Short abstract
Blood monocytes/macrophages infiltrate the brain after ischemic stroke and critically influence brain injury and regeneration. We investigated stroke-induced transcriptomic changes of monocytes/macrophages by RNA sequencing profiling, using a mouse model of permanent focal cerebral ischemia. Compared to non-ischemic conditions, brain ischemia induced only moderate genomic changes in blood monocytes, but triggered robust genomic reprogramming in monocytes/macrophages invading the brain. Surprisingly, functional enrichment analysis of the transcriptome of brain macrophages revealed significant overrepresentation of biological processes linked to neurovascular remodeling, such as angiogenesis and axonal regeneration, as early as five days after stroke, suggesting a previously underappreciated role for macrophages in initiating post-stroke brain repair. Upstream Regulator analysis predicted peroxisome proliferator-activated receptor gamma (PPARγ) as a master regulator driving the transcriptional reprogramming in post-stroke brain macrophages. Importantly, myeloid cell-specific PPARγ knockout (mKO) mice demonstrated lower post-stroke angiogenesis and neurogenesis than wild-type mice, which correlated significantly with the exacerbation of post-stroke neurological deficits in mKO mice. Collectively, our findings reveal a novel repair-enhancing transcriptome in brain macrophages during post-stroke neurovascular remodeling. As a master switch controlling genomic reprogramming, PPARγ is a rational therapeutic target for promoting and maintaining beneficial macrophage functions, facilitating neurorestoration, and improving long-term functional recovery after ischemic stroke.
Keywords: Angiogenesis, fluorescence-activated cell sorting, focal cerebral ischemia, neurogenesis, PPARγ
Introduction
Under steady-state conditions, the central nervous system (CNS) is subject to active immune surveillance, a role fulfilled by parenchymal microglia as well as innate immune sentinel cells existing at the brain–periphery interfaces, including the perivascular space, meninges, and choroid plexus.1 In addition to dynamic surveillance by CNS-resident immune cells, brain injuries can induce the migration of peripheral immune cells into the brain through the blood–brain barrier (BBB), and this immune cell influx propels a cascade of inflammatory responses that determine stroke outcome.2,3 Among the infiltrating immune cells, monocytes/macrophages are rapidly recruited to the injured brain in massive numbers and display remarkable functional plasticity.4 Macrophages can adopt pro-inflammatory or inflammation-resolving phenotypes that potentiate brain injury or facilitate brain repair, respectively.5 In line with their complex, multifaceted functions, studies on the role of monocytes/macrophages after ischemic stroke have generated controversial results. Some studies support a beneficial role of monocytes/macrophages in the post-stroke brain, wherein depletion of monocyte/macrophages or inhibition of their recruitment worsens brain injury and long-term functional deficits after ischemic stroke.6,7 Other studies reported neutral8–10 or improved11 stroke outcomes after monocyte-targeting interventions. The functional role of monocytes/macrophages in the post-stroke brain may therefore depend on the temporal or spatial context of the ischemic injury and other factors. To date, a comprehensive characterization of the mechanistic molecular network through which macrophages acquire certain functional phenotype is lacking, hindering our understanding of the beneficial and/or pathophysiological roles of these cells in the post-stroke brain.
The past decade has witnessed considerable advances in our knowledge of the ontogeny and heterogeneity of brain myeloid cells, largely from unbiased profiling approaches, such as genome-wide transcriptomics and proteomics.12,13 These studies reveal important physiological roles of macrophages at CNS interfaces, such as promotion of vascular sprouting in the CNS and the phagocytosis of cellular debris.1 Although the phenotype of myeloid cells has been extensively interrogated at the whole-genome level under physiological conditions, much less is known of the functional roles of monocytes/macrophages after homeostasis is disrupted in the stroke brain. Monocyte/macrophage populations that circulate in the blood after acute brain injuries may possess unique genomic programs that promote their mobilization and brain-homing behavior and exert a profound influence over brain injury and repair.
To address the aforementioned gaps in the field, we performed genome-wide transcriptome profiling of monocytes/macrophages in the mouse brain and blood after ischemic stroke, using bulk RNA sequencing (RNA-seq). The objectives were two-fold: (1) To unravel the genomic adaptations of monocytes/macrophages following ischemic stroke and their functional implications and (2) to identify potential targets that can be manipulated to promote beneficial macrophage responses and improve stroke outcomes. We show that macrophages in the post-stroke brain have a unique transcriptome that strongly favors the remodeling processes in the neurovascular niche. Furthermore, peroxisome proliferator-activated receptor gamma (PPARγ) dictates this transcriptome and determines the reparative macrophage phenotype, ultimately enhancing recovery of neurological functions after stroke.
Materials and methods
Methodological details beyond the descriptions below are provided in Supplementary Materials. Key resources that are essential to reproduce the results are provided in Supplementary Table 1.
Animals
C57BL/6J, Cx3cr1CreER, and PPARγloxP mice were purchased from the Jackson Laboratory. Myeloid cell-specific PPARγ knockout (PPARγ mKO) mice were obtained by crossing the Cx3cr1CreER and PPARγloxP mice for two generations, using breeding strategies described previously.14 The PPARγ mKO mice (genotype: Cx3cr1CreER/wt; Ppargflox/flox) were viable, fertile, and normal in size and did not exhibit any gross physical or behavioral abnormalities. To induce gene deletion, PPARγ mKO mice received intraperitoneal injections of tamoxifen (75 mg/kg daily for four days). Hemizygous Cx3cr1CreER mice (genotype: Cx3cr1CreER/wt; Ppargwt/wt) served as age- and weight-matched WT control mice for the PPARγ mKO mice and received the same tamoxifen treatment. Mice were subjected to cerebral ischemia 14 days after the first tamoxifen injection. Only male mice were used in this study.
Mice were housed in a temperature and humidity-controlled animal facility with a 12-h light/dark cycle. Food and water were available ad libitum. All animal procedures were approved by the University of Pittsburgh Institutional Animal Care and Use Committee, performed in accordance with the Guide for the Care and Use of Laboratory Animals, and reported in accordance with the ARRIVE guidelines.15 All efforts were made to minimize animal suffering and the number of animals used.
Permanent focal cerebral ischemia
Focal cerebral ischemia was induced in mice (8–12 weeks old, 25–30 g) by permanent occlusion of the left distal middle cerebral artery (MCA) and left common carotid artery (CCA), as described previously.16,17 We refer to this model as distal MCA occlusion (dMCAO). Cortical cerebral blood flow (CBF) was monitored using two-dimensional laser speckle techniques.18 Failure to reduce CBF to 30% or less of baseline levels or death immediately after ischemia led to subject exclusion (<10%). Experimental procedures were performed in a randomized and blinded manner following STAIR guidelines.19
RNA sequencing and data analysis
Monocytes and macrophages were extracted from mouse blood and brain by fluorescence-activated cell sorting (FACS) and subjected to RNA-seq. All RNA-seq data are deposited at GEO (GSE138805). All software and algorithms are provided in Supplementary Table 1.
Neurobehavioral tests
The adhesive removal, foot fault and open field tests were performed as described previously20,21 to assess neurological functions before and up to 21 days after dMCAO.
Statistical analysis
High-throughput sequencing data were analyzed, as described above. Other datasets are presented as mean±SD. Individual data points are plotted where applicable. Statistical comparison of the means between two groups was accomplished by the Student’s t test or the Mann-Whitney U test (both two-tailed). Differences in means among multiple groups were analyzed using one or two-way ANOVA, followed by the Bonferroni/Dunn post hoc correction. Pearson product linear regression analysis was used to correlate histological parameters with neurobehavioral functions. A p value equal to or less than 0.05 was deemed statistically significant. All statistics are summarized in Supplementary Table 2.
Results
Genome-wide transcriptional analysis of monocytes/macrophages in the blood and brain after ischemic stroke in mice
Following ischemic stroke, blood monocytes/macrophages invade the injured brain through the compromised BBB, serving as an important source of neuroinflammation.3 In a mouse model of permanent focal cerebral ischemia induced by dMCAO, we detected massive accumulation of CD11b+CD45high cells in the brain by flow cytometry at five days post-ischemia (Supplementary Figure 1(a) and (b)), an injury stage at which the infiltration of peripheral immune cells peaks.22 A concomitant increase of CD11b+CD45high cells in post-stroke blood was observed (Supplementary Figure 1 (a) and (b)), which may result from their de novo proliferation, exit from the reservoir (e.g. spleen),23,24 or an increased proportion relative to other cell populations. The numbers of resident microglia (CD11b+CD45low cells) in the brain were comparable between dMCAO and sham groups (Supplementary Figure 1(a) and (b)). To obtain a comprehensive understanding of the role of monocytes/macrophages in brain injury and repair, we performed RNA-seq profiling of the CD11b+CD45high cells sorted from the blood under non-ischemic sham conditions, and from the blood and brain following dMCAO (Supplementary Figure 1(c)). FACS-sorted cells were monocyte-lineage cells, as reflected by their high expression of the prototypic monocyte marker Csf1r and low expression of markers for other blood cells and brain cells (Supplementary Figure 1(d)). As microglia represent a special population of macrophages entering the brain during embryonic stage, we examined the expression of established microglia and macrophage markers in FACS-sorted cells (Supplementary Figure 1(e)), including genes held in common by microglia and macrophages (e.g. Itgam, Cx3cr1), systemic macrophage-specific genes (e.g. Cd163, Fabp4), and microglia-specific genes (e.g. Tmem119, P2ry13).25 Cells sorted from sham blood, dMCAO blood, and dMCAO brain all expressed high levels of microglia/macrophage-common genes and macrophage-specific genes (Supplementary Figure 1(e)). Notably, cells sorted from the dMCAO brain expressed higher levels of microglia-specific genes than cells from the blood (Supplementary Figure 1(e)), suggesting that a subset of microglia might be present in the sorted cells as a result of CD45 upregulation in response to stroke. Principal component analysis based on the expression of all mapped genes showed a clustering of samples from the same group, and indicated a close relationship between expression profiles of sham blood and dMCAO blood and relatively more distinct profiles in the dMCAO brain (Supplementary Figure 1(f)).
Ischemic stroke induces moderate transcriptome changes in monocytes/macrophages in the blood
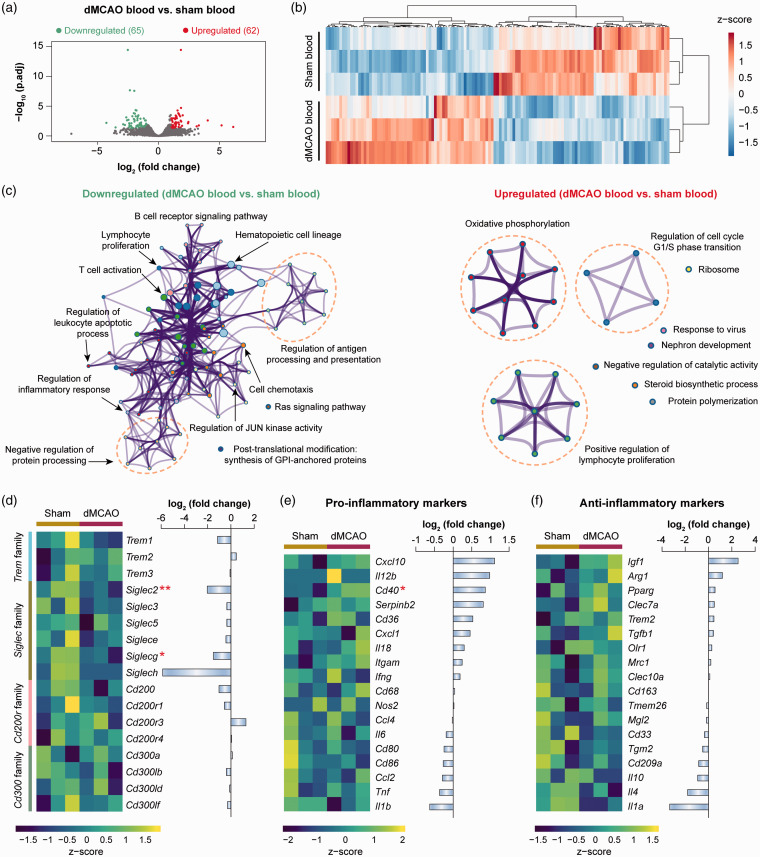
Genomic changes in blood monocytes in response to ischemic stroke were assessed by comparing the RNA-seq expression profiles of cells from dMCAO blood versus sham blood (Figure 1(a)). A total of 127 differentially expressed genes (DEGs; defined as genes with fold change >2 or <−2 and Benjamini-Hochberg adjusted p-value < 0.05) were identified in monocytes from dMCAO versus sham blood (Figure 1(a)). Unsupervised hierarchical clustering of all DEGs illustrated distinct transcriptional signatures between dMCAO blood and sham blood monocytes (Figure 1(b)). To elucidate the functional implications of the DEGs, we performed pathway enrichment analysis on the upregulated and downregulated genes. Nine clusters of functions were enriched in the upregulated DEGs (p-value < 0.01; Figure 1(c) and Supplementary Table 3). The largest cluster was associated with oxidative phosphorylation, which appears to be enhanced in macrophages during M2 phenotype activation.26 In the downregulated DEGs, 12 clusters of functions were overrepresented (Figure 1(c)), encompassing the classical functions of monocytes, such as regulation of antigen processing and presentation and regulation of inflammatory response (Supplementary Table 3).
Figure 1.
Transcriptome changes of blood monocytes after ischemic stroke. Differential gene analysis was performed on RNA-seq data obtained from blood monocytes at five days after dMCAO or sham operation. (a) Volcano plot showing the differentially expressed genes (DEGs; fold change > 2 or < −2, adjusted p-value < 0.05) in monocytes from dMCAO blood versus sham blood. (b) Heatmap with unsupervised hierarchical clustering showing the expression profile of DEGs in dMCAO blood monocytes compared to sham blood monocytes. Each row represents one sample, and each column represents one DEG. The scaled expression value (column z-score) is displayed in a blue-red color scheme with blue and red indicating low and high expression, respectively. (c) Functional enrichment analysis was performed by Metascape on downregulated (left panel) and upregulated (right panel) DEGs in monocytes from dMCAO blood versus sham blood. The significantly overrepresented (p<0.01) ontology terms were grouped into color-coded clusters based on their membership similarities and rendered as network plots. Each node represents an enriched term, and one representative term is shown for each cluster. Terms with a similarity > 0.3 are connected by edges. The complete enrichment results are provided in Supplementary Table 3. (d) Gene expression profiles of selected families of immunoreceptors containing activating and inhibitory members. Left panel: heatmap containing z-scaled gene expression levels. Each column represents one sample. Right panel: summarized gene expression fold changes in monocytes from dMCAO blood versus sham blood. (e, f) Expression profiles of representative genes that are LPS-inducible (pro-inflammatory markers; e) or IL-4-inducible (anti-inflammatory markers; f). Shown are heatmap containing z-scaled expression levels (left panel) and summarized gene expression fold changes (right panel). * and ** indicate adjusted p-value less than 0.05 and 0.01, respectively.
dMCAO: distal middle cerebral artery occlusion.
The strength of immune responses is partly titrated through the signaling of activating and inhibitory receptors on the immune cell surface. Immunoreceptor tyrosine-based activation motifs (ITAMs) and immunoreceptor tyrosine-based inhibition motifs (ITIMs) are two conserved motifs found in the intracellular domain of various signaling proteins and play a pivotal role in the balance between activating and inhibitory signals for immune cells.27 We assessed the expression profiles of a panel of genes encoding immunoreceptors containing ITAMs or ITIMs,12 such as the triggering receptor expressed on myeloid cells (TREM) family, sialic acid-binding immunoglobulin-type lectins (Siglec) family, CD200 receptor family, and CD300 family (Figure 1(d)). In the 17 genes examined, only Siglec2 and Siglecg were significantly downregulated (fold change < −2; adjusted p-value < 0.05) in monocytes from dMCAO versus sham blood (Figure 1(d)), both of which contain ITIMs. Other genes in these receptor families were not differentially expressed between the two monocyte groups (Figure 1(d)). We further determined the immunophenotypes of monocytes from dMCAO blood by assessing the expression levels of genes that could be induced by classical (lipopolysaccharide; LPS) or alternative (interleukin-4; IL-4) activation.12 The results showed that most markers examined were not differentially expressed in monocytes from dMCAO versus sham blood (Figure 1(e) and (f)), suggesting that these cells do not display a specific polarized state. In summary, our data revealed moderate transcriptomic changes in blood monocytes upon ischemic stroke. These cells, however, did not demonstrate features associated with strong activation or inhibition functional state, nor did they possess a polarized pro or anti-inflammatory phenotype.
Monocytes/macrophages bear robust transcriptome changes upon entering the post-ischemic brain
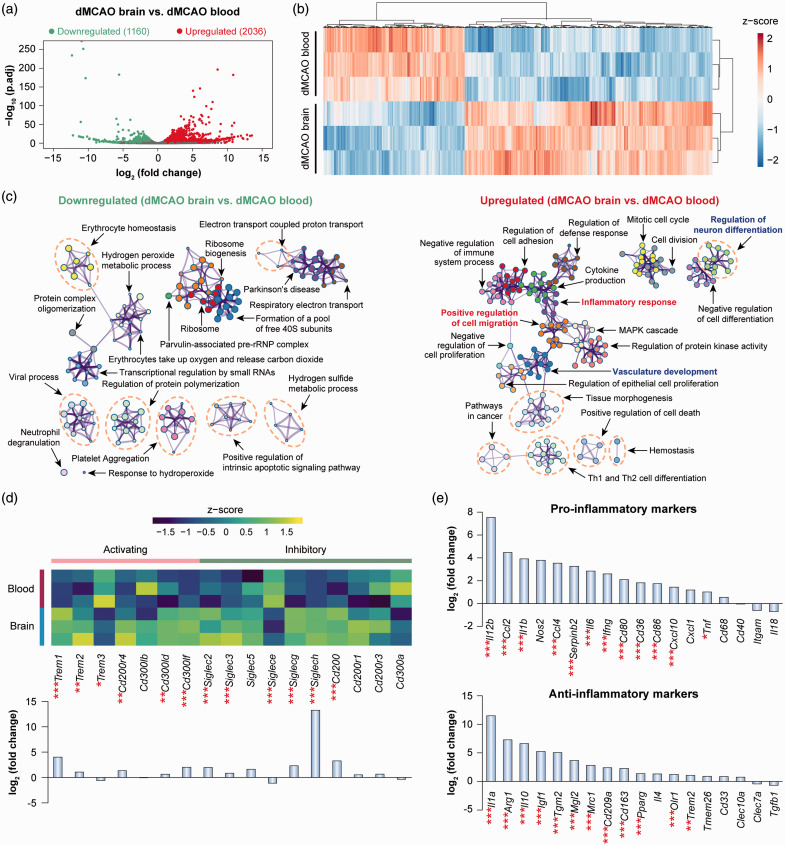
In contrast to the moderate genomic reprogramming occurring in blood monocytes after ischemic stroke, monocytes/macrophages entering the post-stroke brain underwent robust transcriptomic changes. A total of 3196 DEGs (fold change >2 or <−2, adjusted p-value <0.05) were detected in monocytes/macrophages in the dMCAO brain versus blood, of which 2036 were upregulated and 1160 were downregulated (Figure 2(a)). Unsupervised clustering of these DEGs confirmed the distinct transcriptional signatures between dMCAO brain versus blood monocytes/macrophages (Figure 2(b)). Pathway enrichment analysis revealed a total of 218 and 332 ontology terms which were overrepresented (p-value < 0.01) via downregulated and upregulated DEGs, respectively (Figure 2(c) and Supplementary Table 4). A cluster of functions related to cell migration and movement was enriched by the upregulated DEGs in dMCAO brain macrophages (Figure 2(c)), such as positive regulation of cell migration (GO:0030335, p-value = 10−22), positive regulation of cell motility (GO:2000147, p-value = 10−21), and leukocyte chemotaxis (GO:0030595, p-value = 10−15) (Supplementary Table 4), consistent with the intrinsic properties of cells invading the post-ischemic brain. A number of clusters of ontology terms regarding immune and inflammatory responses were overrepresented in upregulated DEGs (Figure 2(c) and Supplementary Table 4), such as inflammatory response (GO:0006954), regulation of innate immune response (GO:0045088), and MAPK cascade (GO:0000165), in line with an established role of these cells in post-stroke neuroinflammation. Notably, such enriched functions encompass both positive regulation (e.g. positive regulation of defense response, positive regulation of toll-like receptor signaling pathway) and negative regulation (e.g. negative regulation of immune system process, negative regulation of cell activation) of inflammatory responses (Supplementary Table 4). Accordingly, the expression of activating and inhibitory immunoreceptors in the brain macrophages demonstrated a mixed profile (Figure 2(d)). We identified both activating and inhibiting receptors that were upregulated in brain macrophages (Figure 2(d)), suggesting a complex and potentially multifaceted role of this cell population. Furthermore, brain macrophages simultaneously upregulated a panel of pro-inflammatory markers as well as anti-inflammatory markers (Figure 2(e)), suggesting that these cells did not possess a specific polarization state. Future studies are warranted to unravel the differential role of macrophages in post-stroke neuroinflammation at various stages after ischemia, and by distinct subpopulations, such as via single-cell RNA-seq approaches.
Figure 2.
Genomic reprogramming of monocytes/macrophages upon invasion of the post-stroke brain. Differential gene analysis was performed on RNA-seq data obtained from monocytes/macrophages in the blood and brain at five days after dMCAO. (a) Volcano plot showing the DEGs (fold change > 2 or < −2, adjusted p-value < 0.05) in macrophages from dMCAO brain versus dMCAO blood. (b) Heatmap with unsupervised hierarchical clustering showing the expression profile of DEGs in macrophages from dMCAO brain compared to dMCAO blood. Each row represents one sample, and each column represents one DEG. The scaled expression value (column z-score) is displayed in a blue-red color scheme. (c) Functional enrichment analysis was performed by Metascape on downregulated (left panel) and upregulated (right panel) DEGs in macrophages from dMCAO brain versus dMCAO blood. The significantly overrepresented (p<0.01) ontology terms were grouped into color-coded clusters based on their membership similarities and rendered as network plots. Shown are the top 20 clusters according to p-values from smallest to largest. The complete enrichment results are provided in Supplementary Table 4. (d) Gene expression profiles of selected families of immunoreceptors containing activating and inhibitory members. Shown are heatmap containing z-scaled gene expression levels (upper panel) and summarized gene expression fold changes in cells from dMCAO brain versus dMCAO blood (lower panel). (e) Expression profiles of representative genes that are LPS-inducible (pro-inflammatory markers) or IL-4-inducible (anti-inflammatory markers). Data are expressed as fold changes in cells from dMCAO brain versus dMCAO blood. *, **, and *** indicate adjusted p-value less than 0.05, 0.01, and 0.001, respectively.
dMCAO: distal middle cerebral artery occlusion.
In addition to the expected macrophage functions related to cell migration and regulation of inflammatory responses, we also discovered that a large number of functions related to brain remodeling and plasticity were enriched in upregulated DEGs in brain macrophages (Figure 2(c) and Supplementary Table 4). These functions can be further categorized into terms related to vascular plasticity (e.g. vasculature development, angiogenesis, endothelial cell proliferation) and neuroplasticity (e.g. regulation of neuron differentiation, regulation of neuron projection development, regulation of axonogenesis). These key findings suggest a previously underappreciated role of brain macrophages in regulating post-stroke neurovascular remodeling.
Genomic reprogramming in brain macrophages favors neurovascular plasticity in the post-ischemic brain
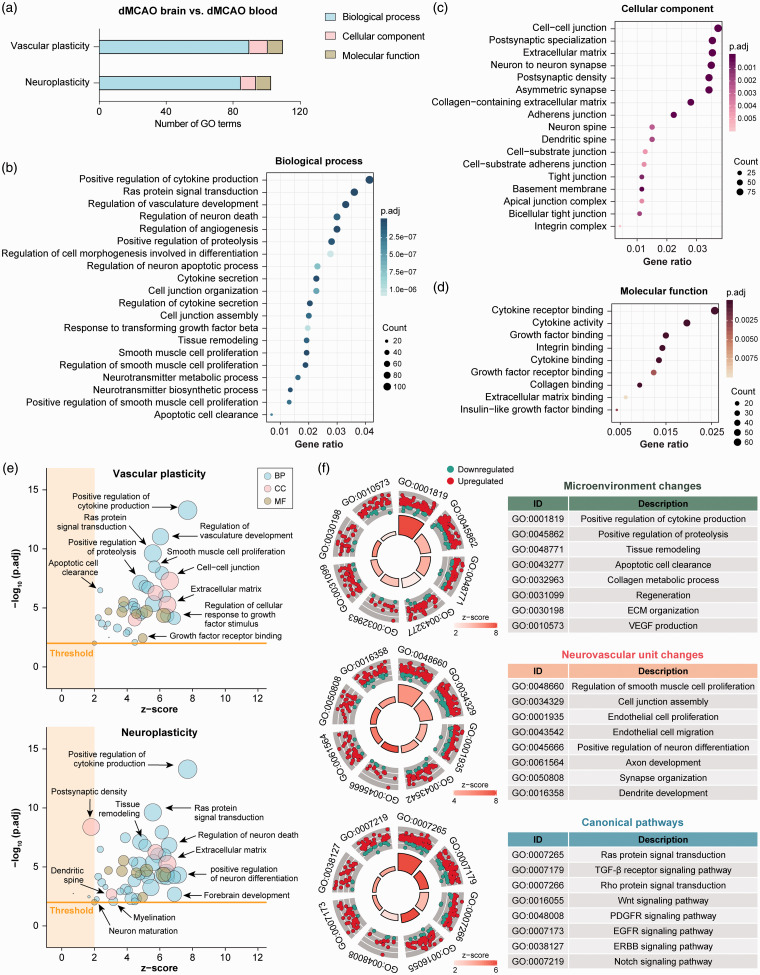
To further characterize the potential role of macrophages in post-stroke neurovascular remodeling and plasticity, DEGs in macrophages in the dMCAO brain versus blood were re-analyzed for gene ontology (GO) enrichment by clusterProfiler on all functions related to vascular plasticity (Supplementary Table 5) and neuroplasticity (Supplementary Table 6). A total of 110 GO terms related to vascular plasticity and 103 terms on neuroplasticity were significantly enriched (Benjamini-Hochberg adjusted p-value < 0.01; Figure 3(a)). These terms belonged to the three GO categories BP (the biological processes culminating from the activities of multiple gene products), CC (the cellular components where gene products are active) and MF (the molecular functions of gene products) (Figure 3(a) to (d)). Various biological processes related to neurovascular remodeling were enriched in the dataset, including regulation of angiogenesis, tissue remodeling, and apoptotic cell clearance (Figure 3(b)). Furthermore, enriched terms under the categories CC and MF also strongly implied that the gene products exert their functions at locations where active neurovascular remodeling occurs (e.g. extracellular matrix, postsynaptic density, and dendritic spine; Figure 3(c)), and that the gene products participate in molecular functions associated with remodeling processes, such as integrin binding, extracellular matrix (ECM) binding, and growth factor receptor binding (Figure 3(d)).
Figure 3.
Transcriptome analysis of macrophages in the post-stroke brain implicates promotion of neurovascular plasticity. Functional enrichment analysis was performed on DEGs in monocytes/macrophages from the dMCAO brain versus dMCAO blood by the R package clusterProfiler. (a) the numbers of significantly overrepresented (adjusted p-value < 0.01) gene ontology (GO) terms in the three categories related to vascular plasticity and neuroplasticity: Biological process (BP), cellular component (CC), and molecular function (MF). (b–d) The top 20 enriched BP (b), CC (c) and MF (d) terms according to the adjusted p-values from smallest to largest. The full list of GO terms is provided in Supplementary Tables 5 and 6. Terms are arranged by the numbers of genes under each term (count). (e) Enriched GO terms related to vascular plasticity or neuroplasticity were filtered according to their gene membership similarities, to reduce the number of redundant terms. Terms with similarity < 0.8 were presented as bubble plots, where the sizes of the bubbles reflect the number of genes under each term. The threshold was set at z-score >2 and adjusted p-value < 0.01. (f) Enriched biological processes were further classified into three clusters: (1) Functions related to microenvironment and extracellular (ECM) changes (upper panel); (2) functions related to changes in the neurovascular unit (middle panel); and (3) canonical pathways (lower panel). Shown are the circular visualizations of eight representative biological processes in each cluster. The DEGs under each biological process were shown as dots which were color-coded according to their direction of change.
dMCAO: distal middle cerebral artery occlusion.
As GO vocabularies are organized as a hierarchy that could contain multiple ascendants or descendants for a term, multiple terms from the same hierarchical path appeared in our enriched GO list on neurovascular plasticity. We reduced the functional redundancies of the enriched terms by deleting all terms that have a gene overlap greater than or equal to 0.8 using the R package GOplot.28 The resulting GO list (Supplementary Table 7) consisted of 41 terms related to vascular plasticity and 50 terms related to neuroplasticity that were predicted to be activated (z-score > 2 and adjusted p-value < 0.01). These ontology terms were largely independent and were associated with various functional aspects of neurovascular plasticity (Figure 3(e)). We further classified the activated biological processes into three functional clusters that are instrumental in the neurovascular remodeling processes after brain injury (Figure 3(f)): (1) Functions related to microenvironment and ECM changes, such as tissue remodeling, apoptotic cell clearance, and ECM organization; (2) Functions related to changes in neurovascular unit components, such as endothelial cell proliferation and migration, and axon and dendrite development; and (3) Canonical pathways involved in angiogenesis and neurogenesis, such as Wnt, EGFR, and Notch signaling pathways.29,30 These biological functions were predicted to be strongly activated (z-score > 2), as visualized by the dominant number of upregulated genes over downregulated genes, under each term (Figure 3(f)). In summary, these data suggest that macrophages in the post-ischemic brain may play critical roles in neurovascular repair and remodeling processes.
Post-stroke brain macrophages upregulate genes strongly linked to neurovascular remodeling and plasticity
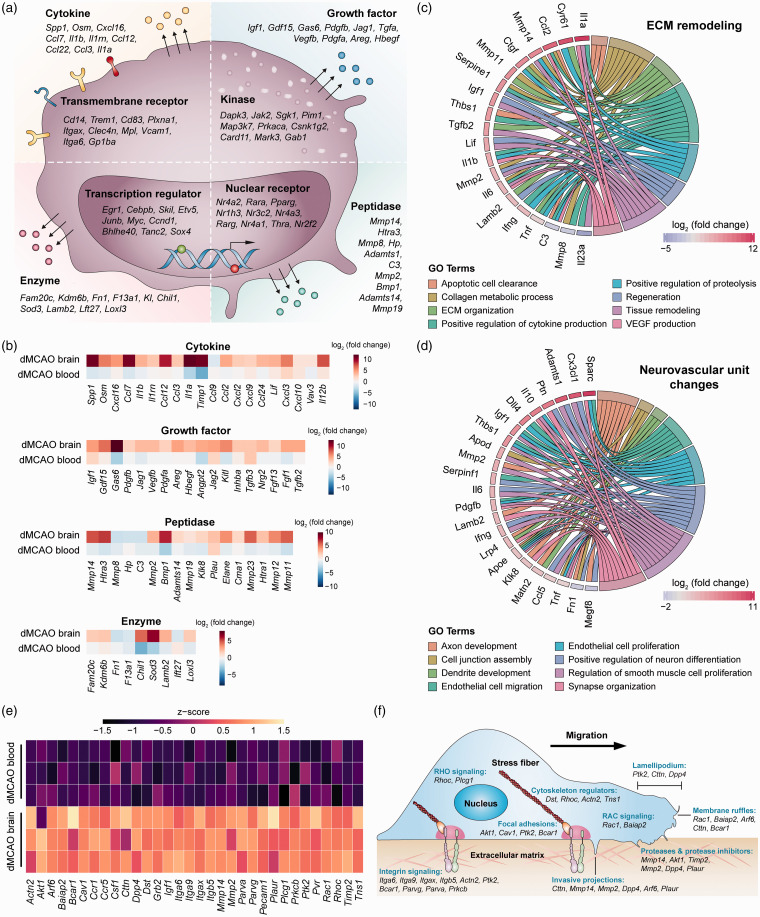
Next, we characterized the transcriptional program associated with neurovascular remodeling at the gene levels. A sum of 1034 DEGs (32% of total DEGs) in dMCAO brain macrophages were associated with enriched GO terms regarding neurovascular plasticity (Supplementary Table 8). To gain further insight into the molecular functions of these genes, we manually annotated them according to the type and subcellular location of the gene products (Figure 4(a)). These products included transcription regulators (e.g. EGR1, SOX4) and ligand-dependent nuclear receptors (e.g. PPARγ, RARα) in the nucleus, kinases in the cytoplasm (e.g. JAK2, MAP3K7), and receptors on the plasma membrane (e.g. CD14, TREM1). Notably, a large number of gene products predicted to be active in the extracellular space were differentially expressed in brain macrophages, including a variety of cytokines, growth factors, peptidases, and enzymes (Figure 4(a)). Cytokines that promote neovascularization, such as osteopontin (Spp1), oncostatin M (Osm), and CXCL2,31–33 were upregulated in macrophages from the dMCAO brain (Figure 4(b)). Growth factors involved in angiogenesis and neurogenesis (e.g. GDF15, VEGF, FGF1),34 and peptidases capable of modulating ECM components (e.g. MMP-14, ELANE) were also upregulated. While all these factors were broadly related to neurovascular remodeling, some of them may participate in different aspects of the remodeling processes. Examples include Il1a, Ccl2, and Mmp14, which were associated with at least two functional subcategories of ECM remodeling (Figure 4(c)). We also identified another panel of genes that could contribute to multiple steps in the restoration of the neurovascular unit, e.g. Igf, Il10, Il6 and Ccl5 (Figure 4(d)).35–37
Figure 4.
Gene-level characterization of macrophage transcriptome changes favoring neurovascular plasticity in the post-stroke brain. DEGs in macrophages from the dMCAO brain versus dMCAO blood were analyzed for their potential influence on post-stroke neurovascular plasticity. (a) DEGs related to neurovascular plasticity were manually annotated according to the type and subcellular localization of gene products. The top 10 genes were shown under each category according to the adjusted p-value, from lowest to highest. The complete gene list is provided in Supplementary Table 8. (b) Heatmaps showing the expression profiles of four clusters of genes whose products are in the extracellular space. Data were expressed as log2 transformation of fold changes in dMCAO brain versus dMCAO blood (first row), and dMCAO blood versus sham blood (second row). (c, d) Genes related to ECM remodeling (c) and neurovascular unit changes (d) were explored for their association with eight functional subcategories. Shown are genes associated with at least two subcategories, displayed as Circos plots. (e) Heatmap of the expression profile of 32 DEGs related to cell movement, cell–cell adhesion, and monocyte–vessel interactions, which were upregulated in macrophages in the dMCAO brain compared to those in the dMCAO blood. (f) The DEGs were manually annotated to functional categories of cell movement and cell–cell interactions.
ECM: extracellular matrix; dMCAO: distal middle cerebral artery occlusion.
In addition to promoting tissue regeneration, recent studies indicate that macrophages may assist with the repair of damaged vessels through direct mechanical contact with the endothelium.38 We identified a panel of genes related to cell movement and adhesion that were highly upregulated after stroke in brain versus blood macrophages (Figure 4(e)). The products of these genes participate in multiple steps essential for canonical cell movement and cell–cell interactions (Figure 4(f)), including cell morphological changes and formation of protrusions (Cttn, Dpp4, Arf6), interaction with the ECM (Mmp2, Mmp14, Timp2, Plaur), formation of focal adhesions (Bcar1, Ptk2, Cav1), and intracellular signaling related to cell polarity (Rhoc, Rac1, Plcg1, Baiap2).39,40 Furthermore, the products of several upregulated genes are known to play a crucial role in monocyte–endothelium interactions,41,42 such as Pecam1, Pvr, Ccr1, and Ccr5 (Figure 4(e)). These genes participate in monocyte trafficking across the blood vessel wall (diapedesis) as well as monocyte–endothelium adhesions during vessel repair. Genes encoding BCAP (Pik3ap1), an essential activator of the PI3K pathway, and the PI3K downstream target Rac1 were significantly upregulated in brain macrophages (Supplementary Table 8). Both PI3K and Rac1 activity are important for vessel repair by macrophages.38 Together, these transcriptomic alterations may underlie the repair-enhancing monocyte/macrophage phenotype in the post-ischemic brain through both contact-dependent and contact-independent mechanisms.
PPARγ is a master switch dictating macrophage reprogramming in the post-stroke brain
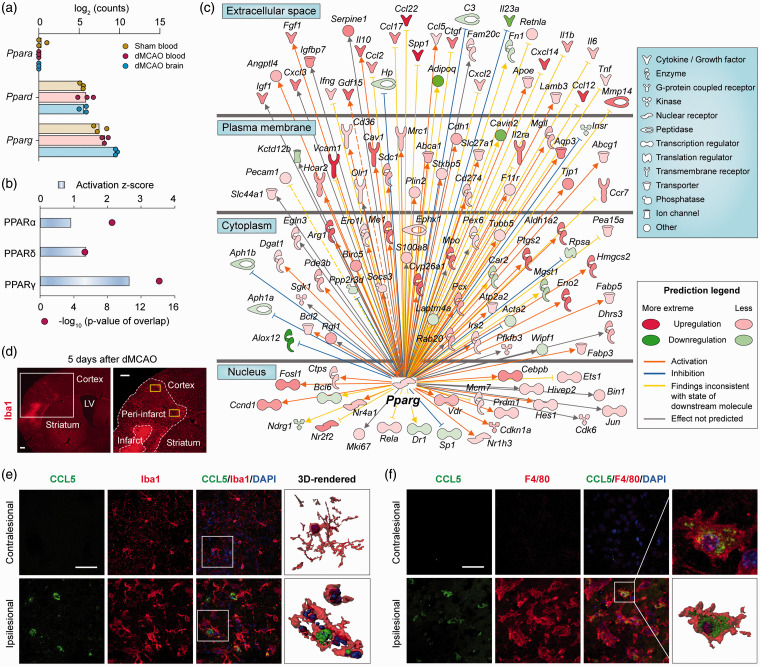
Our data suggest that macrophages reprogram upon entry into the post-stroke brain and might contribute to restorative processes in the neurovascular niche. Next, we sought to identify the master regulators driving their transcriptional reprogramming. PPARs are a family of ligand-activated transcription factors with ubiquitous tissue distributions and a wide variety of target genes and biological functions.43 We examined the expression of all three PPAR-family genes in FACS-sorted cells (Figure 5(a)). Ppara was expressed at negligible levels in all groups, whereas Ppard and Pparg were readily detected in all samples (Figure 5(a)). The expression of Pparg was higher than Ppard in monocytes from sham blood and was further elevated in macrophages from the dMCAO brain. To assess the possibility that PPAR family members are the dominant regulators of differential gene expression in macrophages from dMCAO brain versus blood, we performed Upstream Regulator analysis in IPA.44 Among all PPARs, only PPARγ had an activation z-score larger than 2, indicating robust activation (Figure 5(b)). Accordingly, PPARγ was predicted to be a potent master regulator of the transcriptional program in post-stroke brain macrophages. A total of 120 DEGs were in the molecular network of PPARγ targets, amongst which 87 genes were associated with neurovascular plasticity (Figure 5(c)). The products of these genes include a variety of growth factors (e.g. GDF15, IGF-1, FGF1), cytokines, and peptidase (e.g. osteopontin, IL-10, MMP-14) that exist in the extracellular space, thereby modulating ECM composition and the behavior of other cells to facilitate brain remodeling. At least 12 of the 28 PPARγ-targeted gene products overlapped with the extracellular factors that may participate in neurovascular remodeling (Figure 4(c) and (d)), including Ccl2, Mmp14, Ctgf, Serpine1, Igf1, Il10, and Ccl5. Some of these genes may participate in multiple aspects of neurovascular plasticity (Figure 4(c) and (d)), such as Ccl5, Ccl2, Mmp14, and Il10.
Figure 5.
PPARγ is predicted to regulate the genomic reprogramming of monocytes/macrophages after ischemic stroke. (a) RNA-seq expression profiles of the PPAR family genes Ppara, Ppard and Pparg in monocytes/macrophages from sham blood, dMCAO blood, and dMCAO brain groups. n=3 biological replicates per group. (b, c) Upstream regulator analyses were performed by Ingenuity Pathway Analysis (IPA) on all DEGs in macrophages from dMCAO brain versus dMCAO blood. (b) The activation z-score and p-value of overlap were calculated for each PPAR as a potential upstream regulator. The cutoff values for predicated activation were z-score > 2 and p-value < 0.01. (c) DEGs that are regulated by Pparg are shown in a network view, with annotations on the subcellular localization and types of their products. (d–f) The expression of CCL5 was examined by immunofluorescence staining at five days after dMCAO in the ipsilesional peri-infarct area and the corresponding region in the non-injured contralateral hemisphere. (d) Representative images demonstrate the peri-infarct area defined by Iba1 (red) immunostaining. Yellow rectangles illustrate the regions where images in (e) and (f) were captured. Scale bars: 50 µm. (e, f) Double-label immunostaining of CCL5 with Iba1 (e) or F4/80 (f). Nuclei were counterstained with DAPI. Squares illustrate the regions that were enlarged and 3D-rendered in the fourth column. Scale bars: 50 µm. Robust CCL5 immunosignal was detected in Iba1+ or F4/80+ cells in the ipsilesional cortex, which was absent in the contralesional side.
dMCAO: distal middle cerebral artery occlusion; LV: lateral ventricle.
Using dual immunofluorescence staining of CCL5 and Iba1, we verified stroke-induced CCL5 expression in brain myeloid cells at five days after dMCAO (Figure 5(d)). No CCL5 immunosignal was detected in the non-injured contralesional cortex, where most Iba1+ cells displayed the ramified morphology of resident microglia (Figure 5(e)). In contrast, robust CCL5 immunosignal was observed in Iba1+ cells in the ipsilesional peri-infarct cortex, and these cells demonstrated the amoeboid morphology typical of activated microglia or infiltrated macrophages (Figure 5(e)). Similar findings were obtained for CCL5 colocalization with a more specific macrophage marker F4/80 (Figure 5(f)). CCL5 and F4/80 immunosignal was negligible in the contralateral cortex at five days after dMCAO, whereas massive accumulation of CCL5+F4/80+ cells occurred in the ipsilesional cortex (Figure 5(f)).
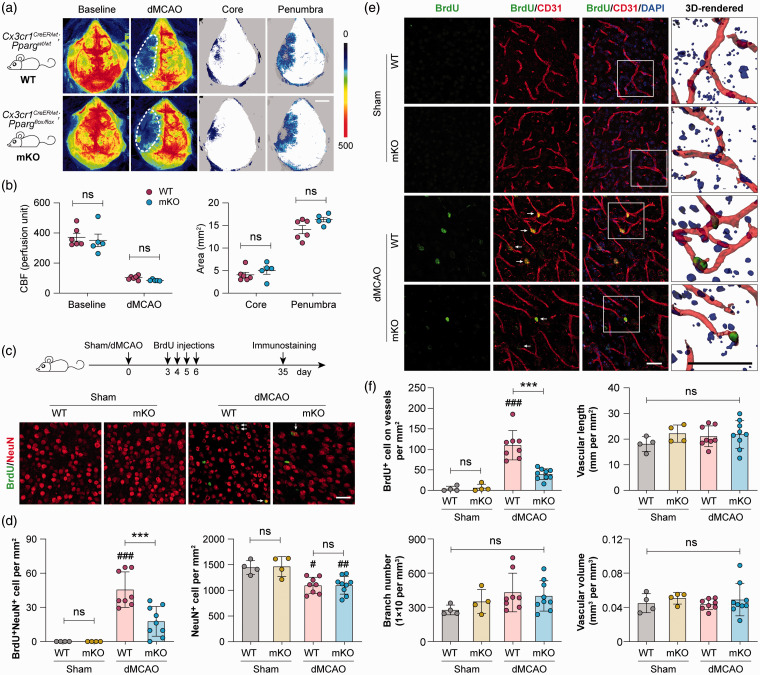
Selective deletion of PPARγ in myeloid-lineage cells hampers post-stroke neurovascular repair
To determine whether macrophage/microglial expression of PPARγ indeed enhances post-stroke neurovascular remodeling, we investigated the effect of conditional PPARγ knockout on two well-characterized brain repair processes – neurogenesis and angiogenesis. To this end, myeloid cell-specific PPARγ knockout (PPARγ mKO) mice were generated by crossing the Cx3cr1CreER mice45 and PPARγloxP mice.46 dMCAO was induced in PPARγ mKO mice and WT control mice 14 days following the first tamoxifen injection. All mice demonstrated similar levels of baseline cortical CBF and dMCAO-induced CBF reductions (Figure 6(a) and (b)), suggesting that PPARγ mKO did not alter the magnitudes of initial ischemic insult. All mice received BrdU injections at 3–6 days after dMCAO to label newly proliferated cells, and neurogenesis and angiogenesis were examined at 35 days after dMCAO by double-label immunostaining of BrdU with the neuronal marker NeuN (Figure 6(c) and (d)) or the endothelial marker CD31 (Figure 6(e) and (f)), respectively. At 35 days after dMCAO, a mild neurogenic response was observed in WT mice, and BrdU and NeuN double-positive cells were observed in the ipsilesional peri-infarct cortex (Figure 6(c)). In PPARγ mKO mice, BrdU+NeuN+ cells were much less frequently detected than in WT mice (Figure 6(c) and (d)), despite that the numbers of BrdU+NeuN+ cells were small in both groups (Figure 6(d)). The numbers of total NeuN+ viable neurons were comparable between PPARγ mKO mice and WT mice under both sham and dMCAO conditions (Figure 6(c) and (d)). There was also no difference in the numbers of BrdU+DCX+ neural progenitor cells between PPARγ mKO mice and WT mice under both sham and dMCAO conditions (data not shown).
Figure 6.
Selective deletion of PPARγ in myeloid-lineage cells impairs neurovascular plasticity after ischemic stroke. Focal cerebral ischemia was induced in myeloid cell-specific PPARγ knockout (PPARγ mKO) mice and wild-type (WT) control mice by dMCAO. (a) Two-dimensional laser speckle images showing cortical cerebral blood flow (CBF) before (baseline) and 30 min after dMCAO. Ischemic core (areas with CBF reduction of >70% of baseline) and penumbra (areas with CBF reduction of 50–70% of baseline) were illustrated by blue dots. Scale bar: 3 mm. (b) Summarized CBF data showing that PPARγ mKO did not significantly alter CBF changes during dMCAO. n= 5–6 mice per group. ns: no significant difference. (c–f) WT and PPARγ mKO mice received BrdU injections at 3–6 days after dMCAO or sham operation to label newly proliferated cells. Neurogenesis and angiogenesis were examined at 35 days after dMCAO. (c) Double-label immunofluorescence of BrdU and the neuronal marker NeuN in the peri-infarct cortex. Arrow: BrdU and NeuN double-positive cell. Scale bar: 50 µm. (d) The numbers of BrdU and NeuN double-positive cells (left panel) and total NeuN-positive cells (right panel) were counted. (e) Double-label immunofluorescence of BrdU and the endothelial marker CD31 in the peri-infarct cortex. Nuclei were counterstained with DAPI. Arrow: BrdU-positive cell on CD31-positive vessels. Squares indicate the regions that were enlarged and 3D-rendered in the fourth column, Scale bars: 50 µm. (f) Summarized data on the numbers of BrdU-positive cells on vessels and vascular branch numbers, lengths and volumes. n=6 mice per sham group. n=8–9 mice per dMCAO group. #p<0.05, ##p<0.01, ###p<0.001 dMCAO vs. sham. ***p<0.001 mKO dMCAO vs. WT dMCAO.
dMCAO: distal middle cerebral artery occlusion; ns: no significant difference.
Similar to the neurogenic responses, a moderate angiogenic response was induced by dMCAO in WT mice. BrdU+ cells on CD31+ microvessels were frequently observed in the peri-infarct cortex of WT mice at 35 days after dMCAO (Figure 6(e)), suggesting the existence of endogenous vascular restorative responses. Remarkably, angiogenesis was largely inhibited in PPARγ mKO mice, which had a significantly smaller number of BrdU+ cells on vessels than WT mice (Figure 6(f)). The gross morphology of microvessels, such as vascular branch number, vascular length, and volume did not differ between WT and PPARγ mKO mice under either sham or dMCAO conditions (Figure 6(f)). Together, these data suggest that ischemia-induced endogenous neurovascular restoration was impaired in the absence of PPARγ in myeloid cells.
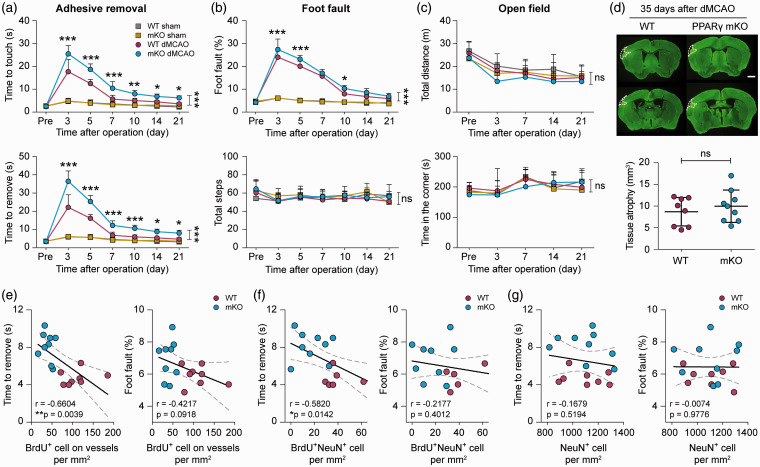
Deficiency of neurovascular plasticity is associated with poor functional recovery in PPARγ mKO mice after ischemic stroke
To determine whether myeloid cell-specific PPARγ knockout influences long-term neurological functions after ischemic stroke—the primary outcome assessed in stroke patients—we performed a battery of neurobehavioral tests in these mice after dMCAO. dMCAO induced prominent sensorimotor deficits in WT versus sham-operated mice, according to the adhesive removal test (Figure 7(a)) and foot fault test (Figure 7(b)). PPARγ mKO mice demonstrated comparable performance with WT control mice after non-ischemic sham operation (Figure 7(a) and (b)). However, PPARγ mKO mice performed significantly worse than WT mice after dMCAO, as reflected by longer latencies to touch and remove the adhesive tape (Figure 7(a)), and increased foot faults during the grid walking task (Figure 7(b)). Locomotor and anxiety-like behavior examined by the open field test were not different between sham- and dMCAO-operated mice (Figure 7(c)). At 35 days after dMCAO, PPARγ mKO mice developed similar degrees of brain atrophy compared to WT mice (Figure 7(d)). Together, these results demonstrate that PPARγ expression in myeloid cells alleviates long-term sensorimotor functional deficits after ischemic stroke.
Figure 7.
Deficient neurovascular plasticity is associated with poor functional recovery in PPARγ mKO mice after ischemic stroke. PPARγ mKO mice and WT control mice were subjected to dMCAO or sham operation. (a–c) Neurological functions were assessed before (pre) and up to 21 days after dMCAO or sham operation, with a battery of behavior tests. (a) Forepaw sensitivity and motor impairments were assessed by the adhesive removal test. Shown are the time the mouse spent to touch the adhesive tape (upper panel) and completely remove the tape (lower panel). (b) Sensorimotor coordination was assessed by the foot fault test. Foot faults were expressed as percentages of total steps (upper panel). The numbers of total steps (lower panel) were comparable among the four groups, indicating similar gross locomotor functions in all mice. (c) General locomotor activity and anxiety-like behavior were assessed by the open field test. Shown are the total distances the mouse traveled (upper panel) and the time the mouse spent in the corner zones (lower panel). (d) Chronic tissue atrophy was assessed at 35 days after dMCAO on MAP2-immunostained coronal brain sections. Representative images of MAP2 immunofluorescence (green) are shown with cortical tissue atrophy demarcated by dashed lines in the left hemisphere. Scale bar: 1 mm. The volumes of tissue atrophy were not different between WT and PPARγ mKO mice. n=6 mice per sham group. n=8–9 mice per dMCAO group. *p<0.05, **p<0.01, ***p<0.001 mKO dMCAO vs. WT dMCAO by one-way ANOVA (individual time point) or two-way repeated measures ANOVA (bracket). (e–g) Pearson correlation between the animals’ performance in the adhesive removal test (time to remove; left panel) and foot fault test (percentage of foot fault; right panel) at 21 days after dMCAO and the number of BrdU-positive cells on vessels (e), BrdU and NeuN double-positive cells (f), and total NeuN-positive cells (g) at 35 days after dMCAO in WT and PPARγ mKO mice. Dashed lines: 95% confidence intervals. n=8 and 9 mice for WT and PPARγ mKO groups, respectively. *p<0.05, **p<0.01.
dMCAO: distal middle cerebral artery occlusion; ns: no significant difference.
Importantly, neurovascular remodeling responses in the post-stroke brain were correlated with behavioral performance. The numbers of BrdU+ cells along vessels were negatively correlated with sensorimotor deficits detected by the adhesive removal test (Figure 7(e), left panel) and showed a trend toward negative correlation (p = 0.09) with the percentage of foot faults (Figure 7(e), right panel) at 21 days after MCAO. The numbers of BrdU+NeuN+ cells showed significant negative correlations (r=−0.5820, p = 0.0142) with the latency to remove the adhesive tape (Figure 7(f)). The total number of NeuN+ cells did not significantly correlate with the animals’ behavioral performance in either test (Figure 7(g)). In summary, these results suggest, but do not prove, that an impairment in post-stroke angiogenesis contributes to poor functional recovery of PPARγ mKO mice.
Discussion
This study compared the transcriptomic profiles of CD11b+CD45high cells in the blood and brain after ischemic stroke and under non-stroke control conditions, using whole-genome RNA-seq profiling. The results demonstrate that post-stroke brain CD11b+CD45high cells have a distinct transcriptome program compared to their counterparts in the blood and support the view that these versatile cells facilitate neurovascular remodeling.
Monocytes are continuously generated in the bone marrow and are released into peripheral blood during adult hematopoiesis. Following ischemic stroke, brain-derived antigens are released into the periphery and trigger prominent monocytosis.47 Not only do monocytes play essential roles in innate immunity, they also profoundly impact the adaptive arm of the peripheral immune system. For example, monocytes contribute to post-stroke immunodepression by suppressing lymphocyte activity through soluble factors, such as CD163 and CD14.48 We did not detect dramatic transcriptomic changes in blood monocytes following ischemic stroke (only 127 DEGs). Furthermore, this mild genomic change did not support an activation or inhibition state for these cells nor did these cells acquire conventional polarization phenotypes. At the subacute stages (3–7 days) after stroke, while peripheral immune responses subside, brain infiltration reaches its peak.22,49,50 It is possible that a fraction of activated monocytes migrates into the post-stroke brain. It should be noted that our analysis of peripheral monocytes is limited to those circulating in the blood, and, therefore, does not represent the full extent of the peripheral immune response after stroke. Future studies are warranted to examine the profiles of monocytes in other peripheral immune organs, such as the spleen and bone marrow, to gain further insights into post-stroke innate immune responses in the periphery.
We discovered dramatic transcriptomic differences between CD11b+CD45high cells in the post-stroke brain and blood. Functions related to the regulation of inflammatory responses and cell migration were enriched, in line with the mobilization and homing of these cells to the brain. Our most noteworthy finding was the pro-neurovascular remodeling effects of brain CD11b+CD45high cells. Monocyte-derived macrophages were long believed to be detrimental in the post-stroke brain by exacerbating neuroinflammation, until evidence in the past decade revealed their beneficial functions in CNS repair. Monocytes can be primed to a neuroprotective phenotype in vitro51 and are important players in reparative activities such as phagocytosis and resolution of inflammation.52,53 In addition to these classical monocyte functions, our study provides whole-genome evidence supporting the existence of repair-enhancing macrophage phenotypes, which may include both contact-dependent and contact-independent mechanisms. First, a large number of genes participating in cell movement and cell–cell interactions were upregulated, consistent with previous findings that macrophages repair damaged vessels through direct mechanical contact with the endothelium.38 Second, a panel of trophic cytokines and growth factors were produced by brain macrophages, which may facilitate neurovascular remodeling and repair. Together, these changes shaped a highly active macrophage phenotype whose influence may extend to other types of cells in the CNS beyond the aforementioned roles. For example, astrocytes critically regulate post-stroke neurorestorative responses and closely interact with infiltrating monocytes/macrophages.54,55 The enhanced cell movement and adhesion functions of brain macrophages may implicate more interactions with astrocytes. Whether these two types of cells play independent, synergistic, or antagonistic roles in brain repair are important topics for future studies.
The robust genomic differences between monocytes/macrophages in the post-stroke brain and blood suggest that the functional roles of brain-specific macrophages are complex, perhaps explaining the discrepancies amongst previous studies investigating the effects of monocyte/macrophage depletion on stroke outcomes. Instead of deleting the cells, we propose an alternative approach to dissect the beneficial versus detrimental molecular functional clusters and to manipulate the upstream regulators of macrophage phenotype. Our analysis predicted PPARγ to be a master switch whose downstream targets are involved in the brain remodeling processes. The beneficial role of PPARγ in the post-stroke brain is likely to be multidimensional. For example, PPARγ is critical in the respiratory burst and resolution of inflammation in macrophages.56–58 Therefore, PPARγ may promote the macrophage phagocytosis and apoptotic cell clearance, which might synergize with the production of anti-inflammatory and trophic factors and facilitate brain remodeling. Many PPARγ downstream genes are known to contribute to the neurovascular remodeling, such as Igf1, Gdf15, Spp1, all of which promote angiogenesis and neurogenesis.31,59 The predictions from our bioinformatics analyses were supported by mechanistic in vivo experiments, whereby myeloid cell-specific PPARγ conditional knockout inhibited the endogenous neurovascular restorative responses and impaired post-stroke functional recovery. Notably, a natural switch of macrophage phenotype from pro‐regenerative to pro-inflammatory roles occurs at subacute stage (seven days) after stroke.60 Persistent activation of PPARγ may be a promising approach to maintain a reparative phenotype of invading macrophages at subacute to chronic stages after ischemic stroke. A limitation of our conditional PPARγ knockout mouse model is that all myeloid cells are targeted by the Cx3cr1 promoter. As a result, we cannot yet differentiate between microglia and macrophage PPARγ. In addition, there are other upstream regulators, besides PPAR, that could be explored as well to develop a strategy simultaneously manipulating multiple functional clusters in myeloid cells.
Endogenous brain remodeling processes play a pivotal role in the spontaneous recovery after stroke.61 However, such endogenous efforts are insufficient to repair the injured brain for satisfactory recovery of function.62 Stimulating the endogenous restorative responses in the post-stroke brain is therefore a promising approach to act early and achieve long-term rehabilitative benefits.62 To this end, we found genomic changes in brain macrophages that favor brain repair processes as early as five days after stroke, consistent with previous observations that angiogenesis after stroke is correlated with increased numbers of macrophages.63 The newly formed microvessels may further facilitate macrophage infiltration and removal of necrotic brain tissues,63 commencing a self-perpetuating restorative cycle. In the present study, post-stroke angiogenesis, but not the number of surviving NeuN+ cells, were correlated with the recovery of neurological functions, suggesting that other factors may determine functional outcomes, including the fine structural and functional integrity of surviving neurons and white matter functionality.64,65 The trophic factors produced by macrophages may also be beneficial to injured white matter. It should be noted that our RNA-seq profiling was performed at a single time point (five days) after dMCAO. This snapshot may not catch all characteristics of the macrophage genome in the post-stroke brain, as the functional roles of macrophages may differ at different injury stages after stroke. Future studies are warranted to examine the full temporal profiles of macrophage genomic changes and the repair responses in the brain at subacute and chronic stages after brain ischemia.
The increasing application of unbiased, high-throughput gene expression profiling approaches has greatly advanced our understanding on the pathophysiology of stroke.66,67 The present study performed bulk RNA-seq on FACS-sorted cells, and therefore had limited capability in differentiating the subpopulations of cells within each sample. Brain CD11b+CD45low cells (primarily resident microglia) can upregulate CD45 after ischemic stroke and become indistinguishable from the CD11b+CD45high cells in CNS border regions or the periphery.68 This may explain our observation that samples from the post-stroke brain expressed higher levels of microglia signature genes. To fully understand the functions of different brain myeloid cells after stroke, future studies need to compare the CD11b+CD45high cells with the CD11b+CD45low cells. Nevertheless, the majority of CD11b+CD45high cells sorted from the post-stroke brain are likely to originate in the periphery, as supported by their high expression of prototypical macrophage markers and markers exclusive for infiltrating immune cells (but not CNS myeloid cells), such as CD44.69 The complex cellular composition of brain myeloid cells may be addressed in future studies employing single-cell RNA-seq. Furthermore, the present study used a permanent ischemia model without acute reperfusion. With recent advances in reperfusion therapies against ischemic strokes,70–72 conclusions drawn from this study should be tested in other stroke models in future research efforts.
In summary, the present study has uncovered novel restorative properties of brain-invading macrophages, which implies that the traditionally negative views of brain inflammation may need to be revised. If invading macrophages facilitate brain tissue repair in human stroke survivors, boosting these natural functions would be expected to raise neurological recovery rates. A considerable advantage of leveraging macrophages clinically to access the difficult-to-reach brain in stroke victims might be their easy accessibility from peripheral blood samples, followed by potential ex vivo manipulation, prior to autologous intravenous injections. The timeframes of the genomic changes in peripheral and brain macrophages in the present study (five days post-stroke) are consistent with the technical feasibility of the latter approach, although the temporal kinetics of the response needs to be confirmed in humans. Brain cells cannot be easily collected from stroke survivors, but our data on peripheral monocyte/macrophages might guide future RNA-seq studies of human blood macrophages after stroke.73 Based on the present findings, human monocytes/macrophage biomarker studies74,75 are warranted to help achieve successful translation of novel stroke recovery therapies.
Supplemental Material
Supplemental material, JCB888630 Supplementary material1 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material2 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material3 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material4 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material5 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material6 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material7 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
This project used the University of Pittsburgh HSCRF Genomics Research Core RNA sequencing service. Data analysis was performed using Ingenuity Pathway Analysis software licensed through the Molecular Biology Information Service of the Health Sciences Library System, University of Pittsburgh. We thank Patricia Strickler for administrative support.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the University of Pittsburgh Medical Center and the American Heart Association. This work was supported by the University of Pittsburgh Medical Center (UPMC) Immune Transplant and Therapy Center grant (to J.C., L.R.W., Y.S. and X.H.) and the American Heart Association grant 17SDG33630130 (to Y.S.). Y.S. was also supported by the Competitive Medical Research Fund of the UPMC Health System and the Pittsburgh Institute of Brain Disorders & Recovery startup funds. J.C. is the Richard King Mellon Professor of Neurology at the University of Pittsburgh and also supported by the Senior Research Career Scientist Award from the Department of Veterans Affairs. L.R.W. is the Henry B. Higman Professor of Neurology at the University of Pittsburgh.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
YS and JC designed the research. RW, YL, QY, SHH, JZ and SL performed the experiments. YS, JC, RW and YL analyzed and/or interpreted the data. Y.S. wrote the paper. XH, RKL, MR, LRW and JC critically revised the paper.
Supplemental material
Supplemental material for this article is available online.
References
- 1.Prinz M, Erny D, Hagemeyer N.Ontogeny and homeostasis of CNS myeloid cells. Nat Immunol 2017; 18: 385–392. [DOI] [PubMed] [Google Scholar]
- 2.Jiang X, Andjelkovic AV, Zhu L, et al. Blood-brain barrier dysfunction and recovery after ischemic stroke. Prog Neurobiol 2018; 163-164: 144–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An C, Shi Y, Li P, et al. Molecular dialogs between the ischemic brain and the peripheral immune system: dualistic roles in injury and repair. Prog Neurobiol 2014; 115: 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Planas AM.Role of immune cells migrating to the ischemic brain. Stroke 2018; 49: 2261–2267. [DOI] [PubMed] [Google Scholar]
- 5.Hu X, Leak RK, Shi Y, et al. Microglial and macrophage polarization-new prospects for brain repair. Nat Rev Neurol 2015; 11: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perego C, Fumagalli S, Zanier ER, et al. Macrophages are essential for maintaining a M2 protective response early after ischemic brain injury. Neurobiol Dis 2016; 96: 284–293. [DOI] [PubMed] [Google Scholar]
- 7.Wattananit S, Tornero D, Graubardt N, et al. Monocyte-derived macrophages contribute to spontaneous long-term functional recovery after stroke in mice. J Neurosci 2016; 36: 4182–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt A, Strecker JK, Hucke S, et al. Targeting different monocyte/macrophage subsets has no impact on outcome in experimental stroke. Stroke 2017; 48: 1061–1069. [DOI] [PubMed] [Google Scholar]
- 9.Laterza C, Wattananit S, Uoshima N, et al. Monocyte depletion early after stroke promotes neurogenesis from endogenous neural stem cells in adult brain. Exp Neurol 2017; 297: 129–137. [DOI] [PubMed] [Google Scholar]
- 10.Michaud JP, Pimentel-Coelho PM, Tremblay Y, et al. The impact of Ly6Clow monocytes after cerebral hypoxia-ischemia in adult mice. J Cereb Blood Flow Metab 2014; 34: e1–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y, Li Y, Jiang L, et al. Macrophage depletion reduced brain injury following middle cerebral artery occlusion in mice. J Neuroinflammation 2016; 13: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabert K, Michoel T, Karavolos MH, et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci 2016; 19: 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Hove H, Martens L, Scheyltjens I, et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci 2019; 22: 1021–1035. [DOI] [PubMed] [Google Scholar]
- 14.Stetler RA, Gao Y, Leak RK, et al. APE1/Ref-1 facilitates recovery of gray and white matter and neurological function after mild stroke injury. Proc Natl Acad Sci USA 2016; 113: E3558–E3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilkenny C, Browne W, Cuthill IC, et al. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 2010; 160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suenaga J, Hu X, Pu H, et al. White matter injury and microglia/macrophage polarization are strongly linked with age-related long-term deficits in neurological function after stroke. Exp Neurol 2015; 272: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pu H, Shi Y, Zhang L, et al. Protease-independent action of tissue plasminogen activator in brain plasticity and neurological recovery after ischemic stroke. Proc Natl Acad Sci USA 2019; 116: 9115–9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, Jiang X, Zhang L, et al. Endothelium-targeted overexpression of heat shock protein 27 ameliorates blood-brain barrier disruption after ischemic brain injury. Proc Natl Acad Sci USA 2017; 114: E1243–E1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher M, Feuerstein G, Howells DW, et al. Update of the stroke therapy academic industry roundtable preclinical recommendations. Stroke 2009; 40: 2244–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Mu H, Liu L, et al. Transient selective brain cooling confers neurovascular and functional protection from acute to chronic stages of ischemia/reperfusion brain injury. J Cereb Blood Flow Metab 2019; 39: 1215–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Zhang W, Gao X, et al. Preconditioning with partial caloric restriction confers long-term protection against grey and white matter injury after transient focal ischemia. J Cereb Blood Flow Metab 2019; 39: 1394–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benakis C, Garcia-Bonilla L, Iadecola C, et al. The role of microglia and myeloid immune cells in acute cerebral ischemia. Front Cell Neurosci 2014; 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E, Yang J, Beltran CD, et al. Role of spleen-derived monocytes/macrophages in acute ischemic brain injury. J Cereb Blood Flow Metab 2014; 34: 1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu ZJ, Chen C, Li FW, et al. Splenic responses in ischemic stroke: new insights into stroke pathology. CNS Neurosci Ther 2015; 21: 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickman SE, Kingery ND, Ohsumi TK, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci 2013; 16: 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer PA, Ravi S, Chacko B, et al. A review of the mitochondrial and glycolytic metabolism in human platelets and leukocytes: implications for their use as bioenergetic biomarkers. Redox Biol 2014; 2: 206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Billadeau DD, Leibson PJ.ITAMs versus ITIMs: striking a balance during cell regulation. J Clin Invest 2002; 109: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter W, Sanchez-Cabo F, Ricote M.GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015; 31: 2912–2914. [DOI] [PubMed] [Google Scholar]
- 29.Clevers H.Wnt/beta-catenin signaling in development and disease. Cell 2006; 127: 469–480. [DOI] [PubMed] [Google Scholar]
- 30.Roca C, Adams RH.Regulation of vascular morphogenesis by Notch signaling. Genes Dev 2007; 21: 2511–2524. [DOI] [PubMed] [Google Scholar]
- 31.Rowe GC, Raghuram S, Jang C, et al. PGC-1alpha induces SPP1 to activate macrophages and orchestrate functional angiogenesis in skeletal muscle. Circ Res 2014; 115: 504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasse M, Pourtau J, Trochon V, et al. Oncostatin M induces angiogenesis in vitro and in vivo. Arterioscler Thromb Vasc Biol 1999; 19: 1835–1842. [DOI] [PubMed] [Google Scholar]
- 33.Mehrad B, Keane MP, Strieter RM.Chemokines as mediators of angiogenesis. Thromb Haemost 2007; 97: 755–762. [PMC free article] [PubMed] [Google Scholar]
- 34.Hermann DM, Zechariah A.Implications of vascular endothelial growth factor for postischemic neurovascular remodeling. J Cereb Blood Flow Metab 2009; 29: 1620–1643. [DOI] [PubMed] [Google Scholar]
- 35.Corey S, Abraham DI, Kaneko Y, et al. Selective endovascular cooling for stroke entails brain-derived neurotrophic factor and splenic IL-10 modulation. Brain Res 2019; 1722: 146380. [DOI] [PubMed] [Google Scholar]
- 36.Gertz K, Kronenberg G, Kalin RE, et al. Essential role of interleukin-6 in post-stroke angiogenesis. Brain 2012; 135: 1964–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neal EG, Acosta SA, Kaneko Y, et al. Regulatory T-cells within bone marrow-derived stem cells actively confer immunomodulatory and neuroprotective effects against stroke. J Cereb Blood Flow Metab 2019; 39: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Wu C, Yang Q, et al. Macrophages mediate the repair of brain vascular rupture through direct physical adhesion and mechanical traction. Immunity 2016; 44: 1162–1176. [DOI] [PubMed] [Google Scholar]
- 39.Lawson CD, Ridley AJ.Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol 2018; 217: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayor R, Etienne-Manneville S.The front and rear of collective cell migration. Nat Rev Mol Cell Biol 2016; 17: 97–109. [DOI] [PubMed] [Google Scholar]
- 41.Gerhardt T, Ley K.Monocyte trafficking across the vessel wall. Cardiovasc Res 2015; 107: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang J, Andjelkovic AV, Zhou N, et al. Is there a central role for the cerebral endothelium and the vasculature in the brain response to conditioning stimuli? Cond Med 2018; 1: 220–232. [PMC free article] [PubMed] [Google Scholar]
- 43.Cai W, Yang T, Liu H, et al. Peroxisome proliferator-activated receptor gamma (PPARgamma): a master gatekeeper in CNS injury and repair. Prog Neurobiol 2018; 163–164: 27–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kramer A, Green J, Pollard J, Jr, et al. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014; 30: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkhurst CN, Yang G, Ninan I, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 2013; 155: 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He W, Barak Y, Hevener A, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA 2003; 100: 15712–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaito M, Araya S, Gondo Y, et al. Relevance of distinct monocyte subsets to clinical course of ischemic stroke patients. PLoS One 2013; 8: e69409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connell GC, Tennant CS, Lucke-Wold N, et al. Monocyte-lymphocyte cross-communication via soluble CD163 directly links innate immune system activation and adaptive immune system suppression following ischemic stroke. Sci Rep 2017; 7: 12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Zhu ZY, Huang TT, et al. The peripheral immune response after stroke – a double edge sword for blood-brain barrier integrity. CNS Neurosci Ther 2018; 24: 1115–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Xuan W, Zhu ZY, et al. The evolving role of neuro-immune interaction in brain repair after cerebral ischemic stroke. CNS Neurosci Ther 2018; 24: 1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Bonilla L, Brea D, Benakis C, et al. Endogenous protection from ischemic brain injury by preconditioned monocytes. J Neurosci 2018; 38: 6722–6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woo MS, Yang J, Beltran C, et al. Cell Surface CD36 Protein in monocyte/macrophage contributes to phagocytosis during the resolution phase of ischemic stroke in mice. J Biol Chem 2016; 291: 23654–23661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim E, Cho S.Microglia and monocyte-derived macrophages in stroke. Neurotherapeutics 2016; 13: 702–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z, Chopp M.Astrocytes, therapeutic targets for neuroprotection and neurorestoration in ischemic stroke. Prog Neurobiol 2016; 144: 103–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frik J, Merl-Pham J, Plesnila N, et al. Cross-talk between monocyte invasion and astrocyte proliferation regulates scarring in brain injury. EMBO Rep 2018; 19: e45294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo B, Wang J, Liu Z, et al. Phagocyte respiratory burst activates macrophage erythropoietin signalling to promote acute inflammation resolution. Nat Commun 2016; 7: 12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo B, Gan W, Liu Z, et al. Erythropoeitin signaling in macrophages promotes dying cell clearance and immune tolerance. Immunity 2016; 44: 287–302. [DOI] [PubMed] [Google Scholar]
- 58.Zhao XR, Gonzales N, Aronowski J.Pleiotropic role of PPARgamma in intracerebral hemorrhage: an intricate system involving Nrf2, RXR, and NF-kappaB. CNS Neurosci Ther 2015; 21: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim DH, Lee D, Chang EH, et al. GDF-15 secreted from human umbilical cord blood mesenchymal stem cells delivered through the cerebrospinal fluid promotes hippocampal neurogenesis and synaptic activity in an Alzheimer's disease model. Stem Cells Dev 2015; 24: 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajan WD, Wojtas B, Gielniewski B, et al. Dissecting functional phenotypes of microglia and macrophages in the rat brain after transient cerebral ischemia. Glia 2019; 67: 232–245. [DOI] [PubMed] [Google Scholar]
- 61.Zhao LR, Willing A.Enhancing endogenous capacity to repair a stroke-damaged brain: an evolving field for stroke research. Prog Neurobiol 2018; 163–164: 5–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hess DC, Borlongan CV.Stem cells and neurological diseases. Cell Prolif 2008; 41(Suppl 1): 94–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manoonkitiwongsa PS, Jackson-Friedman C, McMillan PJ, et al. Angiogenesis after stroke is correlated with increased numbers of macrophages: the clean-up hypothesis. J Cereb Blood Flow Metab 2001; 21: 1223–1231. [DOI] [PubMed] [Google Scholar]
- 64.Dai X, Chen J, Xu F, et al. TGFalpha preserves oligodendrocyte lineage cells and improves white matter integrity after cerebral ischemia. J Cereb Blood Flow Metab. Epub ahead of print 5 March 2019. DOI: 10.1177/0271678X19830791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bastian C, Politano S, Day J, et al. Mitochondrial dynamics and preconditioning in white matter. Cond Med 2018; 1: 64–72. [PMC free article] [PubMed] [Google Scholar]
- 66.Guo S, Tjarnlund-Wolf A, Deng W, et al. Comparative transcriptome of neurons after oxygen-glucose deprivation: Potential differences in neuroprotection versus reperfusion. J Cereb Blood Flow Metab 2018; 38: 2236–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo S, Deng W, Xing C, et al. Effects of aging, hypertension and diabetes on the mouse brain and heart vasculomes. Neurobiol Dis 2019; 126: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim JY, Kim N, Yenari MA.Mechanisms and potential therapeutic applications of microglial activation after brain injury. CNS Neurosci Ther 2015; 21: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korin B, Ben-Shaanan TL, Schiller M, et al. High-dimensional, single-cell characterization of the brain's immune compartment. Nat Neurosci 2017; 20: 1300–1309. [DOI] [PubMed] [Google Scholar]
- 70.Albers GW, Marks MP, Kemp S, et al. Thrombectomy for stroke at 6 to 16 hours with selection by perfusion imaging. N Engl J Med 2018; 378: 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi L, Rocha M, Leak RK, et al. A new era for stroke therapy: integrating neurovascular protection with optimal reperfusion. J Cereb Blood Flow Metab 2018; 38: 2073–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nogueira RG, Jadhav AP, Haussen DC, et al. Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct. N Engl J Med 2018; 378: 11–21. [DOI] [PubMed] [Google Scholar]
- 73.Sharp FR, Xu H, Lit L, et al. The future of genomic profiling of neurological diseases using blood. Arch Neurol 2006; 63: 1529–1536. [DOI] [PubMed] [Google Scholar]
- 74.Liu DZ, Tian Y, Ander BP, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab 2010; 30: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stamova B, Ander BP, Jickling G, et al. The intracerebral hemorrhage blood transcriptome in humans differs from the ischemic stroke and vascular risk factor control blood transcriptomes. J Cereb Blood Flow Metab 2019; 39: 1818–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, JCB888630 Supplementary material1 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material2 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material3 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material4 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material5 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material6 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, JCB888630 Supplementary material7 for RNA sequencing reveals novel macrophage transcriptome favoring neurovascular plasticity after ischemic stroke by Rongrong Wang, Yaan Liu, Qing Ye, Sulaiman H Hassan, Jingyan Zhao, Sicheng Li, Xiaoming Hu, Rehana K Leak, Marcelo Rocha, Lawrence R Wechsler, Jun Chen and Yejie Shi in Journal of Cerebral Blood Flow & Metabolism