Abstract
The 1918 Spanish flu virus has claimed more than 50 million lives. However, the mechanism of its high pathogenicity remains elusive; and the origin of the virus is controversial. The matrix (M) segment regulates the replication of influenza A virus, thereby affecting its virulence and pathogenicity. This study found that the M segment of the Spanish flu virus is a recombinant chimera originating from avian influenza virus and human influenza virus. The unique mosaic M segment might confer the virus high replication capacity, showing that the recombination might play an important role in inducing high pathogenicity of the virus. In addition, this study also suggested that the NA and NS segments of the virus were generated by reassortment between mammalian and avian viruses. Direct phylogenetic evidence was also provided for its avian origin.
1. INTRODUCTION
The 1918 “Spanish Flu” was the most devastating pandemic in modern history (Honigsbaum, 2018). In 11 months between the spring of 1918 and the winter of 1919, it killed approximately 50 million people worldwide (Johnson & Mueller, 2002; Patterson & Pyle, 1991). The morbidity pattern, together with the rapid disease progression to multiorgan failure and death, characterizes the influenza pandemic (Honigsbaum, 2018). So far, its origin, its unusual epidemiologic features, and the basis of its pathogenicity remain elusive (Taubenberger & Morens, 2006).
Its pathogen, a variant of H1N1 influenza A virus (IVA) (Gibbs, Armstrong, & Gibbs, 2001; Kilbourne, 2006), has the eight‐segmented genome housed in an enveloped virion (Noda et al., 2006). Knowing the origin of the virus might be a key for uncovering its epidemiologic features and pathogenicity basis (Reid & Taubenberger, 1999). However, there are two controversies regarding its origin. The first one is whether the virus originated from mammalian IVA reassortment or directly from an avian virus (Antonovics, Hood, & Baker, 2006; Gibbs & Gibbs, 2006; Taubenberger, 2006; Vana & Westover, 2008). And, the second one is whether homologous recombination shaped the formation of the virus (Gibbs et al., 2001; Worobey, Rambaut, Pybus, & Robertson, 2002).
Based on a comparison analysis of the amino acid (AA) sequences of the viral proteins, Taubenberger et al., (2005). found that some proteins have the characteristics of avian viruses, and proposed that the virus was not a reassortant, but an avian virus in origin. However, this interpretation was questioned by several other groups because some genes of the virus were obviously clustered into swine or human branches in IVA phylogenetic trees (Antonovics et al., 2006; Gibbs & Gibbs, 2006; Vana & Westover, 2008). Therefore, the virus is considered a reassortant from mammalian IVAs (Smith et al., 2009).
Besides of genetic reassortment (Taubenberger, 2006), some additional events might also contribute to the formation of the virus and the significant change of its phenotype triggering the pandemic (Basler et al., 2001; Reid, Fanning, Hultin, & Taubenberger, 1999; Webster, 1999). Novel virulent variants of several other viruses have been shown to be generated through homologous recombination (Parrish et al., 2008; Sabir et al., 2016; Worobey, Rambaut, & Holmes, 1999). It was thus proposed that homologous recombination also occurred in the HA gene and resulted in the increased virulence associated with the pandemic (Gibbs et al., 2001). Unfortunately, the recombination event was doubted because of the complete absence of phylogenetic evidence for recombination in HA (Worobey et al., 2002).
Of the eight segments in IVA, the segment 7 encodes the two matrix (M) proteins, M1 and M2 (Lamb, Lai, & Choppin, 1981; Winter & Fields, 1980). The open reading frames (ORFs) of the M1 and M2 genes share the first nine codons at the N‐terminus, and the C‐terminus of M1 overlaps with a region of M2. M1 consists of 252 AAs, while M2 consists of 97 AAs encoded by an alternatively spliced transcript. Lining the inner layer of the viral membrane and contacting the ribonucleoprotein (RNP) core, M1 is highly conserved and is the most abundant protein in viral particles (Reid, Fanning, Janczewski, McCall, & Taubenberger, 2002). M1 regulates the nuclear export of viral RNPs (Bui, Wills, Helenius, & Whittaker, 2000; Martin & Helenius, 1991), restricts viral replication (Liu & Ye, 2002), inhibits viral transcription in the late stages of infection and the switch from replication to viral assembly (Perez & Donis, 1998; Ye, Baylor, & Wagner, 1989) and influences virus assembly and budding (Gomez‐Puertas, Albo, Perez‐Pastrana, Vivo, & Portela, 2000; Helenius, 1992; Latham & Galarza, 2001). Anchored in the viral envelope, M2 serves as a transmembrane ion channel (Lamb, Zebedee, & Richardson, 1985; Sugrue & Hay, 1991) and plays key roles in both virion uncoating and viral budding (Grambas & Hay, 1992; Pinto, Holsinger, & Lamb, 1992).
To clarify the origin of the Spanish flu virus, we re‐dissected the phylogenetic history of the virus and found that its M segment was a mosaic recombined from human and avian influenza viruses, showing that homologous recombination is truly linked with the most devastating pandemic. In addition, we also provided direct phylogenetic evidence for the avian origin of the virus.
2. MATERIALS AND METHODS
2.1. Viruses
The segment sequences of the 1918 IVA isolate A/Brevig Mission/1/1918(H1N1) are from previous reports (Basler et al., 2001; Reid et al., 1999, 2002; Reid, Fanning, Janczewski, Lourens, & Taubenberger, 2004; Reid, Fanning, Janczewski, & Taubenberger, 2000; Taubenberger et al., 2005). Referring to previous studies (Smith et al., 2009; Vana & Westover, 2008), eight segments of classical strains of different species (human, avian, swine, and horse) and A/Brevig Mission/1/1918(H1N1) were respectively concatenated to analyze the potential reassortment or recombination event between viruses of different species in A/Brevig Mission/1/1918(H1N1). Employing basic local alignment search tool (BLAST), reassortment between viruses of different species in these representative genomes was checked through comparing their sequence identity with IVAs deposited in GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Based on sequence identity to the query segment, the host species distribution of the top 100 sbjct viruses is shown in Table S1. Except for PB1 of A/chicken/Rostock/45/1934 (H7N1) that may be associated with human IVA, no other reassortment events were found in these representative viruses. To dissect the phylogenetic history of the M segment of the Spanish flu virus, 23 viruses of different years (from 1902 to 2012) and different species were selected. Information about these viruses was included in virus name.
2.2. Phylogenetic analysis
The sequences of IVAs were aligned with the MUSCLE programme implemented in MEGA 6 (Tamura et al., 2011). The phylogenetic histories of these viruses were inferred using the Maximum Likelihood (ML) or Neighbour‐Joining (NJ) methods based on the best substitution model selected by the model test programme in MEGA 6 (Tamura et al., 2011). The robustness of each lineage was tested using the bootstrap method (≥1,000 replicates). And the monophylogenetic lineage with a bootstrap value ≥70% was considered robust.
2.3. Recombination analysis
Recombination analysis was carried out as per our previous studies (He et al., 2012, 2009). In brief, to distinguish the recombinant, similarity comparison of nucleotide sequences between the putative recombinants and their parents was performed using the sliding window method in the Simplot programme (Lole et al., 1999). The sequence with contradiction identity was considered to be a putative recombinant. Integrating the Fisher's exact test method, we identified the putative breakpoints (p < 0.05) with the maximum chi‐square value of information site.
A set of statistically incongruent phylogenetic trees were recommended as the gold‐standard approach for confirming the presence of recombination (Boni, Jong, Doorn, & Holmes, 2010). Therefore, the recombinants were finally determined through the incongruent phylogenetic histories of different regions delimited by the putative recombination breakpoints. Shimodaira–Hasegawa test was implemented to prove whether phylogenetic trees estimated from different regions were significantly different employing the Tree test programme (http://aix1.uottawa.ca/~sarisbro/).
3. RESULTS
In order to dissect the phylogenetic history of the M segment of the Spanish flu virus, 23 IVA representatives including H1N1 human (n = 11), classical H1N1 swine (n = 2), mixture avian (n = 8) and H7N7 equine (n = 2) were used to infer the origin of the M segment of the virus. According to the phylogenetic history constructed from the complete M segments of these viruses, the Spanish flu virus belongs to the same monophylogentic lineage as the human H1N1 viruses (Figure 1).
Figure 1.
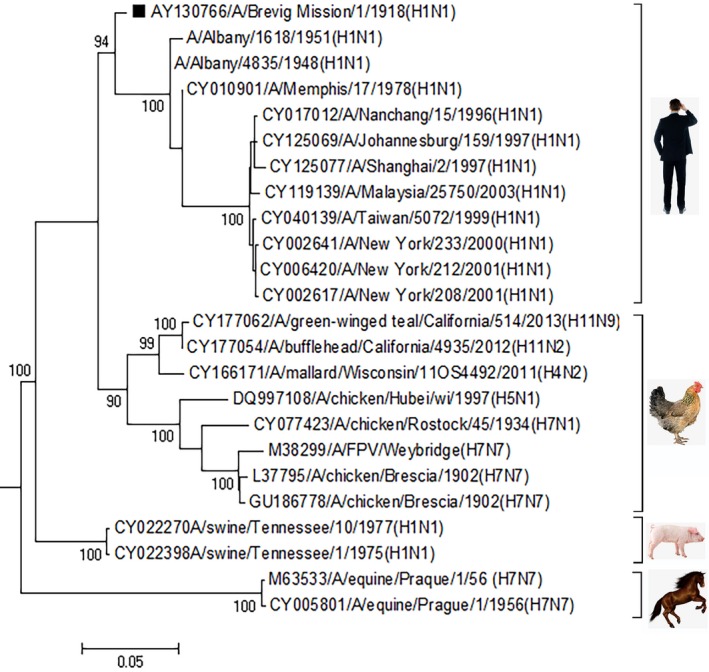
The 1918 “Spanish Flu” virus evolutionary history. The M segment phylogenetic history of influenza viruses was reconstructed using the Maximum Likelihood method. The percentage (>80%) of replicate trees in which influenza virus clustered together in the bootstrap test (1,000 replicates) are shown above the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances, computed using the Kimura 2‐parameter method, are in the units of the number of base substitutions per site [Colour figure can be viewed at http://wileyonlinelibrary.com]
Using the horse flu viruses as the control group and the Spanish flu virus as the query, we compared the M sequence of the Spanish flu virus with those of human and avian flu viruses and discovered the crossover site between human and avian viruses in the M segment of the Spanish flu virus. Before the site, the Spanish flu virus had higher similarity with the human viruses; otherwise it was more like the bird viruses (Figure 2a). Using the Spanish flu virus as the query to compare the M segment polymorphic sites of human and avian flu viruses, this contradiction in sequence similarity signal became clearer (Figure 2a). Interestingly, bootscan analysis gave two putative breakpoints (Figure 2b).
Figure 2.
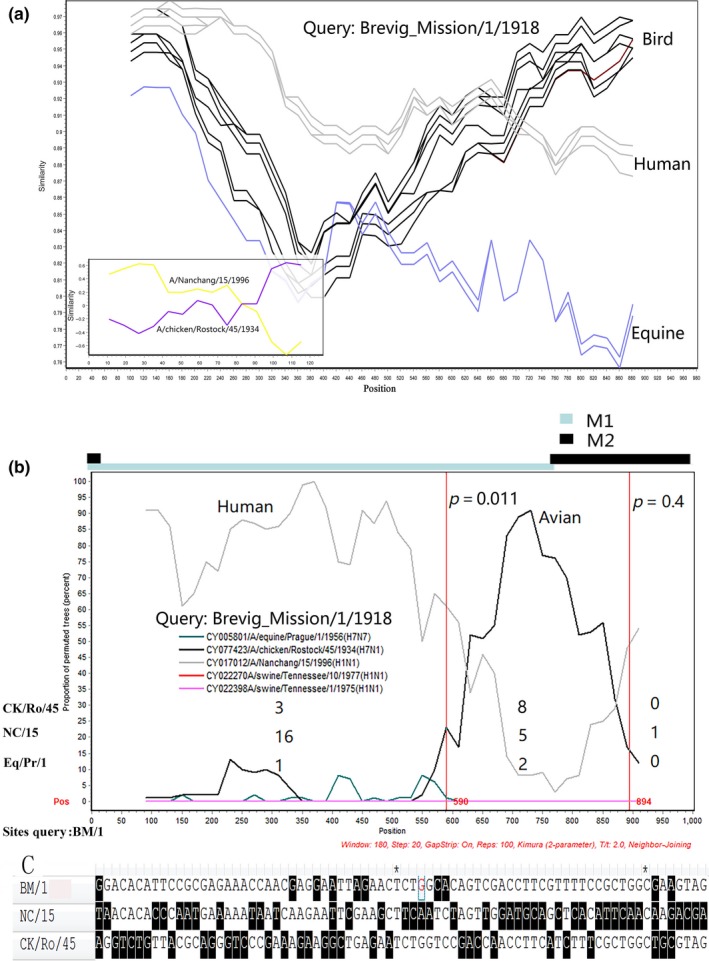
Recombination evidence of the M segment of the 1918 Spanish flu virus. (a) The similarity analysis of the M segment of A/Brevig Mission/1/1918(H1N1) and its parent lineages. The large panel: comparison of the M segment of A/Brevig Mission/1/1918(H1N1), avian branch, human H1N1 subtype and horse branch. The 1918 Spanish flu virus was used as a query. The horse branch was used as the control. The small panel: comparison of the M segment variant sites of A/Brevig Mission/1/1918(H1N1) and two representatives isolates of avian and human branches, A/chicken/Rostock/45/1934 and A/Nanchang/15/1996. The y‐axis indicates the similarity between A/Brevig Mission/1/1918(H1N1) and its parent lineages. The x‐axis indicates the position sites relative to the M1 opening reading frame (ORF) (large box) or variant sites (small box). (b) Bootscan analysis of A/Brevig Mission/1/1918(H1N1) and its parent lineages. The Bootscan analysis was performed using the consensus sequences of different branches. The corresponding positions of the M1 and M2 ORFs are represented by panels of different colours. The vertical lines indicate the putative breakpoints determined by maximization of χ2 as described in Methods. The information sites were analyzed using four isolates, A/Brevig Mission/1/1918 (BM/1), A/chicken/Rostock/45/1934 (Ck/Ro/45), A/Nanchang/15/1996 (NC/15), and A/equine/Prague/1/1956 (Eq/Pr/1). The number in the middle of each panel indicates the same number of information sites as A/Brevig Mission/1/1918. The p‐values from Fisher's Exact Test are listed near the putative breakpoint lines. (c) The alignment of the 1918 virus and its parent lineages’ variant sites surrounding the two putative breakpoints (after the position 437 of M 1 ORF). Two asterisks represent putative recombination breakpoints [Colour figure can be viewed at http://wileyonlinelibrary.com]
These results suggested that the M segment of the Spanish flu virus might be a recombinant from human and avian flu viruses. Then, choosing one representative isolate of human, avian and horse flu viruses (Figure 2b), respectively, we employed Fisher's exact test to analyze information sites so as to determine the robustness of the potential recombination breakpoints. Of the two putative breakpoints with Maximum chi‐square value, the first one (position 590 in the M1 ORF) was statistically significant (p < 0.05), whereas the second one (position 894) was not because there were not enough information sites after it.
To further determine this recombination event, we reconstructed the phylogenetic history of the M segment using different regions delimited by the putative breakpoints. Before the first breakpoint, the Spanish flu virus was clustered into the human H1N1 lineage with 100% bootstrap support (Figure 3a). Inside the two breakpoints; however, it was nested in the clade of the bird viruses with 93% bootstrap value (Figure 3b). The topologies of the two trees were significantly different (Shimodaira–Hasegawa test, p < 0.0001). Notably, this difference was only attributed to the Spanish flu virus.
Figure 3.
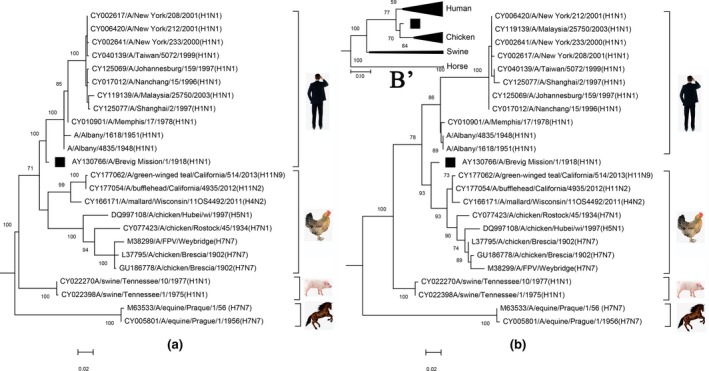
Phylogenetic histories of different M segment regions. (a) Phylogenetic history inferred from all positions of the M segment region 1–589. The first nucleotide of the M1 ORF is defined as the position 1. (b) Phylogenetic history inferred from all positions of the M segment region 590–894. (b’). Phylogenetic history reconstructed using the codon position 3 of the M1 and M2 ORFs in the region 590–894. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2‐parameter model. The rate variation model allowed some sites to be evolutionarily invariable ([+I], 49.1% sites in A and 66.0% sites in b). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site [Colour figure can be viewed at http://wileyonlinelibrary.com]
In order to distinguish whether the cluster of the Spanish flu virus into the avian branch was due to homologous recombination or convergent evolution adapting to avian host, we reconstructed its phylogenetic history using the third nucleotides of codons in the M1 and M2 ORFs between the two breakpoints (Figure 3b’). The topologies of the two trees were identical (Figure 3b and b’). Therefore, it is more likely that homologous recombination caused the Spanish flu virus to jump into the avian branch rather than convergent evolution adapting to avian host.
The genetic material provided by the avian parent to the Spanish flu virus is approximately 305 nucleotides (nt) in the M segment, which encodes for 56 AAs at the M1 C‐terminus and 60 AAs at the M2 N‐terminus. The M1 protein region from the avian virus is different from that of the human virus at two AAs (T218A, D231N). This could lead to rapid changes in virion composition since M1 is the most abundant protein in viral particles. For M2, the recombination might cause a significant substitution up to five AAs (Figure 4c). The M2 genetic region from the avian virus encoded the complete pH‐gated proton channel of the virus (Figure 4a and b).
Figure 4.
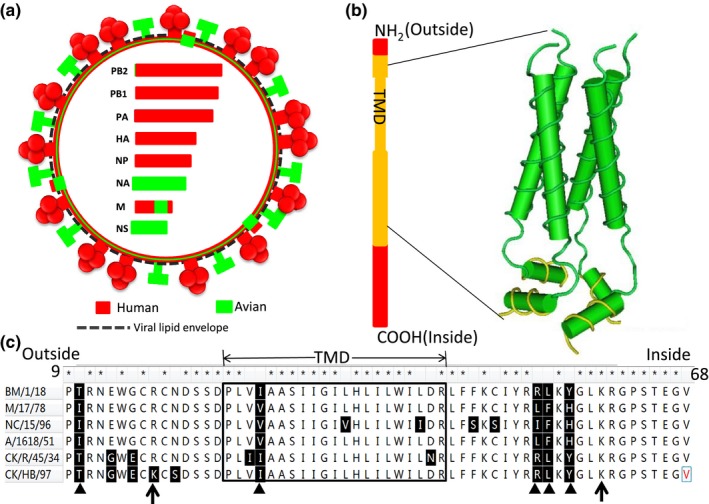
Schematic diagram of the Spanish flu virus with the recombinant M segment. (a) The schematic diagram of the Spanish flu virus structure. Segments of different origins are indicated with different colours. The encoded proteins are described using the identical colour respectively. The recombinant M segment is shown with two different colours, red and green. (b) The schematic diagram of the mosaic M2 protein of the Spanish flu virus and the 3D structure diagram of its pH‐gated proton channel encoded by genetic materials originating from avian influenza virus. TMD indicates the region of transmembrane domain, and is framed in the box. The amino acids (AAs) in the inside helix are shown in yellow. The 3D structure of the proton channel is deviated from the previous report (PDB ID: 2RLF). (c) The AA comparison of the Spanish flu virus and its parent lineage representatives in the recombination region of M2. The asterisks indicate the identical AAs between the Spanish flu virus and its parents. The triangles show the unique AAs of bird lineage and the Spanish flu virus. The two black arrows indicate the start and end points of the pH‐gated proton channel. The AAs masked with light yellow constitute the inside helix of the channel. BM/1/18, A/Brevig_Mission/1/1918(H1N1); NC/15/97, A/Nanchang/15/1996(H1N1); M/17/78, A/Memphis/17/1978(H1N1), A/1618/51, A/Albany/1618/1951(H1N1) CK/R/45/34, A/chicken/Rostock/45/1934(H7N1); CK/HB/97, A/chicken/Hubei/wi/1997(H5N1) [Colour figure can be viewed at http://wileyonlinelibrary.com]
In addition, according to the sequence similarity and bootscan analysis of the concatenated IVA genome and phylogenetic reconstruction of NA and NS, the NA and NS segments of the Spanish flu virus might be re‐assorted from avian IVAs (Figure S1). Within the HA segments, there is also a crossover site in the sequence similarity plot (Figure S1A). However, there is no phylogenetic evidence that the similarity crossover of the HA gene is due to recombination (Figure S1C and D), which is consistent with the previous report (Worobey et al., 2002).
4. DISCUSSION
Establishing the genetic basis for interspecies transmission of IVAs is paramount for predicting the potential human danger associated with new pandemic strains. The latest influenza pandemic was also caused by H1N1 virus in 2009. The 2009 virus is a triple reassortant virus of swine origin that contains gene segments from swine, human and avian influenza viruses (Garten et al., 2009), but it does not have the high virulence as the Spanish flu virus. Unlike the 2009 virus, the Spanish flu virus is considered not to have acquired genetic material from avian IVA via reassortment or recombination (Antonovics et al., 2006; Gibbs & Gibbs, 2006; Smith et al., 2009; Vana & Westover, 2008). However, the Spanish flu virus has multiple proteins carrying the unique AAs of the avian branch, suggesting its progenitors had infected avian before its outbreak; and thus, the mutations adaptive to birds might have been fixed in virus genome during circulation in avian host (Basler et al., 2001; Reid et al., 1999, 2004, 2000; Taubenberger et al., 2005). Interestingly, in this study, by introducing a bird IVA lineage isolated in the early 20th century into the dataset, we found that the NA and NS segments of the Spanish flu virus are more likely from avian IVA. Moreover, approximately 300 nt fragment of its M segment is also inherited from avian IVA. These results provided direct evidence for the avian origin of the virus.
The two major advantages of recombination over mutation are that recombination accelerates the rate at which advantageous genetic combinations are produced and allows more efficient removal of deleterious mutations (Simon‐Loriere & Holmes, 2011). This could also result in the change of host tropism and virulent phenotypes, as occurred with the human immunodeficiency virus (HIV) (Lemey, Rambaut, & Pybus, 2006) and Middle East respiratory syndrome coronavirus (Sabir et al., 2016). Even for negative RNA viruses, recombination has also been responsible for outbreaks of bovine ephemeral fever virus in cattle (He et al., 2016), bat rabies virus in skunk and raccoon (Ding, Xu, Sun, He, & He, 2017) and severe fever with thrombocytopenia syndrome bunyavirus in human (He & Ding, 2012). For the Spanish flu virus, Gibbs et al., (2001). proposed its HA was recombined from human and swine IVAs, and the recombination played the key role in the outbreak of the virus. However, this recombination evidence was thought to be invalid because the phylogenetic histories of HA were not different in the regions inside and outside the proposed recombination breakpoints (Worobey et al., 2002). Here, we provided robust phylogenetic evidence that the M segment was a chimera inherited from a human virus and a bird virus, showing that homologous recombination does be the important genetic mechanism driving the formation of the Spanish flu virus, although recombination might infrequently occur between IVAs.
The avian HA protein binds preferentially to 2,3‐linked sialic acids, whereas human HA protein binds preferentially to 2,6‐linked sialic acids. This determines the host difference (Matrosovich & Klenk, 2003). However, outbreaks of avian IVAs, such as H5N1 and H7N9, have been found in human being (Nga et al., 2019), suggesting that avian IVA can potential infect human. Several studies have also shown that HA of H7N9 can bind both human‐ and avian‐type receptors (Belser et al., 2013; Watanabe et al., 2013; Zhang et al., 2013). Therefore, it is not strange that the Spanish flu virus is a mosaic from avian and human IVAs.
Based on this study and previous reports (Smith et al., 2009; Taubenberger et al., 2005; Vana & Westover, 2008), we would like to propose a hypothesis about the origin of the Spanish flu virus. Its progenitor (a human flu virus) occasionally infected birds. During circulation in avian host, the virus obtained the NA and NS segments via reassortment and the 300 nt M segment through recombination from avian IVAs, and later jumped back to humans and caused the devastating pandemic. However, it is difficult to trace when the recombination event in the M segment took place because the real parents of the Spanish flu virus cannot be identified. Therefore, we are not sure to what extent this recombination affected the virus.
Recent studies have showed that both the HA and PB2 proteins of the Spanish flu virus can enhance viral replication. Moreover, PB2 can also increase the virulence of the virus (Malaspinas, Malaspinas, Evans, & Slatkin, 2012; Qi et al., 2018). Here, we found that the recombination has shaped a new M segment that is significantly different from any avian and mammalian IVAs, which may give the virus a unique biological phenotype. It has been known that the disease course and pathological damage caused by the virus are associated with the capability of replicating to titers higher than those of other strains (Wolbach, 1919). The M protein is the regulator of IVA growth (Yasuda, Bucher, & Ishihama, 1994; Yasuda, Toyoda, Nakayama, & Ishihama, 1993). Small changes in M can have large effects on replication phenotype (Reid et al., 2002). M can also affect the virulence of the virus (Brown, Liu, Kit, Baird, & Nesrallah, 2001; Smeenk, Wright, Burns, Thaker, & Brown, 1996). Through the recombination, the Spanish flu virus acquired the seven unique AAs from the avian IVA in M1 and M2. Previous studies have shown that the mosaic proteins might have given the Spanish flu virus a high replication capacity. The recombinant M segments of its offspring strains PR34 (A/Puerto Rico/8/34 (H1N1)) and WSN33 (A/WSN/33 (H1N1)) (Figure S2) have been shown to confer high‐growth characteristics in single‐gene reassortant strains (Yasuda et al., 1994), suggesting that the recombination may give the Spanish flu virus a high replication power, and thus, influence its pathogenicity.
The 1957 Asian flu and 1968 Hong Kong flu pandemics also involved IVAs of avian and human origins (Kawaoka, Krauss, & Webster, 1989). To see whether their M segments are the direct descendants of the 1918 Spanish flu virus, we also analyzed the M segment phylogenetic history of the three viruses based on the recombination event. It turned out that the M segments of 1957 and 1968 viruses might not be directly descended from the 1918 virus since the recombination signal of the 1918 virus was not present in their M segments (Figure S2).
In conclusion, this study showed that the M segment of the Spanish flu virus was a recombinant originating from human and avian IVAs, while the NA and NS segments were generated by reassortment between mammalian and avian IVAs, providing direct evidence of its avian origin. Moreover, the recombination might be associated with the high replication capacity of the virus and thus play an important role in its high pathogenicity.
Supporting information
He C‐Q, He M, He H‐B, Wang H‐M, Ding N‐Z. The matrix segment of the “Spanish flu” virus originated from intragenic recombination between avian and human influenza A viruses. Transbound Emerg Dis. 2019;66:2188–2195. 10.1111/tbed.13282
Funding information
This work was supported by grants from Key R & D project of Shandong Province (2017GNC10125) and the University and Institute Independent Innovation Programme of Jinan (201303031).
REFERENCES
- Antonovics, J. , Hood, M. E. , & Baker, C. H. (2006). Molecular virology: was the 1918 flu avian in origin? Nature, 440, E9; discussion E9–10. [DOI] [PubMed] [Google Scholar]
- Basler, C. F. , Reid, A. H. , Dybing, J. K. , Janczewski, T. A. , Fanning, T. G. , Zheng, H. , … Taubenberger, J. K. (2001). Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proceedings of the National Academy of Sciences of the United States of America, 98, 2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser, J. A. , Gustin, K. M. , Pearce, M. B. , Maines, T. R. , Zeng, H. , Pappas, C. , … Tumpey, T. M. (2013). Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature, 501, 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni, M. F. , de Jong, M. D. , van Doorn, H. R. , & Holmes, E. C. (2010). Guidelines for identifying homologous recombination events in influenza A virus. PLoS ONE, 5, e10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E. G. , Liu, H. , Kit, L. C. , Baird, S. , & Nesrallah, M. (2001). Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: Identification of functional themes. Proceedings of the National Academy of Sciences of the United States of America, 98, 6883–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui, M. , Wills, E. G. , Helenius, A. , & Whittaker, G. R. (2000). Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. Journal of Virology, 74, 1781–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, N. Z. , Xu, D. S. , Sun, Y. Y. , He, H. B. , & He, C. Q. (2017). A permanent host shift of rabies virus from Chiroptera to Carnivora associated with recombination. Scientific Reports, 7, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten, R. J. , Davis, C. T. , Russell, C. A. , Shu, B. , Lindstrom, S. , Balish, A. , … Cox, N. J. (2009). Antigenic and genetic characteristics of swine‐origin 2009 A(H1N1) influenza viruses circulating in humans. Science, 325, 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, M. J. , Armstrong, J. S. , & Gibbs, A. J. (2001). Recombination in the hemagglutinin gene of the 1918 "Spanish flu". Science, 293, 1842–1845. [DOI] [PubMed] [Google Scholar]
- Gibbs, M. J. , & Gibbs, A. J. (2006). Molecular virology: was the 1918 pandemic caused by a bird flu? Nature, 440, E8; discussion E9–10. [DOI] [PubMed] [Google Scholar]
- Gomez‐Puertas, P. , Albo, C. , Perez‐Pastrana, E. , Vivo, A. , & Portela, A. (2000). Influenza virus matrix protein is the major driving force in virus budding. Journal of Virology, 74, 11538–11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grambas, S. , & Hay, A. J. (1992). Maturation of influenza A virus hemagglutinin–estimates of the pH encountered during transport and its regulation by the M2 protein. Virology, 190, 11–18. [DOI] [PubMed] [Google Scholar]
- He, C. Q. , & Ding, N. Z. (2012). Discovery of severe fever with thrombocytopenia syndrome bunyavirus strains originating from intragenic recombination. Journal of Virology, 86, 12426–12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C. Q. , Ding, N. Z. , Mou, X. , Xie, Z. X. , Si, H. L. , Qiu, R. , … Cao, R. N. (2012). Identification of three H1N1 influenza virus groups with natural recombinant genes circulating from 1918 to 2009. Virology, 427, 60–66. [DOI] [PubMed] [Google Scholar]
- He, C. Q. , Liu, Y. X. , Wang, H. M. , Hou, P. L. , He, H. B. , & Ding, N. Z. (2016). New genetic mechanism, origin and population dynamic of bovine ephemeral fever virus. Veterinary Microbiology, 182, 50–56. [DOI] [PubMed] [Google Scholar]
- He, C. Q. , Xie, Z. X. , Han, G. Z. , Dong, J. B. , Wang, D. , Liu, J. B. , … Li, G. R. (2009). Homologous recombination as an evolutionary force in the avian influenza A virus. Molecular Biology and Evolution, 26, 177–187. [DOI] [PubMed] [Google Scholar]
- Helenius, A. (1992). Unpacking the incoming influenza virus. Cell, 69, 577–578. [DOI] [PubMed] [Google Scholar]
- Honigsbaum, M. (2018). Spanish influenza redux: Revisiting the mother of all pandemics. Lancet, 391, 2492–2495. [DOI] [PubMed] [Google Scholar]
- Johnson, N. P. , & Mueller, J. (2002). Updating the accounts: Global mortality of the 1918–1920 "Spanish" influenza pandemic. Bulletin of the History of Medicine, 76, 105–115. [DOI] [PubMed] [Google Scholar]
- Kawaoka, Y. , Krauss, S. , & Webster, R. G. (1989). Avian‐to‐human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. Journal of Virology, 63, 4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourne, E. D. (2006). Influenza pandemics of the 20th century. Emerging Infectious Diseases, 12, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, R. A. , Lai, C. J. , & Choppin, P. W. (1981). Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: Colinear and interrupted mRNAs code for overlapping proteins. Proceedings of the National Academy of Sciences of the United States of America, 78, 4170–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, R. A. , Zebedee, S. L. , & Richardson, C. D. (1985). Influenza virus M2 protein is an integral membrane protein expressed on the infected‐cell surface. Cell, 40, 627–633. [DOI] [PubMed] [Google Scholar]
- Latham, T. , & Galarza, J. M. (2001). Formation of wild‐type and chimeric influenza virus‐like particles following simultaneous expression of only four structural proteins. Journal of Virology, 75, 6154–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey, P. , Rambaut, A. , & Pybus, O. G. (2006). HIV evolutionary dynamics within and among hosts. AIDS Reviews, 8, 125–140. [PubMed] [Google Scholar]
- Liu, T. , & Ye, Z. (2002). Restriction of viral replication by mutation of the influenza virus matrix protein. Journal of Virology, 76, 13055–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole, K. S. , Bollinger, R. C. , Paranjape, R. S. , Gadkari, D. , Kulkarni, S. S. , Novak, N. G. , … Ray, S. C. (1999). Full‐length human immunodeficiency virus type 1 genomes from subtype C‐infected seroconverters in India, with evidence of intersubtype recombination. Journal of Virology, 73, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspinas, A. S. , Malaspinas, O. , Evans, S. N. , & Slatkin, M. (2012). Estimating allele age and selection coefficient from time‐serial data. Genetics, 192, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, K. , & Helenius, A. (1991). Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell, 67, 117–130. [DOI] [PubMed] [Google Scholar]
- Matrosovich, M. , & Klenk, H. D. (2003). Natural and synthetic sialic acid‐containing inhibitors of influenza virus receptor binding. Reviews in Medical Virology, 13, 85–97. [DOI] [PubMed] [Google Scholar]
- Nga, V. T. , Ngoc, T. U. , Minh, L. B. , Ngoc, V. T. N. , Pham, V. H. , Nghia, L. L. , … Chu, D. T. (2019). Zoonotic diseases from birds to humans in Vietnam: Possible diseases and their associated risk factors. European Journal of Clinical Microbiology & Infectious Diseases, 38, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Noda, T. , Sagara, H. , Yen, A. , Takada, A. , Kida, H. , Cheng, R. H. , & Kawaoka, Y. (2006). Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature, 439, 490–492. [DOI] [PubMed] [Google Scholar]
- Parrish, C. R. , Holmes, E. C. , Morens, D. M. , Park, E. C. , Burke, D. S. , Calisher, C. H. , … Daszak, P. (2008). Cross‐species virus transmission and the emergence of new epidemic diseases. Microbiology and Molecular Biology Reviews : MMBR, 72, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, K. D. , & Pyle, G. F. (1991). The geography and mortality of the 1918 influenza pandemic. Bulletin of the History of Medicine, 65, 4–21. [PubMed] [Google Scholar]
- Perez, D. R. , & Donis, R. O. (1998). The matrix 1 protein of influenza A virus inhibits the transcriptase activity of a model influenza reporter genome in vivo. Virology, 249, 52–61. [DOI] [PubMed] [Google Scholar]
- Pinto, L. H. , Holsinger, L. J. , & Lamb, R. A. (1992). Influenza virus M2 protein has ion channel activity. Cell, 69, 517–528. [DOI] [PubMed] [Google Scholar]
- Qi, W. , Jia, W. , Liu, D. , Li, J. , Bi, Y. , Xie, S. , … Liao, M. (2018). Emergence and adaptation of a novel highly pathogenic H7N9 influenza virus in birds and humans from a 2013 human‐infecting Low‐Pathogenic Ancestor. Journal of Virology, 92 10.1128/JVI.00921-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, A. H. , Fanning, T. G. , Hultin, J. V. , & Taubenberger, J. K. (1999). Origin and evolution of the 1918 "Spanish" influenza virus hemagglutinin gene. Proceedings of the National Academy of Sciences of the United States of America, 96, 1651–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, A. H. , Fanning, T. G. , Janczewski, T. A. , Lourens, R. M. , & Taubenberger, J. K. (2004). Novel origin of the 1918 pandemic influenza virus nucleoprotein gene. Journal of Virology, 78, 12462–12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, A. H. , Fanning, T. G. , Janczewski, T. A. , McCall, S. , & Taubenberger, J. K. (2002). Characterization of the 1918 "Spanish" influenza virus matrix gene segment. Journal of Virology, 76, 10717–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, A. H. , Fanning, T. G. , Janczewski, T. A. , & Taubenberger, J. K. (2000). Characterization of the 1918 "Spanish" influenza virus neuraminidase gene. Proceedings of the National Academy of Sciences of the United States of America, 97, 6785–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, A. H. , & Taubenberger, J. K. (1999). The 1918 flu and other influenza pandemics: "over there" and back again. Laboratory Investigation, 79, 95–101. [PubMed] [Google Scholar]
- Sabir, J. S. , Lam, T. T. , Ahmed, M. M. , Li, L. , Shen, Y. , Abo‐Aba, S. E. , … Guan, Y. (2016). Co‐circulation of three camel coronavirus species and recombination of MERS‐CoVs in Saudi Arabia. Science, 351, 81–84. [DOI] [PubMed] [Google Scholar]
- Simon‐Loriere, E. , & Holmes, E. C. (2011). Why do RNA viruses recombine? Nature Reviews. Microbiology, 9, 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk, C. A. , Wright, K. E. , Burns, B. F. , Thaker, A. J. , & Brown, E. G. (1996). Mutations in the hemagglutinin and matrix genes of a virulent influenza virus variant, A/FM/1/47‐MA, control different stages in pathogenesis. Virus Research, 44, 79–95. [DOI] [PubMed] [Google Scholar]
- Smith, G. J. , Bahl, J. , Vijaykrishna, D. , Zhang, J. , Poon, L. L. , Chen, H. , … Guan, Y. (2009). Dating the emergence of pandemic influenza viruses. Proceedings of the National Academy of Sciences of the United States of America, 106, 11709–11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue, R. J. , & Hay, A. J. (1991). Structural characteristics of the M2 protein of influenza A viruses: Evidence that it forms a tetrameric channel. Virology, 180, 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. , & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger, J. K. (2006). The origin and virulence of the 1918 "Spanish" influenza virus. Proceedings of the American Philosophical Society, 150, 86–112. [PMC free article] [PubMed] [Google Scholar]
- Taubenberger, J. K. , & Morens, D. M. (2006). 1918 Influenza: The mother of all pandemics. Emerging Infectious Diseases, 12, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger, J. K. , Reid, A. H. , Lourens, R. M. , Wang, R. , Jin, G. , & Fanning, T. G. (2005). Characterization of the 1918 influenza virus polymerase genes. Nature, 437, 889–893. [DOI] [PubMed] [Google Scholar]
- Vana, G. , & Westover, K. M. (2008). Origin of the 1918 Spanish influenza virus: A comparative genomic analysis. Molecular Phylogenetics and Evolution, 47, 1100–1110. [DOI] [PubMed] [Google Scholar]
- Watanabe, T. , Kiso, M. , Fukuyama, S. , Nakajima, N. , Imai, M. , Yamada, S. , … Kawaoka, Y. (2013). Characterization of H7N9 influenza A viruses isolated from humans. Nature, 501, 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, R. G. (1999). 1918 Spanish influenza: The secrets remain elusive. Proceedings of the National Academy of Sciences of the United States of America, 96, 1164–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, G. , & Fields, S. (1980). Cloning of influenza cDNA ino M13: The sequence of the RNA segment encoding the A/PR/8/34 matrix protein. Nucleic Acids Research, 8, 1965–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbach, S. B. (1919). Comments on the pathology and bacteriology of fatal influenza cases, as observed at Camp Devens Massachusetts. Johns Hopkins Hospital Bulletin, 30, 104. [Google Scholar]
- Worobey, M. , Rambaut, A. , & Holmes, E. C. (1999). Widespread intra‐serotype recombination in natural populations of dengue virus. Proceedings of the National Academy of Sciences of the United States of America, 96, 7352–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey, M. , Rambaut, A. , Pybus, O. G. , & Robertson, D. L. (2002): Questioning the evidence for genetic recombination in the 1918 "Spanish flu" virus. Science, 296, 211 discussion 211. [DOI] [PubMed] [Google Scholar]
- Yasuda, J. , Bucher, D. J. , & Ishihama, A. (1994). Growth control of influenza A virus by M1 protein: Analysis of transfectant viruses carrying the chimeric M gene. Journal of Virology, 68, 8141–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, J. , Toyoda, T. , Nakayama, M. , & Ishihama, A. (1993). Regulatory effects of matrix protein variations on influenza virus growth. Archives of Virology, 133, 283–294. [DOI] [PubMed] [Google Scholar]
- Ye, Z. P. , Baylor, N. W. , & Wagner, R. R. (1989). Transcription‐inhibition and RNA‐binding domains of influenza A virus matrix protein mapped with anti‐idiotypic antibodies and synthetic peptides. Journal of Virology, 63, 3586–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Shi, J. , Deng, G. , Guo, J. , Zeng, X. , He, X. , … Chen, H. (2013). H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science, 341, 410–414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


