Abstract
Ascomycete Sclerotinia sclerotiorum (Lib.) de Bary is one of the most damaging soilborne fungal pathogens affecting hundreds of plant hosts, including many economically important crops. Its genomic sequence has been available for less than a decade, and it was recently updated with higher completion and better gene annotation. Here, we review key molecular findings on the unique biology and pathogenesis process of S. sclerotiorum, focusing on genes that have been studied in depth using mutant analysis. Analyses of these genes have revealed critical players in the basic biological processes of this unique pathogen, including mycelial growth, appressorium establishment, sclerotial formation, apothecial and ascospore development, and virulence. Additionally, the synthesis has uncovered gaps in the current knowledge regarding this fungus. We hope that this review will serve to build a better current understanding of the biology of this under-studied notorious soilborne pathogenic fungus.
Keywords: soilborne fungal pathogen, Sclerotinia sclerotiorum, fungal pathogenesis, sclerotial formation, apothecial development, fungal growth
1. Introduction
Ascomycete Sclerotinia sclerotiorum (Lib.) de Bary is a soilborne pathogenic fungus, which was first described as Peziza sclerotiorum by Libert [1]. In 1945, Whetzel [2] established S. sclerotiorum (Lib.) de Bary as the major species of Sclerotinia, as it is the best-known and most economically important pathogen of the genus. It was proposed as the representative species for the genus Sclerotinia in the 1970s [3,4]. The Special Committee on Fungi and Lichens recommended the generic name Sclerotinia, and so did the General Committee of the International Association of Plant Taxonomists later on [5].
S. sclerotiorum belongs to the family Sclerotiniaceae [2] of the class Leotiomycetes. Sclerotiniaceae also includes two other very closely related species, S. minor and S. trifoliorum. All three species produce vegetative resting structures, termed sclerotia, and mushroom-like reproductive structures, named apothecia, during their life cycles. However, S. minor forms small sesame-sized sclerotia, whereas S. sclerotiorum and S. trifoliorum can form large sclerotia as big as peas. They all lack an obvious conidial stage. S. sclerotiorum has the broadest host range, which includes both herbaceous and succulent plants, and even some woody ornamentals and monocots. S. trifoliorum was isolated from vegetable legumes, while S. minor has been found on sunflowers, tomatoes, carrots, peanuts, and lettuce.
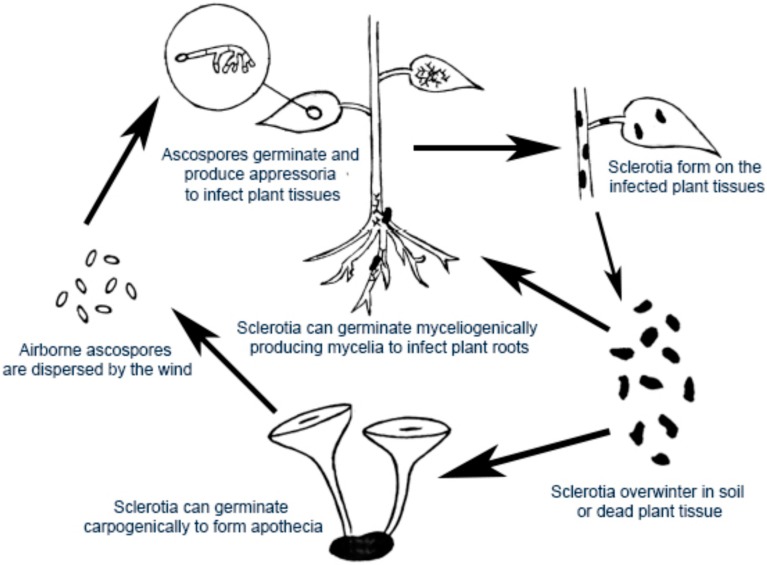
S. sclerotiorum is a highly damaging pathogen with diverse infection modes and a double feeding lifestyle of both biotroph and necrotroph (Figure 1) [6,7]. It attacks host plants either by means of ascospores that can be discharged forcibly upwards from apothecia into the air, or by mycelium arising from infected tissue or from germinated sclerotia [8]. When ascospores land on susceptible host tissue, they can germinate under favorable conditions and start a new cycle of infection. To date, the microconidia of S. sclerotiorum have been observed amongst the mycelium and on the surfaces of sclerotia. However, no evidence of their functionality has been identified. Under moist and cool conditions, this fungus rapidly grows inside the infected host tissues and develops symptoms of browning, water-soaking, and a white, cotton-like mycelium, which leads to necrosis, stunting, premature ripening, and wilting of the host [8]. Therefore, the diseases that it causes have been given names, including stem rot, drop, crown rot, cottony rot, watery soft rot, blossom blight, and Sclerotinia blight. Upon killing the host, the fungus can saprophytically grow on the dead plant tissue. Sclerotia are later abundantly formed on the host surface and cavities, in plant debris, and in soil, where they are able to remain dormant for up to 10 years [9].
Figure 1.
Life cycle of Sclerotinia sclerotiorum.
S. sclerotiorum has an extremely broad host range, which consists of more than 600 plant species, including almost all dicotyledonous and some monocotyledonous plants [10,11]. It can infect many economically important crops, such as canola; legumes such as soybean and peanut; sunflower; various vegetables such as lettuce and tomato; and monocotyledonous plants such as tulip and onion [10,12]. It is therefore not surprising that it causes significant economic losses globally every year [13]. For example, in China, oilseed rape yield losses that are caused by SSR (Sclerotinia stem rot) usually range from 10 to 20% and they may be up to 80% for severe SSR outbreak seasons [14,15,16]. In The United States, annual losses that are caused by this pathogen have exceeded $200 million [12].
As with most other fungal pathogens, chemical control is the most commonly used method for controlling the diseases that are caused by S. sclerotiorum, although it is environmentally unfriendly and can induce resistance from the pathogen. Chemical control also becomes less effective once the sclerotia are formed. Obviously, the incidence of disease would be greatly reduced if sclerotia could be either prevented from forming or destroyed in soil and plant debris. Flooding and crop rotation are good options that accelerate the decay of sclerotia [17]. However, flooding is impractical in most areas, and crop rotation has been found to be less effective as a control measure [18,19], because of the long survival periods of sclerotia in soil. Measures, like widening row spacing, and using wire trellis supports to raise foliage from the ground have been reported to be helpful [18,20]. However, factors, such as type of crop, method of cultivation, and environmental conditions, always affect the effectiveness of each control method.
For major crop diseases, the most welcoming and environmentally friendly control method is through the use of resistant cultivars. However, strong host single-gene resistance has not been found against S. sclerotiorum [12], which makes it difficult to improve resistance using classical breeding methods [7]. Even with mild resistance, the traits seem to be multigenic. Future development of integrated approaches may help to control the spread of the fungus with better understanding of the biology of this notorious pathogen and better QTL mapping strategies.
In this review, we focus on recent key molecular findings on the unique biology and pathogenesis process of S. sclerotiorum. A brief summary of the genome of the pathogen will also be discussed. The genomic sequences of S. sclerotiorum have been available for less than a decade [21]. However, it was recently updated with higher completion and better gene annotation [22]. We hope that this review will serve to build a better current understanding of the biology of this notorious soilborne pathogenic fungus.
2. The Features of the S. sclerotiorum Genome
2.1. Genome Sequences
The genome of S. sclerotiorum strain 1980, originally isolated from beans in western Nebraska USA, was first sequenced in 2011 using Sanger technology with 9.1x coverage. As approximately 1.6 Mb of sequences were not covered by the 38.0 Mb scaffolds, and thus missed from the assembly, the final size of S. sclerotiorum genome was estimated to be approximately 38.3 Mb, which consists of 16 linkage groups likely corresponding to 16 chromosomes (GenBank accession numbers: AAGT01000000) [21]. Most of the uncovered regions are located in the middle of chromosomes, which probably correspond to centromeres. The genome was predicted to contain 14,522 genes, with average GC contents of 41.8%. After removing genes encoding small proteins that are less than 100 amino acids in length or without evidence of expression (from ESTs and/or microarray signals), 11,860 predicted proteins with high confidence were deduced [21].
Initial analyses of the genome sequences revealed a large number of virulence related genes that were possessed by the S. sclerotiorum 1980 strain, including genes encoding cell wall degrading enzymes (CWDEs) and biosynthesis genes of phytotoxins and other secondary metabolites [21]. Amselem et al. also identified a significant amount of repetitive transposable elements (TEs), which comprises approximately 7.7% of the whole genome. This led to a postulation that the S. sclerotiorum genome experienced a recent major remodeling that was associated with a dramatic expansion of TEs. The TE expansion in S. sclerotiorum might also have an impact on its genome organization and function of gene inactivation, modification, or expression regulation [21].
Derbyshire et al. recently sequenced the genome sequence of the same S. sclerotiorum 1980 strain to near completion in 2017 using PacBio technology. It was annotated using extensive RNA-Seq data and manual curation. Therefore, this version of the whole-genome sequence became the choice of standard for Sclerotinia. They identified 70 candidate effector genes, and found a significant association between the positions of these secreted proteins and regions with a high relative RIP (repeat-induced point mutation) index, which suggested that S. sclerotiorum exhibits a subtly enhanced mutation rate of secreted proteins in specific genomic compartments as a result of transposition and RIP activity [23].
More recently, Derbyshire et al. sequenced the genomes of 25 field isolates of S. sclerotiorum that were collected from four different continents—Australia, Africa (north and south), Europe, and North America (Canada and the Northern United States). They conducted SNP-based analyses on population structure and selective sweeps [24,25]. These 25 isolates can be grouped into two major populations, where population 1 consists of 11 isolates from Canada, the USA, and France, and population 2 includes nine isolates from Australia and one from Morocco. A single candidate selective sweep was identified in the Australian and Moroccan group, which covers less than 0.001% of the genome. It is reasonable to speculate that a slow evolution rate and extensive self-fertilization of S. sclerotiorum may have led to a striking absence of strong selective sweeps, as there is only one gene within this 10 Kb region, which encodes a major facilitator superfamily transporter that was only negligibly expressed at late stages of Brassica napus infection [22]. In support of such genome conservation, S. sclerotiorum strains from even a tropical and a temperate region, although segregated in the populations, still exhibit similar genetic structure [26].
2.2. Transcriptomic and Secretomic Analysis
The availability of the high-quality genome sequences facilitated comparative transcriptomic and secretomic analysis of this notorious pathogen, which in turn facilitated biological investigations. Seifbarghi et al. (2017) was able to delineate gene expression patterns that signified transitions between pathogenic phases of S. sclerotiorum, namely, host penetration, ramification, and necrotrophic stages, through RNA-Seq analysis focusing on events occurring through the early (1 h) to the middle (48 h) stages of infection. These expression data provide evidence for the occurrence of a brief biotrophic phase soon after host penetration [27].
432 proteins were identified in S. sclerotiorum in an interspecies comparative analysis of the predicted secretomes of S. sclerotiorum and Botrytis cinerea. Among them, 16% of the encoding genes reside in small gene clusters that are distributed over 13 of the 16 predicted chromosomes [28]. These candidate genes provide a reservoir for future reverse genetics analyses to delineate the pathogenesis mechanisms in the S. sclerotiorum-host plant interactions.
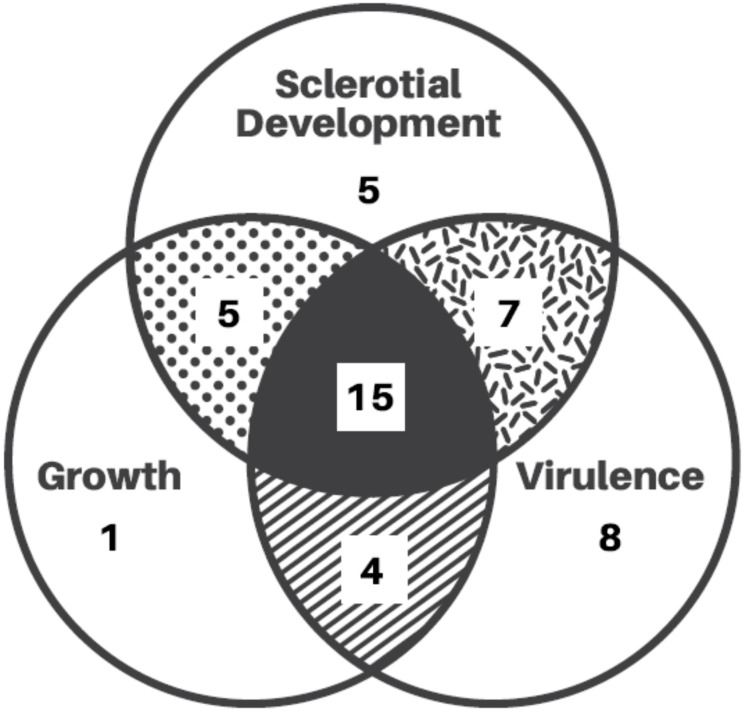
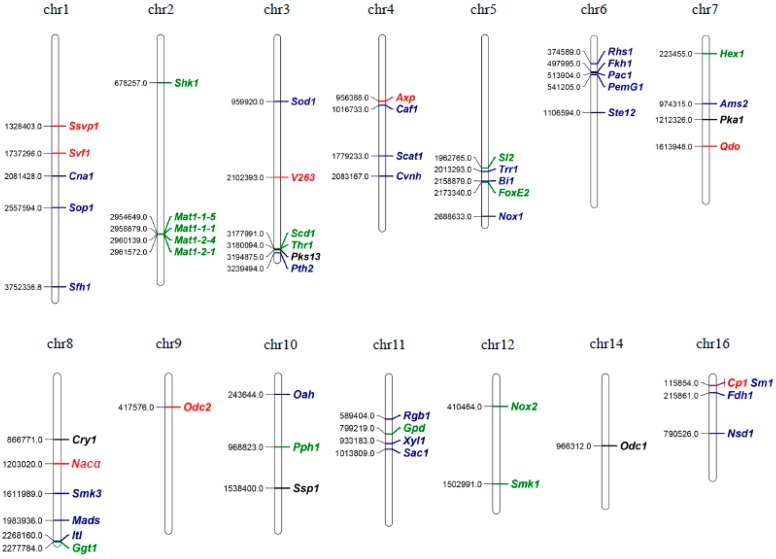
Mutant analysis is paramount to establish a causal relationship between a gene and a studied biological process, as omics experiments only provide vague association clues. Hence, the rest of this review will summarize all of the genes studied in S. sclerotiorum so far using such convincing mutant analysis methods. A Venn diagram of the functions of these encoded proteins is provided for readers’ overview (Figure 2). We also constructed a chromosomal map for these genes to detect possible clustering events that are often associated with virulence factors (Figure 3). However, little clustering is observed, except for genes contributing to mating and melanisation, reflecting the under-studied nature of this fungus.
Figure 2.
A Venn diagram summary of the genes that have been studies with mutant analysis (detailed in Table 1).
Figure 3.
Map positions of the genes in Table 1. The numbers on the left of each chromosome represent the locations of these genes. Genes labelled in green are involved in the development of S. sclerotiorum, including hyphal growth, sclerotial formation, apothecial formation and etc. Genes labelled in red mainly play roles in virulence of S. sclerotiorum. Genes labelled in blue are involved in both while the ones labelled in black are involved in other biological processes. The chromosomal map was drawn using ‘MapChart’ software.
3. Molecular Dissection of S. sclerotiorum Biology
Here we adopt the gene/mutant/protein nomenclature that is most commonly used by Sclerotinia researchers, as there are many discrepancies in S. sclerotiorum literature. As an example, Abc1 protein is encoded by wild-type gene that is italicized as Abc1. Mutant is denoted as abc1. Exceptions are specifically explained. Another consideration to keep in mind is the presence of huge variations in the field isolates and local hosts used in these studies (Table 1), which likely contribute to some of the discrepancies that were observed in the phenotypes of mutants of the same genes.
Table 1.
List of all Sclerotinia sclerotiorum genes studied so far using mutant analysis. The genes are ordered in regards to their chromosomal positions (Figure 3).
| Gene Code (New) | Gene Code (Old) | Strains | Mutant Name | Gene Full Name | Mutant Type | Mutant Phenotypes | Secretion Signal | Tested Hosts | Other Functions | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyphal Growth | Sclerotial Formation | Oxalate Production | Virulence | Appressoria Formation | Induce Host HR/Resistance | ||||||||||
| sscle_01g003850 | SS1G_02068 | Ep-1PNA367, Heilongjiang, China | ssvp1 | Small secreted virulence-related protein | RNAi | - | NA | NA | + | NA | + | Yes | Canola (Zhongyou 821) | HR induction | [30] |
| sscle_01g004990 | SS1G_01919 | 1980, Nebraska, USA | svf1 | Survival factor 1 homologue | RNAi | - | NA | - | + | + | NA | No | Arabidopsis and canola (Zhongyou 821) | Cell wall integrity, ROS production | [31] |
| sscle_01g006030 | SS1G_01788 | 1980, Nebraska, USA | cna1 | Catalytic subunit calcineurin-encoding gene | RNAi | + | + | - | + | - | NA | No | Arabiposis and tomato (Bonny Best) | Hyphal elongation | [32] |
| sscle_01g007450 | SS1G_01614 | Ep-1PNA367, Heilongjiang, China | sop1 | Microbial opsin homolog gene | RNAi | + | + | NA | + | NA | NA | No | Arabidopsis | Stress responses | [33] |
| sscle_01g011030 | SS1G_01151 | 1980, Nebraska, USA | sfh1 | GATA-box and SNF5 domains containing transcription factor | RNAi | + | + | NA | + | + | NA | No | Soybean, common bean and tomato | ROS accumulation | [34] |
| sscle_02g013550 | SS1G_12694 | HA61, Jiangsu, China | shk1 | Histidine kinases | Deletion | + | + | NA | - | NA | NA | No | Rapeseed, strawberry, tomato and cucumber | Oxidative stresses, glycerol accumulation | [35] |
| sscle_02g020240 | SS1G_04003 | 1980, Nebraska, USA | mat1-1-5 | Mating-type gene | Deletion | - | - | NA | - | NA | NA | No | Tomato | Apothecial development | [29] |
| sscle_02g020250 | SS1G_04004 | 1980, Nebraska, USA | mat1-1-1 | Mating-type gene | Deletion | - | - | NA | - | NA | NA | No | Tomato | Apothecial development | [29] |
| sscle_02g020260 | SS1G_04005 | 1980, Nebraska, USA | mat1-2-4 | Mating-type gene | Deletion | - | - | NA | - | NA | NA | No | Tomato | Apothecial development, ascospore production | [29] |
| sscle_02g020270 | SS1G_04006 | 1980, Nebraska, USA | mat1-2-1 | Mating-type gene | Deletion | + | - | NA | - | NA | NA | No | Tomato | Apothecial development | [29] |
| sscle_03g025030 | SS1G_00699 | WMA1, Washington, USA | sod1 | Cu/Zn superoxide dismutase | T-DNA | - | - | - | + | NA | NA | No | Pea (Guido) | Detoxification of host ROS | [36] |
| sscle_03g025030 | SS1G_00699 | 1980, Nebraska, USA | sod1 | Cu/Zn superoxide dismutase | Deletion | + | + | + | + | NA | NA | No | Tomato (Garden Peach) and tobacco | Oxidative stress tolerance, repression of host ROS | [37] |
| sscle_03g028510 | SS1G_00263 | Isolated from an infected canola stem, Alberta, Canada | v263 | Hypothetical secreted protein | Deletion | - | NA | NA | + | NA | NA | Yes | Canola | [38] | |
| sscle_03g031470 | SS1G_13314 | Isolated from diseased rapeseed, Alberta, Canada | scd1 | Scytalone dehydratase | Deletion | + | + | NA | - | NA | NA | No | Rapeseed (Westar) | Hyphal branching | [39] |
| sscle_03g031480 | SS1G_13315 | Isolated from diseased rapeseed, Alberta, Canada | thr1 | Trihydroxynaphthalene reductase | Deletion | + | + | NA | - | NA | NA | No | Rapeseed (Westar) | Hyphal branching | [39] |
| sscle_03g031520 | SS1G_13322 | 1980, Nebraska, U.S.A.; UF1, Florida; WMA, Washington, USA | pks13 | Polyketide synthase | Deletion by CRISPR | - | - | NA | - | + | NA | No | Soybean, canola, tomato (Better Boy), faba bean (Windsor) and pea (Sugar Daddy) | Pigmentation of compound appressoria | [40] |
| sscle_03g031670 | SS1G_13339 | 274, Nebraska, USA | pth2 | Peroxysomal carnitine acetyl transferase | Deletion | + | + | + | + | + | NA | No | Soybean | Oxalic acid accumulation | [41] |
| sscle_04g034810 | SS1G_02462 | Isolated from an infected canola stem, Alberta, Canada | axp | Arabinofuranosidase/β-xylosidase | Deletion | NA | NA | NA | + | NA | NA | Yes | Canola | [42] | |
| sscle_04g034960 | SS1G_02486 | Sunf-M, Inner Mongolia, China | caf1 | Secreted protein with a putative Ca2+-binding EF-hand motif | RNAi, T-DNA | + | + | - | + | + | + | Yes | Arabidopsis, rapeseed, pak choi cabbage, hot pepper, tomato, cucumber and soybean | [43] | |
| sscle_04g037170 | SS1G_02784 | 1980, Nebraska, USA | scat1 | Type A catalase | Deletion | + | + | NA | + | NA | NA | No | Tomato (Garden Peach) | Modulation of ROS | [44] |
| sscle_04g038020 | SS1G_02904 | Ep-1PNA367, Heilongjiang, China | cvnh | Secreted protein | RNAi | + | + | NA | + | NA | NA | Yes | Arabidopsis | [45] | |
| sscle_05g046240 | SS1G_05917 | SUN-F-M, Inner Mongolia, China | sl2 | Cell wall protein | RNAi | - | + | NA | - | NA | NA | Yes | Rapeseed and Arabidopsis | Cellular integrity | [46] |
| sscle_05g046390 | SS1G_05899 | 1980, Nebraska, USA | trr1 | Thioredoxin reductase | RNAi | NA | + | NA | + | NA | NA | No | Arabidopsis and tobacco | Oxidative stress tolerance | [47] |
| sscle_05g046790 | SS1G_05839 | 1980, Nebraska, USA | bi1 | Bax inhibitor-1 protein | RNAi | + | - | - | + | NA | NA | No | Arabidopsis and tomato | Stress responses, hyphal tip branching | [48] |
| sscle_05g046830 | SS1G_05834 | JRUF1, Florida, USA | foxe2 | Forkhead-box transcription factor family gene | Deletion, T-DNA | - | - | NA | - | - | NA | No | Tomato | Apothecial development | [49] |
| sscle_05g048220 | SS1G_05661 | 1980, Nebraska, USA | nox1 | NADPH oxidase | RNAi | - | + | + | + | NA | NA | No | Tomato (Rutger) | ROS regulation | [50] |
| sscle_06g049430 | SS1G_07404 | 1980, Nebraska, USA | rhs1 | Rearrangement hot spot repeat-containing protein | RNAi | + | + | - | + | + | NA | Yes | Arabidopsis and canola (Zhongshuang 9) | [51] | |
| sscle_06g049780 | SS1G_07360 | 1980, Nebraska, USA | fkh1 | Atypical forkhead (FKH)-box-containing protein | RNAi | + | + | NA | + | NA | NA | No | Tomato | Cellular integrity | [52] |
| sscle_06g049830 | SS1G_07355 | 1980, Nebraska, USA | pac1 | pH-Responsive transcription factor | Deletion | - | + | + | + | NA | NA | No | Arabidopsis and tomato (Bonnie Best) | [53] | |
| sscle_06g049890 | SS1G_07345 | NGA4, Anhui, China | pemg1 | Elicitor-homologous protein | RNAi | + | NA | - | + | + | NA | No | Oilseed rape and soybean | Negative regulator of growth and virulence | [54] |
| sscle_06g051560 | SS1G_07136 | UF1, Florida, USA | ste12 | Downstream transcription factor of MAPK pathway | RNAi | + | + | NA | + | + | NA | No | Bush bean and tomato | [13] | |
| sscle_07g055970 | SS1G_03527 | SUN-F-M, Inner Mongolia, China | hex1 | Woronin body major protein | RNAi | - | + | NA | NA | NA | NA | No | Cellular integrity | [46] | |
| sscle_07g058030 | SS1G_03252 | UF1, Florida, USA | ams2 | Cell-cycle-regulated GATA transcription factor | RNAi | + | - | NA | + | + | NA | No | Common bean | Chromosome segregation, number and distribution of sclerotia | [55] |
| sscle_07g058620 | SS1G_03171 | 1980, Nebraska, USA | pka1 | Protein kinase A | Deletion | - | - | - | NA | NA | NA | No | PKA activity | [56] | |
| sscle_07g059700 | MK992913 | 1980, Nebraska, USA | qdo | Quercetin dioxygenase gene | Deletion | - | NA | NA | + | NA | NA | Yes | Arabidopsis | Flavonol degradation | [57] |
| sscle_08g064670 | SS1G_05163 | 1980, Nebraska, USA | cry1 | Cryptochrome family CRY-DASH ortholog | Deletion | - | - | NA | - | NA | NA | No | Arabidopsis and tomato | Sclerotial mass, response to UV | [58] |
| sscle_08g065550 | SS1G_05284 | NGA4, Anhui, China | nacα | Nascent polypeptide-associated complex α-subunit | RNAi | - | - | - | + | NA | NA | No | Oilseed rape and tobacco | [59] | |
| sscle_08g066770 | SS1G_05445 | 1980, Nebraska, USA | smk3 | Slt2 ortholog | Deletion | + | + | NA | + | + | NA | No | Canola (Westar) | Cuticle penetration, cell wall integrity | [60] |
| sscle_08g067830 | SS1G_05588 | 1980, Nebraska, U.S.A. | mads | MADS-box proteins | RNAi | + | - | NA | + | NA | NA | No | Tomato | [61] | |
| sscle_08g068500 | SS1G_14133 | Ep-1PNA367, Heilongjiang, China | itl | Integrin-like protein | RNAi | + | + | NA | + | NA | + | Yes | Arabidopsis and canola | Hyphal branching, suppression of host defense | [62] |
| sscle_08g068530 | SS1G_14127 | 1980, Nebraska, USA | ggt1 | γ-Glutamyl transpeptidase | Deletion | - | + | NA | - | + | NA | Yes | Tomato | [63] | |
| sscle_09g069850 | SS1G_10796 | 1980, Nebraska, USA | odc2 | Oxalate decarboxylases | Deletion | - | - | + | + | + | NA | Yes | Common bean (Bush Blue Lake 47), soybean (Harosoy), tomato (Bonnie Best) and celery | [64] | |
| sscle_10g075560 | SS1G_08218 | WMA1, Washington, USA | oah | Oxaloacetate acetylhydrolase | T-DNA | - | - | + | + | NA | NA | No | Faba bean (Broad Windsor), pea (Guido), green bean (Great North Tara) and soybean (Skylla) | [65] | |
| sscle_10g075560 | SS1G_08218 | 1980, Nebraska, USA | oah1 | Oxaloacetate acetylhydrolase | Deletion | - | + | + | + | + | NA | No | Tomato (Bonnie Best), common bean (Bush Blue Lake 47), soybean (Harosoy), canola, Arabidopsis and sunflower | [66] | |
| sscle_10g075560 | SS1G_08218 | 1980, Nebraska, U.S.A.; UF1, Florida; WMA, Washington, USA | oah1 | Oxaloacetate acetylhydrolase | Deletion by CRISPR | - | + | + | + | + | NA | No | Soybean, canola, tomato (Better Boy), faba bean (Windsor), and pea (Sugar Daddy) | [40] | |
| sscle_10g077630 | SS1G_08489 | 1980, Nebraska, USA | pph1 | Type 2A Ser/Thr phosphatase catalytic subunit PP2Ac | RNAi | + | + | NA | NA | NA | NA | No | [67] | ||
| sscle_10g079320 | SS1G_14065 | 1980, Nebraska, USA | ssp1 | Development-specific protein | Deletion | - | - | NA | NA | NA | NA | No | Resistance to glycoside-containing antibiotics | [68] | |
| sscle_11g082700 | SS1G_07871 | 1980, Nebraska, USA | rgb1 | Type 2A Ser/Thr phosphatase B subunit | RNAi | - | + | NA | + | + | NA | No | Arabidopsis and tomato (Bonny Best) | MAPK pathway | [67] |
| sscle_11g083230 | SS1G_07798 | SUN-F-M, Inner Mongolia, China | gpd | glyceraldehyde-3-phosphate dehydrogenase | RNAi | NA | + | NA | NA | NA | NA | No | [46] | ||
| sscle_11g083680 | SS1G_07749 | 1980, Nebraska, USA | xyl1 | Endo-β-1,4-xylanase | Deletion | + | + | NA | + | NA | NA | Yes | Canola and Arabidopsis | Apothecia formation | [69] |
| sscle_11g083950 | SS1G_07715 | 1980, Nebraska, USA | sac1 | Adenylate cyclase | Deletion | + | + | - | + | NA | NA | No | Tomato (Bonnie Best) | Apothecia production, cAMP-signaling | [70] |
| sscle_12g087830 | SS1G_11172 | 1980, Nebraska, USA | nox2 | NADPH oxidase | RNAi | - | + | - | - | NA | NA | No | Tomato (Rutger) | ROS regulation | [50] |
| sscle_12g090900 | SS1G_11866 | 1980, Nebraska, USA | smk1 | ERK -type MAP kinase | RNAi | + | + | NA | NA | NA | NA | No | pH-dependent regulation | [71] | |
| sscle_14g099710 | SS1G_08814 | 1980, Nebraska, USA | odc1 | Oxalate decarboxylases | Deletion | - | NA | NA | - | NA | NA | Yes | Common bean (Bush Blue Lake 47), soybean (Harosoy) and tomato (Bonnie Best) | [64] | |
| sscle_16g107670 | SS1G_10096 | 1980, Nebraska, USA | cp1 | Cerato-platanin protein | Deletion | - | - | NA | + | NA | + | Yes | Arabidopsis and tobacco | Interaction with PR1 | [72] |
| sscle_16g107670 | SS1G_10096 | FXGD2, Anhui, China | sm1 | Cerato-platanin protein | RNAi | + | + | - | + | NA | NA | Yes | Tobacco, oilseed rape and soybean | [73] | |
| sscle_16g107930 | SS1G_10135 | 1980, Nebraska, USA | fdh1 | Formaldehyde dehydrogenase | Deletion | - | + | NA | + | + | NA | No | Common bean and tomato | Osmotic oxidative stress resistance | [74] |
| sscle_16g109570 | ANQ80447 | 1980, Nebraska, USA | nsd1 | GATA-type Ivb zinc-finger transcription factor | Deletion | - | - | NA | + | + | NA | No | Tomato (Bonnie Best), celery and tomato | Ascogonia formation, apothecium development | [75] |
The assembly genome for the new version gene code is ASM185786v1 published in 2017 and ASM14694v1 for the old version published in 2011. ‘+’ in the table represents that the corresponding phenotype of mutant is altered compared to wild type strain, while ‘-’ means that the phenotype remains unchanged. NA means not assessed. The secretion signals were found using ‘SignalP-5.0’. Unless specified, all deletions were generated with homologous recombination. The Latin names of host species used for pathogenicity test of mutants are: Apium graveolens (celery), Arabidopsis thaliana (Arabidopsis), Brassica napus (canola, oilseed rape, rapeseed), Brassica rapa subsp. chinensis (pak choi cabbage), Capsicum frutescens (hot pepper), Cucumis sativus (cucumber), Fragaria ananassa (strawberry), Glycine max (soybean), Helianthus annuus (sunflower), Lycopersicon solanum (tomato), Nicotiana benthamiana (tobacco), Phaseolus vulgaris (bush bean, common bean, green bean), Pisum sativum (pea), Vicia faba (faba bean). The ecotype of A. thaliana mentioned in the table is Columbia-0. The specific local cultivars of host species are shown in brackets.
3.1. Regulation of Mycelial Growth and Virulence
During the last two decades, the availability of whole genome sequences and improved transformation as well as knockdown/knockout methods have greatly facilitated the molecular study of S. sclerotiorum. Most of the genes studied so far that affect growth are relatively conserved genes that also affect differentiation, such as sclerotial and apothecial development (Figure 2), which will be discussed in the next sections. In this part, we specifically discuss genes only affecting hyphal growth rate and virulence, but not differentiation, during development, as mycelial growth precedes processes of virulence and differentiation.
It is not surprising that the mutants here with major growth defects often exhibit virulence attenuation, although there are certainly exceptions such as mat1-2-1, which exhibits slower growth, aberrant apothecial morphogenesis, but normal virulence [29] (discussed more later).
The RNAi knockdown of highly conserved transcription factors Ams2 or Mads lead to reduced mycelial growth rate and virulence, which suggested that they are early growth regulators. Ams2 is a GATA-box domain transcription factor. Ams2 is required for proper expression of histone and cell cycle related genes, similar as in Schizosaccharomyces pombe [55]. Mads is orthologous to yeast Mcm1 with a MADS-box domain, which is essential for viability, cell cycle, mating, mini-chromosome maintenance, recombination, and stress tolerance [61]. Although mutants of these early growth factors severely affect growth, they can still form sclerotia, which indicated that the later stages of fungal differentiation pathways, such as sclerotial formation and apothecial development, might be less dependent on these early growth factors.
BAX inhibitor-1 (Bi1) is a highly conserved protein that is found in both eukaryotes and prokaryotes, exhibiting apoptosis-inhibiting activity. RNAi knockdown mutants in S. sclerotiorum showed more aerial hyphal growth and reduced virulence [48]. The mutants also exhibit reduced tolerance to chemical stress, as its gene expression is induced by various stresses. Bi1 is likely a stress specific regulator dedicated to both biotic and abiotic stress responses, due to its minor roles in growth and development.
Unlike the previously described genes, PemG1 serves an opposite function in regards to growth and virulence. The RNAi mutants of PemG1 exhibit faster mycelial growth and enhanced virulence, indicating that it is a negative regulator in these processes. Consistently, more infection cushions, more CWDE activities, and less susceptibility to salt stress were observed [54]. However, the exact mechanism on how this fungal specific protein negatively regulates growth and virulence is unclear.
3.2. Signaling Events Leading to Sclerotial Formation
Sclerotia of S. sclerotiorum initiate with small primordia and rapidly develop into white compact hyphal masses. They mature with dehydration and pigmentation once they cease growth. The melanised outer surface of the sclerotium is mostly responsible for resisting adverse conditions [8]. We will first discuss the genes affecting both growth and sclerotial formation, which are likely very upstream components that are shared by both processes. As expected for broad regulators, these mutants show not only defective growth and sclerotial malformation, but also almost always loss of virulence and other pleiotropic defects (Table 1).
The melanisation of sclerotia is an important process for the long-term survival of Sclerotinia species in nature. The deletion of melanin biosynthesis genes scytalone dehydratase (Scd1) or trihydroxynaphthalene reductase (Thr1) resulted in slower mycelial growth, reduced number, size, and pigmentation of sclerotia [39], which suggested that melanisation is critical for sclerotial maturation and perhaps feedback promotion of growth. However, unlike in other pathogenic fungi, the virulence of these knockout strains is not affected, suggesting that melanin does not contribute to pathogenesis in Sclerotinia as it does for other fungi, like Magnaporthe species.
Knocking out some other conserved genes in S. sclerotiorum can also lead to diverse defects. For example, the peroxisomal carnitine acetyl transferase (CAT) encoding gene in S. sclerotiorum is Pth2. The CAT activity is required for acetyl-CoA transport, which is needed for fatty acid metabolism. The pth2 knockout mutants show aberrant growth, non-sclerotial formation, and reduced oxalic acid (OA) levels and virulence [41]. In another instance, the silencing of integrin-like gene Itl leads to altered colony morphology, smaller and irregular shaped sclerotia, and hypovirulence [62]. Although Cvnh homologs seem to only exist in Sclerotinia and Botrytis, the CVNH domain is a conserved domain. The silencing of this gene lead to reduced growth rate, non-sclerotial formation and lower virulence [45].
Phosphorylation and de-phosphorylation events are critical for almost all signal transductions. Therefore, it is not surprising that a number of kinases and phosphatases are involved in both growth and sclerotial formation. Mutation of two-component histidine kinase Shk1 leads to slower and altered hyphal growth and failed sclerotial formation [35]. Although the mutant is also sensitive to osmotic stress and shows heightened resistance to fungicides, it exhibits normal virulence. Shk1 likely works upstream of the MAPK cascade to control these processes. Smk1 and Smk3 are two MAPK kinases that have been studied in S. sclerotiorum so far. Mutants of these two kinases show similar growth defects and non-sclerotial formation [60,71], suggesting that they may function downstream of Shk1.
cAMP could act upstream of the MAPKs, as exogenous application of cAMP inhibits Smk1 expression and MAPK phosphorylation [71]. Type 2A phosphoprotein phosphatase Pph1 possibly functions downstream of the MAPK. The mutants of pph1 show arrested growth [67]. Consistently, PP2A R2 B regulatory subunit rgb1 mutants show aberrant sclerotial formation and failed penetration during infection [67].
Reactive oxygen species (ROS) play diverse roles in development and virulence in fungi. This is supported by the mutant analysis of type I catalase Scat1. The deletion of Scat1 lead to slower radial growth, higher number of small sclerotia that are not properly melanised, and hypovirulence [44]. Calcium is another common factor that is involved in signaling. The study of Cna1 and Caf1 supports its contribution in S. sclerotiorum biology. Calcineurin Ser/Thr protein phosphatase Cna1 is conserved in eukaryotes, whose activity is dependent on calcium and calmodulin. Antisense cna1 mutants show altered growth, sclerotial development, and hypovirulence [32]. Caf1 is a secreted protein with a putative Ca2+-binding EF-hand motif, which seems to be conserved in fungi. caf1 T-DNA mutants exhibit slightly slower growth, deformed sclerotia, and hypovirulence [43].
Conserved transcription factors are another large group of genes that are responsible for signaling. Sfh1 is a GATA transcription factor, but with an SNF5 domain. It is a member of the housekeeping RSC (Remodels Structure of Chromatin) complex, which is an ATP-dependent chromatin remodeler that is essential for cell cycle [34]. Its diverse functions explain its involvement in growth, sclerotial development, and virulence. Transcription factor Ste12 likely acts downstream of the MAP kinase cascades. In ste12 RNAi lines, slow growth, smaller sclerotia, and fewer appressoria, leading to hypovirulence, were observed [13]. Forkhead (FKH) box (FOX) family transcription factors are important regulators of primary metabolism, cell cycle, and morphogenesis in animals and fungi. fkh1 RNAi mutants show slower growth, non-sclerotial formation, hypovirulence, and defective stress responses [52], suggesting that it is a transcriptional regulator that is shared by multiple pathways.
CWDEs have been speculated to play key roles in fungal pathogenesis. In support of this, the deletion of Xyl1 that encoded an endo-β-1, 4-xylanase leads to a loss of pathogenicity. Interesting, growth retardation, less sclerotial formation, and dense hyphal branching were also observed in the mutant [69], suggestive of a role of xylanase in development as well.
Opsins are conserved light-sensitive proteins involved in circadian rhythms. sop1 RNAi lines show reduced growth, non-sclerotial formation, hypovirulence, and heightened sensitivities to osmotic and fungicide stresses [33]. These observations suggest that light may have diverse roles in various processes in Sclerotinia through opsins.
Rearrangement hotspot (Rhs) repeat-containing proteins are widely distributed in bacteria and eukaryotes. Rhs1 is a secreted protein and its orthologs seem to only be present in Sclerotinia and Botrytis species. Rhs1 RNAi mutants show slightly slower colony growth, fewer and larger sclerotial formation, and hypovirulence [51]. These data suggest that Rhs1is likely a key virulence factor for Sclerotinia and it also slightly contributes to growth and sclerotial development. Cerato-platanins are fungal secreted elicitors that can trigger plant immunity and cell death. Sm1 can cause host hypersensitive response. Interestingly, besides virulence, RNAi mutants of Sm1 also exhibit slower growth, impaired sclerotial development, and heightened sensitivities to different stresses [73], suggesting a broader role of the elicitor. However, these defects were not detected in a separate study in deletion mutant cp1 mutating the same gene [72]. Future careful transgenic complementation experiments are needed to solve these discrepancies.
The existence of mutants that have normal mycelial growth, but do not form sclerotia, suggest that later stages of sclerotial development are separated from growth. Although most of these mutants still show attenuated virulence, some exhibit normal pathogenicity, such as sl2 and ggt1 (ɤ-glutamyl transpeptidase). RNAi knockdown of either Sl2, GAPDH (Gpd), or Hex1 lead to altered sclerotial formation or melanisation, similar to its interacting proteins glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Hex [46]. However, the virulence of sl2 is not affected, suggesting that sclerotial formation and virulence can be separately controlled by distinct downstream factors. As a fungal specific secreted protein localizing to fungal cell wall, how Sl2 affects sclerotial development is unclear. Ggt is a highly conserved enzyme that contributes to redox homeostasis. The deletion mutants of ggt1 show much higher GSH+GSSG and hydrogen peroxide accumulation, and separation of the cortex layer of mature sclerotia from the medulla [63]. In addition, the mutants are defective in compound appressoria production, only affecting the penetration stage of infection. These mutant phenotypes suggest that GSH, GSSG, or GSH/GSSG ratio contribute to the regulation of sclerotia maturation and appressoria development.
The rest of the genes studied so far that affect sclerotial formation, but not growth, all show attenuated virulence. They likely act upstream of Sl2 and Ggt. ROS increase was observed in sclerotial initials and infection cushions in S. sclerotiorum, likely due to the combined activities of NADPH oxidases Nox1 and Nox2. Similar to exogenous application of NADPH inhibitor or ROS scavenger, the RNAi mutants of either Nox1 or Nox2 exhibit failed sclerotial development [50], indicating that ROS is required for sclerotial initiation. Interestingly, reduced OA production and virulence was only observed in nox1, but not nox2, which suggests that these two genes are not fully redundant, likely due to their different expression patterns. Consistently, when the thioredoxin reductase (Trr1) gene that is partly responsible for ROS detoxification is silenced, less and larger sclerotia are formed [47]. However, virulence is attenuated in Trr1 silenced strains, suggesting that an optimum ROS is needed for virulence.
Low pH was known to promote sclerotial proliferation. The pH-sensitive transcription factor Pac1 is responsible for ambient pH-induced gene expression. Consistently, the deletion of Pac1 caused aberrant sclerotial formation and reduced OA levels, thus leading to hypovirulence [53]. As higher levels of OA reduce pH and may affect ROS homeostasis, proper balance among ROS, OA, and pH is likely paramount for the biology of this fungal pathogen.
3.3. Control of Apothecial and Ascospore Development
The apothecium consisting of a stalk (stipe) and a saucer-shaped disc is characteristic of the Sclerotinia genus (Figure 1). Asci develop on the upper surface of apothecia and when they mature, the ascospores are dispersed [8]. As mating-type (Mat) genes have generally been established as key regulators of sexual development of ascomycetes, it is not surprising that they affect apothecial development in the homothallic S. sclerotiorum. Similar to other fungi, the S. sclerotiorum Mat locus contains a cluster of four Mat genes encoding transcription factors, with MAT1-1s encoding an alpha-domain and Mat1-2s encoding high mobility group (HMG) proteins. By deletion knockout analysis [29], each of the four Mat genes were analyzed in regards to their contributions to regular mycelial growth, virulence, sclerotial formation, and apothecial development. While these Mats contribute little to vegetative growth, development, and pathogenesis, Mat1-1-5, Mat1-1-1, and Mat1-2-1 are essential for early, and Mat1-2-4 is critical for late sexual stages, including apothecial initiation and ascospore development. As transcription factors, it is also not surprising that these Mat gene mutants exhibit altered expression of the Mat locus and putative pheromone and pheromone receptor genes, explaining their potential regulatory mechanism and specific phenotypes.
Similar to the Mats, another gene that specifically contributes to apothecial development is FoxE2 [49]. FoxE2 is a member of the forkhead box (FOX) transcription factor (TF) family. FOX proteins play diverse roles in morphogenesis, development, pathogenicity, and stress responses in animals and fungi. Knockout analysis of FoxE2 in S. sclerotiorum showed that it does not affect mycelial growth, sclerotial formation, or virulence. The striking mutant phenotypic similarity between the Mats and FoxE2 brings the question on the relationship among these transcription factors. It could be possible that a transcriptional network that includes these transcription factors needs to be orchestrated for proper sexual stage development in S. sclerotiorum.
As a UV-A photoreceptor, Cry1 was suspected to be involved in apothecial development, as its expression was induced by UV during this specific developmental phase. However, no obvious defects in apothecial development were detected in the cry1 deletion mutant [58], suggesting that it is not a major regulator, or there exists redundancy masking its effects on photomorphogenesis.
All other mutants in the literature with defects in apothecial development show pleiotropic defects, including sac1 (Adenylate cyclase) [70] and nsd1 (GATA-type IVb zinc-finger transcription factor) [75], both encoding conserved proteins. cAMP is a well-known molecule with diverse roles for many biological processes in plant fungal pathogens. Although protein kinase A catalytic subunit gene (Pka1) was hypothesized to mediate cAMP signaling, the pka1 single mutant exhibits non-detectable defects, likely due to genetic redundancy of Pka2 [56]. In comparison, with dramatic cAMP level reduction, the sac1 mutant displays slower growth, abolished pathogenicity, and failed apothecial development [70]. In other fungi, Nsd1 serves as transcription factor for development and environmental responses. In nsd1 knockout mutants, altered hyphal growth, smaller sclerotia, failed appressorium formation leading to failed pathogenicity, and failed apothecial initiation were observed [75]. These indicate that Nsd1 is likely a general transcription factor that regulates many processes in S. sclerotiorum, or a specific regulator that regulates a general downstream factor that is shared by many pathways. Recently, the NO homeostasis regulating glutathione-dependent formaldehyde dehydrogenases (Fdh1) encoding gene was revealed to be one of the targets of Nsd1 [74]. The deletion mutants of fdh1 exhibit slower mycelial growth, smaller sclerotia, and defective compound appressoria, leading to failed penetration into host tissue, being suggestive of the keys roles NO plays in these processes. The high similarity between the fdh1 and nsd1 mutant phenotypes indicates that Fdh1 could be the main target of Nsd1 during growth and development.
3.4. Regulation of Fungal Pathogenesis of the Fungus
Following a short biotrophic phase after infection, S. sclerotiorum causes tissue maceration and necrosis leading to rapid cell death and host cell wall degradation during fungus colonization. Toxins and CWDEs are believed to play critical roles in promoting these processes. The study of axp mutants deleting a secreted arabinofuranosidase/β-xylosidase precursor reflects the importance of CWDEs in the virulence of S. sclerotiorum. Attenuated virulence was observed in canola. Similarly, the critical contribution of ROS in virulence is corroborated in sod1 mutants affecting Cu/Zn superoxide dismutase [36,37]. OA is another key factor that has long been hypothesized to contribute to the pathogenicity of this pathogen. Readers are referred to two excellent reviews that have summarized the historic and comprehensive studies of the pathogenicity and virulence of S. sclerotiorum [65,66].
During infection, S. sclerotiorum produces large amounts of OA. Earlier physiological and pharmacological studies have highlighted its importance. In addition to virulence, UV mutants where the exact mutations were not identified could not produce OA and failed to form sclerotia [76], also suggesting a link between OA and development. It was not until the availability of genetically defined OA deficient mutants that the contribution of OA towards Sclerotinia biology was partly settled. In odc2 RNAi strain where the gene encoding oxalate decarboxylase in charge of catabolism of OA is silenced, less and non-functional appressoria were formed, leading to hypovirulence [64]. These data support the involvement of developmentally regulated OA accumulation in penetration-dependent infection. In recently generated oah (oxaloacetate acetylhydrolase) mutants through the deletion or CRISPR, where OA biosynthesis is blocked, failed appresorium formation, hypovirulence, and delayed sclerotial formation was observed [40]. However, the deletion or T-DNA mutants of oah in a separate study show pH-dependent virulence alteration and normal growth and development. These discrepancies can likely be resolved through more careful examination of other mutants that are deficient in OA biosynthesis.
Many pathogen effectors are secreted into the host to dampen host immunity and/or promote virulence. In recent years, effector biology has been focusing on the study of secreted proteins, as these effector candidates can be relatively easily identified through secretome and RNA-seq analysis. Many secreted proteins in S. sclerotiorum have diverse roles in both development and virulence (Table 1 and discussions in the previous sections). Examples of effectors only affecting virulence in S. sclerotiorum include Ssvp1, V263 and Qdo (quercetin dioxygenase). Less conserved Ssvp1 [30] and V263 [38] are both small secreted proteins that contribute to virulence. On the other hand, more conserved Qdo catalyzes the cleavage of the flavonol carbon skeleton [57], revealing the critical contributions of degrading host anti-microbial flavonol in fungal virulence.
Besides secreted proteins, there are also non-secreted regulators that are solely dedicated to pathogen penetration and virulence regulation. The examples in S. sclerotiorum include Svf1 (Survival factor 1) [31], Pks13 (polyketide synthase 13) [40], and Nacα (nascent-polypeptide-associated complex alpha subunit) [59]. In yeast, Svf1 promotes survival under oxidative stress conditions. Therefore, it likely assists in coping with ROS during infection in Sclerotinia. Pks13 is likely involved in melanin biosynthesis in appressoria, as its CRISPR mutant exhibits albino compound appressoria without affecting virulence. Nacα is a highly conserved protein with transcriptional co-activator activity. Interesting, Nacα negatively impacts the expression of polygalacturonase-encoding genes, which likely contributes to the enhanced virulence phenotypes of the mutant.
4. Summary and Future Perspectives
Over the decade, many genes that have been involved in pathogen development and pathogenesis have been characterized in S. sclerotiorum (Table 1). Much has been unveiled for the pathogen growth, development, and differentiation signaling pathways. Accordingly, more evidence is supporting the two-phase infection model, where the pathogen suppresses host basal defense prior to killing and degrading host cells [11]. In the short biotrophic phase, the pathogen uses well-orchestrated strategies to overcome host immunity. It might spatially achieve this via the production of OA, ROS, CWDEs, and effectors in compound appressoria or primary invasive hyphae.
Among almost 15,000 genes encoded in its genome, less than 60 have been studied so far using careful mutant analysis. This explains the under-studied nature of this notorious pathogen and the lack of connections in the signaling pathways of its different special biological processes. Besides the traditional one-gene-at-a-time reverse genetics analysis, more robust methods, such as genome-wide CRISPR, should be developed to enhance the study of this economically important fungus.
Due to the short biotrophic period, as well as the switching to necrotrophic phase, identifying and assigning the temporal, spatial, and tissue-specific functions to virulence factors are real challenges. Future development of cell biology and biochemical tools tailored for S. sclerotiorum will offer opportunities for solving many mysteries of the fungus. Considering this specific form of pathogen-host interaction system, challenges also exist ahead in applying quantitative disease resistance breeding and phenotypic screens for stage-specific defenses. More collaboration among the molecular pathologists and breeders will be critical to solve the host resistance difficulties of the pathogen.
Acknowledgments
The authors would like to apologize for works that are not cited due to space limitations. We greatly appreciate the constructive suggestions from the three anonymous reviewers, which helped us improve the quality of this review. We would like to thank National Natural Science Foundation of China (Grants 31971836) and Scientific Research Fund of Hunan Provincial Education Department (Grants 17A095) for funds to S.X., and Canadian foundation of Innovation (CFI), NSERC-Discovery and NSERC-CREATE-PRoTECT programs for financial supports to X.L. Y.X. is partly supported by a scholarship from China Scholarship Council (CSC).
Author Contributions
S.X., Y.X. and X.L. wrote the review. Y.X. made Table 1 and Figure 3. R.H. made Figure 1, and J.Z. made Figure 2. Y.X. and L.Q. collected references. All authors have read and agreed to the published version of the manuscript.
Funding
National Natural Science Foundation of China (Grants 31971836) and Scientific Research Fund of Hunan Provincial Education Department (Grants 17A095) to SX; Canadian foundation of Innovation (CFI), NSERC-Discovery and NSERC-CREATE-PRoTECT funds to XL. YX is partly supported by a scholarship from China Scholarship Council (CSC).
Conflicts of Interest
The authors declare no conflict of interests.
References
- 1.Libert M.A. Plante Crytogamicae Arduennae (Exsiccati) 1837. No. 326. Published by the Author. [Google Scholar]
- 2.Whetzel H.H. A synopsis of the genera and species of the Sclerotiniaceae, a family of stromatic inoperculate discomycetes. Mycologia. 1945;37:648–714. doi: 10.1080/00275514.1945.12024025. [DOI] [Google Scholar]
- 3.Buchwald N.F., Neergaard P.J. A plea for the retention of Sclerotinia sclerotiorum as type species for the genus Sclerotinia Fckl. emend. Friesia. 1972;10:96–99. [Google Scholar]
- 4.Buchwald N.F., Neergaard P.J. Proposal to conserve Sclerotinia Fckl, with S. Sclerotiorum (Lib.) de bary as type species. Taxon. 1976;25:199–200. doi: 10.2307/1220457. [DOI] [Google Scholar]
- 5.Kohn L.M. Delimitation of the economically important plant pathogenic Sclerotinia species. Phytopathology. 1979;69:881–886. doi: 10.1094/Phyto-69-881. [DOI] [Google Scholar]
- 6.Kabbage M., Yarden O., Dickman M.B. Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic Lifestyle. Plant Sci. 2015;233:53–60. doi: 10.1016/j.plantsci.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z., Bao L.L., Zhao F.Y., Tang M.Q., Chen T., Li Y., Wang B.X., Fu B., Fang H., Li G.Y., et al. BnaMPK3 is a key regulator of defense responses to the devastating plant pathogen Sclerotinia sclerotiorum in oilseed rape. Front. Plant Sci. 2019;10:1–18. doi: 10.3389/fpls.2019.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willetts H., Wong J.J. The biology of Sclerotinia sclerotiorum, S. Trifoliorum, and S. Minor with emphasis on specific nomenclature. Bot. Rev. 1980;46:101–165. doi: 10.1007/BF02860868. [DOI] [Google Scholar]
- 9.Harper G.E., Frampton C.M., Stewart A. Factors influencing survival of sclerotia of Sclerotium cepivorum in New Zealand soils. N. Z. J. Crop Hortic. Sci. 2002;30:29–35. doi: 10.1080/01140671.2002.9514196. [DOI] [Google Scholar]
- 10.Boland G.J., Hall R. Index of plant hosts of Sclerotinia sclerotiorum. Plant Sci. 1994;16:93–108. doi: 10.1080/07060669409500766. [DOI] [Google Scholar]
- 11.Liang X., Rollins J.A. Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology. 2018;108:1128–1140. doi: 10.1094/PHYTO-06-18-0197-RVW. [DOI] [PubMed] [Google Scholar]
- 12.Bolton M.D., Thomma B.P.H.J., Nelson B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006;7:1–16. doi: 10.1111/j.1364-3703.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 13.Xu T., Li J., Yu B., Liu L., Zhang X., Liu J., Montes H.M. Transcription factor SsSte12 was involved in mycelium growth and development in Sclerotinia sclerotiorum. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.02476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mei J., Qian L., Disi J.O., Yang X., Li Q., Li J., Frauen M., Cai D., Qian W. Identification of resistant sources against Sclerotinia sclerotiorum in brassica species with emphasis on B. Oleracea. Euphytica. 2011;177:393–399. doi: 10.1007/s10681-010-0274-0. [DOI] [Google Scholar]
- 15.Li G.Q., Huang H.C., Miao H.J., Erickson R.S., Jiang D.H., Xiao Y.N. Biological control of Sclerotinia diseases of rapeseed by aerial applications of the mycoparasite Coniothyrium minitans. Eur. J. Plant Pathol. 2006;114:345–355. doi: 10.1007/s10658-005-2232-6. [DOI] [Google Scholar]
- 16.Yang S.J. An investigation on the host range and some ecological aspects on the Sclerotinia disease of the Rape plant. Acta Phytopathol. Sin. 1959;5:111–122. [Google Scholar]
- 17.Moore W.J. Flooding as a means of destroying the sclerotia of Sclerotinia sclerotiorum. Phytopathology. 1949;39:920–927. [Google Scholar]
- 18.Schwartz H., Steadman J.J. Factors affecting sclerotium populations of, and apothecium production by, Sclerotinia sclerotiorum. Phytopathology. 1978;68:383–388. doi: 10.1094/Phyto-68-383. [DOI] [Google Scholar]
- 19.Stoner W.N., Moore W.D. Lowland rice farming, a possible cultural control for Sclerotinia sclerotiorum in the Everglades. Plant Dis. Rep. 1953;37:181–186. [Google Scholar]
- 20.Coyne D., Steadman J., Anderson F.J. Effect of modified plant architecture of great northern dry bean varieties (Phaseolus vulgaris) on white mold severity, and components of yield. Plant Dis. Rep. 1974;58:379–382. [Google Scholar]
- 21.Amselem J., Cuomo C.A., Van Kan J.A., Viaud M., Benito E.P., Couloux A., Coutinho P.M., De Vries R.P., Dyer P.S., Fillinger S., et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charles M., Id D., Denton-giles M., Id J.K.H., Chang S. A whole genome scan of SNP data suggests a lack of abundant hard selective sweeps in the genome of the broad host range plant pathogenic fungus Sclerotinia sclerotiorum. PLoS ONE. 2019;14:e0214201. doi: 10.1371/journal.pone.0214201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derbyshire M., Denton-giles M., Hegedus D., Seifbarghi S., Rollins J., Van J., Seidl M.F., Faino L., Mbengue M., Navaud O., et al. The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 2017;9:593–618. doi: 10.1093/gbe/evx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermisson J., Pennings P.S. Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messer P.W., Petrov D.A. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 2013;28:659–669. doi: 10.1016/j.tree.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del Ponte E.M., Eduardo S., Lehner M.S., Ju T.J.D.P., Mizubuti G., Pethybridge S.J. Independently founded populations of Sclerotinia sclerotiorum from a tropical and a temperate region have similar genetic structure. PLoS ONE. 2017;12:e0173915. doi: 10.1371/journal.pone.0173915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seifbarghi S., Borhan M.H., Wei Y., Coutu C., Robinson S.J., Hegedus D.D. Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genom. 2017;18:266. doi: 10.1186/s12864-017-3642-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heard S., Brown N.A., Hammond-kosack K. An interspecies comparative analysis of the predicted secretomes of the necrotrophic plant pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS ONE. 2015;10:e0130534. doi: 10.1371/journal.pone.0130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doughan B., Rollins J.A., Avery S.V. Characterization of MAT gene functions in the life cycle of Sclerotinia sclerotiorum reveals a lineage-specific MAT gene functioning in apothecium morphogenesis. Fungal Biol. 2016;120:1105–1117. doi: 10.1016/j.funbio.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Lyu X., Shen C., Fu Y., Xie J., Jiang D., Li G., Cheng J. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 2016;12:e1005435. doi: 10.1371/journal.ppat.1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y., Du J., Wang Y., Zhang M., Huang Z., Cai J., Fang A., Yang Y., Qing L., Bi C., et al. Survival factor 1 contributes to the oxidative stress response and is required for full virulence of Sclerotinia sclerotiorum. Mol. Plant Pathol. 2019;20:895–906. doi: 10.1111/mpp.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harel A., Bercovich S., Yarden O. Calcineurin is required for sclerotial development and pathogenicity of Sclerotinia sclerotiorum in an oxalic acid–independent manner. Mol. Plant Microbe Interact. 2006;19:682–693. doi: 10.1094/MPMI-19-0682. [DOI] [PubMed] [Google Scholar]
- 33.Lyu X., Shen C., Fu Y., Xie J., Jiang D., Li G. The microbial opsin homolog sop1 is involved in Sclerotinia sclerotiorum development and environmental stress response. Front. Microbiol. 2016;6:1–14. doi: 10.3389/fmicb.2015.01504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L., Wang Q., Sun Y., Zhang Y., Zhang X., Liu J. Sssfh1, a gene encoding a putative component of the RSC chromatin remodeling complex, is involved in hyphal growth, reactive oxygen species accumulation, and pathogenicity in Sclerotinia sclerotiorum. Front. Microbiol. 2018;9:1–14. doi: 10.3389/fmicb.2018.01828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan Y., Ge C., Liu S., Wang J., Zhou M. A two-component histidine kinase shk1 controls stress response, sclerotial formation and fungicide resistance in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2013;14:708–718. doi: 10.1111/mpp.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L., Chen W. Random T-DNA mutagenesis identifies a Cu/Zn superoxide dismutase gene as a virulence factor of Sclerotinia sclerotiorum. Mol. Plant Microbe Interact. 2013;26:431–441. doi: 10.1094/MPMI-07-12-0177-R. [DOI] [PubMed] [Google Scholar]
- 37.Veluchamy S., Williams B., Kim K., Dickman M.B. Physiological and molecular plant pathology the CuZn superoxide dismutase from Sclerotinia sclerotiorum is involved with oxidative stress tolerance, virulence, and oxalate production. Physiol. Mol. Plant Pathol. 2012;78:14–23. doi: 10.1016/j.pmpp.2011.12.005. [DOI] [Google Scholar]
- 38.Liang Y., Yajima W., Davis M.R., Kav N.N.V., Stephen E. Disruption of a gene encoding a hypothetical secreted protein from Sclerotinia sclerotiorum reduces its virulence on canola (Brassica Napus) disruption of a gene encoding a hypothetical secreted protein from Sclerotinia sclerotiorum reduces its virulence on Canola (Brassica napus) Can. J. Plant Pathol. 2013;35:46–55. [Google Scholar]
- 39.Liang Y.U.E., Xiong W.E.I., Steinkellner S., Feng J.I.E. Deficiency of the melanin biosynthesis genes SCD1 and THR1 affects sclerotial development and vegetative growth, but not pathogenicity, in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2017;19:1–10. doi: 10.1111/mpp.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J.T., Zhang Y.H., Zhang Y.C., Yu P.L., Pan H.Y., Rollins J.A. Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum. MBio. 2018;9:e00567-18. doi: 10.1128/mBio.00567-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberti D., Rollins J.A., Dobinson K.F. Peroxysomal carnitine acetyl transferase influences host colonization capacity in Sclerotinia sclerotiorum. Mol. Plant Microbe Interact. 2013;26:768–780. doi: 10.1094/MPMI-03-13-0075-R. [DOI] [PubMed] [Google Scholar]
- 42.Yajima W., Liang Y., Kav N.N.V., Sclerotinia A., Bary D. Gene disruption of an arabinofuranosidase/β-xylosidase precursor decreases Sclerotinia sclerotiorum virulence on canola tissue. Mol. Plant Microbe Interact. 2009;22:783–789. doi: 10.1094/MPMI-22-7-0783. [DOI] [PubMed] [Google Scholar]
- 43.Xiao X., Xie J., Cheng J., Li G., Yi X., Jiang D., Fu Y. Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant Microbe Interact. 2014;27:40–55. doi: 10.1094/MPMI-05-13-0145-R. [DOI] [PubMed] [Google Scholar]
- 44.Yarden O., Veluchamy S., Dickman M.B., Kabbage M. Physiological and molecular plant pathology Sclerotinia sclerotiorum catalase SCAT1 affects oxidative stress tolerance, regulates ergosterol levels and controls pathogenic development. Physiol. Mol. Plant Pathol. 2014;85:34–41. doi: 10.1016/j.pmpp.2013.12.001. [DOI] [Google Scholar]
- 45.Lyu X., Shen C., Fu Y., Xie J., Jiang D., Li G., Cheng J. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci. Rep. 2015;5:15565. doi: 10.1038/srep15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y., Jiang D., Xie J., Cheng J., Li G., Yi X., Fu Y. Ss-Sl2, a novel cell wall protein with PAN modules, is essential for sclerotial development and cellular integrity of Sclerotinia sclerotiorum. PLoS ONE. 2012;7:e34962. doi: 10.1371/journal.pone.0034962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J., Wang Y., Du J., Huang Z., Fang A., Yang Y. Sclerotinia sclerotiorum thioredoxin reductase is required for oxidative stress tolerance, virulence, and sclerotial development. Front. Microbiol. 2019;10:1–9. doi: 10.3389/fmicb.2019.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y., Xiao J., Yang Y., Bi C., Qing L., Tan W. Physiological and molecular plant pathology Ss-Bi1 encodes a putative BAX Inhibitor-1 protein that is required for full virulence of Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 2015;90:115–122. doi: 10.1016/j.pmpp.2015.04.005. [DOI] [Google Scholar]
- 49.Wang L., Liu Y., Liu J., Zhang Y., Zhang X., Pan H. The Sclerotinia sclerotiorum FoxE2 gene is required for apothecial development. Mycology. 2016;106:484–490. doi: 10.1094/PHYTO-08-15-0181-R. [DOI] [PubMed] [Google Scholar]
- 50.Kim H., Chen C., Kabbage M., Dickman M.B. Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases. Appl. Environ. Microbiol. 2011;77:7721–7729. doi: 10.1128/AEM.05472-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Y., Xiao J., Zhu W., Yang Y., Mei J., Bi C., Qian W.E.I., Qing L., Tan W. Ss-Rhs1, a secretory Rhs repeat-containing protein, is required for the virulence of Sclerotinia sclerotiorum. Mol. Plant Pathol. 2017;18:1052–1061. doi: 10.1111/mpp.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan H., Yu G., Liu Y., Zhang X., Liu J., Zhang Y., Rollins J.A., Sun F., Pan H. An atypical forkhead-containing transcription factor SsFKH1 is involved in sclerotial formation and is essential for pathogenicity in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2017;18:963–975. doi: 10.1111/mpp.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rollins J.A., Pathology P., Hall F. The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant Microbe Interact. 2003;16:785–795. doi: 10.1094/MPMI.2003.16.9.785. [DOI] [PubMed] [Google Scholar]
- 54.Pan Y., Xu Y., Li X., Yao C., Gao Z. Physiological and molecular plant pathology SsPemG1 encodes an elicitor-homologous protein and regulates pathogenicity in Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 2015;92:70–78. doi: 10.1016/j.pmpp.2015.08.010. [DOI] [Google Scholar]
- 55.Liu L., Wang Q., Zhang X., Liu J., Zhang Y. Ssams2, a gene encoding GATA transcription factor, is required for appressoria formation and chromosome segregation in Sclerotinia sclerotiorum. Front. Microbiol. 2018;9:1–12. doi: 10.3389/fmicb.2018.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jurick W.M., Dickman M.B., Rollins J.A. Characterization and functional analysis of a CAMP-dependent protein kinase a catalytic subunit gene (Pka1) in Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 2004;64:155–163. doi: 10.1016/j.pmpp.2004.07.004. [DOI] [Google Scholar]
- 57.Chen J., Ullah C., Reichelt M., Gershenzon J., Hammerbacher A. Sclerotinia sclerotiorum circumvents flavonoid defenses by catabolizing flavonol glycosides and aglycones. Plant Physiol. 2019;180:1975–1987. doi: 10.1104/pp.19.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veluchamy S., Rollins J.A. A CRY-DASH-type photolyase/cryptochrome from Sclerotinia sclerotiorum mediates minor UV-A-specific effects on development. Fungal Genet. Biol. 2008;45:1265–1276. doi: 10.1016/j.fgb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Li X.L., Guo M., Xu D.F., Chen F.X., Zhang H.J., Pan Y., Li M.M., Gao Z. The nascent-polypeptide-associated complex alpha subunit regulates the polygalacturonases expression negatively and influences the pathogenicity of Sclerotinia sclerotiorum. Mycologia. 2015;107:1130–1137. doi: 10.3852/14-250. [DOI] [PubMed] [Google Scholar]
- 60.Bashi Z.D., Gyawali S., Bekkaoui D., Coutu C., Lee L., Poon J., Rimmer S.R., Khachatourians G.G., Hegedus D.D. The Sclerotinia sclerotiorum Slt2 mitogen-activated protein kinase ortholog, SMK3, is required for infection initiation but not lesion expansion. Can. J. Microbiol. 2016;850:836–850. doi: 10.1139/cjm-2016-0091. [DOI] [PubMed] [Google Scholar]
- 61.Qu X., Yu B., Liu J., Zhang X., Li G., Zhang D. MADS-Box transcription factor SsMADS is involved in regulating growth and virulence in Sclerotinia sclerotiorum. Int. J. Mol. Sci. 2014;15:8049–8062. doi: 10.3390/ijms15058049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu W., Wei W., Fu Y., Cheng J., Xie J., Li G., Yi X. A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE. 2013;8:e53901. doi: 10.1371/journal.pone.0053901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li M., Liang X., Rollins J.A. Sclerotinia sclerotiorum-glutamyl transpeptidase (Ss-Ggt1) is required for regulating glutathione accumulation and development of sclerotia and compound appressoria. Mol. Plant Microbe Interact. 2012;25:412–420. doi: 10.1094/MPMI-06-11-0159. [DOI] [PubMed] [Google Scholar]
- 64.Liang X., Moomaw E.W., Rollins J.A. Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 2015;16:825–836. doi: 10.1111/mpp.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu L., Xiang M., White D., Chen W. pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 2015;17:2896–2909. doi: 10.1111/1462-2920.12818. [DOI] [PubMed] [Google Scholar]
- 66.Liang X., Liberti D., Li M., Kim Y., Hutchens A., Wilson R.O.N., Rollins J.A. Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 2015;16:559–571. doi: 10.1111/mpp.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Erental A., Harel A., Yarden O. Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum. Mol. Plant Microbe Interact. 2007;20:944–954. doi: 10.1094/MPMI-20-8-0944. [DOI] [PubMed] [Google Scholar]
- 68.Li M., Rollins J.A. The development-specific Ssp1 and Ssp2 genes of Sclerotinia sclerotiorum encode lectins with distinct yet compensatory regulation. Fungal Genet. Biol. 2010;47:531–538. doi: 10.1016/j.fgb.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 69.Yu Y., Xiao J., Du J., Yang Y., Bi C., Qing L. Disruption of the gene encoding endo-β-1, 4-xylanase affects the growth and virulence of Sclerotinia sclerotiorum. Front. Microbiol. 2016;7:1–9. doi: 10.3389/fmicb.2016.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ii W.M.J., Rollins J.V.A. Deletion of the adenylate cyclase (Sac1) geneaffects multiple developmental pathways and pathogenicity in Sclerotinia sclerotiorum. Fungal Genet. Biol. 2007;44:521–530. doi: 10.1016/j.fgb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 71.Chen C., Harel A., Gorovoits R., Yarden O., Dickman M.B. MAPK regulation of sclerotial development in Sclerotinia sclerotiorum is linked with pH and CAMP sensing. Mol. Plant Microbe Interact. 2004;17:404–413. doi: 10.1094/MPMI.2004.17.4.404. [DOI] [PubMed] [Google Scholar]
- 72.Yang G., Tang L., Gong Y., Xie J., Fu Y., Jiang D., Li G., Collinge D.B., Chen W., Cheng J. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018;217:739–755. doi: 10.1111/nph.14842. [DOI] [PubMed] [Google Scholar]
- 73.Pan Y., Wei J., Yao C., Reng H., Gao Z. Plant science SsSm1, a cerato-platanin family protein, is involved in the hyphal development and pathogenic process of Sclerotinia sclerotiorum. Plant Sci. 2018;270:37–46. doi: 10.1016/j.plantsci.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 74.Zhu G.L., Yu G., Zhang Y.H., Rollins J.A., Li J., Pan H. The formaldehyde dehydrogenase SsFdh1 is regulated by and functionally cooperates with the GATA transcription. MSystems. 2019;4:1–19. doi: 10.1128/mSystems.00397-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan H., Rollins J.A. The GATA-Type IVb zinc-finger transcription factor SsNsd1 regulates asexual—Sexual development and appressoria formation in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2017;9:1679–1689. doi: 10.1111/mpp.12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Godoy G., Steadman J.R., Dickman M.B., Dam R. Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on phaseolus vulgaris. Physiol. Mol. Plant Pathol. 1990;37:179–191. doi: 10.1016/0885-5765(90)90010-U. [DOI] [Google Scholar]