Abstract
Long noncoding RNAs (lncRNA) expression profiles change in the ischemic brain after stroke, but their roles in specific cell types after stroke have not been studied. We tested the hypothesis that lncRNA modulates brain injury by altering macrophage functions. Using RNA deep sequencing, we identified 73 lncRNAs that were differentially expressed in monocyte-derived macrophages (MoDMs) and microglia-derived macrophages (MiDMs) isolated in the ischemic brain three days after stroke. Among these, the lncRNA, GM15628, is highly expressed in pro-inflammatory MoDMs but not in MiDMs, and are functionally related to its neighbor gene, lymphocyte cytosolic protein 1 (LCP1), which plays a role in maintaining cell shape and cell migration. We termed this lncRNA as Macrophage contained LCP1 related pro-inflammatory lncRNA, Maclpil. Using cultured macrophages polarized by LPS, M(LPS), we found that downregulation of Maclpil in M(LPS) decreased pro-inflammatory gene expression while promoting anti-inflammatory gene expression. Maclpil inhibition also reduced the migration and phagocytosis ability of MoDMs by inhibiting LCP1. Furthermore, adoptive transfer of Maclpil silenced M(LPS), reduced ischemic brain infarction, improved behavioral performance and attenuated penetration of MoDMs in the ischemic hemisphere. We conclude that by blocking macrophage, Maclpil protects against acute ischemic stroke by inhibiting neuroinflammation.
Keywords: Ischemic stroke, lncRNA, macrophages, neuroinflammation, Inflammatory
Introduction
Stroke is one of the leading causes of human death, which results in 6.2 million deaths each year worldwide.1 Currently, tissue plasminogen activator (tPA) and endovascular therapy are the two main treatments. However, both treatments are only applicable to a small number of stroke patients, due to the short therapeutic time window and other restrictive conditions.2 Therefore, it is critical to develop novel and effective treatments to a larger spectrum of stroke patients.
Targeting neuroinflammation has great potential for stroke treatment. During the acute phase, immune cells actively migrate into the brain lesion and accentuate brain injury. Various immune cells including T cells, B cells, NK cells, and macrophages have been shown to be involved in stroke-induced neuroinflammation.3–5 Among these cell types, macrophages vastly outnumber all other leukocytes in the ischemic brain. They receive signals from other leukocytes as well as affect other leukocytes by releasing inflammatory factors, thus they are a central player in stroke-induced neuroinflammation.6 Macrophages derive from both brain residential microglia and peripheral monocytes. Our previous data showed that monocytes-derived macrophages (MoDMs) are a double-edged sword; they exacerbate severe brain injury in the acute phase, but promote functional brain recovery in the chronic phase.7,8 Nevertheless, the most recent studies suggest that microglia and microglia-derived macrophages (MiDMs) are beneficial in reducing brain injury.9 Considering that MoDMs and MiDMs differ in their roles in ischemic brain injury induced by stroke, we aim to leverage their differences to facilitate the development of effective therapies.10
Long non-coding RNAs (lncRNAs) are untranslated regulatory RNAs with more than 200 nt in length. The lncRNA expression profile was significantly altered after ischemic stroke, both in animal models and patients' blood samples.11,12 lncRNAs were shown as a key mediator in gene expression and function regulation in ischemic stroke.13–16 After ischemia, lncRNA H19 levels increased in the ischemic brain, and were shown to promote neuroinflammation in ischemic stroke by driving histone deacetylase 1-dependent M1 microglial polarization in BV2 cells lines.14 However, one limitation is that previous studies analyzed the lncRNA in the whole brain tissue as a bulk assay. Considering the fact that lncRNAs have a high cell type specificity, we aimed to identify the specific profiles of lncRNAs in MoDMs and MiDMs in acute brain injury after stroke and to manipulate them for stroke therapy.
In our current study, we screened the lncRNA expression patterns from FACS sorted MoDMs and MiDMs after ischemia, identified the lncRNA, Maclpil, that is highly expressed in MoDMs but not MiDMs. This lncRNA is functionally related to its neighbor gene, lymphocyte cytosolic protein 1 (LCP1). We studied Maclpil functions using both in vitro macrophage cultures and in vivo mouse stroke models, and demonstrated that inhibition of Maclpil in MoDMs protects against brain injury induced by acute stroke in an experimental mouse model.
Materials and methods
Animals
Male C57BL/6 mice (8–10 weeks, stock number 000664) were purchased from Jackson Laboratories (Maine, USA). Five mice were housed in each cage at the Stanford animal facility under a 12:12-h light-dark cycle (lights on at 07:00 a.m.), at a temperature of 65–75 °F and 40–60% humidity, with freely available food and water. All animal experiments were performed under the protocols based on the NIH Guidelines for Care and Use of Laboratory Animals. All experiments were performed in accordance with the ARRIVE (Animal Research Reporting In Vivo Experiments) guidelines,17 and approved by the Stanford Institutional Animal Care and Use Committee (IACUC). Animals were randomly assigned to groups; behavior tests and treatments were conducted in a blinded fashion – the researcher performing the surgeries was unaware of the animal groups. A total of 177 male mice were included with 36 mice excluded in this study. The criteria to exclude the animals from the study were: (1) had no neurological deficits after stroke; (2) brains had evidence of surgical subarachnoid hemorrhage (no more than two mice were excluded in each group); (3) did not fit the base line of the behavior test before surgery for that test.
Focal cerebral ischemia
Focal ischemia in male mice (22 to 25 g) was induced by middle cerebral artery occlusion (MCAO) for 45 min, which generated infarction in both the cortex and the striatum, as we previously described.18,19 Anesthesia was induced by 5% isoflurane, and maintained with 2% isoflurane throughout the surgery. Core body temperatures were monitored with a rectal probe and maintained at 37 ℃.
Infarct size measurement
Three days after surgery, mice brains were collected and infarct size of the ischemic region was measured as previously described.20 In brief, four to five blocks of 2 mm thick coronal sections were prepared and stained with 2% 2,3,5-triphenyltetrazolium chloride (TTC) for 10 min and fixed with 4% paraformaldehyde overnight. Infarct size was measured using the National Institutes of Health Image J software.21 It was normalized to the contralateral hemisphere and expressed as a ratio according to the following formula: (area of non-ischemic hemisphere – area of non-ischemic tissue in the ischemic hemisphere)/area of non-ischemic hemisphere.20
RNA deep sequencing in FACS sorted MoDMs and MiDMs
To conduct RNA deep sequencing, brain leukocytes were isolated from the ischemic brains. The detailed procedures are listed below:
Brain mononuclear cell isolation and FACS sorting: Three days after stroke, the brain was dissected after trans-cardinal perfusion with 50 ml cold PBS. Mononuclear cells were collected as described previously.3,22 In brief, ischemic hemispheres were homogenized and filtered through a 70 μm cell strainer. Cell pellets were suspended in 37% Percoll (GE Healthcare, Pittsburgh, USA), then 2 ml of 70% Percoll was loaded under the cell suspension. After 30 min of centrifugation at 2000 r/min, the interphase cells were collected. After another wash with 1% BSA in PBS, cells were first stained live/dead aqua for 10 min, then co-stained for 30 min on ice with anti-mouse CD45 (clone 30-F11, PE-Cy7), anti-mouse CD11b (clone M1/70, FITC), anti-mouse Ly6G (clone 1A8 Alexa Flour 647), anti-mouse CD19 (clone 1D3/CD19, PE) and anti-mouse CD3 (clone 17A2, APC) in darkness. All the antibodies were purchased from BioLegend®, California, USA. Cells were then washed with 1% BSA in PBS, centrifuged, and resuspended in 200 µl 1% BSA in PBS. MoDMs and MiDMs were first excluded from T cells, B cells and neutrophils by using the CD3, CD19, and Ly6G markers and identified as CD45hiCD11b+ and CD45lowCD11b+, respectively. For each RNA sample sequenced, five animals were pooled together, and the total cell number was one million cells for MoDMs, and two millions for microglia or MiDMs.
RNA extraction and quantitation: Total RNA was extracted from sorted microglia/MiDMs and MoDMs by using TRIzol® (Life Technologies, California, USA) according to the manufacturer instructions. After centrifuging and removing ethanol, the RNA pellets were dried at room temperature and re-suspended in 20 μl RNase-free water. RNA concentration was determined by using a NanoDrop (NanoDrop Technologies, Wilmington, Delaware, USA), and RNA integrity was assessed by Agilent Bioanalyzer 2100 system with a small RNA chip (Agilent Technologies, California, USA).
RNA library construction and deep sequencing: RNA libraries were created from each sample using the TruSeq Stranded mRNA HT Sample Preparation Kit (Illumina, Inc., Hayward, California, USA; Catalog: RS-122-2103). The first step in the workflow involved two cycles of oligo (dT) magnetic bead purification of poly (A)+ mRNA. (Life Technologies Corporation, Carlsbad, California, USA). The purified mRNA was then fragmented into ∼200 nucleotides using divalent cations at an elevated temperature. The cleaved RNA fragments were subsequently converted into double-stranded cDNA by using RNase H and Escherichia coli DNA polymerase. Through an end-repair reaction, non-templated A residues were added to the 3′ ends of the double-stranded cDNA and special Illumina adapters were ligated onto the cDNA. The products of these reactions were purified and enriched by PCR to generate the final cDNA library. Library quality was assessed on the Agilent Bioanalyzer 2100 system (Agilent, Santa Clara, California, USA). Finally, the purified cDNA was directly used for cluster generation and single-end sequenced in 50 bp using an Illumina HiSeq 2000 platform (Illumina, Inc., San Diego, California, USA).
RNA-seq data analysis
Reads were formatted into Fastq Format using Bam2fastx, then mapped to the mouse genome using Tophat.23 The estimate gene expression was analyzed using Cufflinks.24 Each RefSeq gene's expression was summarized and normalized bioconductor R package. The identified lncRNA must have transcripts of at least 200 bp and not translate into protein product. The pipeline of RNA-seq analysis was summarized in Supplementary Figure 1(b).
Primary monocyte isolation and macrophage culture
Monocytes from C57BL/6J mouse bone marrow were first treated with RBC lysis buffer (eBioscience, Inc., San Diego, California, USA; Catalog: 00-4300-54). To culture monocyte-derived macrophages, the cells were planted into 24-well plates at 37 ℃ in a 5% CO2 incubator. After 12–24 h, cell media was exchanged to 10–20 ng/ml M-CSF recombinant mouse protein (Thermo Fisher Scientific, Waltham, Massachusetts, USA, Catalog: PMC2044) for six days to differentiate peripheral monocytes from the neutral state macrophage, M(M-CSF). M(M-CSF) can be further polarized after 24 h to the pro-inflammatory M(LPS) using 1 μg/ml Lipopolysaccharides (LPS; Sigma-Aldrich, St. Luis, Missouri, USA, Catalog: L2880), and anti-inflammatory M(IL-4) using 2 μg/ml Recombinant Murine IL-4 (PeproTech Sciences, Inc., Ontario, Canada; Catalog: 214-14).
Maclpil and LCP1 knockdown
SiRNA for LCP1 knockdown were purchased from Ambion by Life Technologies (ThermoFisher Scientific, Waltham, Massachusetts, USA; siRNA ID150197), and the control scramble siRNA was purchased from QIAGEN (Qiagen Inc., Germantown, Maryland, USA; Catalog: 1027292). This siRNA negative control was conjugated with AlexaFlor 488, thus the transfection efficiency can be detected using AlexaFlor 488 signal using FACS, which was around 80%. Three siRNAs were initially designed to target Maclpil by using ThermoFisher Scientific's online tool (ThermoFisher Scientific, Waltham, Massachusetts, USA). After comparing the outcomes, siRNA(sequence: AAUGCACAGAGACAUUAGCdTdT) was chosen to perform the experiment. The siRNA was synthesized at the Stanford University Protein and Nucleic Acid (PAN) Facility. The siRNAs were transfected into M(LPS) using the HiperFect Transfection Reagent (Qiagen Inc., Germantown, Maryland, USA, Catalog: 301704) to produce Maclpil-silenced M(LPS) (MSM). In brief, 4 × 104 cells were planted in each well of a 24-well plate; 10 nM siRNA was then mixed with 4.5 μl HiPerFect Reagent for 15 min at room temperature and added into the cell culture following the instructions.
Adoptive transfer of macrophages
The primary monocytes were obtained from bone marrow and then polarized into macrophages, and transfected with siRNA. The cells (6 × 106 cells) as prepared above and control macrophages, were collected and adoptively transferred (IV injection) into mice after MCAO. To track whether the siRNA transferred cells infiltrate into the ischemic brain, donor monocytes were isolated from B6.SJL-Ptprca Pepcb/ BoyJ (CD45.1+) mice, cultured into macrophages and transfected with siRNA, then IV injected into the recipient mice (CD45.2+) after stroke. Immune cells were isolated from the ischemic brain, spleen, blood, bone marrow and lymph nodes 18 and 50 h after transfer, and the CD45.1+ cells were analyzed by FACS.
In vitro cell migration and phagocytosis assay
The cultured MoDM was collected and plated in a 24-well transwell plate (Transwell® Inserts, Sterile, Corning®, Massachusetts, USA, Catalog: 3492). Chemokines CCL2 (MCP1, BioLegend®, California, USA, Catalog: 578402) were added into the lower chamber at 2 ng/ml.25 After 4 h of incubation at 5% CO2, 37 ℃, cell numbers from the upper and lower chambers were collected and counted using trypan blue for live/dead staining. To test the phagocytosis assay, the cultured macrophages were plated in a 6-well plate at 80% density. Latex beads – rabbit IgG-FITC complex – were added directly to the cell culture to a final dilution of 1:250 (phagocytosis assay Kit, IgG FITC, Cayman Chemical Co., Michigan, USA, Catalog: 500290) and cultured cells at 5% CO2, 37 ℃. The cells, which engulf the beads, were detected as FITC+ cells using FACS.
Immunofluorescence staining
Brains were collected after PBS perfusion and immediately frozen, then cut into 20 μm thick coronal sections on a cryostat (Leica, Wetzlar, Germany), collected on premium charged microscope slides, and stored at −80 ℃. Sections were first treated with 4% PFA for 30 min at 37 ℃, then washed with PBS twice, each for 10 min; then treated with 0.3% triton-100 in PBS for 30 min, and blocked by 10% BSA for 1 h at room temperature. Primary antibodies including Rat anti-mouse CD11b antibody (BD biosciences, USA, Catalog: 553309), mouse anti-mouse CD68 antibody (Catalog: ab955, Abcam, USA), and rabbit anti-mouse iNOS (Catalog: ab15323, Abcam, USA) were added on each slide in a 1:100 dilution at 4 ℃ and incubated overnight. After three washes, the sections were incubated with the secondary antibody, Alexa 488-conjugated goat anti-mouse antibody (1:200; Invitrogen, Carlsbad, CA, USA, Catalog: A-11001) and Alexa 594-for 60 min at room temperature. After washing with PBS, stained sections were mounted with DAPI.
Real-time PCR
RNA was purified using RNeasy Mini Kit (Qiagen Inc., Germantown, Maryland, USA; Catalog: 74104) and reverse transcripted to cDNA using the RT2 Easy First Strand Kit (Qiagen Inc., Germantown, Maryland, USA; Catalog: 330421). Real-time PCR (RT-PCR) was conducted using Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific, Waltham, Massachusetts, USA, Catalog: 4367659) and Rox Reference Dye (Thermo Fisher Scientific, Waltham, Massachusetts, USA, Catalog: 1223-012) on the Stratagene Mx3000P QPCR System. GAPDH and 18S RNA were used as reference genes. All primers were synthesized at the Protein and Nucleic Acid (PAN) facility at Stanford University (primer sequences are listed in Supplementary Table 1).
Neurobehavioral examination
Neurobehavioral tests were carried out by an investigator who was blinded to the surgery. This assay is based on a modified neurological severity score (mNSS) system to present a comprehensive assessment of neurological functions including motor, sensory, balance, and reflex tests.26 The higher the scores, the more severe the injury. More specifically, the mNSS ranges from 0 to 14, in which 0 represents normal and 14 represents the highest degree of neurological deficiency. For the motor assay, after holding up the tail of the mouse, the bend and limb torsion was observed (score 0–3). The posture of walking on the floor was also checked (score 0–3). For the balance test, the mice were placed on a beam to test whether the mouse could keep its balance on the beam, if its limbs fell off the beam, and its ability to walk across the beam (score 0–6). For the sensory and reflex tests, pinna and corneal reflexes were examined, respectively (score 0–2). For the behavior assays of the pole tests, mice were trained three times, from five days before surgery. After the MCAO surgery, a test was performed on day 3, day 5, day 7 based on previously described methods.27
Statistical analysis
All results are presented as mean ± SD. Statistical significance of differences among groups was determined by two-tailed unpaired and paired Student's t-test. For experiments with one treatment and more than three groups, one-way ANOVA was applied. For experiments with more than one treatment or variants, two-way ANOVA was performed followed by Tukey's multiple comparisons test using PRISM 7.0 (GraphPad Software, La Jolla, California, USA, www.graphpad.com).
Results
RNA sequencing showed distinct lncRNA expression patterns in MoDMs vs. MiDMs in the ischemic brain after stroke
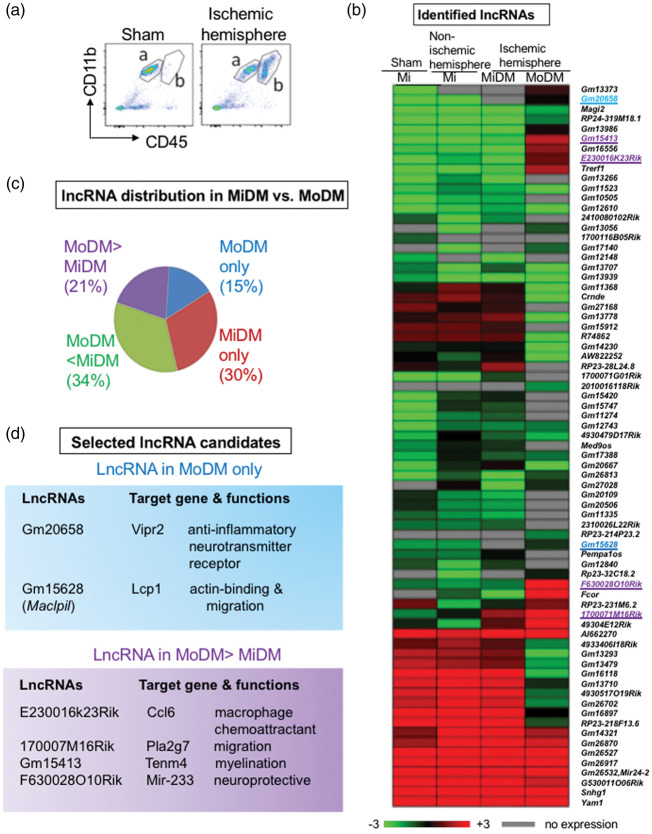
Previous studies have shown that CD11b and CD45 are well defined cell surface markers used to identify MoDMs and MiDMs.7,28,29 In this study, MoDMs and MiDMs were sorted by FACS as CD45hiCD11b+ and CD45lowCD11b+, respectively, after excluding T cells, B cells and neutrophils by using the CD3, CD19, and Ly6G markers (Figure 1(a)). Because our purpose in this study is to compare relative lncRNA expression levels in MoDMs and MiDMs, and thus only microglia (without monocytes) from sham animals was used as a control, which sets a baseline to compare lncRNA expression in MoDMs and MiDMs. As a result, the deep sequencing identified 73 lncRNAs that are differentially expressed in MiDMs and MoDMs three days after stroke. (Figure 1(b), Supplementary Figure 1(a) and (b)). Among those, 21% of lncRNA expression levels were higher in MoDMs, while 34% were higher in MiDMs. In addition, 15% of lncRNAs were specifically expressed in MoDMs but not in MiDMs, while 30% were expressed in MiDMs but not in MoDMs (Figure 1(c)).
Figure 1.
MoDMs and MiDMs purified from the ischemic hemisphere have distinct lncRNAs expression profiles. (a) Representative FACS gating strategies. After excluding T cells (CD3+), B cells (CD19+) and neutrophils (Ly6G+) from live singlets, MoDMs and Mi/MiDMs are gated as CD45hiCD11b+ (a) and CD45lowCD11b+ (b), respectively. (b) The heatmap represents RNA deep sequencing results. After ischemia, a total of 73 lncRNAs in MiDMs and MoDMs were identified. Four cell groups were studied: microglia (Mi) in the sham group, microglia in the non-ischemic hemisphere, MiDMs and MoDMs in the ischemic hemisphere. (c) Percentage of lncRNA distributions in MoDMs and MiDMs in the ischemic hemisphere. (d) Six lncRNAs highly expressed in MoDMs were selected for further screening: their names, neighbor genes, and functions are listed. These six lncRNAs are also highlighted in the heatmap. Gm20658 and Gm15628 (highlighted in blue) were expressed in MoDMs but not in MiDMs. Gm15413, E230016k23Rik, F630028O10Rik, 170007M16Rik (highlighted in purple) had higher expressions in MoDMs than in MiDMs after stroke.
Our hypothesis is that lncRNAs modulate brain injuries by altering MoDMs functions, as we recently reported that MoDMs play critical roles in brain injury.7 Therefore, six lncRNAs that match the following criteria were selected for further studies. First, they are highly or only expressed in MoDMs; second, the lncRNAs may affect macrophage functions by affecting their neighbor genes that were analyzed using NIH genetic sequence database GenBank® to check the location and function of their neighbor genes,30 (Supplementary Figure 2(a), Pipeline for candidate lncRNA selection). Based on the screen result, only two lncRNAs expressed in MoDMs and four lncRNAs had higher expression levels in MoDMs compared to MiDMs were selected as candidates. During the acute stroke phase, the pro-inflammatory macrophages are the major components. Thus, we further screened the six candidate lncRNAs to select only one that is anti-inflammatory for final study. The candidate lncRNAs and their target genes are listed (Figure 1(d), Supplementary Table 2).
Maclpil expression is closely related to pro-inflammatory macrophage polarization
Macrophage polarization is controlled by the tissue microenvironment and extrinsic factors.31 The polarization status of macrophages is relatively plastic; they can be converted from pro-inflammatory to anti-inflammatory depending on the stimuli and signals.31 Our previous study showed that pro-inflammatory macrophages are the dominant type during the acute phase of ischemia.7 Therefore, we polarized the monocytes in vitro, and screened the six candidate lncRNA expression levels in the pro- and anti-inflammatory macrophages (Supplementary Figure 2(b)). The results showed that only the lncRNA Maclpil (Gm15628) expression was significantly higher in M(LPS) compared to M(IL-4) (Supplementary Figure 2(c)). Then, we quantified the Maclpil expressions at different time points after adding LPS, and found that its expression was gradually increased, which was similar to the changes in M(LPS) marker genes iNOS (Figure 2(a)), suggesting that Maclpil is functionally related to pro-inflammatory macrophage polarization.
Figure 2.
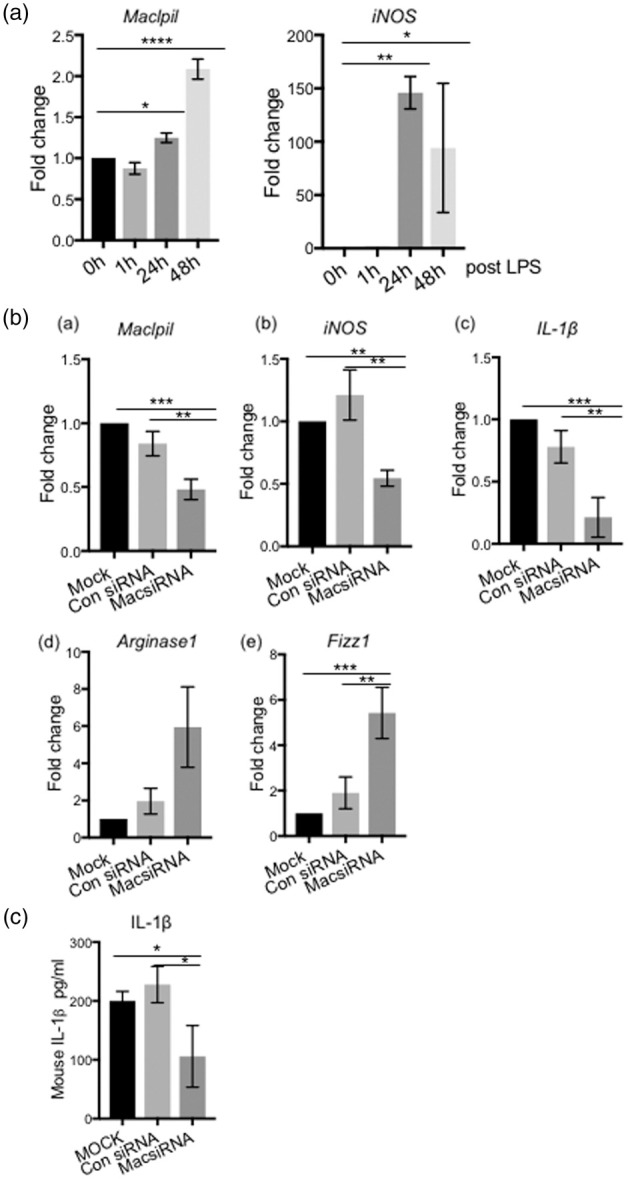
The lncRNA Maclpil (GM15628) expression is linked to the pro-inflammatory macrophage M(LPS) polarization in vitro. Bone marrow-derived monocytes (BMDM) were cultured and differentiated into macrophages, then polarized to pro-inflammatory macrophages by adding LPS in vitro. (a) Real-time qPCR shows the Maclpil and iNOS expression time courses at the transcriptional level in BMDM at 0 h, 1 h, 24 h, and 48 h after LPS addition to the cultured macrophages. The values represent Mean ±SD, one-way ANOVA, *, **, **** p < 0.05, p < 0.01, p < 0.0001, respectively. (b) The effects of Maclpil knockdown by siRNA on inflammatory gene expressions in M(LPS) at 48 h after LPS addition. (a) The RT-qPCR results show that transfecting a Maclpil-specific siRNA in M(LPS) resulted in reductions of Maclpil RNA expressions. Maclpil reduction resulted in the decrease of the pro-inflammatory genes, iNOS and IL-1ß (b–c), but increases in the anti-inflammatory genes, Argnase1 and Fizz1 (e–f). (c) ELISA results show that Maclpil knockdown resulted in the reduction in IL-1β proteins in cell supernatant of cultured M(LPS) measured at 48 h after LPS addition. The experiments were repeated three times. n = 3 in each group, one-way ANOVA, *,**, *** p < 0.05, p < 0.01, p < 0.001.
Next, we designed siRNA to knockdown Maclpil expression (Figure 2(b)). Downregulation of Maclpil by siRNA resulted in reductions in the pro-inflammatory genes iNOS and IL-1ß (Figure 2(b), (b) and (c) ), but increased the anti-inflammatory M(IL-4) marker gene expressions Arginase1 and Fizz1 (Figure 2(b), (d) and (e)). ELISA analyses further confirmed that Maclpil knockdown resulted in reduced IL-1β protein expressions (Figure 2(c)).
Maclpil modulates macrophage migration and phagocytosis by regulating its actin-relevant neighbor gene LCP1
Migration and phagocytosis are highly related to cell cytoskeleton structures.32,33 LCP1 is a key protein involved in both phagocytosis and migration function via actin filaments and actin-based structures.34 Maclpil is located intragenically with LCP1 (Supplementary Figure 3(a)), and thus we hypothesized that LCP1 might be involved in the decline of phagocytosis and migration induced by the downregulation of Maclpil.
To confirm the hypothesis, we first checked STRING (http://string-db.org/), an online database to summarize the network of predicted associations for a protein.35 LCP1 was shown to have strong links to cell skeleton proteins such as Actg1 and myosin (Supplementary Figure 3(b)). RT-PCR results showed that when Maclpil was knocked down by siRNA in cultured M(LPS), both Maclpil and LCP1 expressions were inhibited (Figure 3(a)). In vitro cell migration test showed that when Maclpil or LCP1 (Figure 3(a), Supplementary Figure 3(c) for LCP1) was knocked down, fewer MoDMs migrated into the lower chambers, while more MoDMs stayed in the upper chambers (Figure 3(b)). To test the phagocytosis ability, FITC+-labeled latex–rabbit IgG beads were added into the in vitro cultured MoDMs. When LCP1 or Maclpil expression was blocked, fewer FITC+ beads were engulfed by M(LPS) (Figure 3(c)).
Figure 3.
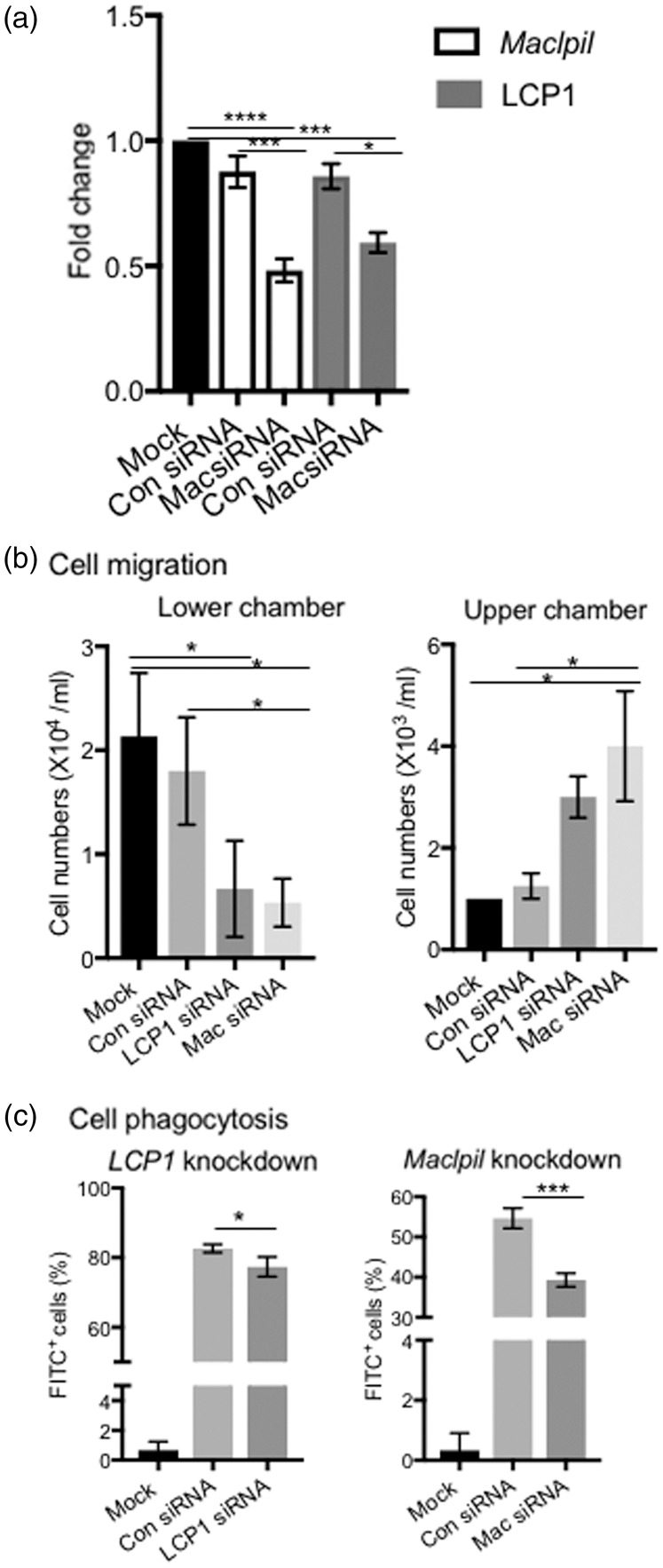
Maclpil modulates migration and phagocytosis by its actin-relevant neighbor gene LCP1. (a) RT-PCR results show that when Maclpil was knocked down by siRNA in cultured M(LPS), both Maclpil and LCP1 expressions were inhibited for 48 h after LPS addition. ****, p < 0.0001, one-way ANONVA, n = 3. (b) In vitro transwell cell migration assay. The knockdown of LCP1 or Maclpil decreased the cell migration from the upper chamber to the lower chamber. Mock control (cells with transfection reagent only, without adding siRNA). (c) In vitro cell phagocytosis assay. FACS results showed that when LCP1 or Maclpil was knocked down by siRNA, the numbers of cells phagocytosed FITC+ latex beads were significantly reduced. Mock control (cells with transfection reagent only, without adding FITC+ beads). *, ***, p < 0.05, p < 0.001, one-way ANOVA.
Silencing Maclpil in M(LPS) protects against ischemic stroke in vivo
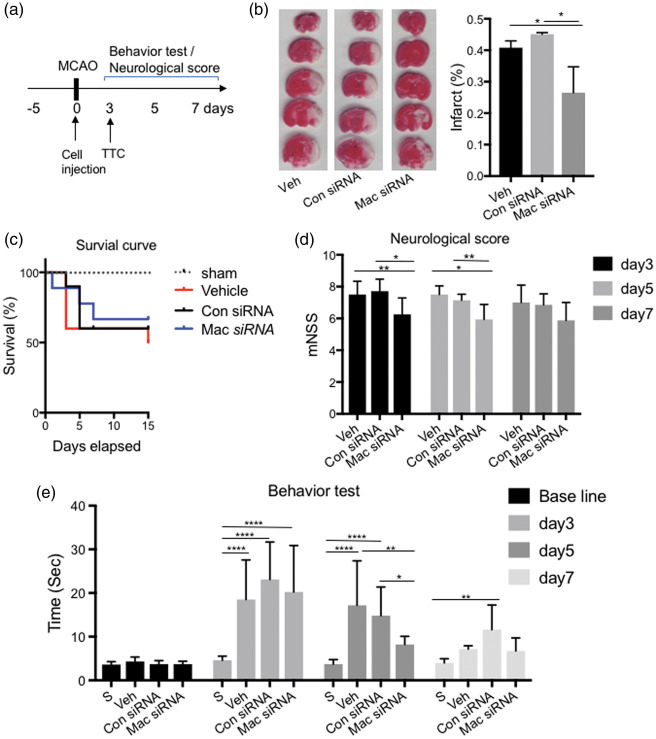
To test whether blocking Maclpil in MoDMs protected against stroke in vivo, macrophages transfected with Maclpil siRNA (6 × 106 cells), i.e. Maclpil-silenced macrophages (MSM), were IV injected into mice after stroke onset (Figure 4(a)). Infarct sizes were measured by 2,3,5-TTC staining three days after stroke. The results showed that mice adoptively transferred with MSM had significantly reduced infarct sizes (Figure 4(b)), and a higher survival ratio (Figure 4(c)). The MSM group had significantly higher neurological scores on day 3 and day 5 (Figure 4(d)) and better behavior performances compared to mice receiving pro-inflammatory macrophages transfected with control siRNA, using the pole test27 (Figure 4(e)).
Figure 4.
IV injection of Maclpil-silenced M(LPS) (MSM) protected against acute ischemic stroke. (a) Schematic diagram of experimental design. Behavioral training started five days before surgery. Cells were injected immediately after MCAO surgery (Day 0). Mice were euthanized for TTC staining on day 3 after stroke onset. Behavior test and neurological scores were tested from day 3 to day 7. (b) Representative TTC staining and the bar graph show the statistical results of infarct sizes as percentages measured at three days after ischemic stroke. n = 6–8 mice/group. (c) Survival curve from day 0 to day 15 after MCAO. n = 10–12 mice/group. (d) Neurological score from day 3 to day 7, n = 10–12 mice/group. One-way ANOVA, *,** p < 0.05, p < 0.01. (e) The behavioral assessment was conducted by using the pole test, showing the average walking time from the top to the bottom of the vertical pole. n = 10–12 mice/group. *, **, **** p < 0.05, p < 0.01, p < 0.0001. Two-way ANOVA followed by Tukey's multiple comparisons test (within each column).
Maclpil sliced macrophages inhibited neuroinflammation and relieved the peripheral immunosuppression
To understand how this modified macrophage functions after stroke, we tracked whether the Maclpil-silenced CD45.1 macrophages that were adoptively transferred into CD45.2 mice infiltrated into the ischemic brain (Supplementary Figure 5). CD45.1+ cell numbers were analyzed by FACS in the ischemic brain, spleen, blood, bone marrow, and lymph nodes. The results showed that Maclpil-silenced CD45.1+ macrophages were increased in the ischemic brain at 18 h post-transfer, but few were identified at 50 h. While these modified macrophages had largely accumulated in the spleen at both 18 h and 50 h, few were detected in the blood, bone marrow, and lymph nodes (Supplementary Figure 5).
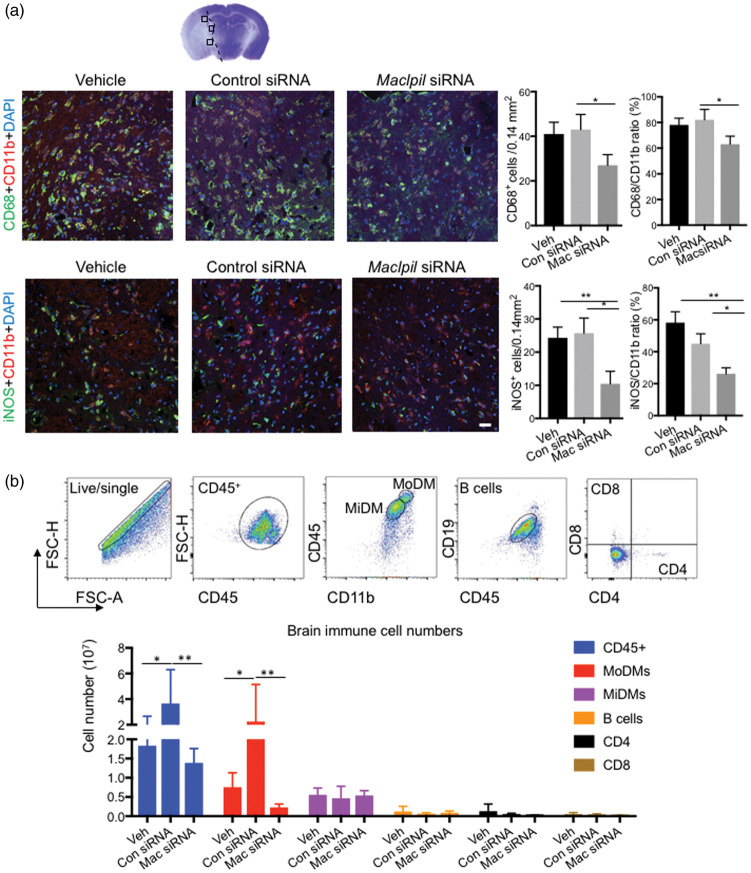
We then analyzed the effects of MSM on neuroinflammatory response in the ischemic brain. The immune-fluorescence staining showed that stroke resulted in strong expressions of CD68+ and iNOS in control mice, but MSM significantly attenuated CD68+ and iNOS+ cell numbers and their percentages in total microglia/macrophages (Figure 5(a)). The FACS results confirmed that MSM blocked MoDMs infiltration into the ischemic brain three days after stroke (Figure 5(b), Supplementary Figure 5(a)), but it did not significantly affect microglia, B cells and T cells. When we checked the immune cells in the peripheral organs, MSM significantly attenuated reductions in the total cell numbers and most leukocyte subtypes in the spleen. A similar trend is also observed in the peripheral blood, although significant differences were not observed in the total numbers and some leukocyte subsets (Supplementary Figure 4, Supplementary Figure 5). Previous studies showed that stroke could induce profound immunosuppression.36 In this study, we showed that injections of MSM could relieve this immunosuppression.
Figure 5.
The effects of MSM adoptive transfer on leukocyte accumulations in the ischemic brain. (a) Confocal immunofluorescent staining for CD11b, CD68, and iNOS, and counter stained with DAPI. The top panel shows a representative brain section stained with cresol/violet, on which three indicated regions are identified for cell number quantification. The middle panel shows the representative staining of CD11b+ (red) and CD68+ (green), and the bottom panel shows the representative staining of CD11b+(red) and iNOS+ (green) in vehicle, control siRNA, and MSM groups at 72 h after stroke. The bar graphs in the right show CD68+ and iNOS+ cell numbers and their percentages in total microglia/macrophage (CD11b+ cells). Scale bar: 20 μm. n = 5–6 mice/group. *, **p < 0.05, p < 0.01. (b) FACS analysis of the ischemic brain immune cells. The upper pictures show the representative gating methodology in the brain to identify MoDMs, MiDMs, B cells, CD4 and CD8 T cells. Bar graphs of the statistical results for each cell types. n = 5 mice/group, two-way ANOVA, * p < 0.05, **p < 0.01.
Discussion
In the current study, we showed that MoDMs and MiDMs had distinctive lncRNA expression profiles after ischemic stroke. LncRNA Maclpil was specifically expressed in MoDMs three days after ischemia. Downregulation of Maclpil converted macrophages from pro-inflammatory to anti-inflammatory phenotypes, decreased their migration and phagocytosis ability by inhibiting its neighbor gene LCP1. Adoptive transfer of MSM reduced brain infarction by inhibiting neuroinflammation.
LncRNA expression profiles had significantly altered after ischemic stroke in the mouse model.15,16,37 Some lncRNAs were reported to play direct roles on ischemic outcomes. For instance, lncRNA MEG3 regulated ischemic neuronal death38; silencing RMST had a protective function against ischemic stroke induced by MCAO.39 In stroke patients, a total of 299 lncRNAs expressed differently in 266 whole-blood RNA samples.12 However, a major limitation of these studies was using bulk tissue rather than specific cell types, which lack the information of lncRNA cell specificity. In our current study, we tested the hypothesis that lncRNAs have distinct expression profiles in MoDMs and MiDMs, and specifically regulate cell functions. This is based on previous lncRNA studies. First, lncRNA expression had cell type specificity.40,41 Using the in situ hybridization data from the Allen Brain Atlas, the expression signatures of lncRNAs were shown to have a strong relationship with specific cell types.42,43 lncRNA expressions were reported to have significant differences between gray and white matter, as these two brain components contain different cell populations.44 The highly distinct lncRNAs expression profiles were reported in the hippocampus and the pre-frontal cortex in the mouse brain based on the transcriptome assay data.45 Second, LncRNAs also play critical roles in gene expression regulation. For instance, lncRNA rhabdomyosarcoma 2-associated transcript (Rmst) interacted with its target gene SOX2 to regulate neurogenesis and blocking Rmst from extending the initiation of neural differentiation.46 The neural-specific lncRNA Pinky (Pnky) interacted with a splicing regulator, PTBP1, to regulate neurogenesis from the embryonic and postnatal neural stem cell (NSC) populations.47 Inspired by these findings, our current study provides novel insights about lncRNA specificity in macrophages after ischemic stroke.
In this study, using the FACS sorting technique, we obtained the separated MoDMs and MiDMs, which enabled us to study the lncRNAs cell type specificity. Using RNA deep sequencing, 73 lncRNAs were identified to be differentially expressed in MoDMs and MiDMs isolated in the ischemic brain three days after stroke. As we recently reported that MoDMs play critical roles in neuroinflammation and brain injuries after stroke,7 we aimed to identify lncRNAs that may protect against stroke by modulating MoDMs. Therefore, we selected and screened six lncRNAs that are specifically expressed in MoDMs, and their neighbor genes that are involved in inflammation, and found that the lncRNA Maclpil (GM15628) is critical for MoDMs functions. We confirmed that Maclpil is mainly expressed in MoDMs rather than in MiDMs. Inhibiting Maclpil in M(LPS) decreased pro-inflammatory gene expressions, while it increased some pro-inflammatory gene expressions, suggesting that Maclpil knockdown can reverse the functional macrophage phenotypes.
We have recently reported that adoptive transfer of anti-inflammatory MoDMs significantly improved recovery in mice during the chronic phase after stroke.7 In our current study, we further demonstrated that the adoptive transfer of MSM attenuates infarct sizes and improves behavioral performance. We showed that MSM was identified in the ischemic brain at 18 h but not at 50 h post-transfer. Nevertheless, a large amount of these modified macrophages were accumulated in the spleen at the both time points. It is likely that MSM directly affects brain tissues through their presence in the ischemic brain, and also indirectly affect brain injury via the peripheral immune organs. This protective effect is linked with a reduced inflammatory response compared to its control group, as we found that MSM attenuated CD68+ and iNOS+ cell numbers, and their percentages in total microglia/macrophages in the ischemic hemisphere. It not only inhibited neuroinflammation in the ischemic brain, but also attenuated lymphopenia in the peripheral circulation, which is consistent with our previous studies that neuroinflammation is proportional with immunosuppression in the peripheral circulation.29,48
The underlying mechanism of the lncRNA Maclpil regulating macrophage functions remains unknown. Nevertheless, our results showed that Maclpil downregulation decreased LCP1 expression, which is linked with the reversal of pro-inflammatory macrophage to anti-inflammatory phenotypes. In addition, we demonstrated that Maclpil inhibition induced LCP1 downregulation, which attenuated macrophage phagocytosis and migration abilities. LCP1 is an actin-related cytoskeleton protein needed to maintain cell shape. We speculate that LCP1 downregulation may result in altered cell skeletons thus leading to cell shape changes, and results in macrophage polarization status change from the pro-inflammatory to anti-inflammatory phenotypes. This speculation is supported by previous studies. First, cell shape modulation has been shown to determine macrophage polarization.49 For instance, the anti-inflammatory M(IL-4) phenotype shape is longer, while the pro-inflammatory M(LPS) shape is more rounded (Supplementary Figure 2(b)). McWhorter et al.49 showed that only the engineered cell culture substrates that elongate macrophages can promote an anti-proinflammatory phenotype in the absence of cytokine stimulation.49 In addition, LCP1 is functionally correlated to cell skeleton components, which affect cell shape and migration.50 LCP1 is also called the actin-bundling protein L-plastin (LPL), which binds to actin filaments to increase the stability of the actin-based structure, which has been shown to be closely related to immune cell functions.34 Second, LCP1 is functionally related to immune responses. LCP1 is functionally related to various immune response related proteins, including leukocyte specific transcript 1 (LST1), and interleukin 2 receptor (IL2RG).50 LCP1 can activate T cells by phosphorylating serine together with cytokine stimulation.51 LCP1 knock down by siRNA in T cells diminished actin polymerization during synapse formation, and synapse size.52 LCP1−/− T cells did not sufficiently produce TCR-mediated cytokines (e.g. IFN-γ and IL-17) and had lower proliferation rates than normal T cells did.53 Neutrophils isolated from LCP1−/− mice showed respiratory burst deficiencies.54 Third, LCP1 is also required for leukocyte trafficking, and working together with Coronin A facilitates chemokine induced cell trafficking.51 LCP1 downregulation decreased the motility towards chemokine CCL20 in Jurkat T cells.55
In summary, we have demonstrated that MoDMs and MiDMs have specific lncRNA expression profiles in the ischemic hemisphere after stroke, and we identified that the lncRNA Maclpil is only expressed in MoDMs. Our in vitro studies showed that Maclpil knockdown alters functional macrophage phenotypes by converting pro-inflammatory macrophages into anti-inflammatory phenotypes, and in vivo studies demonstrated that adoptive transfer of macrophages with silenced Maclpil robustly reduced infarction and improved behavioral performance. In conclusion, Maclpil knockdown in macrophages protects against brain injuries, which is linked with the inhibition of its target gene LCP1.
Supplemental Material
Supplemental Material for Silencing the lncRNA Maclpil in pro-inflammatory macrophages attenuates acute experimental ischemic stroke via LCP1 in mice by Yan Wang, Ying Luo, Yang Yao, Yuhua Ji, Liangshu Feng, Fang Du, Xiaoya Zheng, Tao Tao, Xuan Zhai, Yaning Li, Pei Han, Baohui Xu and Heng Zhao in Journal of Cerebral Blood Flow & Metabolism
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This project is supported by National Institutes of Health grant (R01NS064236 to H.Z.).
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors' contributions
YW and HZ conceived and designed the project. YW, YL and XY Z conducted the experiments. YL, YY, LSF, DF and XZ preformed the MCAO surgery and YW and YL did the behavior test. YHJ sorted the cells and did sequencing. TT and LS F injected cells; YW did FACS. PH and BHX contributed ideas and technical support. YW and HZ wrote the manuscript.
Supplemental Material
Supplemental Material for this article is available online.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics – 2015 update: a report from the American Heart Association. Circulation 2015; 131: e29–322. [DOI] [PubMed] [Google Scholar]
- 2.Menon BK, Saver JL, Goyal M, et al. Trends in endovascular therapy and clinical outcomes within the nationwide Get With The Guidelines-Stroke registry. Stroke 2015; 46: 989–995. [DOI] [PubMed] [Google Scholar]
- 3.Xiong X, Gu L, Zhang H, Xu B, et al. The protective effects of T cell deficiency against brain injury are ischemic model-dependent in rats. Neurochem Int 2013; 62: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu L, Xiong X, Wei D, et al. T cells contribute to stroke-induced lymphopenia in rats. PLoS One 2013; 8: e59602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin WN, Ducruet AF, Liu Q, et al. Activation of JAK/STAT3 restores NK-cell function and improves immune defense after brain ischemia. FASEB J 2018; 32: 2757–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalogeris T, Baines CP, Krenz M, et al. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol 2012; 298: 229–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang W, Zhai X, Han D, et al. CCR2-dependent monocytes/macrophages exacerbate acute brain injury but promote functional recovery after ischemic stroke in mice. Theranostics 2018; 8: 3530–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wattananit S, Tornero D, Graubardt N, et al. Monocyte-derived macrophages contribute to spontaneous long-term functional recovery after stroke in mice. J Neurosci 2016; 36: 4182–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anttila JE, Whitaker KW, Wires ES, et al. Role of microglia in ischemic focal stroke and recovery: focus on Toll-like receptors. Prog Neuropsychopharmacol Biol Psychiatry 2017; 79: 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kronenberg G, Uhlemann R, Richter N, et al. Distinguishing features of microglia- and monocyte-derived macrophages after stroke. Acta Neuropathol 2018; 135: 551–568. [DOI] [PubMed] [Google Scholar]
- 11.He W, Wei D, Cai, et al. Altered long non-coding RNA transcriptomic profiles in ischemic stroke. Hum Gene Ther 2018; 29: 719–732. [DOI] [PubMed] [Google Scholar]
- 12.Dykstra-Aiello C, Jickling GC, et al. Altered expression of long noncoding rnas in blood after ischemic stroke and proximity to putative stroke risk loci. Stroke 2016; 47: 2896–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta SL, Kim T, Vemuganti R. Long noncoding RNA FosDT promotes ischemic brain injury by interacting with REST-associated chromatin-modifying proteins. J Neurosci 2015; 35: 16443–16449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Zhao H, Fan Z, et al. Long noncoding RNA H19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1-dependent M1 microglial polarization. Stroke 2017; 48: 2211–2221. [DOI] [PubMed] [Google Scholar]
- 15.Dharap A, Pokrzywa C, Vemuganti R. Increased binding of stroke-induced long non-coding RNAs to the transcriptional corepressors Sin3A and coREST. ASN Neuro 2013; 5: 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dharap A, Nakka VP, Vemuganti R. Effect of focal ischemia on long noncoding RNAs. Stroke 2012; 43: 2800–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilkenny C, Browne W, Cuthill IC, et al. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 2010; 160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu L, Xiong X, Zhang H, et al. Distinctive effects of T cell subsets in neuronal injury induced by cocultured splenocytes in vitro and by in vivo stroke in mice. Stroke 2012; 43: 1941–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong X, Xie R, Zhang H, et al. PRAS40 plays a pivotal role in protecting against stroke by linking the Akt and mTOR pathways. Neurobiol Dis 2014; 66: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi T, Steinberg GK, Zhao H. Lithium treatment reduces brain injury induced by focal ischemia with partial reperfusion and the protective mechanisms dispute the importance of akt activity. Aging Dis 2012; 3: 226–233. [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim JK, Obara CJ, Rivollier A, et al. Chemokine receptor Ccr2 is critical for monocyte accumulation and survival in West Nile virus encephalitis. J Immunol 2011; 186: 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van den Bossche J, Neele AE, Hoeksema MA, et al. Macrophage polarization: the epigenetic point of view. Curr Opin Lipidol 2014; 25: 367–373. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Chopp M, Chen J, et al. Intrastriatal transplantation of bone marrow nonhematopoietic cells improves functional recovery after stroke in adult mice. J Cereb Blood Flow Metab 2000; 20: 1311–1319. [DOI] [PubMed] [Google Scholar]
- 27.Balkaya M, Kröber JM, Rex A, et al. Assessing post-stroke behavior in mouse models of focal ischemia. J Cereb Blood Flow Metab 2013; 33: 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponomarev ED, Veremeyko T, Barteneva N, et al. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat Med 2011; 17: 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joo SP, Xie W, Xiong X, et al. Ischemic postconditioning protects against focal cerebral ischemia by inhibiting brain inflammation while attenuating peripheral lymphopenia in mice. Neuroscience 2013; 243: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res 2013; 41: D36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray PJ. Macrophage polarization. Annu Rev Physiol 2017; 79: 541–566. [DOI] [PubMed] [Google Scholar]
- 32.Cui S, Qian J, Bo P. Inhibitive effect on phagocytosis of Candida albicans induced by pretreatment with quercetin via actin cytoskeleton interference. J Tradit Chin Med 2013; 33: 804–809. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Jiao Y, Fan EK, et al. Aging-impaired filamentous actin polymerization signaling reduces alveolar macrophage phagocytosis of bacteria. J Immunol 2017; 199: 3176–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morley SC. The actin-bundling protein L-plastin: a critical regulator of immune cell function. Int J Cell Biol 2012; 2012: 935173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43: D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Offner H, Vandenbark AA, Hurn PD. Effect of experimental stroke on peripheral immunity: CNS ischemia induces profound immunosuppression. Neuroscience 2009; 158: 1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Yuan L, Zhang X, et al. Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol 2016; 277: 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan H, Rao J, Yuan J, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate ischemic neuronal death by targeting miR-21/PDCD4 signaling pathway. Cell Death Dis 2017; 8: 3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou XX, Cheng H. Long non-coding RNA RMST silencing protects against middle cerebral artery occlusion (MCAO)-induced ischemic stroke. Biochem Biophys Res Commun 2018; 495: 2602–2608. [DOI] [PubMed] [Google Scholar]
- 40.Ward M, McEwan C, Mills JD, et al. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J Hum Transcr 2015; 1: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012; 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mercer TR, Dinger ME, Sunkin SM, et al. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 2008; 105: 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007; 445: 168–176. [DOI] [PubMed] [Google Scholar]
- 44.Mills JD, Kavanagh T, Kim WS, et al. Unique transcriptome patterns of the white and grey matter corroborate structural and functional heterogeneity in the human frontal lobe. PLoS One 2013; 8: e78480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadakkuzha BM, Liu XA, McCrate J, et al. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front Cell Neurosci 2015; 9: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng SY, Bogu GK, Soh BS, et al. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 2013; 51: 349–359. [DOI] [PubMed] [Google Scholar]
- 47.Ramos AD, Andersen RE, Liu SJ, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 2015; 16: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong X, Gu L, Wang Y, et al. Glycyrrhizin protects against focal cerebral ischemia via inhibition of T cell activity and HMGB1-mediated mechanisms. J Neuroinflammation 2016; 13: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McWhorter FY, Wang T, Nguyen P, et al. Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci U S A 2013; 110: 17253–17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorenshteyn D, Zaslavsky E, Fribourg M, et al. Interactive big data resource to elucidate human immune pathways and diseases. Immunity 2015; 43: 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freeley M, O'Dowd F, Paul T, et al. L-plastin regulates polarization and migration in chemokine-stimulated human T lymphocytes. J Immunol 2012; 188: 6357–6370. [DOI] [PubMed] [Google Scholar]
- 52.Wabnitz GH, Lohneis P, Kirchgessner H, et al. Sustained LFA-1 cluster formation in the immune synapse requires the combined activities of L-plastin and calmodulin. Eur J Immunol 2010; 40: 2437–2449. [DOI] [PubMed] [Google Scholar]
- 53.Wang C, Morley SC, Donermeyer D, et al. Actin-bundling protein L-plastin regulates T cell activation. J Immunol 2010; 185: 7487–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H, Mocsai A, Zhang H, et al. Role for plastin in host defense distinguishes integrin signaling from cell adhesion and spreading. Immunity 2003; 19: 95–104. [DOI] [PubMed] [Google Scholar]
- 55.Lin SL, Chien CW, Han CL, et al. Temporal proteomics profiling of lipid rafts in CCR6-activated T cells reveals the integration of actin cytoskeleton dynamics. J Proteome Res 2010; 9: 283–297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Silencing the lncRNA Maclpil in pro-inflammatory macrophages attenuates acute experimental ischemic stroke via LCP1 in mice by Yan Wang, Ying Luo, Yang Yao, Yuhua Ji, Liangshu Feng, Fang Du, Xiaoya Zheng, Tao Tao, Xuan Zhai, Yaning Li, Pei Han, Baohui Xu and Heng Zhao in Journal of Cerebral Blood Flow & Metabolism