) culture medium (Invitrogen, Grand Island, New York, USA) supplemented with 10% FBS, 1% L‐GLUTAMINE 200 mM, 1% penicillin G (100 U/mL), streptomycin (100 μg/mL) and 10% tryptose phosphate. The Vero cells were maintained at 37 °C and 5% CO2. C6/36, an Aedes albopictus cell line, was cultured in L‐15 medium supplemented with 10% FBS, l‐glutamine, 10% tryptose phosphate, 1% penicillin G (100 U/mL) and streptomycin (100 μg/mL) at 28 °C in the absence of CO2. Suspensions of U937 cells (monocytic lineage) were cultured in RPMI 1640 medium, supplemented with 0.29 g/L L‐GLUTAMINE, 2 g/L sodium bicarbonate, 25 mM/L HEPES, 1% penicillin G (100 U/mL), streptomycin (100 μg/mL) and 10% FBS (Invitrogen) and maintained at 37 °C and 5% CO2.
Virus stock
Parental DENV‐2 New Guinea C strain was recovered from the brains of 1‐ to 2‐day‐old suckling Swiss mice. This virus was passaged several times in C6/36 cells contained in 75 cm2 culture flasks with virus diluted in 1 mL of L‐15–2% FBS. After 1 hr, 14 mL of L‐15 supplemented with 10% FBS was added and the cells cultured for 7 days. Cells culture supernatants were then harvested and centrifuged at 2000g for 5 min to remove cell debris. The suspensions were kept in ice to prevent further microbial loss. The virus suspensions were adjusted to 20% FBS and then divided into aliquots and stored frozen at −70 °C. Virus stock and cell culture supernatants used in the present study were free of lipopolysaccharides and mycoplasma.
Dengue virus titration
Virus production was titrated by plaque assay using Vero cells. Vero cells were seeded in 12‐well (6 × 105 cells/well) plate in L‐15 medium with 10% FBS for 48 hr at 37 °C. The medium was removed and decimal serial dilutions of virus stock or supernatant of U937 cells treated with CLQ prepared in L‐15 medium with 2% FBS were added (0.1 mL/well) to the cells, which were then incubated for 2 hr at 37 °C. Subsequently, L‐15 medium containing 5% FBS and 3% carboxymethyl‐cellulose (1 mL/well) (overlay) was added and the plates incubated at 37 °C for 7 days. The overlay was removed on Day 7 and the cells fixed with a solution of 10% formaldehyde in PBS. After 2 hr at room temperature, the formaldehyde solution was removed and the cells washed twice with PBS and stained (15 min) with a 1% crystal violet solution in 20% ethanol. Plaques of cell lysis were counted and the virus concentration expressed as PFU per mL.
Chloroquine cytotoxic assay
Cytotoxicity induced by CLQ was assayed in U937 cells to determine the ideal concentration for the experiments. Suspension of U937 cells were seeded in 24‐well (1 × 104 cells/well) plates. After an incubation period of approximately 72 hr, the culture medium was replaced with RPMI medium containing 2% FBS and different concentrations of CLQ (Sigma‐Aldrich, St Louis, MO, USA). After incubation periods of 1, 6, 12, 24, 48, 72 and 96 hr with the drugs, the U937 cells were removed and their viability confirmed by the Trypan blue exclusion method (Invitrogen).
Indirect immunofluorescence
Indirect immunofluorescence assays were performed to determine the presence of dengue‐2 viral infection in U937 cells cultures treated with CLQ. To prepare the slides for immunofluorescence, cell suspensions were centrifuged at 1000 rpm for 5 min. The cells were then washed twice with PBS, placed on slides (10 µL/well), fixed in cold acetone for 10 min and stored at −20 °C. The slides were incubated with DENV‐2‐specific hyperimmune mouse ascitic fluid diluted 1:100 in PBS with 3% FBS at 37 °C for 30 min in a humidified atmosphere, and then rinsed four times in PBS. The samples were then similarly incubated with a 1:100 dilution of fluorescein isothiocyanate‐labeled goat anti‐mouse antibodies (Sigma‐Aldrich) in Evans blue at 37 °C for 30 min, rinsed four times in PBS and air dried. After washing, the coverslips were mounted in buffered glycerin, examined at 200–1000× and photographed using an inverted fluorescence microscope. Fluorescent foci that were located within a 0.7 cm2 area of each well were counted.
Treatment of DENV‐2 infected cells with CLQ
Suspensions of U937 cells (1 × 106 cells/mL) were seeded onto 24‐well plates infected with DENV‐2 (1 × 106 PFU/mL) at a MOI of 1. At 1 hr post‐infection, the inoculums were removed. U937 cells were incubated in the presence of different concentrations of CLQ (0, 0.05, 0.5, 5 and 50 μg/mL) at the following defined time intervals in different experiments: (i) concomitantly, 1 hr after infection; (ii) concomitantly, 1 hr after infection, and at 24‐hr intervals for 7 days; and (iii) concomitantly, 1 hr after infection and at 12‐hr intervals for 7 days. Mock controls for DENV‐2 replication contained no CLQ. Infected cell supernatants were collected at 0, 6, 12, 24, 48, 72, 96, 120, 144 and 168 hr post‐infection, cleared by centrifugation, and stored in aliquots at −70 °C until use in real‐time PCR and plaque assays. The experiments were carried out in duplicate and the results are shown as the mean values obtained from three individual experiments.
Viral RNA extraction
Viral RNA was extracted from 140 μL of each supernatant sample using a QIAamp Viral RNA kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions.
Total RNA extraction: TRIzol method (guanidinium thiocyanate‐phenol‐chloroform extraction)
Total RNA from DENV‐2 infected or uninfected U937 cells, treated and untreated CLQ (1 hr after infection and at 24–24 hr intervals) was extracted after 1, 2, 12, 48 and 72 hr post‐infection using the Trizol reagent (Invitrogen) according to the manufacturer's instructions.
qRT‐PCR assay
Real time q‐PCR was performed to determine the number of DENV‐2 RNA copies present in each supernatant, as described previously 13. Briefly, to construct a standard curve, the number of DENV‐2 particles produced by infected cells was measured as the number of RNA copies quantitated by qRT‐PCR in serially diluted infected cell supernatants. Each qRT‐PCR contained 12.5 μL of Sybr Green Master Mix reagent (Applied Biosystems, Foster, CA, USA), 0.5 μL of RNase inhibitor, 0.13 μL of MultiScribe (50 U/μL; Life Technologies, Carlsbad, CA, USA), 0.5 μL of primers (20 nM) DV2U (5′‐AAGGTGAGATGAAGCTGTAGTCTC‐3′), and DVL1 (5′‐CATTCCATTTTCTGGCGTTCT‐3′) 14 specifically designed to anneal to the DENV‐2 3′‐untranslated region (3′‐UTR), 5.87 μL of diethylpyrocarbonate water and 5 μL of RNA to a final volume of 25 μL. The amplification protocol consisted of the following steps: 48 °C for 30 min, 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 sec, and finally 60 °C for 1 min. The same protocol was used to quantify the DENV‐2 RNA copies present in the supernatants of CLQ‐treated and untreated U937 cells infected with DENV‐2, collected at defined post‐infection intervals, as described above.
Relative quantification of cytokines (IFN‐α, IFN‐β, IFN‐γ, TNF‐α, IL‐6, IL‐12 and IL‐10) in relation to the transcripts of β‐actin gene (endogenous control) was performed using PCR reactions in real time using a QuantiTect SYBR Green PCR kit (Qiagen). The reactions were performed in a final volume of 25 µL containing 1 µL of cDNA, 12.5 µL of 2× QuantiTect SYBR Green PCR Master Mix (HotStarTaq DNA polymerase, QuantiTect SYBR Green PCR buffer, SYBR Green I and ROX fluorophore used as a passive reference for normalization levels of fluorescence) and 0.5 µL of oligonucleotide sense and antisense (20 nm) (Table 1) under the following conditions: 95 °C for 15 min, followed by 40 cycles of 94 °C for 15 sec, 60 °C for 30 sec, and 72 °C for 30 sec.
Table 1.
Primers for quantitative real‐time PCR
| Primers | Nucleotide sequences (5′ → 3′) |
|---|---|
| β‐actin | (forward) GGACTTCGAGCAAGAGATGG |
| (reverse) AGCACTGTGTTGGCGTACAG | |
| IL‐6 | (forward) CCAGGAGCCCAGCTATGAAC |
| (reverse) CCCAGGGAGAAGGCAACTG | |
| TNF‐α | (forward) CCTACCAGACCAAGGTCAAC |
| (reverse) AGGGGGTAATAAAGGGATTG | |
| IL‐12 | (forward) TGCAAAGCTTCTGATGGATCC |
| (reverse) AAAATCCGGTTCTTCAAGGGA | |
| IFN‐α | (forward) GATGGCAACCAGTTCCAGAAG |
| (reverse) AAAGAGGTTGAAGATCTGCTGGAT | |
| IFN‐β | (forward) TTGTGCTTCTCCACTACAGC |
| (reverse) CTGTAAGTCTGTTAATGAAG | |
| IFN‐γ | (forward) TTTTAATGCAGGTCATTCAGATGT |
| (reverse) AAGTTTGAAGTAAAAGGAGACAATTTGG | |
| IL‐10 | (forward) CGAGATGCCTTCAGCAGAGTG |
| (reverse) TCATCTCAGAACAAGGCTTGGC |
Complementary DNA synthesis
The extracted RNA was converted into cDNA through reactions with reverse transcriptase enzyme M‐MLV (Invitrogen) according to the manufacturer's protocol. To prepare the cDNA, concentrations of 400 ηg of total RNA were used for all samples.
Statistical analysis
Statistical analysis was used to assess differences between viral yield at time‐defined intervals from infected cells in contact with CLQ and control cells (without the drug). Data were entered into the GraphPad Prism software, version 6.0 and submitted to one‐way ANOVA (nonparametric test) analysis followed by the Bonferroni test. Cytokine mRNA quantification by real‐time PCR was tested using Student's t‐test. for all analyses, values of P < 0.05 were considered statistically significant.
RESULTS
Cytotoxicity of CLQ in U937 cells
Chloroquine was highly cytotoxic to U937 cells when they were treated with a concentration ≥500 μg/mL, whereas no significant cytotoxicity was observed when the cells were treated with a concentration ≤50 μg/mL (Fig. 1). Based on these data, CLQ was used in concentrations ≤50 μg/mL in these experiments.
Figure 1.
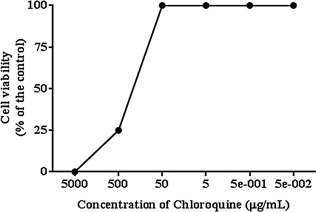
The effect of CLQ on the cytotoxicity in U937 cells.
Concentrations equal or higher than 500 μg/mL (CLQ) were highly cytotoxic to U937 cells, while concentrations equal or lower than 50 μg/mL (CLQ) did not induce significant cytotoxicity.
Effect of CLQ on DENV‐2 replication as assessed by qRT‐PCR
To determine whether CLQ inhibits DENV‐2 replication, suspensions of U937 cells were infected with DENV‐2 and incubated with 2% FBS RPMI medium containing different concentrations of the drug; then virus production was quantified by qRT‐PCR and plaque assay. Results of qRT‐PCR showed that CLQ inhibited DENV‐2 replication in U937 cells in a dose‐dependent manner (Fig. 2a). Viral replication in U937 cells was significantly less after addition of ≥ 5 μg/mL of CLQ 1 hr after infection than in untreated cells; this inhibition was maintained up to 24 hr after infection (Fig. 2). The same results were obtained when virus production was assessed by plaque assay, the results showing an excellent correlation with those of qRT‐PCR (Fig. 2b).
Figure 2.
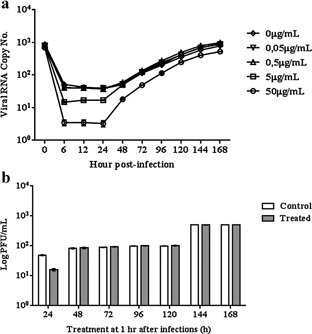
Action of CLQ on DENV‐2 replication in U937 cells.
(a) The viral RNA present in the culture supernatants of U937cells infected with DENV‐2, treated and untreated CLQ post‐infection, was extracted and analyzed by qRT‐PCR. (b) Culture supernatants of U937cells infected with DENV‐2, treated and untreated CLQ post‐infection, were analyzed by plaque assay. The results represent the average values of the viral RNA copy number (P < 0.001).
Because the duration of viral inhibition effect induced by CLQ may have been related to the timing of drug administration, the antiviral effect of CLQ was further analyzed in U937 cells treated with the drug 1 hr after infection and at 24‐ and 12‐hr intervals post‐infection. qRT‐PCR showed that treated cells produced significantly less virus than untreated cells up to the 7th day after infection (Figs 3a and 4). Again, assessment by plaque assay was in agreement with that by qRT‐PCR (Fig. 3b).
Figure 4.
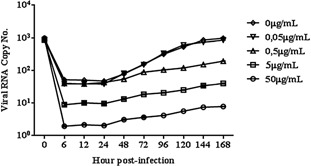
Action of CLQ added at 12‐hr intervals on DENV‐2 replication in U937 cells.
The viral RNA present in the culture supernatants of U937 cells infected with DENV‐2, treated and untreated CLQ (every 12 hr post‐infection), was extracted and analyzed by qRT‐PCR. The results represent the average values of the viral RNA copy number (P < 0.05).
Figure 3.
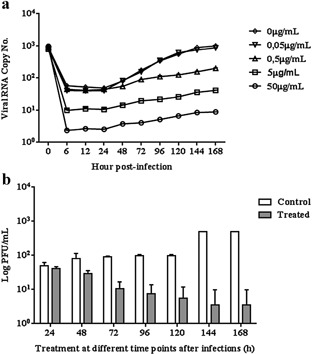
Action of CLQ added at 24‐hr intervals on DENV‐2 replication in U937 cells.
(a) The viral RNA present in the culture supernatants of U937 cells infected with DENV‐2, treated and untreated CLQ (every 24 hr post‐infection), was extracted and analyzed by qRT‐PCR. (b) Culture supernatants of U937 cells infected with DENV‐2, treated and untreated CLQ (every 24 hr post‐infection), was analyzed by plaque assay. The results represent the average values of the viral RNA copy number (P < 0.05).
Effect of CLQ on DENV‐2 replication assessed by IFA
U937 cells infected or uninfected with dengue‐2 virus in the presence and absence of CLQ added 1 hr after infection and thereafter at 24 hr intervals yielded significantly less virus than untreated cells up to the sixth day after infection (Fig. 5).
Figure 5.
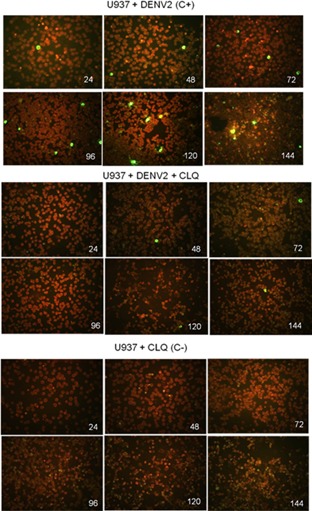
Indirect immunofluorescence assay.
Detection of DENV‐2 virus in U937 cells infected or uninfected, treated and untreated CLQ (every 24 hr post‐infection). (Original magnification 200–1,000×).
Analysis of cytokines by real‐time PCR
To determine whether CLQ interfered with IFN‐α, IFN‐β, IFN‐γ, TNF‐α, IL‐6, IL‐12 and IL‐10 release pretranslation, the effect of this drug on cytokines’ mRNA expression was investigated by real‐time PCR in U937 cells infected or uninfected with DENV‐2 and treated and untreated with CLQ (Fig. 6). In infected U937 cells, treatment with CLQ downregulated expression of the proinflammatory cytokines, IFN‐β, IFN‐γ, TNF‐α, IL‐6 and IL‐12 compared with the positive control (Fig. 6b–f). In the case of IFN‐α, expression was inhibited at hour 72 post‐infection (Fig. 6a). There was no statistically significant difference in expression of the anti‐inflammatory cytokine IL‐10 compared with the positive control (Fig. 6g).
Figure 6.
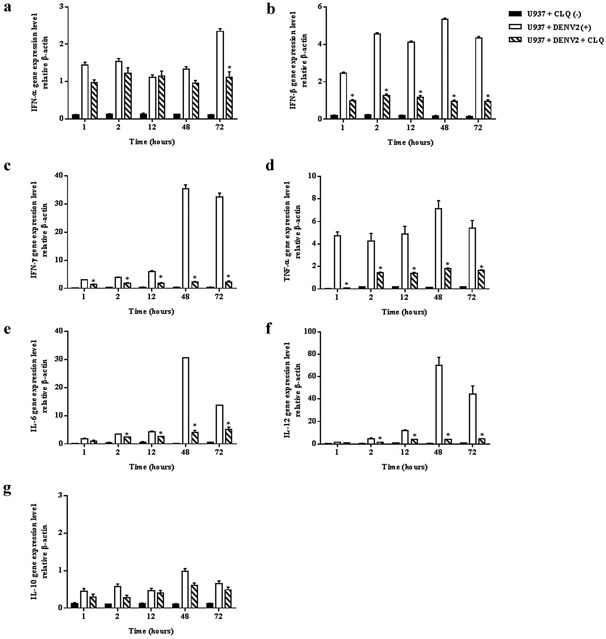
Relative quantification of cytokines by real‐time PCR in U937 cells infected or not with DENV‐2 in the presence or absence of CLQ added at 24‐hr intervals.
IFN‐α (a), IFN‐β (b), IFN‐γ (c), TNF‐α (d), IL‐6 (e), IL‐12 (f), and IL‐10 (g) mRNA expression in DENV‐2 infected or uninfected U937 cells, treated and untreated CLQ were analyzed by quantitative RT‐PCR, and normalized to mouse beta actin gene. Values were submitted to t test in which *P < 0.05 vs. U937 cells infected with DENV‐2s.
DISCUSSION
In addition to its antimalarial effect, CLQ may interfere with the replication cycle of many viruses by interacting with viral entry mediated by endosomes or in the late stages of replication of enveloped viruses 15.
Chloroquine, an 9‐aminoquinoline, was first synthesized in Germany by Bayer in 1934 and emerged during the first half of the 20th century as an effective substitute for quinine, the previous drug of choice against malaria 16, 17. It has multiple mechanisms of action; these differ according to the pathogen. It is a weak base which increases the pH of acidic vesicles and is concentrated in acidic organelles such as endosomes, lysosomes, vesicles and the Golgi by a mechanism involving a proton pump which requires energy, increasing the pH.
During the last decades, two major mechanisms of action of CLQ have been well described: alkalization of acidic vesicles, thereby interfering with endosome‐mediated viral entry, and alteration of post‐translational modifications of proteins newly synthesized in cells infected by virus 18.
We demonstrate here the antiviral effect of CLQ on U937 cells infected with DENV‐2. Our findings are in agreement with those reported by Farias et al., who showed that when Vero cells infected with DENV‐2 are treated with CLQ at 1 hr post‐infection and at different time intervals (24/12 hr), viral replication decreases in a dose dependent manner 19.
In vitro studies have demonstrated that CLQ has a strong antiviral effect on viruses such such as hepatitis B virus, herpes simplex virus type 1, influenza virus, human immunodeficiency virus, severe acute respiratory syndrome (coronavirus) and Epstein‐Barr virus 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30. Our in vitro study has demonstrated the therapeutic action of CLQ after exposure to the dengue‐2 virus (1 hr after infection). Recent studies have shown that CLQ treatment inhibits DENV‐2‐induced mTRAIL relocalization and IFN‐α production by plasmacytoid dendritic cells 31. One study showed that patients with dengue treated with CLQ have an improved quality of life and are able to resume their daily activities 32.
Tricou et al. evaluated the effect of CLQ in adult patients with dengue and found that treatment with CLQ had no effect on viremia or NS1 antigenemia 33. Their study had some limitations. They measured viremia by quantitative RT‐PCR as a surrogate and well‐characterized marker of infection, though they recognized this is not the same as a quantitative biological assay of infectious virus 33. In our in vitro study, we measured viral yield by quantitative RT‐PCR and plaque assay.
Regarding the amount of cytokines in cell culture (U937) as assessed by real‐time PCR, we found that infected U937 cells treated with CLQ contained significantly less IFN‐β, IFN‐γ, TNF‐α, IL‐6, and IL‐12 than the positive control (Fig. 6b–f). CLQ reduced IFN‐α production 72 hr post‐infection (6a). The drug reduced production of proinflammatory cytokines by interfering with mRNA synthesis of IL‐12, IFN‐γ, IL‐6, TNF‐α, IFN‐β and IFN‐α.
Van den borne et al. showed that CLQ decreases production of proinflammatory cytokines such as INF‐γ, TNF‐α and IL‐6 in peripheral blood mononuclear cells stimulated with LPS 34. Chloroquine inhibits the release of TNF‐α in macrophages by inhibiting synthesis of TNF‐α mRNA 35, 36. Jang et al. showed that CLQ interferes with production of the cytokines TNF‐α, IL‐1β and IL‐6 from monocytes/macrophages stimulated with LPS 37. Chloroquine reportedly decreases expression of TNF‐α receptors on the surfaces of human histiocytic U937 cells by delaying its transport to cell surface 38. Gandini et al. showed that CLQ treatment completely blocks IFN‐α secretion by plasmacytoid dendritic cells 31. Silva et al. showed that CLQ‐mediated blocking of this process partially inhibits West Nile virus‐induced IFN‐α production by plasmacytoid dendritic cells cultures 39.
Our results corroborate those of such studies, demonstrating that CLQ interferes with expression of proinflammatory cytokines.
We postulated that the reduction in proinflammatory cytokines in infected cells treated with CLQ could be due to the drug's anti‐inflammatory effects, particularly because CLQ is a TNF‐α inhibitor 40. Another possible mechanism of the effect of CLQ in cells infected with dengue virus could be via its antiviral action, which decreases the viral load and thus the degree of immune system activation.
In conclusion, the therapeutic use as CLQ, which is considered a safe and effective drug and is used to treat many diseases, including malaria, could be expanded because we have shown in this study that the drug has a significant antiviral effect on the replication of DENV‐2 in cells culture. Chloroquine is a well known immunomodulatory agent 15. These data indicate that CLQ inhibits IFN‐α, IFN‐β, IFN‐γ, TNF‐α, IL‐6 and IL‐12 genes expression in U937cells infected with DENV‐2 and treated with CLQ.
DISCLOSURE
All authors declare that they have no conflict of interests.
ACKNOWLEDGMENTS
This study was supported by the São Paulo Foundation for Research (FAPESP; Grant no. 05/04450‐4). K.J.S. Farias was supported by a FAPESP fellowship (Grant no. 06/55789‐4).
REFERENCES
- 1. Karabatsos N.( 1985) International Catalog of Arboviruses Including Certain Other Viruses of Vertebrates. 3rd edn. Fort Collins, CO: American Society of Tropical Medicine & Hygiene, p. 1147. [Google Scholar]
- 2. Gibbons R.V., Vaughn D.W. ( 2002) Dengue: An escalating problem. Br Med J 324: 1563–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rigau‐pérez J.G., Clark G.G., Gubler J.D., Reiter P., Sanders J.E., Vorndam V. ( 1998) Dengue and dengue haemorrhagic fever. Lancet 353: 971–7. [DOI] [PubMed] [Google Scholar]
- 4. Monath T.P., Burke D.S.( 2001) Flaviviruses. Fields Virology. Vol. 33, 4th edn. Philadelphia: Lippincott Willians & Wilkins, pp. 1043–1126. [Google Scholar]
- 5. Rey F.A. ( 2003) Dengue virus envelope glycoprotein structure: New insight into its interactions during viral entry. Proc Natl Acad Sci U S A 100: 6899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers T.C., Hahn C.S., Galler R., Rice C.M. ( 1990) Flavivirus genome organization, expression, and replication. Annu Rev Microbiol 44: 649–88. [DOI] [PubMed] [Google Scholar]
- 7. Rice C.M. ( 1996) Flaviviridae: The viruses and their replication. Fields Virology. In: Fields BN, Knipe DM, Howley PMed. 3rd edn. New York: Lippincott‐Raven, pp. 931–959. [Google Scholar]
- 8. Tassaneetrithep B., Burgess T.H., Granelli‐piperno A., Trumpfheller C., Finke J., Sun W., Eller M.A., Pattanapanyasat K., Sarasombath S., Birx D.L., Steinman R.M., Schlesinger S., Marovich M.A. ( 2003) DC‐SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med 197: 823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lozach P.Y., Burleigh L., Staropoli I., Navarro‐sanchez E., Harriague J., Virelizier J.L., Rey F.A., Despres P., Arenzana‐seisdedos F., Amara A. ( 2005) Dendritic cell‐specific intercellular adhesion molecule 3‐grabbing non‐integrin (DCSIGN)‐mediated enhancement of dengue virus infection is independent of DC‐SIGN internalization signals. J Biol Chem 280: 23698–708. [DOI] [PubMed] [Google Scholar]
- 10. Miller J.L., Dewet B.J., Martinez‐pomares L., Radcliffe C.M., Dwek R.A., Rudd P.M., Gordon S. ( 2008) The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog 4: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marianneau P., Steffan A.M., Royer C., Drouet M.T., Kirn A., Deubel V. ( 1998) Differing infection patterns of dengue and yellow fever viruses in a human hepatoma cell line. J Infect Dis 178: 1270–8. [DOI] [PubMed] [Google Scholar]
- 12. de Duve C. ( 1983) Lysosomes revisited. Eur J Biochem 137: 391–7. [DOI] [PubMed] [Google Scholar]
- 13. Ruiz A.C.G., Nascimento R.T., de Paula S.O., da Fonseca B.A.L. ( 2006) SYBR green and TaqMan real‐time PCR assays are equivalent for the diagnosis of dengue virus type 3 infections. J Med Virol 78: 760–3. [DOI] [PubMed] [Google Scholar]
- 14. Houng H.S.H., Chen R.C., Vaughn D.W., Kanesathasan N. ( 2001) Development of a fluorogenic RT‐PCR system for quantitative identification of dengue virus serotypes 1‐4 using conserved and serotype‐specific 3(noncoding sequences. J Virol Methods 95: 19–32. [DOI] [PubMed] [Google Scholar]
- 15. Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. ( 2003) Effects of chloroquine on viral infections: An old drug against today's diseases?. Lancet Infect Dis 3: 722–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bruce‐chwatt L.J. ed. ( 1981) Chemotherapy of Malaria. 2nd edn. WHO Monograph Series 27. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 17. Wellems T.E., Plowe C.V. ( 2001) Chloroquine‐resistant malaria. J Infect Dis 184: 770–6. [DOI] [PubMed] [Google Scholar]
- 18. Rolain J.M., Colson P., Raoult D. ( 2007) Recycling of chloroquine and its hydroxyl analogue to face bacterial, fungal and viral infections in the 21st century. Int J Antimicrob Agents 30: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farias K.J.S., Machado P.R.L., Fonseca B.A.L. ( 2013) Chloroquine inhibits dengue virus type 2 replication in Vero cells but not in C6/36 cells. Sci World J 2013: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Civitico G., Wang Y.Y., Luscombe C., Bishop N., Tachedjian G., Gust I., Locarnini S. ( 1990) Antiviral strategies in chronic hepatitis B virus infection: II. Inhibition of duck hepatitis B virus in vitro using conventional antiviral agents and supercoiled‐DNA active compounds. J Med Virol 31: 90–7. [DOI] [PubMed] [Google Scholar]
- 21. Offensperger W.B., Offensperger S., Walter E., Blum H.E., Gerok W. ( 1991) Inhibition of duck hepatitis B virus infection by lysosomotropic agents. Virology 183: 415–8. [DOI] [PubMed] [Google Scholar]
- 22. Koyama A.H., Uchida T. ( 1984) Inhibition of multiplication of herpes simplex virus type 1 by ammonium chloride and chloroquine. Virology 138: 332–5. [DOI] [PubMed] [Google Scholar]
- 23. Ooi E.E., Chew J.S., Loh J.P., Chua R.C. ( 2006) In vitro inhibition of human influenza A virus replication by chloroquine. Virol J 3: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sperber K., Kalb T.H., Stecher V.J., Banerjee R., Mayer L. ( 1993) Inhibition of human immunodeficiency virus type 1 replication by hydroxychloroquine in T cells and monocytes. AIDS Res Hum Retroviruses 9: 91–8. [DOI] [PubMed] [Google Scholar]
- 25. Pardridge W.M., Yang J., Diagne A. ( 1998) Chloroquine inhibits HIV‐1 replication in human peripheral blood lymphocytes. Immunol Lett 64: 45–47. [DOI] [PubMed] [Google Scholar]
- 26. Savarino A., Lucia M.B., Rastrelli E., Rutella S., Golotta C., Morra E., Tamburrini E., Perno C.F., Boelaert J.R., Sperber K., Cauda R. ( 2004) Anti‐HIV effects of chloroquine: Inhibition of viral particle glycosylation and synergism with protease inhibitors. J Acquir Immune Defic Syndr 35: 223–32. [DOI] [PubMed] [Google Scholar]
- 27. Keyaerts E., Vijgen L., Maes P., Neyts J., Ranst M.V. ( 2004) In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Biophys Res Commun 323: 264–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. ( 2005) Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Clercq E. ( 2006) Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev Anti Infect Ther 4: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller N., Hutt‐fletcher L.M. ( 1992) Epstein‐Barr virus enters B cells and epithelial cells by different routes. J Virol 66: 3409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gandini M., Gras C., Azeredo E.L., Pinto L.M., Smith N., Despres P., da Cunha R.V., de Souza L.J., Kubelka C.F., Herbeuval J.P. ( 2013) Dengue virus activates membrane TRAIL relocalization and IFN‐α production by human plasmacytoid dendritic cells in vitro and in vivo. PLoS Negl Trop Dis 7: e2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borges M.C., Castro L.A., da Fonseca B.A.L. ( 2013) Chloroquine use improves dengue‐related symptoms. Mem Inst Oswaldo Cruz. Mem Inst Oswaldo Cruz, Rio de Janeiro 108 (5): 596–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tricou V., Minh N.N., Van T.P., Lee S.J., Farrar J., Wills B., Tran H.T., Simmons C.P. ( 2010) A randomized controlled trial of chloroquine for the treatment of dengue in Vietnamese adults. PLoS Negl Trop Dis 4: e785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van den borne B.E., Dijkmans B.A., de Rooij H.H., le Cessie S., Verweij C.L. ( 1997) Chloroquine and hydroxychloroquine equally affect tumor necrosis factor‐alpha, interleukin 6, and interferon‐gamma production by peripheral blood mononuclear cells. J Rheumatol 24: 55–60. [PubMed] [Google Scholar]
- 35. Weber S.M., Levitz S.M. ( 2000) Chloroquine interferes with lipopolysaccharide‐induced TNF‐alpha gene expression by a nonlysosomotropic mechanism. J Immunol 165: 1534–40. [DOI] [PubMed] [Google Scholar]
- 36. Weber S.M., Chen J.M., Levitz S.M. ( 2002) Inhibition of mitogen‐activated protein kinase signaling by chloroquine. J Immunol 168: 5303–9. [DOI] [PubMed] [Google Scholar]
- 37. Jang C.H., Choi J.H., Byun M.S., Jue D.M.( 2006) Chloroquine inhibits production of TNF‐alpha, IL‐1beta and IL‐6 from lipopolysaccharide‐stimulated human monocytes/ macrophages by different modes. Rheumatology (Oxford) 45: 703–10. [DOI] [PubMed] [Google Scholar]
- 38. Jeong J.Y., Choi J.W., Jeon K.I., Jue D.M. ( 2002) Chloroquine decreases cell‐ surface expression of tumour necrosis factor receptors in human histiocytic U‐937 cells. Immunology 105: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silva M.C., Guerrero‐Plata A., Gilfoy F.D., Garofalo R.P., Mason P.W. ( 2007) Differential activation of human monocyte‐derived and plasmacytoid dendritic cells by West Nile virus generated in different host cells. J Virol 81: 13640–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jeong J.Y., Jue D.M. ( 1997) Chloroquine inhibits processing of tumor necrosis factor in lipopolysaccharide‐stimulated RAW 264. 7 macrophages. J Immunol 158: 4901–7. [PubMed] [Google Scholar]


