Abstract
Members of the genus Curvularia are melanin-producing dematiaceous fungi of increasing clinical importance as causal agents of both local and invasive infections. This study contributes to the taxonomical and clinical knowledge of this genus by describing two new Curvularia species based on isolates from corneal scrapings of South Indian fungal keratitis patients. The phylogeny of the genus was updated based on three phylogenetic markers: the internal transcribed spacer (ITS) region of the ribosomal RNA gene cluster as well as fragments of the glyceraldehyde-3-phosphate dehydrogenase (gpdh) and translation elongation factor 1-α (tef1α) genes. The maximum likelihood phylogenetic tree constructed from the alignment of the three concatenated loci revealed that the examined isolates are representing two new, yet undescribed, Curvularia species. Examination of colony and microscopic morphology revealed differences between the two species as well as between the new species and their close relatives. The new species were formally described as Curvularia tamilnaduensis N. Kiss & S. Kocsubé sp. nov. and Curvularia coimbatorensis N. Kiss & S. Kocsubé sp. nov. Antifungal susceptibility testing by the broth microdilution method of CLSI (Clinical & Laboratory Standards Institute) revealed that the type strain of C. coimbatorensis is less susceptible to a series of antifungals than the C. tamilnaduensis strains.
Keywords: Curvularia, keratitis, taxonomy, antifungal susceptibility, Curvularia coimbatorensis, Curvularia tamilnaduensis
1. Introduction
The fungal genus Curvularia (Ascomycota, Pleosporales, Pleosporaceae) comprises of dematiaceous, melanin-producing molds with various lifestyles including saprophytism, plant endophytism [1], plant parasitism [2], and human pathogenicity [3].
The genus-level identification of Curvularia was performed traditionally by the examination of pigmentation, as well as the morphology of the septate conidia and hyphae [3]. The first sequence-based species-level identification attempts targeted the internal transcribed spacer (ITS) region of the ribosomal RNA gene cluster, which alone, however, proved inappropriate, either for the purposes of exact diagnosis [4] or for the phylogenetic resolution of the genus and the clarification of its relationship to the closely related genera Bipolaris, Cochliobolus, and Drechslera [3]. Multilocus sequence typing (MLST) involving fragments of the nuclear ribosomal large subunit RNA (LSU) as well as the glyceraldehyde-3-phosphate dehydrogenase (gpdh) and translation elongation factor 1-α (tef1a) genes in addition to ITS had resulted in the recently accepted phylogenetic concept of the genus Curvularia [5], which was applied in more recent works [6,7,8]. Recently, the genus involves more than 100 described species, which can be divided into six clades (americana, eragrostidis, hominis, lunata, spicifera, and trifolii) according to Madrid et al. [7] based on MLST of four loci (ITS, LSU, gpdh, and the RNA polymerase II subunit rpb2).
Krizsán et al. [3] reviewed the clinical importance of the genus Curvularia, and identified Curvularia australiensis, Curvularia geniculata, Curvularia hawaiiensis, Curvularia lunata, Curvularia pallescens, and Curvularia spicifera as the species most frequently isolated from clinical samples. Further members of the genus with confirmed clinical relevance include Curvularia americana, Curvularia chlamydospora, Curvularia hominis, Curvularia muehlenbeckiae, Curvularia pseudolunata [7], Curvularia brachyspora [9], Curvularia senegalensis [10,11], Curvularia clavata [12], Curvularia tuberculata [13], and Curvularia inaequalis [14,15,16]. A Curvularia infection in humans is designated as curvulariosis, a subtype of phaeohyphomycoses (i.e., fungal infections caused by dematiaceous fungi) [3]. The resulting diseases include deep and disseminated infections [3,17,18,19], infections complicating peritoneal dialysis [14,20,21], respiratory infections including sinusitis and bronchopulmonary mycosis [3,10,22], urinary tract infections [23], as well as localized infections affecting the skin, nail [4,24,25], and the eye. Among eye infections, the involvement of Curvularia spp. is most frequent in keratitis—a suppurative, ulcerative disease of the cornea, but endophthalmitis and chronic dacryocystitis cases have also been reported [3,26].
In this study, we describe two new species of the genus Curvularia, the type strains of which were isolated from corneal scraping samples derived from South Indian patients diagnosed with fungal keratitis.
2. Results
2.1. Strain Selection and Case Details
About two thirds of the dematiaceous fungi isolated from corneal ulcers in the Aravind Eye Hospital, Coimbatore, Tamil Nadu, India belong to the genus Curvularia (unpublished data). The four strains involved in this study were selected retrospectively based on the inability of reliable species-level identification of some Curvularia isolates by ITS sequence analysis. Details available of the cases are presented in Table 1. All four patients were diagnosed with fungal corneal ulcer. The corneal scrapings from the ulcers were in all cases positive for fungal filaments in direct microscopy (both 10% KOH and Gram staining). None of the cases had a history of contact lens wear. History of falling dust (2) and mud (1) into the eye was recorded as predisposing factors. Based on the typical clinical picture and the KOH report, topical antifungal therapy was started with natamycin (5% suspension) and econazole drops (2%) every half an hour, along with homatropine (1%) administered three times a day. Unfortunately, the patients were lost to follow up after one or two visits.
Table 1.
Case details of the fungal keratitis infections.
| Strain | Age | Sex | Clinical Diagnosis | Corneal Scraping | Therapy | Outcome |
|---|---|---|---|---|---|---|
| SZMC 22225 | 80 | Male | Fungal corneal ulcer | 11 July 2012 | NAT, ECZ, HTR | Lost to follow up after two visits |
| SZMC 22226 | 66 | Male | Fungal corneal ulcer | 2 March 2013 | NAT, ECZ, HTR | Lost to follow up after one visit |
| SZMC 26758 | 40 | Male | Fungal corneal ulcer | 21 March 2011 | NAT, ECZ, HTR | Lost to follow up after one visit |
| SZMC 26759 | NA | NA | Fungal corneal ulcer | NA | NA | Lost to follow up |
NAT: natamycin (5%); ECZ: econazole (2%), HTR: homatropine (1%); NA: data not available.
2.2. Updated Phylogeny of the Genus Curvularia
Table 2 shows the strains and sequences involved in the phylogenetic analysis of the genus Curvularia, including four isolates derived from cases of fungal keratitis diagnosed and treated in the Aravind Eye Hospital, Coimbatore, Tamil Nadu, India. The tef1α dataset consisted of 902 characters of nucleotide alignment without binary characters. The gpdh dataset contained 684 characters with 601 characters of nucleotide alignment and 63 binary characters derived from indel coding. The length of the ITS alignment was 1193 characters long, containing 896 bp of nucleotide data and 297 binary characters.
Table 2.
Sequences used for the phylogenetic analysis.
| Curvularia Species | Strain | GenBank Accession Number | ||
|---|---|---|---|---|
| ITS | tef1a | gpdh | ||
| Bipolaris maydis | CBS 136.29 T | KJ909780 | KM093794 | KM034846 |
| Curvularia aeria | BRIP 61232b | KX139029 | KU552155 | KU552162 |
| Curvularia affinis | CBS 154.34 T | KJ909782 | KM196566 | KM230401 |
| Curvularia ahvazensis | SCUA-1bi T | KJ415539 | MG428686 | MG428693 |
| Curvularia akaii | CBS 317.86 | JX256420 | KM196569 | KM230402 |
| Curvularia akaiiensis | BRIP 16080 T | HE861833 | KJ415453 | KJ415407 |
| Curvularia alcornii | MFLUCC 10-0703 T | JX256424 | JX266589 | JX276433 |
| Curvularia americana | UTHSC 08-3414 T | KJ415540 | - | HF565488 |
| Curvularia asiatica | MFLUCC 10-0711 T | KJ415541 | JX266593 | JX276436 |
| Curvularia australiensis | BRIP 12044 T | KJ415542 | KJ415452 | KJ415406 |
| Curvularia australis | BRIP 12521 T | MH414892 | KJ415451 | KJ415405 |
| Curvularia bannonii | BRIP 16732 T | MH414894 | KJ415450 | KJ415404 |
| Curvularia beasleyi | BRIP 10972 T | MH414911 | MH433654 | MH433638 |
| Curvularia beerburrumensis | BRIP 12942 T | KP400638 | MH433657 | MH433634 |
| Curvularia boeremae | IMI 164633 T | KJ415543 | - | MH433641 |
| Curvularia borreriae | MFLUCC 11-0422 | KJ922372 | KM196571 | KP419987 |
| Curvularia bothriochloae | BRIP 12522 T | KJ909765 | KJ415449 | KJ415403 |
| Curvularia brachyspora | CBS 186.50 | HG778984 | KM230405 | KM061784 |
| Curvularia buchloës | CBS 246.49 T | MF490814 | KM196588 | KM061789 |
| Curvularia carica-papayae | CBS 135941 T | HG779021 | - | HG779146 |
| Curvularia chiangmaiensis | CPC 28829 T | MH275055 | MF490857 | MF490836 |
| Curvularia chlamydospora | UTHSC 07-2764 T | KU552205 | - | HG779151 |
| Curvularia chonburiensis | MFLUCC 16-0375 T | MH414897 | - | MH412747 |
| Curvularia clavata | BRIP 61680b | AF081447 | KU552159 | KU552167 |
| Curvularia coatesiae | BRIP 24261 T | MH414898 | MH433659 | MH433636 |
| Curvularia coicis | CBS 192.29 T | LT631357 | JN601006 | AF081410 |
| Curvularia colbranii | BRIP 13066 T | LT631310 | MH433660 | MH433642 |
| Curvularia comoriensis | CBS 110673 | KJ415544 | - | LT715841 |
| Curvularia crassiseptum | CBS 503.90 T | HG778985 | - | LT715882 |
| Curvularia crustacea | BRIP 13524 T | MF490815 | KJ415448 | KJ415402 |
| Curvularia cymbopogonis | CBS 419.78 | KJ415545 | - | HG779129 |
| Curvularia dactyloctenicola | CPC 28810 T | LT631356 | MF490858 | MF490837 |
| Curvularia dactyloctenii | BRIP 12846 T | JN192375 | KJ415447 | KJ415401 |
| Curvularia deightonii | CBS 537.70 | MH414899 | - | LT715839 |
| Curvularia ellisii | CBS 193.62 T | HG778986 | JN601007 | JN600963 |
| Curvularia eragrosticola | BRIP 12538 T | KJ909781 | MH433661 | MH433643 |
| Curvularia eragrostidis | CBS 189.48 | HG778987 | - | HG779154 |
| Curvularia geniculata | CBS 187.50 | JN192376 | KM230410 | KM083609 |
| Curvularia gladioli | CBS 210.79 | KJ415546 | - | HG779123 |
| Curvularia graminicola | BRIP 23186a T | KJ415547 | JN601008 | JN600964 |
| Curvularia harveyi | BRIP 57412 T | KJ415548 | KJ415446 | KJ415400 |
| Curvularia hawaiiensis | BRIP 11987 T | KJ415549 | KJ415445 | KJ415399 |
| Curvularia heteropogonicola | BRIP 14579 T | HG779011 | KJ415444 | KJ415398 |
| Curvularia heteropogonis | CBS 284.91 T | JN192380 | JN601013 | JN600969 |
| Curvularia hominis | CBS 136985 T | KJ922375 | - | HG779106 |
| Curvularia homomorpha | CBS 156.60 T | HG778991 | JN601014 | JN600970 |
| Curvularia inaequalis | CBS 102.42 T | MH861533 | KM196574 | KM061787 |
| Curvularia intermedia | CBS 334.64 | MH414900 | - | HG779155 |
| Curvularia ischaemi | CBS 630.82 T | MH855025 | - | LT715790 |
| Curvularia kenpeggii | BRIP 14530 T | MH414901 | MH433662 | MH433644 |
| Curvularia kusanoi | CBS 137.29 | JX256429 | JN601016 | LT715862 |
| Curvularia lamingtonensis | BRIP 12259 T | JF812154 | MH433663 | MH433645 |
| Curvularia lunata | CBS 730.96 T | MH414902 | JX266596 | JX276441 |
| Curvularia malina | CBS 131274 T | HE792934 | KR493095 | KP153179 |
| Curvularia mebaldsii | BRIP 12900 T | MF139088 | MH433664 | MH433647 |
| Curvularia micropus | CBS 127235 | KJ909770 | - | LT715859 |
| Curvularia microspora | GUCC6272 T | MG846737 | MF139115 | MF139106 |
| Curvularia miyakei | CBS 197.29 T | KP400647 | KM196568 | KM083611 |
| Curvularia mosaddeghii | IRAN 3131C T | KJ415550 | MH392152 | MH392155 |
| Curvularia muehlenbeckiae | CBS 144.63 T | MH414910 | KM196578 | KP419996 |
| Curvularia neergaardii | BRIP 12919 T | KJ415551 | KJ415443 | KJ415397 |
| Curvularia neoindica | IMI 129790 T | MF490816 | MH433667 | MH433649 |
| Curvularia nicotiae | BRIP 11983 T | JN601033 | KJ415442 | KJ415396 |
| Curvularia nodosa | CPC 28800 T | KP400650 | MF490859 | MF490838 |
| Curvularia nodulosa | CBS 160.58 | JN192384 | JN601019 | JN600975 |
| Curvularia oryzae | CBS 169.53 T | KJ922380 | KM196590 | KP645344 |
| Curvularia ovariicola | CBS 470.90 T | MH275056 | JN601020 | JN600976 |
| Curvularia pallescens | CBS 156.35 T | KJ415552 | KM196570 | KM083606 |
| Curvularia pandanicola | MFLUCC 15-0746 T | HG778995 | MH412763 | MH412748 |
| Curvularia papendorfii | CBS 308.67 T | MH414905 | KJ415441 | KJ415395 |
| Curvularia perotidis | CBS 350.90 T | KY905678 | KM230407 | HG779138 |
| Curvularia petersonii | BRIP 14642 T | MH414906 | MH433668 | MH433650 |
| Curvularia pisi | CBS 190.48 T | KJ415553 | KY905697 | KY905690 |
| Curvularia platzii | BRIP 27703b T | KJ922373 | MH433669 | MH433651 |
| Curvularia portulacae | BRIP 14541 T | KJ922376 | KJ415440 | KJ415393 |
| Curvularia prasadii | CBS 143.64 T | MF490819 | KM230408 | KM061785 |
| Curvularia protuberata | CBS 376.65 T | HE861842 | KM196576 | KM083605 |
| Curvularia pseudobrachyspora | CPC 28808 T | HE861838 | MF490862 | MF490841 |
| Curvularia pseudolunata | UTHSC 09-2092 T | JN192386 | - | HF565459 |
| Curvularia pseudorobusta | UTHSC 08-3458 | MH414907 | - | HF565476 |
| Curvularia ravenelii | BRIP 13165 T | KJ415555 | JN601024 | JN600978 |
| Curvularia reesii | BRIP 4358 T | KJ909783 | MH433670 | MH433637 |
| Curvularia richardiae | BRIP 4371 T | KX139030 | KJ415438 | KJ415391 |
| Curvularia robusta | CBS 624.68 T | KJ415556 | KM196577 | KM083613 |
| Curvularia rouhanii | SCUA-2bi-2 T | HG779001 | MG428687 | MG428694 |
| Curvularia ryleyi | BRIP 12554 T | KY905679 | KJ415437 | KJ415390 |
| Curvularia senegalensis | CBS 149.71 | KJ415558 | - | HG779128 |
| Curvularia soli | CBS 222.96 T | MH414904 | KY905698 | KY905691 |
| Curvularia sorghina | BRIP 15900 T | KP400655 | KJ415435 | KJ415388 |
| Curvularia sp. | BRIP 17068b | KP400654 | MH433666 | MH433648 |
| Curvularia sp. | AR5117 | HE861826 | KP735698 | KP645349 |
| Curvularia sp. | MFLUCC 120177 | JN192387 | KP735697 | KP645348 |
| Curvularia sp. | UTHSC 8809 | MH414908 | - | HF565477 |
| Curvularia spicifera | CBS 274.52 | KJ909777 | JN601023 | JN600979 |
| Curvularia sporobolicola | BRIP 23040b T | MH275057 | MH433671 | MH433652 |
| Curvularia subpapendorfii | CBS 656.74 T | HG779023 | KM196585 | KM061791 |
| Curvularia thailandicum | MFLUCC 15-0747 T | JN192388 | MH412764 | MH412749 |
| Curvularia trifolii | CBS 173.55 | KJ415559 | - | HG779124 |
| Curvularia tripogonis | BRIP 12375 T | KC424596 | JN601025 | JN600980 |
| Curvularia tropicalis | BRIP 14834 T | JX256433 | KJ415434 | KJ415387 |
| Curvularia tsudae | ATCC 44764 T | HG779024 | KC503940 | KC747745 |
| Curvularia tuberculate | CBS 146.63 T | MF490822 | JX266599 | JX276445 |
| Curvularia uncinate | CBS 221.52 T | HG779026 | - | HG779134 |
| Curvularia variabilis | CPC 28815 T | KP400652 | MF490865 | MF490844 |
| Curvularia verruciformis | CBS 537.75 | MH414909 | - | HG779133 |
| Curvularia verruculosa | CBS 150.63 | MH275058 | KP735695 | KP645346 |
| Curvularia warraberensis | BRIP 14817 T | AF071338 | MH433672 | MH433653 |
| Curvularia xishuangbannaensis | KUMCC 17-0185 T | KJ909780 | MH412765 | MH412750 |
| Curvularia gudauskasii | DAOM 165085 | KX139029 | KM093794 | AF081393 |
| Curvularia tamilnaduensis sp. nov. | SZMC 22226 T * | MN628311 | MN628303 | MN628307 |
| SZMC 26758 * | MN628308 | MN628300 | MN628304 | |
| SZMC 26759 * | MN628309 | MN628301 | MN628305 | |
| Curvularia coimbatorensis sp. nov. | SZMC 22225 T * | MN628310 | MN628302 | MN628306 |
T type strain; * Strains examined during the present study. Sequences derived from the present study are set in bold.
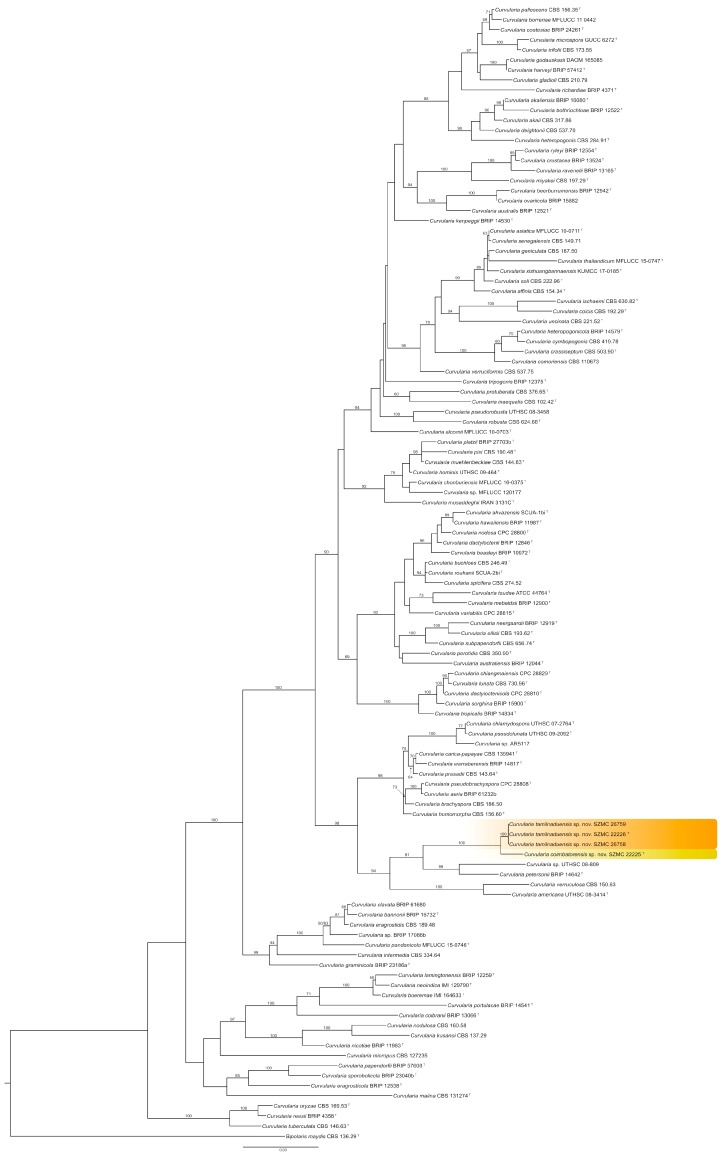
On the phylograms obtained from each of the three loci, the four keratitis isolates of this study were resolved as two new species with over 80% of confidence values (data not shown), one of them represented by the single isolate SZMC 22225, while the other one by isolates SZMC 22226, SZMC 26758, and SZMC 26758. As the individual inferences were largely congruent, the three loci were concatenated and partitioned. The phylogenetic tree obtained from the concatenated dataset is shown in Figure 1.
Figure 1.
Maximum likelihood phylogeny of the genus Curvularia inferred from the concatenated internal transcribed spacer (ITS), translation elongation factor 1-α (tef1a), and glyceraldehyde-3-phosphate dehydrogenase (gpdh) sequences. The isolates examined in this study are shown as the new species Curvularia tamilnaduensis and Curvularia coimbatorensis (highlighted in color). Sequences of the reference Curvularia strains were collected from the GenBank Nucleotide database (Table 1). Bootstrap support values greater than 60% are shown above the branches. Bipolaris maydis CBS 136.29 was used to root the tree. Abbreviations of culture collections: BRIP: Plant Pathology Herbarium, Queensland, Australia; CBS: Westerdijk Fungal Biodiversity Institute culture collection, The Netherlands; CPC: Cultures of Pedro Crous, housed at Westerdijk Fungal Biodiversity Institute; DAOM: Canadian National Mycological Herbarium, Ottawa, Canada; GUCC: Guizhou University Culture Collection, Guizhou, China; IMI: CABI Bioscience, Eggham, UK; IRAN: Iranian Fungal Culture Collection, Iranian Research Institute of Plant Protection, Tehran, Iran; KUMCC: Culture Collection of Kunming Institute of Botany, Kunming, China; MFLUCC: Mae Fah Luang Culture Collection, Chiang Rai, Thailand; SCUA: Collection of Fungal Cultures, Department of Plant Protection, Shahid Chamran University of Ahvaz, Iran; SZMC: Szeged Microbiology Collection, Szeged, Hungary; UTHSC: University of Tennessee Health Science Center, Memphis, USA. T: type strain.
2.3. Taxonomy and Related Information
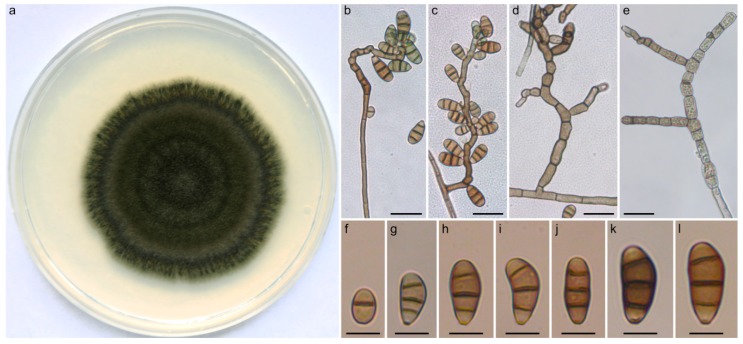
Curvularia coimbatorensis N. Kiss & S. Kocsubé sp. nov. (Figure 2). MycoBank accession number: MB 833656. The etymology is referring to the city in Tamil Nadu, South India where the type strain was isolated.
Figure 2.
Morphological features of Curvularia coimbatorensis SZMC 2225. (a) Colony morphology on PDA (potato dextrose agar) medium after 7 days at 25 °C; (b,c) conidiophores with septate conidia; (d) branching conidiophores; (e) swollen cells; (f–l) septate conidia. Scale bars: (b–e) 20 µm; (f–l) 10 µm.
Vegetative hyphae septate, subhyaline to brown, branched, smooth, 3–4 µm in width. Colonies on PDA reaching approximately 4–6 cm in diameter after 7 days at 25 °C, surface funiculose, margin fimbriate, olivaceous black to olivaceous grey, velutinous with sparse aerial mycelium. Conidiophores erect, often branched, in most cases uniformly brown, sometimes pale brown at apex, seminematous, septate, flexuous, in most cases geniculate towards the apex, up to 210 µm long, 3–4 µm wide, basal cells sometimes swollen. Conidiogenous cells integrated, terminal, or intercalary with sympodial proliferation, smooth, brown, mono- or polytretic. Chlamydospores not observed. Conidia ellipsoidal to clavate to obovoid, asymmetrical with paler end cells, usually curved at the third cell from the base, (13-)16–18(-23) × (7-)8–9(-10) µm, 3-distoseptate, hila slightly protuberant, thickened and darkened.
Specimens examined: India, Coimbatore, human corneal scraping from corneal ulcer, 2012, (holotype: freeze dried culture specimen in the Szeged Microbiological Collection (SZMC) at the Department of Microbiology, Faculty of Science and Informatics, University of Szeged, Hungary, SZMC 22225, includes ex-type culture).
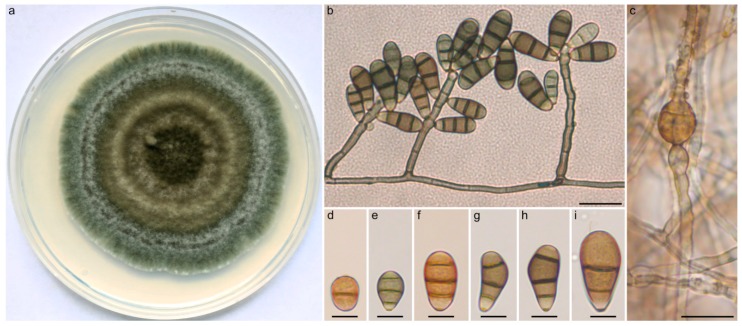
Curvularia tamilnaduensis N. Kiss & S. Kocsubé sp. nov. (Figure 3). MycoBank accession number: MB 833657. The etymology is referring to the state of South India where the type strain and the other two examined strains were isolated.
Figure 3.
Morphological features of Curvularia tamilnaduensis SZMC 2226. (a) Colony morphology on PDA medium after 7 days at 25 °C; (b) conidiophores with septate conidia; (c) subglobose intercalary chlamydospore; (d–f) septate conidia. Scale bars: (b,c) 20 µm; (d–i) 10 µm.
Vegetative hyphae septate, subhyaline to brown, branched, smooth walled, but often heavily asperulate, 2–3 µm in width. Colonies on PDA reaching approximately 6–7 cm in diameter after 7 days at 25 °C, surface lanose, aerial mycelium abundant, margin fimbriate, olivaceous green. Conidiophores erect, usually unbranched, in most cases uniformly brown, sometimes with paler tip, seminematous, septate, slightly flexuous, rarely geniculate towards the apex, up to 125 µm long, 2.5–4 µm wide. Conidiogenous cells integrated, terminal or intercalary, smooth, pale brown to brown, mono- or polytretic, proliferating sympodially. Chlamydospores present, subglobose, terminal and intercalary, 8–22 µm in diameter. Conidia ellipsoidal to clavate to obovoid, asymmetrical with paler basal and apical cells, usually curved at the third cell from the base which is darker than the other cells, (15-)20–23(-28) × (7-)8–10(-11) µm, (2-)3-distoseptate with non-protuberant, thickened, and darkened hila.
Specimens examined: India, Coimbatore, human corneal scraping from corneal ulcer, 2013, (holotype: freeze dried culture specimen in the Szeged Microbiological Collection (SZMC) at the Department of Microbiology, Faculty of Science and Informatics, University of Szeged, Hungary, SZMC 22226, includes ex-type culture); India, Coimbatore, human corneal scraping from corneal ulcer, 2011, (SZMC 26758); India, Coimbatore, human corneal scraping from corneal ulcer, 2011–2013, (SZMC 26759).
2.4. Antifungal Susceptibilities of Curvularia Strains Isolated from Fungal Keratitis
The minimum inhibitory concentrations (MIC) of nine antifungal agents towards C. coimbatorensis SZMC 22225, C. tamilnaduensis SZMC 22226, SZMC 26758, and SZMC 26759, as well as the type strains of C. australiensis (CBS 172.57), C. hawaiiensis (CBS 173.57), and C. spicifera (CBS 274.52) are shown in Table 3. The MIC of natamycin was 2 µg mL−1 for both new species and all other strains tested, while substantial differences between them could be observed in the case of clotrimazole, econazole, miconazole, and terbinafine, with the type strain of C. coimbatorensis having 4, 8, 4, and 4–8 times higher values, respectively. Among the tested isolates, the type strain of C. spicifera proved to be the less susceptible to clotrimazole, econazole, fluconazole, ketoconazole, and miconazole. Notable strain-to-strain variations between the C. tamilnaduensis strains could be observed only in the case of itraconazole and ketoconazole with detected MIC ranges of 0.03–0.25 and 0.06–0.25, respectively.
Table 3.
Antifungal susceptibilities of the Curvularia coimbatorensis and Curvularia tamilnaduensis strains in comparison with the type strains of Curvularia australiensis, Curvularia hawaiiensis, and Curvularia spicifera determined by the CLSI (Clinical & Laboratory Standards Institute) broth microdilution method (minimum inhibitory concentrations (MIC) values in µg mL−1).
| Strain | Antifungal Agent | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMB | CLT | ECN | FLC | ITC | KTC | MCZ | NTM | TRB | |
| C. australiensis CBS 172.57 T | 0.25 | 0.25 | 0.125 | 16 | 0.03 | 0.25 | 0.25 | 2 | 0.25 |
| C. hawaiiensis CBS 173.57 T | 0.25 | 0.06 | 0.06 | 4 | 0.03 | 0.06 | 0.125 | 2 | 0.25 |
| C. spicifera CBS 274.52 T | 0.5 | 4 | 2 | >32 | 0.25 | 2 | 2 | 2 | 1 |
| C. coimbatorensis SZMC 22225 T | 0.5 | 0.5 | 1 | 32 | 0.25 | 0.25 | 1 | 2 | 1 |
| C. tamilnaduensis SZMC 22226 T | 1 | 0.125 | 0.125 | 8 | 0.03 | 0.06 | 0.25 | 2 | 0.25 |
| C. tamilnaduensis SZMC 26758 | 0.5 | 0.125 | 0.125 | 16 | 0.03 | 0.25 | 0.25 | 2 | 0.125 |
| C. tamilnaduensis SZMC 26759 | 1 | 0.125 | 0.125 | 16 | 0.25 | 0.25 | 0.25 | 2 | 0.25 |
T: type strain; AMB: amphotericin B; CLT: clotrimazole; ECN: econazole; FLC: fluconazole; ITC: itraconazole; KTC: ketoconazole; MCZ: miconazole; NTM: natamycin; TRB: terbinafine.
3. Discussion
The phylogenetic tree obtained from the concatenated dataset of three loci presents an update about the phylogeny of Curvularia, which is mostly in agreement with the recently published phylogenies of this genus (Figure 1). C. ischaemi formed a clade with C. coicis, which is in contradiction with the results of Tan et al. [8] and Tibpromma et al. [27], where C. ischaemi formed a sister clade to C. gladioli, but in agreement with the phylogram obtained by Madrid et al. [7] and Manamgoda et al. [28]. Our analysis placed C. perotidis as a sister clade to C. australiensis, however, other studies [7,8,27,29] suggested that this species is closer to C. spicifera. The placement of C. variabilis was also different from previously published articles [8,29]. According to the analyses of Tan et al. [8] and Marin-Felix et al. [29], C. variabilis forms a clade with C. hawaiiensis, C. nodosa, C. dactyloctenicola, and C. beasleyi, however, in this study we found C. variabilis as a sister clade of C. tsudae and C. mebaldsii. The same authors found C. tripogonis, C. pseudorobusta, C. robusta, C. alcornii, C. protuberata, and C. inaequalis as members of two distinct monophyletic clades, while our results indicate that these species are closely related and paraphyletic, however, none of the topologies have strong statistical supports. The observed slight differences between the previous inferences and our analyses did not affect the validity of any of the previously described species, and some of them might be the result of the slightly broader taxon sampling.
One of the newly described species, C. coimbatorensis is only known from the type specimen isolated from corneal ulcer. Phylogenetic analysis based on three loci placed C. coimbatorensis as a sister clade to the other newly described species C. tamilnaduensis. The two species are closely related, but can be distinguished by tef1a, gpdh, and ITS sequences, with percentage identities of 99%, 98%, and 99%, respectively. C. petersonii [8] is also closely related and can be distinguished by all three loci (98% in tef1a, 93% in gpdh and 96% in ITS). C. coimbatorensis differs from C. tamilnaduensis in colony morphology, the lack of chlamydospores, and the size of conidia. C. petersonii is very similar in colony morphology, however, has significantly shorter (up to 110 µm) and only slightly geniculate conidiophores bearing narrower (5-)5.5–6(-7) conidia [8]. C. coimbatorensis has longer conidiophores.
The phylogenetic analysis based on three loci placed the other newly described species, C. tamilnaduensis as a sister clade to the recently described species C. petersonii. C. tamilnaduensis can be reliably distinguished from the ex-type of C. petersonii by tef1a, gpdh and ITS sequences with percentage identities of 99%, 95%, and 96%, respectively. The two species also differ by morphology, as C. petersonii has not been reported to produce chlamydospores and has different conidial dimensions (17–19 × 5.5–6) [8]. C. americana [7] and C. verruculosa [30] are also related species with considerable amount of genetic distances and none of these species have been reported before to have chlamydopores. C. americana has 4(-5)-distoseptate and wider (7–15 µm) conidia, while C. verruculosa has mostly 3-distoseptate conidia, but also wider (12–17 µm) than those of C. tamilnaduensis.
The antifungal susceptibilities of the examined strains of C. coimbatorensis and C. tamilnaduensis to amphotericin B, clotrimazole, econazole, fluconazole, itraconazole, ketoconazole, miconazole, natamycin, and terbinafine were within the MIC ranges reported for other clinically relevant Curvularia species in the study of Guarro et al. [11] and the review of Krizsán et al. [3]. The type strain of C. coimbatorensis proved to be less susceptible than the strains of C. tamilnaduensis to all antifungals except for natamycin. For itraconazole and ketoconazole our results are in agreement with the study of Guarro et al. [11], who reported that amphotericin B, itraconazole, miconazole and ketoconazole are highly effective against a series of Curvularia species known from fungal keratitis (C. brachyspora, C. clavata, C. geniculata, C. lunata, C. pallescens, C. senegalensis, and C. verruculosa).
4. Materials and Methods
4.1. Curvularia Strains, Culture Conditions, and Morphological Examination
The Curvularia strains involved in this study derived from corneal scrapings from fungal corneal ulcers of keratitis patients attending the Aravind Eye Hospital and Postgraduate Institute of Ophthalmology, Coimbatore, India. All cases were initially screened by experienced ophthalmologists, and the corneal scrapings were collected following the clinical diagnosis of fungal keratitis. The samples were initially processed microbiologically for the isolation of the causative agents as described earlier [31]. The corneal scrapings of all patients were subjected to Gram stain, Giemsa stain, and 10% KOH wet mount. Culture methods involved direct inoculation of specimens onto 5% sheep blood agar, chocolate agar, non-nutrient agar, potato dextrose agar, thioglycolate broth, and brain–heart infusion broth. The microbial cultures were considered positive only if the growth of the same organism was demonstrated on two or more solid media, or there was confluent growth at the site of inoculation on one solid medium with consistent direct microscopic findings. The isolates were deposited in the Szeged Microbiology Collection (SZMC, Szeged, Hungary) under the accession numbers SZMC 22225, SZMC 22226, SZMC 26758, and SZMC 26759. Colony morphology of the isolates was examined on PDA (BioLab, Budapest, Hungary) medium after 7 days of incubation at 25 °C under normal day/night light conditions. Micromorphological characters were examined with a Leica DMI 4000B (Leica, Wetzlar, Germany) microscope equipped with a Leica DFC 295 camera. Microscopic features were examined in lactic acid (100% v/v) on glass slides. Conidiophores were studied in the same mounting fluid with the transparent tape method. Conidiophores and conidia were measured using the software ImageJ v2.52a (National Institute of Mental Health, Bethesda, MD, USA). Size ranges of the conidia were derived from 50 measurements. Lengths and widths are given as (minimum value) mean size minus SD-mean size plus SD (maximum value).
4.2. DNA Extraction, Amplification, Sequencing, and Phylogenetic Analysis
Genomic DNA was isolated from the examined Curvularia strains SZMC 22225, SZMC 22226, SZMC 26758, and SZMC 26759 with the Masterpure™ Yeast DNA Purification Kit (Epicentre Biotechnologies, Madison, WI, USA) according to the manufacturer’s instructions. Fragments of tef1a and gpdh were amplified as described previously [5,32,33]. The ITS region of the ribosomal RNA gene cluster was amplified according to White et al. [34]. Sequencing of the amplicons was carried out on a 3500 Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA) by the sequencing service of the Biological Research Centre, Szeged, Hungary. Resulting sequences were deposited in the GenBank Nucleotide database (www.ncbi.nlm.nih.gov) under the accession numbers shown in Table 2.
Sequences of the four clinical isolates were aligned with publicly available sequences of 108 previously described Curvularia species, as well as Bipolaris maydis as the outgroup (Table 2). Phylogenetic analyses were conducted using three loci (tef1α, gpdh and ITS). Sequences of all three loci were aligned with the phylogeny-aware sequence alignment tool Canopy v0.1.4 using RAxML as tree estimator and PRANK [35] with the -F option as the aligner with 10 iterations and seed decomposition strategy. Alignments of the three loci were concatenated and partitioned by region. The tef1α sequences formed one partition while in the case of gpdh sequences the dataset was partitioned to exons and introns. The ITS dataset was divided to rDNA and ITS1-ITS2 regions. Alignments of gpdh and ITS datasets contained high number of indels with important phylogenetic signal, therefore gaps were coded as absence/presence characters by SequenceMatrix v1.8 [36] using the simple indel coding algorithm [37]. The two indel matrices were concatenated and added as a single partition to the dataset. Maximum likelihood analysis was performed using RAxML-NG v0.9.0 [38] under the GTR model with gamma-distributed rate heterogeneity using empirical base frequencies. As indel-based datasets do not contain constant sites, the ascertainment bias correction described by Lewis [39] was used for this partition. Statistical support of the best ML tree was obtained with 1000 thorough bootstrap replicates.
4.3. Antifungal Susceptibility Testing
In vitro antifungal susceptibility tests were carried out according to the CLSI M38-A2 broth microdilution method [40]. Nine antifungal agents: amphotericin B, clotrimazole, econazole, fluconazole, itraconazole, ketoconazole, miconazole, natamycin and terbinafine (Sigma-Aldrich, Budapest, Hungary) were examined. Microtiter plates were incubated at 35 °C for 72 h. Plates were evaluated both spectrophotometrically with a Spectrostar Nano microplate reader (BMG Labtech, Ortenberg, Germany) and by visual examination.
5. Conclusions
The present study demonstrates, that although the phylogeny of the genus Curvularia is resolved and well established, further expansion can be expected both in the list of described Curvularia species and in the known spectrum of clinically relevant members of the genus. The collection of further keratitis isolates from the genus Curvularia and gaining data about their antifungal susceptibilities are therefore tasks of increasing importance. Furthermore, comparing the infectivity of various Curvularia species causing keratitis—including the recently described ones—in animal keratitis models would be an intriguing topic for future research.
Acknowledgments
The authors wish to thank Venkatapathy Narendran (Aravind Eye Hospital and Postgraduate Institute of Ophthalmology, Coimbatore, Tamil Nadu, India), Coimbatore Subramanian Shobana (Department of Microbiology, PSG College of Arts and Science, Coimbatore, Tamil Nadu, India) and Kanesan Panneer Selvam (Department of Microbiology, M.R Government Arts College, Mannargudi, Tamil Nadu, India) for constantly supporting the research efforts on fungal keratitis within the frames of the Indo-Hungarian Fungal Keratitis Research Group.
Author Contributions
Conceptualization, S.K., L.K., T.P. and C.V.; methodology, N.K., A.M., M.H. and S.K.; software, S.K.; validation, R.R., P.M., M.H., C.V., M.V. and S.K.; formal analysis, K.K., T.P. and M.V.; investigation, N.K., A.M., P.M., K.K., M.H., M.V. and S.K.; resources, R.R., P.M., A.M., T.P. and C.V.; data curation, N.K., S.K., K.K., L.K. and M.H.; writing—original draft preparation, N.K., S.K., L.K.; writing—review and editing, N.K., S.K., L.K., P.M., T.P. and C.V.; visualization, N.K., M.H. and S.K.; supervision, S.K.; project administration, S.K., T.P., L.K., and P.M.; funding acquisition, S.K., T.P., L.K. and P.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants NKFI PD-116609 (National Research, Development and Innovation Office, Hungary), GINOP-2.3.2-15-2016-00035 (Széchenyi 2020 Programme) and also supported by the COST action HUPLANTcontrol (Control of Human Pathogenic Micro-organisms in Plant Production Systems, CA16110). LK is grantee of the János Bolyai Research Scholarship (Hungarian Academy of Sciences) and the Bolyai Plus Scholarship (New National Excellence Programme). TP and MH are supported by the grants LP2016-8/2016 and by the FIKP program (TUDFO/4738-1/2019 ITM) of the Ministry of Human Capacities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bengyella L., Iftikhar S., Nawaz K., Fonmboh D.J., Yekwa E.L., Jones R.C., Njanu Y.M.T., Roy P. Biotechnological application of endophytic filamentous Bipolaris and Curvularia: A review on bioeconomy impact. World J. Microbiol. Biotechnol. 2019;35:69. doi: 10.1007/s11274-019-2644-7. [DOI] [PubMed] [Google Scholar]
- 2.Kusai N.A., Azmi M.M.Z., Zulkifly S., Yusof M.T., Zainudin N.A.I.M. Morphological and molecular characterization of Curvularia and related species associated with leaf spot disease of rice in Peninsular Malaysia. Rend. Lincei Sci. Fis. Nat. 2016;27:205–214. doi: 10.1007/s12210-015-0458-6. [DOI] [Google Scholar]
- 3.Krizsán K., Papp T., Manikandan P., Shobana C.S., Chandrasekaran M., Vágvölgyi C., Kredics L. Clinical Importance of the Genus Curvularia. In: Razzaghi-Abyaneh M., Shams-Ghahfarokhi M., Rai M., editors. Medical Mycology: Current Trends and Future Prospects. CRC Press; Boca Raton, FL, USA: 2016. pp. 147–204. [DOI] [Google Scholar]
- 4.Yanagihara M., Kawasaki M., Ishizaki H., Anzaw K., Udagawa S., Mochizuki T., Sato Y., Tachikawa N., Hanakawa H. Tiny keratotic brown lesions on the interdigital web between the toes of a healthy man caused by Curvularia species infection and a review of cutaneous Curvularia infections. Mycoscience. 2010;51:224–233. doi: 10.1007/S10267-009-0030-2. [DOI] [Google Scholar]
- 5.Manamgoda D.S., Cai L., McKenzie E.H.C., Crous P.W., Madrid H., Chukeatirote E., Shivas R.G., Tan Y.P., Hyde K.D. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012;56:131–144. doi: 10.1007/s13225-012-0189-2. [DOI] [Google Scholar]
- 6.Paredes K., Capilla J., Sutton D.A., Mayayo E., Fothergill A.W., Guarro J. Virulence of Curvularia in a murine model. Mycoses. 2013;56:512–515. doi: 10.1111/myc.12064. [DOI] [PubMed] [Google Scholar]
- 7.Madrid H., da Cunha K.C., Gené J., Dijksterhuis J., Cano J., Sutton D.A., Guarro J., Crous P.W. Novel Curvularia species from clinical specimens. Persoonia. 2014;33:48–60. doi: 10.3767/003158514X683538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan Y.P., Crous P.W., Shivas R.G. Cryptic species of Curvularia in the culture collection of the Queensland Plant Pathology Herbarium. MycoKeys. 2018;35:1–25. doi: 10.3897/mycokeys.35.25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcus L., Vismer H.F., van der Hoven H.J., Gove E., Meewes P. Mycotic keratitis caused by Curvularia brachyspora (Boedjin). A report of the first case. Mycopathologia. 1992;119:29–33. doi: 10.1007/BF00492227. [DOI] [PubMed] [Google Scholar]
- 10.Travis W.D., Kwon-Chung K.J., Kleiner D.E., Geber A., Lawson W., Pass H.I., Henderson D. Unusual aspects of allergic bronchopulmonary fungal disease: Report of two cases due to Curvularia organisms associated with allergic fungal sinusitis. Hum. Pathol. 1991;22:1240–1248. doi: 10.1016/0046-8177(91)90106-Y. [DOI] [PubMed] [Google Scholar]
- 11.Guarro J., Akiti T., Horta R.A., Morizot Leite-Filho L.A., Gené J., Ferreira-Gomes S., Aguilar C., Ortoneda M. Mycotic keratitis due to Curvularia senegalensis and in vitro antifungal susceptibilities of Curvularia spp. J. Clin. Microbiol. 1999;37:4170–4173. doi: 10.1128/jcm.37.12.4170-4173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Y.M., Huang W.M., Li S.F., Wu G.F., Li W., Chen R.Y. Cutaneous phaeohyphomycosis of foot caused by Curvularia clavata. Mycoses. 2009;52:544–546. doi: 10.1111/j.1439-0507.2008.01646.x. [DOI] [PubMed] [Google Scholar]
- 13.Vasikasin V., Nasomsong W., Srisuttiyakorn C., Mitthamsiri W., Oer-Areemitr N., Changpradub D. Disseminated phaeohyphomycosis caused by Curvularia tuberculata in a previously healthy man. Mycopathologia. 2019;184:321–325. doi: 10.1007/s11046-019-00323-0. [DOI] [PubMed] [Google Scholar]
- 14.Pimentel J.D., Mahadevan K., Woodgyer A., Sigler L., Gibas C., Harris O.C., Lupino M., Athan E. Peritonitis due to Curvularia inaequalis in an elderly patient undergoing peritoneal dialysis and a review of six cases of peritonitis associated with other Curvularia spp. J. Clin. Microbiol. 2005;43:4288–4292. doi: 10.1128/JCM.43.8.4288-4292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Posteraro B., Scarano E., La Sorda M., Torelli R., De Corso E., Mulé A., Paludetti G., Fadda G., Sanguinetti M. Eosinophilic fungal rhinosinusitis due to the unusual pathogen Curvularia inaequalis. Mycoses. 2010;53:84–88. doi: 10.1111/j.1439-0507.2008.01671.x. [DOI] [PubMed] [Google Scholar]
- 16.Cruz R., Barthel E., Espinoza J. Allergic rhinosinusitis by Curvularia inaequalis (Shear) Boedijn. Rev. Chil. Infectol. 2013;30:319–322. doi: 10.4067/S0716-10182013000300008. (In Spanish) [DOI] [PubMed] [Google Scholar]
- 17.Flanagan K.L., Bryceson A.D. Disseminated infection due to Bipolaris australiensis in a young immunocompetent man: Case report and review. Clin. Infect. Dis. 1997;25:311–313. doi: 10.1086/514528. [DOI] [PubMed] [Google Scholar]
- 18.Filizzola M.J., Martinez F., Rauf S.J. Phaeohyphomycosis of the central nervous system in immunocompetent hosts: Report of a case and review of the literature. Int. J. Infect. Dis. 2003;7:282–286. doi: 10.1016/S1201-9712(03)90108-1. [DOI] [PubMed] [Google Scholar]
- 19.Gadgil N., Kupferman M., Smitherman S., Fuller G.N., Rao G. Curvularia brain abscess. J. Clin. Neurosci. 2013;20:173–175. doi: 10.1016/j.jocn.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Vachharajani T.J., Zaman F., Latif S., Penn R., Abreo K.D. Curvularia geniculata fungal peritonitis: A case report with review of literature. Int. Urol. Nephrol. 2005;37:781–784. doi: 10.1007/s11255-004-0628-4. [DOI] [PubMed] [Google Scholar]
- 21.Diskin C.J., Stokes T.J., Dansby L.M., Radcliff L., Carter T.B. Case report and review: Is the tendency for Curvularia tubular obstruction significant in pathogenesis? Perit. Dial. Int. 2008;28:678–679. [PubMed] [Google Scholar]
- 22.Saenz R.E., Brown W.D., Sanders C.V. Allergic bronchopulmonary disease caused by Bipolaris hawaiiensis presenting as a necrotizing pneumonia: Case report and review of literature. Am. J. Med. Sci. 2001;321:209–212. doi: 10.1097/00000441-200103000-00012. [DOI] [PubMed] [Google Scholar]
- 23.Robson A.M., Craver R.D. Curvularia urinary tract infection: A case report. Pediatr. Nephrol. 1994;8:83–84. doi: 10.1007/BF00868275. [DOI] [PubMed] [Google Scholar]
- 24.Safdar A. Curvularia—Favorable response to oral itraconazole therapy in two patients with locally invasive phaeohyphomycosis. Clin. Microbiol. Infect. 2003;9:1219–1223. doi: 10.1111/j.1469-0691.2003.00791.x. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez M., Noyola D.E., Rossmann S.N., Edwards M.S. Cutaneous phaeohyphomycosis caused by Curvularia lunata and a review of Curvularia infections in pediatrics. Pediatr. Infect. Dis. J. 1999;18:727–731. doi: 10.1097/00006454-199908000-00016. [DOI] [PubMed] [Google Scholar]
- 26.Dave V.P., Joseph J., Pathengay A., Pappuru R.R., Das T. Clinical presentations, diagnosis, and management outcomes of Curvularia endophthalmitis and a review of literature. Retina. 2018 doi: 10.1097/IAE.0000000000002375. [DOI] [PubMed] [Google Scholar]
- 27.Tibpromma S., Hyde K.D., Bhat J.D., Mortimer P.E., Xu J., Promputtha I., Doilom M., Yang J.B., Tang A.M.C., Karunarathna S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys. 2018;33:25–67. doi: 10.3897/mycokeys.33.23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manamgoda D.S., Rossman A.Y., Castlebury L.A., Crous P.W., Madrid H., Chukeatirote E., Hyde K.D. The genus Bipolaris. Stud. Mycol. 2014;79:221–288. doi: 10.1016/j.simyco.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marin-Felix Y., Senwanna C., Cheewangkoon R., Crous P.W. New species and records of Bipolaris and Curvularia from Thailand. Mycosphere. 2017;8:1556–1574. doi: 10.5943/mycosphere/8/9/11. [DOI] [Google Scholar]
- 30.Sivanesan A. Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. Mycol. Pap. 1987;158:1–261. [Google Scholar]
- 31.Mythili A., Babu Singh Y.R., Priya R., Shafeeq Hassan A., Manikandan P., Panneerselvam K., Narendran V., Shobana C.S. In vitro and comparative study on the extracellular enzyme activity of molds isolated from keratomycosis and soil. Int. J. Ophthalmol. 2014;7:778–784. doi: 10.3980/j.issn.2222-3959.2014.05.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoch C.L., Crous P.W., Groenewald J.Z., Boehm E.W.A., Burgess T.I., de Gruyter J., de Hoog G.S., Dixon L.J., Grube M., Gueidan C., et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009;64:1–15. doi: 10.3114/sim.2009.64.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berbee M., Pirseyedi M., Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977. doi: 10.1080/00275514.1999.12061106. [DOI] [Google Scholar]
- 34.White T.J., Bruns T.D., Lee S., Taylor J.W. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White J.W., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 35.Löytynoja A. Phylogeny-aware alignment with PRANK. Meth. Mol. Biol. 2014;1079:155–170. doi: 10.1007/978-1-62703-646-7_10. [DOI] [PubMed] [Google Scholar]
- 36.Vaidya G., Lohman D.J., Meier R. SequenceMatrix: Concatenation software for the fast assembly of multigene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 37.Simmons M.P., Ochoterena H. Gaps as characters in sequence-based phylogenetic analysis. Syst. Biol. 2000;49:369–381. doi: 10.1093/sysbio/49.2.369. [DOI] [PubMed] [Google Scholar]
- 38.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis P.O. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 2001;50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- 40.Clinical and Laboratory Standards Institute . Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 3rd ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2008. Approved Standard, CLSI Document M27-A3. [Google Scholar]