SUMMARY
Positive‐stranded RNA viruses, like many other viruses, have evolved to exploit the host cellular machinery to their own advantage. In eukaryotic cells, the ubiquitin‐proteasome system (UPS) that serves as the major intracellular pathway for protein degradation and modification plays a crucial role in the regulation of many fundamental cellular functions. A growing amount of evidence has suggested that the UPS can be utilized by positive‐sense RNA viruses. The UPS eliminates excess viral proteins that prevent viral replication and modulates the function of viral proteins through post‐translational modification mediated by ubiquitin or ubiquitin‐like proteins. This review will discuss the current understanding of how positive RNA viruses have evolved various mechanisms to usurp the host UPS to modulate the function and stability of viral proteins. In addition to the pro‐viral function, UPS‐mediated viral protein degradation may also constitute a host defense process against some positive‐stranded RNA viral infections. This issue will also be discussed in the current review. Copyright © 2012 John Wiley & Sons, Ltd.
Abbreviations used
- TMV
tobacco mosaic virus
- TYMV
turnip yellow mosaic virus
- TBSV
tomato bushy stunt virus
- HAV
hepatitis A virus
- HCV
hepatitis C virus
- EMCV
encephalomyocarditis virus
- SARS‐CoV
severe acute respiratory syndrome coronavirus
- RdRp
RNA‐dependent RNA polymerase
- NSP4
non‐structural protein 4
- NS5B
non‐structural 5B protein
- NS2
non‐structural 2 protein 2
- E protein
envelope protein
- N protein
nucleocapsid protein
- MP
movement protein
- CP
coat protein
INTRODUCTION
Positive‐strand RNA viruses cover more than one‐third of all virus genera and infect a wide range of hosts, for example, the plants, animals, and humans. Examples of this class of viruses include poliovirus, coxsackievirus, EMCV, HCV, HAV, SARS‐CoV, West Nile virus, and dengue fever virus 1. Positive‐strand RNA viruses, either enveloped or non‐enveloped, consist of a single positive‐strand of RNA acting as mRNA to direct the synthesis of viral proteins. Viral structural proteins make up the viral capsid, and non‐structural proteins, such as viral proteases and RNA‐dependent RNA polymerases, function to process the viral polyprotein and catalyze the synthesis of the viral progeny genomes, respectively.
Similar to other viral pathogens, positive‐strand RNA viruses utilize and subvert the host cellular machinery to support their life cycle. Among the many host pathways that can be modulated by positive strand RNA viruses, the ubiquitin‐proteasome system (UPS), the major intracellular protein degradation pathway, has recently received considerable attention because the interaction between viruses and the UPS has been found to play important roles in many aspects of the viral life cycle 2, 3. For positive‐strand RNA viruses, it has been shown that an appropriate stoichiometric ratio of viral structural to non‐structural proteins is essential for effective viral replication 4, 5, 6. There is evidence that too much of certain viral non‐structural proteins are disadvantageous for certain viruses to successfully replicate 6. To ensure a proper ratio of viral proteins, some positive‐strand RNA viruses employ the host regulatory mechanisms, for example, the UPS, to degrade excess viral proteins to prevent interference of these proteins with the viral replication. In addition, post‐translational modification of some viral proteins mediated by ubiquitin and ubiquitin‐like proteins of the UPS has also been demonstrated as an important means to regulate viral protein function without altering protein stability 2.
The objective of this review is to present an overview of current knowledge on how positive‐strand RNA viruses interact and subvert the host UPS, a central component of the host protein degradation system, to maintain optimal levels of viral proteins and to modify the functions of virus‐encoded proteins. The possibility that UPS‐mediated viral protein degradation may also constitute a host defense process against some positive‐stranded RNA viral infections will be briefly discussed as well. How positive‐stranded RNA viruses utilize the UPS to modify the level/function of host proteins to generate a favorable environment for their infection will not be the focus of the current review (please refer to other reviews on this topic 2, 7, 8).
THE UBIQUITIN‐PROTEASOME SYSTEM
Ubiquitin‐dependent and ubiquitin‐independent proteasomal degradation
In eukaryotic cells, the best‐known function of the UPS is to degrade misfolded/damaged proteins or intracellular regulatory proteins that are involved in a variety of cellular activities, including cell‐cycle regulation, membrane protein trafficking, transcription, antigen presentation, and signal transduction 9, 10. Protein degradation via the UPS starts with the covalent attachment of ubiquitin to a target protein (a process referred to as ubiquitylation) (Figure 1). This process consists of multi‐step ATP‐dependent enzyme reactions. First, the ubiquitin is activated by ubiquitin‐activating enzyme E1 to form an E1‐ubiquitin thioester intermediate. The activated ubiquitin is then transferred to a ubiquitin‐conjugating enzyme E2 that subsequently transfers the ubiquitin to the target protein either directly or with the help of a ubiquitin ligase enzyme E3 by forming an isopeptide bond between the carboxyl terminus of ubiquitin and the ε‐amino group of a lysine residue on the target protein 11. The lysine residue of the conjugated ubiquitin is attached by another ubiquitin, consecutively, resulting in the formation of a poly‐ubiquitin chain. The ubiquitin‐tagged substrate protein is then delivered to the 20S proteasome, and the polypeptide is hydrolyzed into short oligopeptides and the free ubiquitin is recycled through the action of de‐ubiquitinating enzymes 12, 13. De‐ubiquitinating enzyme plays important roles in UPS as it proofreads ubiquitin‐protein conjugates, processes inactive ubiquitin precursors, and keeps sustainable proteolysis by maintaining sufficient ubiquitin within the cell 14. The 20S proteasome consists of two 7 subunit‐composed outer α‐rings and two inner β‐rings. The function of the α‐ring is to translocate the target protein to the core of the β‐rings. The β‐rings possess the catalytic property to conduct the action of the degradation. The 20S proteasome is normally inactive and inaccessible to protein substrates. Proteasome activators (PA), such as PA700 (also known as 19S proteasome), PA28 (also known as REG or 11S proteasome), and PA200, bind and activate the 20S proteasome 15. Although the function of PA200 remains to be fully characterized, PA700 has been shown to mainly mediate ubiquitin‐dependent proteasomal degradation, whereas PA28 facilitates substrate degradation in a ubiquitin and ATP‐independent manner 15 (Figure 1).
Figure 1.
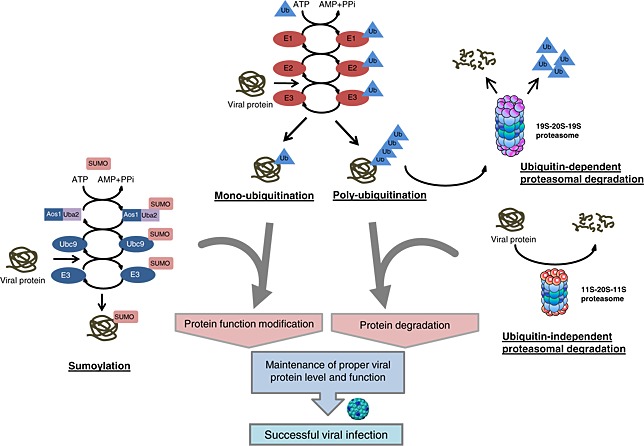
Schematic illustration of post‐translational modification of viral proteins by the ubiquitin‐proteasome system. Post‐translational modification mediated by mono‐ubiquitylation or sumoylation is necessary for the regulation of viral protein functions. Proteasomal degradation mediated through either ubiquitin‐dependent or ubiquitin‐independent mechanisms controls the expression levels of viral proteins. The harmony of these modifications is required to maintain the appropriate level and function of viral proteins, which are crucial for successful infection
Protein modification by ubiquitin and ubiquitin‐like proteins
Unlike protein modification by poly‐ubiquitylation that mostly influences the stability of substrate protein, attachment of a single ubiquitin, a process called mono‐ubiquitylation, regulates the function of target proteins without targeting them for degradation (Figure 1). Mono‐ubiquitylation has been shown to alter protein sub‐cellular localization, regulate transcriptional and enzymatic activities, and change binding affinities to their partners 16, 17.
In addition to ubiquitin, several ubiquitin‐like (UBL) proteins have been found to also function as protein modifiers to regulate a variety of cellular functions, including transcription, DNA repair, signal transduction, and cell cycle control, by post‐translational modification of target proteins. The best‐characterized family of UBL proteins is the small ubiquitin‐like modifiers (SUMO), including SUMO‐1, SUMO‐2, and SUMO‐3 18, 19. Protein modification by sumoylation is directed by an enzymatic cascade parallel to that involved in ubiquitylation (Figure 1). SUMO is activated in an ATP‐dependent manner by an E1‐activating enzyme consisting of a heterodimer of the Aos1 and Uba2 proteins. After activation, SUMO is transferred to the SUMO E2‐conjugating enzyme Ubc9. Three classes of SUMO E3‐ligases have been reported, RanBP2, PIAS and the Polycomb protein Pc2. The sumoylation target is a lysine that occurs in the consensus motif ΨKXE where Ψ is a hydrophobic amino acid and X is any residue. Like mono‐ubiquitylation, sumoylation does not target proteins for degradation, but instead, regulates protein function and sub‐cellular localization 20, 21. A growing body of evidence suggests that sumoylation is particularly important in the regulation of transcription 22. The role of sumoylation in controlling the function of viral proteins by post‐translational modification has also been increasingly recognized 18, 23.
Another well‐studied UBL molecule is interferon‐stimulated gene 15 (ISG15). Expression of ISG15 is highly induced upon viral infection and interferon stimulation 24. Protein modification by ISG15, termed ISGylation, occurs in a mode similar to ubiquitylation and sumoylation that requires the sequential action of E1 (UBE1L), E2 (UbcH8), and E3 enzymes 24. The process of ISG15 conjugation can be reversed by de‐ISGylating enzymes 24. Function of ISGylation has not been fully understood, but it is thought to alter the biological activities of the target proteins or compete for ubiquitylation and play significant roles in many cellular functions 25. Although ISGylation has been mainly associated with host defense response against infection of many viruses, including dengue virus 26, West Nile virus 26, Sindbis virus 27, and Japanese encephalitis virus 28, there were reports that the host ISG15 conjugations system can also be exploited by HCV to facilitate viral production 29, 30, 31. The exact mechanism for the pro‐viral function of ISGylation remains unclear; however, it is speculated that ISGylation of HCV proteins or host proteins critical for HCV life‐cycle promotes HCV production by altering the function or preventing target proteins from ubiquitin‐mediated degradation 31.
RNA‐DEPENDENT RNA POLYMERASE
RNA‐dependent RNA polymerase (RdRp) catalyzes the synthesis of new viral RNA genomes from the original viral RNA 32. Appropriate balance of viral RdRp concentration in infected cells was found to be a key factor affecting the membrane rearrangements, RNA replication, and RNA recombination efficiencies 33, 34, 35. Transgenic overexpression of the 3D RdRp of the Theiler's murine encephalomyelitis virus has been shown to have antiviral effects against the infection of this virus itself and other viruses 6, 36. Although the detailed mechanisms remain to be elucidated, changes of the stoichiometry have been proposed to be one mechanism responsible for such antiviral properties of overexpressed 3D 37. These studies imply that maintaining an appropriate level of RdRp is important for viral growth 6. Indeed, the expression levels of RdRp of Sindbis virus, TYMV, HCV, and HAV have been reported to be regulated via UPS‐regulated protein proteolysis 5, 19, 38, 39, 40. It was shown that the RNA polymerase (NSP4) of Sindbis virus is degraded by the proteasome through the ubiquitin‐dependent N‐end rule pathway 38. The N‐end rule is a highly conserved degradation mechanism relating the stability of a protein with the nature of its N‐terminal residue 41. TYMV RdRp was also shown to be degraded by the proteasome at a late stage of viral infection 5. This UPS‐mediated TYMV RdRp degradation requires prerequisite phosphorylation at two residues (threonine‐64 and serine‐80) localized within the putative N‐terminal PEST sequence (P, proline; E, glutamic acid; S, serine; T, threonine), a known protein degradation motif 19, 42.
The RdRps of HAV and HCV are another two examples of UPS‐mediated degradation 40. The HAV 3D polymerase and its precursors were observed to be present at low levels in infected cells 43 and was demonstrated to be poly‐ubiquitinated for rapid proteasomal degradation 40. Non‐structural 5B (NS5B) protein, the RdRp of HCV, was reported to bind to a cellular ubiquitin‐like protein 39. Overexpression of this protein, but not mutant lacking the NS5B‐binding domain, increases the poly‐ubiquitylation of NS5B and significantly decreases its stability 39.
What is the significance of RdRp degradation during viral infection? Low amounts of RdRp may be required for regulating the switch between the negative‐strand and the positive‐strand viral RNA synthesis during viral replication 5. It is also possible that maintaining a low concentration of RdRp is a strategy for virus to preserve its genome integrity 5. Furthermore, it is conceivable that keeping a low quantity of viral proteins may be beneficial for virus to escape host immune surveillance.
As discussed earlier, in addition to UPS‐mediated rapid turnover of viral proteins, viral protein modification by ubiquitin and/or UBLs also plays a key role in the regulation of viral protein function. It has been recently shown that coxsackieviral 3D RdRp can be post‐translationally modified by mono‐ubiquitylation 44. Although the specific E3 ligase responsible for 3D ubiquitylation and the exact ubiquitylation site remain undetermined, this modification appears to be required for its function in regulating transcription of viral genome 44. Besides ubiquitylation, it has also been found that the 3D polymerase of coxsackievirus can be sumoylated with SUMO‐1, SUMO‐2, and SUMO‐3 (unpublished data). DNA alignment reveals one highly conserved sequence Valine‐Lysine‐Aspartic acid‐Glutamic acid (VKDE), which could be found in the 3D of several different species of enteroviral 3D polymerases, that matches the ΨKXE consensus motif for sumoylation, suggesting that sumoylation may also be required for the regulation of 3D transcriptional activity.
AUTOPROTEASE (VIRUS‐ENCODED AUTOCATALYTIC PROTEASE)
Autoproteases are virus‐encoded proteases that not only proteolytically process viral polyprotein to yield individual structural and non‐structural proteins but also mediate the cleavage of many host proteins essential for transcription/translation and maintenance of cellular structures 45, 46. The levels of mature 3C protease of EMCV and HAV, both belonging to the picornavirus family, have been found to decrease rapidly after they reach maximum proteolytic activity, about halfway through the infectious cycle 47, 48, 49, 50. It was later demonstrated that low concentrations of 3C in infected cells are largely attributed to increased protein turnover through ubiquitin‐mediated proteasome degradation 48, 51, 52, 53. In the presence of proteasome inhibitors, poly‐ubiquitinated 3C protein accumulated and the rate of 3C degradation was significantly reduced 48, 52. Evidence also suggests that EMCV and HAV exploit the same ubiquitin enzymes for 3C ubiquitylation as 3C proteases of these two viruses were shown to compete with each other for ubiquitin conjugation 48. Further investigation identified the amino acid sequences 34LLVRGRTLVV43 and 32LGVKDDWLLV41 that serve as the protein destruction signals for recognition and ubiquitylation of EMCV and HAV 3C proteases, respectively, by the host ubiquitin‐conjugating system 40, 51, 52, 53. Mutations within these sequences lead to increased stability of 3C proteases. The E3 ubiquitin ligase, E3α, has been shown to recognize these destruction signals of EMCV and HAV 3C proteases and catalyze the conjugation of ubiquitin to them 40, 51, 53. The 3C protein of another virus in the picornavirus family, enterovirus 71 (EV71) has also been reported to be regulated by ubiquitin‐mediated degradation 54. It was found that sumoylation of EV71 3C at lysine 52 promotes its ubiquitylation and subsequent degradation 54.
Virus‐encoded proteases are required for successful virus replication via cleaving viral polyprotein precursors and can trigger host cell apoptosis by activating pro‐apoptotic mediators and suppressing host protein translation and transcription 55, 56. Apoptosis at late stages of viral replication can promote viral progeny release. But premature cell death will perturb viral replication before the virus has completed its life cycle. It is therefore speculated that keeping a small amount of 3C protease is necessary, at least at the early stage of viral infection, to prevent premature cell death and allow efficient viral replication. However, the potential significance of UPS targeting viral protease for degradation in host antiviral defense cannot be completely ruled out.
In the study of the regulation of HCV proteases, it was found that both enzymatically inactive non‐structural 2/3 (NS2/3) protein and cleaved NS2 protein of HCV are rapidly degraded during the course of viral infection, underlining the importance of tight regulation of these proteins during the viral life cycle 57, 58, 59. The degradation of NS2 was shown to be regulated in a phosphorylation‐dependent manner mediated by casein kinase 2 on serine 164 residue 57. Notably, ubiquitylation appears not to be required for NS2 degradation as NS2 lysine‐to‐arginine mutagenesis does not affect its stability and expression levels 57, 59. A non‐proteasomal degradation pathway, for example, lysosome‐mediated proteolysis, has also been suggested to be involved in the rapid turnover of NS2 59.
Not all proteases in the family of positive‐stranded RNA viruses are regulated by the UPS. The 3C proteases of rhinovirus and poliovirus, similar to those of EMCV and HAV, are present in low concentrations in infected cell 35, 60, 61. However, it was found that poliovirus 3C is not conjugated with ubiquitin and remains stable during the viral life cycle 48. Further studies are warranted to elucidate the mechanisms involved in maintaining low levels of polioviral 3C protease. Possible mechanisms may include blockage of ribosomal read‐through and differential processing of polyproteins, leading to reduced production of 3C protease. A recent report has suggested that pseudoknot structures of the SARS‐CoV RNA can stop the translation from upstream to downstream encoded viral proteins 62.
Some viral proteases have been shown to possess de‐ubiquitinating enzyme activity. It was reported that the papain‐like cysteine proteases of coronavirus 63, 64, 65, 66, 67, 68, hepatitis E virus 69, and foot‐and‐mouth disease virus 70 have structural similarity to the cellular de‐ubiquitinating enzymes and are able to efficiently hydrolyze ubiquitinated substrates. Although the viral and cellular targets remain largely unknown, the de‐ubiquitinating activity of these proteases appears to play a significant role in viral life cycle and in blockage of host innate immunity.
STRUCTURAL PROTEIN
Although the structural proteins need to be expressed at higher levels than non‐structural proteins 71, the levels and functions of some viral structural proteins can also be regulated by the UPS. The core protein of HCV is a structural protein that not only packages the viral genomic RNA but also modulates multiple cellular functions, including apoptosis, cell proliferation, cell transformation, and signal transduction, and contributes to HCV pathogenesis 72, 73. The stability of HCV core protein has been reported to be regulated by the proteasome in both ubiquitin‐dependent and ubiquitin‐independent manner 74, 75, 76, 77, 78, 79, 80. It was demonstrated that the E3 ligase, E6‐associated protein (E6AP), binds to the core protein of HCV and promotes its ubiquitylation and subsequent degradation by the proteasome 78. Ubiquitin‐independent proteasomal degradation mediated by PA28γ has also been shown to facilitate the degradation of HCV core protein 76, 79. PA28γ interacts directly with the core protein of HCV in the nucleus and regulates its degradation 76. In animal experiments, this interaction has been demonstrated to play an important role in HCV pathogenesis as PA28γ knockout disrupts the progression of steatosis, hepatocarcinoma, and insulin resistance induced by HCV core protein 74, 75. Moreover, recent evidence revealed a role for PA28γ‐mediated and E6AP‐mediated degradation of HCV core protein in the regulation of HCV propagation 77.
Recently discovered F protein, a frame‐shift product of the core protein of HCV, has also been reported to be directly degraded by the 20S proteasome through a mechanism independent of ubiquitin 81, 82. The function of HCV F protein remains unclear. Available data suggest a role for F protein in HCV pathogenesis by regulating the expression and activity of several pro‐inflammatory cytokines, transcriptional factors, and oncogenes 5, 83, 84. HCV F protein was found to be very labile with a short half‐life (~10 min) and degraded by the proteasome 81, 85. Further experimentation revealed that the degradation of F protein is ubiquitin‐independent as a lysine‐less F protein mutant remains unstable and its protein levels do not appears to be affected in a cell line with a temperature‐sensitive E1 82. It was demonstrated that HCV F protein binds to the α3 subunit of the 20S proteasome and is degraded directly by the 20S proteasome in vitro 82. The functional significance of F protein degradation in viral replication (pro‐viral strategy versus host defense mechanism) warrants future investigation.
West Nile virus capsid protein is another example of a viral structural protein being the target of the UPS 86, 87. The Makorin ring finger protein 1, a member of the Makorin family of proteins, was identified as the E3 ligase facilitating the ubiquitylation and consequent degradation of this capsid protein by the proteasome 86. In addition to its role in nucleocapsid assembly, the West Nile capsid protein is also involved in viral pathogenesis by inducing apoptosis 88, 89. Overexpression of Makorin ring finger protein 1 has been shown to result in reduced apoptosis triggered by West Nile virus infection, whereas depletion of this protein promotes viral cytotoxicity 86. It remains unclear whether such modification and degradation of the capsid protein is a host defense mechanism or a viral strategy to prevent premature cell death.
The envelope (E) protein of SARS‐CoV has been reported to be ubiquitinated in vitro and in cells, likely through its interaction with the N‐terminal ubiquitin‐like domain‐1 of non‐structural protein (NSP3) 90. The coat protein of TMV has also been shown to be modified by mono‐ubiquitylation 91. However, the functional consequence of these post‐translational modifications in the regulation of the viral life cycle and viral pathogenesis is not known and requires further investigation.
Besides ubiquitylation, UBL‐mediated modification has also been reported for viral structural proteins. The nucleocapsid (N) protein, a structural protein of SARS‐CoV, was demonstrated to undergo post‐translational modification by sumoylation 92, 93. Using a yeast two‐hybrid system, Ubc9, the E2‐conjugating enzyme for sumoylation, was identified as the cellular protein interacting with the SARS‐CoV N protein 92. It was further demonstrated that N protein is covalently modified mainly at lysine 62 residue by SUMO 94. Further investigation using wild‐type N protein and a sumoylation mutant revealed that sumoylation of this protein increases its homo‐oligomerization that may play a role in controlling viral replication cycle 94. Swine fever virus core protein was also found to interact with the intracellular sumoylation pathway 95. It binds to Ubc9 and SUMO‐1 and disruption of these interactions results in attenuated viral virulence, suggesting a regulatory role of SUMO modification in viral infectivity 95. Dengue virus is a member of the virus family Flaviviridae. Its envelope protein that is responsible for the virus attachment and entry to host cells has been shown to interact directly with SUMO E2 enzyme Ubc9. Further investigation demonstrates that overexpression of Ubc9 reduces the production of infectious virus, suggesting a role for SUMO modification in attenuating viral infectivity 96.
OTHER PROTEINS
Movement protein (MP) is a non‐structural protein encoded by plant viruses to facilitate cell to cell movement. TMV MP was previously observed to be only transiently expressed during virus infection 97. Further investigation demonstrates that TMV MP is poly‐ubiquitinated and subsequently degraded by the proteasome and inhibition of proteasome function leads to accumulation of MP preferentially on the perinuclear ER 98. The exact role of UPS‐mediated degradation of MP in TMV infection remains to be elucidated. It is speculated that such modification of MP plays a critical role in regulating virus spread by attenuating its damage on ER structure 98.
Ubiquitin‐conjugating enzyme Cdc34p has been identified as one of the host proteins binding to the p33 replication protein of Tombusvirus 99. It was shown that p33 can be ubiquitinated both in vitro and in vivo, and overexpression of Cdc34p increases, whereas down‐regulation of Cdc34p reduces Tombusvirus replication 99. Further investigation identified the lysine residues required for p33 mono‐ubiquitylation and demonstrated the functional significance of p33 ubiquitylation in Tombusvirus replication 100.
The interaction between the host UPS and positive‐stranded RNA viruses is summarized in Table 1.
Table 1.
Degradation and functional modification of positive‐strand RNA viral proteins by the host ubiquitin‐proteasome system
| Family | Virus | Target protein | UPS‐mediated modification | Function (confirmed or proposed) | References |
|---|---|---|---|---|---|
| Picornaviruses | HAV | RdRp(3D) | Poly‐ubiquitination and degradation | Regulate viral RNA synthesis | Losick et al. 40 |
| 3C protease | Poly‐ubiquitination and degradation | Prevent premature cell death | Gladding et al. 48 | ||
| Lawson et al. 53 | |||||
| Losick et al. 40 | |||||
| EMCV | 3C protease | Poly‐ubiquitination and degradation | Prevent premature cell death | Lawson et al. 51 | |
| Lawson et al. 52 | |||||
| Lawson et al. 53 | |||||
| Enterovirus | 3C protease | Poly‐ubiquitination and degradation | Prevent premature cell death | Chen et al. 54 | |
| Sumoylation | Promote poly‐ubiquitination of 3C | Chen et al. 54 | |||
| Coxsackivirus | RdRp (3D) | Mono‐ubiquitination | Modulate 3D polymerase function | Si et al. 44 | |
| Sumoylation | Modulate 3D polymerase function | [unpublished] | |||
| Flaviviruses | HCV | RdRp (NS5B) | Poly‐ubiquitination and degradation | Regulate viral RNA synthesis | Gao et al. 2 |
| NS2 protease | Ubiquitin‐independent degradation | ||||
| Non‐proteasomal degradation | Prevent premature cell death | Franck et al. 57 | |||
| Welbourn et al. 59 | |||||
| Core protein | Ubiquitination and degradation | Regulate viral propagation | Moriishi et al. 75 | ||
| Shirakura et al. 78 | |||||
| Ubiquitin‐independent degradation | Regulate viral propagation and HCV pathogenesis | Miyamoto et al. 74 | |||
| Moriishi et al. 75 | |||||
| Moriishi et al. 76 | |||||
| Moriishi et al. 77 | |||||
| Suzuki et al. 79 | |||||
| F protein | Ubiquitin‐independent degradation | Unknown | Xu et al. 81 | ||
| Yuksek et al. 82 | |||||
| West Nile virus | Capsid protein | Poly‐ubiquitination and degradation | Prevent premature cell death | Ko et al. 86 | |
| Oh et al. 87 | |||||
| Swine fever virus | Core protein | Sumoylation | Promote viral propagation | Gladue et al. 95 | |
| Dengue virus | E protein | Sumoylation | Attenuate viral infectivity | Chiu et al. 96 | |
| Togaviruses | Sindbis Virus | RdRp (NSP4) | Poly‐ubiquitination and degradation | Regulate viral RNA synthesis | de Groot et al. 38 |
| Tymoviruses | TYMV | RdRp (3D) | Poly‐ubiquitination and degradation | Regulate viral RNA synthesis | Camborde et al. 5 |
| Hericourt et al. 19 | |||||
| Tobamoviruses TMV | MP | Poly‐ubiquitination and degradation | Regulate viral spread | Reichel et al. 98 | |
| CP | Mono‐ubiquitination | Unknown | Dunigan et al. 91 | ||
| Tombusvirus | TBSV | p33 | Mono‐ubiquitination | Enhance p33 binding activity | Li et al. 99 |
| Promote viral replication | Barajas et al. 100 | ||||
| Coronaviruses | SARS‐CoV | E Protein | Poly‐ubiquitination and degradation | Unknown | Alvarez et al. 90 |
| N protein | Sumoylation | Promote its homo‐oligomerization | Fan et al. 92 | ||
| Li et al. 93 | |||||
| Li et al. 94 |
CONCLUSION
Current evidence, reviewed herein, strongly supports a notion that the maintenance of appropriate levels of certain viral proteins within infected cells is crucial for successful viral reproduction. Autoproteases at high concentrations have been suggested to trigger apoptosis of the host cells by cleaving cellular proteins 55, 56, whereas high amounts of RdRps have been shown to perturb the process of appropriate viral packaging and even become antiviral when expressed at high levels 6, 36. Growing studies have suggested that positive‐stranded RNA viruses manipulate the host UPS for the degradation of excess viral proteins that disturb efficient viral growth and for the modulation of viral protein function through ubiquitin‐mediated or UBL‐mediated modification (Table 1 and Figure 1). These findings will not only be able to explain the differential expression of various viral proteins but also provide a drug target through a thorough understanding of the mechanism regulating the levels and activities of viral proteins.
CONFLICT OF INTEREST
The authors have no competing interest.
ACKNOWLEDGEMENT
This work was supported by the Canadian Institutes of Health Research (to HL).
REFERENCES
- 1. Van Regenmortel MHV. Virus Taxonomy. Academic Press: San Diego, Calif: 2000. [Google Scholar]
- 2. Gao G, Luo H. The ubiquitin‐proteasome pathway in viral infections. Canadian Journal of Physiology and Pharmacology 2006; 84(1): 5–14. [DOI] [PubMed] [Google Scholar]
- 3. Luo H, Wong J, Wong B. Protein degradation systems in viral myocarditis leading to dilated cardiomyopathy. Cardiovascular Research 2010; 85(2): 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banerjee R, Weidman MK, Echeverri A, Kundu P, Dasgupta A. Regulation of poliovirus 3C protease by the 2C polypeptide. Journal of Virology 2004; 78(17): 9243–9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Camborde L, Planchais S, Tournier V, Jakubiec A, Drugeon G, Lacassagne E, Pflieger S, Chenon M, Jupin I. The ubiquitin‐proteasome system regulates the accumulation of turnip yellow mosaic virus RNA‐dependent RNA polymerase during viral infection. The Plant Cell 2010; 22(9): 3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kerkvliet J, Papke L, Rodriguez M. Antiviral effects of a transgenic RNA‐dependent RNA polymerase. Journal of Virology 2011; 85(1): 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Randow F, Lehner PJ. Viral avoidance and exploitation of the ubiquitin system. Nature Cell Biology 2009; 11(5): 527–534. [DOI] [PubMed] [Google Scholar]
- 8. Viswanathan K, Fruh K, Defilippis V. Viral hijacking of the host ubiquitin system to evade interferon responses. Current Opinion in Microbiology 2010; 13(4): 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rechsteiner M. Natural substrates of the ubiquitin proteolytic pathway. Cell 1991; 66(4): 615–618. [DOI] [PubMed] [Google Scholar]
- 10. Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin‐like proteins as multifunctional signals. Nature Reviews Molecular Cell Biology 2005; 6(8): 599–609. [DOI] [PubMed] [Google Scholar]
- 11. Glickman MH, Ciechanover A. The ubiquitin‐proteasome proteolytic pathway: destruction for the sake of construction. Physiological Reviews 2002; 82(2): 373–428. [DOI] [PubMed] [Google Scholar]
- 12. Pickart CM. Ubiquitin in chains. Trends in Biochemical Sciences 2000; 25(11): 544–548. [DOI] [PubMed] [Google Scholar]
- 13. Pickart CM. Ubiquitin enters the new millennium. Molecular Cell 2001; 8(3): 499–504. [DOI] [PubMed] [Google Scholar]
- 14. Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochimica et Biophysica Acta 2004; 1695(1–3): 189–207. [DOI] [PubMed] [Google Scholar]
- 15. Mao I, Liu J, Li X, Luo H. REGgamma, a proteasome activator and beyond? Cellular and Molecular Life Sciences 2008; 65(24): 3971–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature Cell Biology 2006; 8(4): 339–347. [DOI] [PubMed] [Google Scholar]
- 17. Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Current Opinion in Cell Biology 2004; 16(2): 119–126. [DOI] [PubMed] [Google Scholar]
- 18. Boggio R, Chiocca S. Viruses and sumoylation: recent highlights. Current Opinion in Microbiology 2006; 9(4): 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hericourt F, Blanc S, Redeker V, Jupin I. Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA‐dependent RNA polymerase domain expressed in a baculovirus‐insect cell system. Biochemical Journal 2000; 349(Pt 2): 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geiss‐Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nature Reviews Molecular Cell Biology 2007; 8(12): 947–956. [DOI] [PubMed] [Google Scholar]
- 21. Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes & Development 2004; 18(17): 2046–2059. [DOI] [PubMed] [Google Scholar]
- 22. Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Reports 2003; 4(2): 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hilgarth RS, Murphy LA, Skaggs HS, Wilkerson DC, Xing H, Sarge KD. Regulation and function of SUMO modification. Journal of Biological Chemistry 2004; 279(52): 53899–53902. [DOI] [PubMed] [Google Scholar]
- 24. Skaug B, Chen ZJ. Emerging role of ISG15 in antiviral immunity. Cell 2010; 143(2): 187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang D, Zhang DE. Interferon‐stimulated gene 15 and the protein ISGylation system. Journal of Interferon and Cytokine Research 2011; 31(1): 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai J, Pan W, Wang P. ISG15 facilitates cellular antiviral response to dengue and West Nile virus infection in vitro . Virology Journal 2011; 8: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giannakopoulos NV, Arutyunova E, Lai C, Lenschow DJ, Haas AL, Virgin HW. ISG15 Arg151 and the ISG15‐conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. Journal of Virology 2009; 83(4): 1602–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsiao NW, Chen JW, Yang TC, Orloff GM, Wu YY, Lai CH, Lan YC, Lin CW. ISG15 over‐expression inhibits replication of the Japanese encephalitis virus in human medulloblastoma cells. Antiviral Research 2010; 85(3): 504–511. [DOI] [PubMed] [Google Scholar]
- 29. Broering R, Zhang X, Kottilil S, Trippler M, Jiang M, Lu M, Gerken G, Schlaak JF. The interferon stimulated gene 15 functions as a proviral factor for the hepatitis C virus and as a regulator of the IFN response. Gut 2010; 59(8): 1111–1119. [DOI] [PubMed] [Google Scholar]
- 30. Chen L, Li S, Mcgilvray I. The ISG15/USP18 ubiquitin‐like pathway (ISGylation system) in hepatitis C virus infection and resistance to interferon therapy. The International Journal of Biochemistry & Cell Biology 2011; 43(10): 1427–1431. [DOI] [PubMed] [Google Scholar]
- 31. Chen L, Sun J, Meng L, Heathcote J, Edwards AM, Mcgilvray ID. ISG15, a ubiquitin‐like interferon‐stimulated gene, promotes hepatitis C virus production in vitro: implications for chronic infection and response to treatment. Journal of General Virology 2010; 91(Pt 2): 382–388. [DOI] [PubMed] [Google Scholar]
- 32. Ahlquist P. RNA‐dependent RNA polymerases, viruses, and RNA silencing. Science 2002; 296(5571): 1270–1273. [DOI] [PubMed] [Google Scholar]
- 33. Jaag HM, Stork J, Nagy PD. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology 2007; 368(2): 388–404. [DOI] [PubMed] [Google Scholar]
- 34. Pantaleo V, Rubino L, Russo M. The p36 and p95 replicase proteins of Carnation Italian ringspot virus cooperate in stabilizing defective interfering RNA. Journal of General Virology 2004; 85(Pt 8): 2429–2433. [DOI] [PubMed] [Google Scholar]
- 35. Schwartz M, Chen J, Lee WM, Janda M, Ahlquist P. Alternate, virus‐induced membrane rearrangements support positive‐strand RNA virus genome replication. Proceedings of the National Academy of Sciences of the United States of America 2004; 101(31): 11263–11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kerkvliet J, Zoecklein L, Papke L, Denic A, Bieber AJ, Pease LR, David CS, Rodriguez M. Transgenic expression of the 3D polymerase inhibits Theiler's virus infection and demyelination. Journal of Virology 2009; 83(23): 12279–12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kerkvliet J, Edukulla R, Rodriguez M. Novel roles of the picornaviral 3D polymerase in viral pathogenesis. Advances in Virology 2010; 2010: 368068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Groot RJ, Rumenapf T, Kuhn RJ, Strauss EG, Strauss JH. Sindbis virus RNA polymerase is degraded by the N‐end rule pathway. Proceedings of the National Academy of Sciences of the United States of America 1991; 88(20): 8967–8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao L, Tu H, Shi ST, Lee KJ, Asanaka M, Hwang SB, Lai MM. Interaction with a ubiquitin‐like protein enhances the ubiquitination and degradation of hepatitis C virus RNA‐dependent RNA polymerase. Journal of Virology 2003; 77(7): 4149–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Losick VP, Schlax PE, Emmons RA, Lawson TG. Signals in hepatitis A virus P3 region proteins recognized by the ubiquitin‐mediated proteolytic system. Virology 2003; 309(2): 306–319. [DOI] [PubMed] [Google Scholar]
- 41. Bachmair A, Finley D, Varshavsky A. In vivo half‐life of a protein is a function of its amino‐terminal residue. Science 1986; 234(4773): 179–186. [DOI] [PubMed] [Google Scholar]
- 42. Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 1986; 234(4774): 364–368. [DOI] [PubMed] [Google Scholar]
- 43. Updike WS, Tesar M, Ehrenfeld E. Detection of hepatitis A virus proteins in infected BS‐C‐1 cells. Virology 1991; 185(1): 411–418. [DOI] [PubMed] [Google Scholar]
- 44. Si X, Gao G, Wong J, Wang Y, Zhang J, Luo H. Ubiquitination is required for effective replication of coxsackievirus B3. PloS One 2008; 3(7): e2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blair WS, Semler BL. Self‐cleaving proteases. Current Opinion in Cell Biology 1991; 3(6): 1039–1045. [DOI] [PubMed] [Google Scholar]
- 46. Lloyd RE. Translational control by viral proteinases. Virus Research 2006; 119(1): 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gauss‐Muller V, Jurgensen D, Deutzmann R. Autoproteolytic cleavage of recombinant 3C proteinase of hepatitis A virus. Virology 1991; 182(2): 861–864. [DOI] [PubMed] [Google Scholar]
- 48. Gladding RL, Haas AL, Gronros DL, Lawson TG. Evaluation of the susceptibility of the 3C proteases of hepatitis A virus and poliovirus to degradation by the ubiquitin‐mediated proteolytic system. Biochemical and Biophysical Research Communication 1997; 238(1): 119–125. [DOI] [PubMed] [Google Scholar]
- 49. Lawson TG, Smith LL, Palmenberg AC, Thach RE. Inducible expression of encephalomyocarditis virus 3C protease activity in stably transformed mouse cell lines. Journal of Virology 1989; 63(12): 5013–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oberst MD, Gollan TJ, Gupta M, Peura SR, Zydlewski JD, Sudarsanan P, Lawson TG. The encephalomyocarditis virus 3C protease is rapidly degraded by an ATP‐dependent proteolytic system in reticulocyte lysate. Virology 1993; 193(1): 28–40. [DOI] [PubMed] [Google Scholar]
- 51. Lawson TG, Gronros DL, Evans PE, Bastien MC, Michalewich KM, Clark JK, Edmonds JH, Graber KH, Werner JA, Lurvey BA, Cate JM. Identification and characterization of a protein destruction signal in the encephalomyocarditis virus 3C protease. Journal of Biological Chemistry 1999; 274(14): 9904–9980. [PubMed] [Google Scholar]
- 52. Lawson TG, Gronros DL, Werner JA, Wey AC, Digeorge AM, Lockhart JL, Wilson JW, Wintrode PL. The encephalomyocarditis virus 3C protease is a substrate for the ubiquitin‐mediated proteolytic system. Journal of Biological Chemistry 1994; 269(45): 28429–28435. [PubMed] [Google Scholar]
- 53. Lawson TG, Sweep ME, Schlax PE, Bohnsack RN, Haas AL. Kinetic analysis of the conjugation of ubiquitin to picornavirus 3C proteases catalyzed by the mammalian ubiquitin‐protein ligase E3alpha. Journal of Biological Chemistry 2001; 276(43): 39629–39637. [DOI] [PubMed] [Google Scholar]
- 54. Chen SC, Chang LY, Wang YW, Chen YC, Weng KF, Shih SR, Shih HM. Sumoylation‐promoted enterovirus 71 3C degradation correlates with a reduction in viral replication and cell apoptosis. Journal of Biological Chemistry 2011; 286(36): 31373–31384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barco A, Feduchi E, Carrasco L. Poliovirus protease 3C(pro) kills cells by apoptosis. Virology 2000; 266(2): 352–360. [DOI] [PubMed] [Google Scholar]
- 56. Chau DH, Yuan J, Zhang H, Cheung P, Lim T, Liu Z, Sall A, Yang D. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis 2007; 12(3): 513–524. [DOI] [PubMed] [Google Scholar]
- 57. Franck N, Le Seyec J, Guguen‐Guillouzo C, Erdtmann L. Hepatitis C virus NS2 protein is phosphorylated by the protein kinase CK2 and targeted for degradation to the proteasome. Journal of Virology 2005; 79(5): 2700–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Welbourn S, Green R, Gamache I, Dandache S, Lohmann V, Bartenschlager R, Meerovitch K, Pause A. Hepatitis C virus NS2/3 processing is required for NS3 stability and viral RNA replication. Journal of Biological Chemistry 2005; 280(33): 29604–29611. [DOI] [PubMed] [Google Scholar]
- 59. Welbourn S, Jirasko V, Breton V, Reiss S, Penin F, Bartenschlager R, Pause A. Investigation of a role for lysine residues in non‐structural proteins 2 and 2/3 of the hepatitis C virus for their degradation and virus assembly. Journal of General Virology 2009; 90(Pt 5): 1071–1080. [DOI] [PubMed] [Google Scholar]
- 60. Aschauer B, Werner G, Mccray J, Rosenwirth B, Bachmayer H. Biologically active protease 3C of human rhinovirus 1A is expressed from a cloned cDNA segment in Escherichia coli . Virology 1991; 184(2): 587–594. [DOI] [PubMed] [Google Scholar]
- 61. Thomas AA, Voorma HO, Boeye A. Relationship between synthesis and cleavage of poliovirus‐specific proteins. Journal of Virology 1983; 48(1): 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tholstrup J, Oddershede LB, Sorensen MA. mRNA pseudoknot structures can act as ribosomal roadblocks. Nucleic Acids Research 2012; 40(1): 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain‐like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. Journal of Virology 2005; 79(24): 15189–15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, Baker SC. Proteolytic processing and deubiquitinating activity of papain‐like proteases of human coronavirus NL63. Journal of Virology 2007; 81(11): 6007–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez‐Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. Deubiquitinating and interferon antagonism activities of coronavirus papain‐like proteases. Journal of Virology 2010; 84(9): 4619–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lindner HA, Fotouhi‐Ardakani N, Lytvyn V, Lachance P, Sulea T, Menard R. The papain‐like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. Journal of Virology 2005; 79(24): 15199–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME, Baker SC, Ghosh AK, Mesecar AD. A noncovalent class of papain‐like protease/deubiquitinase inhibitors blocks SARS virus replication. Proceedings of the National Academy of Sciences of the United States of America 2008; 105(42): 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD. Severe acute respiratory syndrome coronavirus papain‐like protease: structure of a viral deubiquitinating enzyme. Proceedings of the National Academy of Sciences of the United States of America 2006; 103(15): 5717–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Karpe YA, Lole KS. Deubiquitination activity associated with hepatitis E virus putative papain‐like cysteine protease. Journal of General Virology 2011; 92(Pt 9): 2088–2092. [DOI] [PubMed] [Google Scholar]
- 70. Wang D, Fang L, Li P, Sun L, Fan J, Zhang Q, Luo R, Liu X, Li K, Chen H, Chen Z, Xiao S. The leader proteinase of foot‐and‐mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. Journal of Virology 2011; 85(8): 3758–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bell YC, Semler BL, Ehrenfeld E. Requirements for RNA replication of a poliovirus replicon by coxsackievirus B3 RNA polymerase. Journal of Virology 1999; 73(11): 9413–9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Khaliq S, Jahan S, Pervaiz A. Sequence variability of HCV Core region: important predictors of HCV induced pathogenesis and viral production. Emerging Infectious Diseases 2011; 11(3): 543–556. [DOI] [PubMed] [Google Scholar]
- 73. Watashi K, Shimotohno K. The roles of hepatitis C virus proteins in modulation of cellular functions: a novel action mechanism of the HCV core protein on gene regulation by nuclear hormone receptors. Cancer Science 2003; 94(11): 937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Miyamoto H, Moriishi K, Moriya K, Murata S, Tanaka K, Suzuki T, Miyamura T, Koike K, Matsuura Y. Involvement of the PA28gamma‐dependent pathway in insulin resistance induced by hepatitis C virus core protein. Journal of Virology 2007; 81(4): 1727–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Moriishi K, Mochizuki R, Moriya K, Miyamoto H, Mori Y, Abe T, Murata S, Tanaka K, Miyamura T, Suzuki T, Koike K, Matsuura Y. Critical role of PA28gamma in hepatitis C virus‐associated steatogenesis and hepatocarcinogenesis. Proceedings of the National Academy of Sciences of the United States of America 2007; 104(5): 1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Moriishi K, Okabayashi T, Nakai K, Moriya K, Koike K, Murata S, Chiba T, Tanaka K, Suzuki R, Suzuki T, Miyamura T, Matsuura Y. Proteasome activator PA28gamma‐dependent nuclear retention and degradation of hepatitis C virus core protein. Journal of Virology 2003; 77(19): 10237–10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Moriishi K, Shoji I, Mori Y, Suzuki R, Suzuki T, Kataoka C, Matsuura Y. Involvement of PA28gamma in the propagation of hepatitis C virus. Hepatology 2010; 52(2): 411–420. [DOI] [PubMed] [Google Scholar]
- 78. Shirakura M, Murakami K, Ichimura T, Suzuki R, Shimoji T, Fukuda K, Abe K, Sato S, Fukasawa M, Yamakawa Y, Nishijima M, Moriishi K, Matsuura Y, Wakita T, Suzuki T, Howley PM, Miyamura T, Shoji I. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. Journal of Virology 2007; 81(3): 1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Suzuki R, Moriishi K, Fukuda K, Shirakura M, Ishii K, Shoji I, Wakita T, Miyamura T, Matsuura Y, Suzuki T. Proteasomal turnover of hepatitis C virus core protein is regulated by two distinct mechanisms: a ubiquitin‐dependent mechanism and a ubiquitin‐independent but PA28gamma‐dependent mechanism. Journal of Virology 2009; 83(5): 2389–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Suzuki R, Tamura K, Li J, Ishii K, Matsuura Y, Miyamura T, Suzuki T. Ubiquitin‐mediated degradation of hepatitis C virus core protein is regulated by processing at its carboxyl terminus. Virology 2001; 280(2): 301–309. [DOI] [PubMed] [Google Scholar]
- 81. Xu Z, Choi J, Lu W, Ou JH. Hepatitis C virus f protein is a short‐lived protein associated with the endoplasmic reticulum. Journal of Virology 2003; 77(2): 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yuksek K, Chen WL, Chien D, Ou JH. Ubiquitin‐independent degradation of hepatitis C virus F protein. Journal of Virology 2009; 83(2): 612–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fiorucci M, Boulant S, Fournillier A, Abraham JD, Lavergne JP, Paranhos‐Baccala G, Inchauspe G, Bain C. Expression of the alternative reading frame protein of Hepatitis C virus induces cytokines involved in hepatic injuries. Journal of General Virology 2007; 88(Pt 4): 1149–1162. [DOI] [PubMed] [Google Scholar]
- 84. Wu WB, Shao SW, Zhao LJ, Luan J, Cao J, Gao J, Zhu SY, Qi ZT. Hepatitis C virus F protein up‐regulates c‐myc and down‐regulates p53 in human hepatoma HepG2 cells. Intervirology 2007; 50(5): 341–346. [DOI] [PubMed] [Google Scholar]
- 85. Roussel J, Pillez A, Montpellier C, Duverlie G, Cahour A, Dubuisson J, Wychowski C. Characterization of the expression of the hepatitis C virus F protein. Journal of General Virology 2003; 84(Pt 7): 1751–1759. [DOI] [PubMed] [Google Scholar]
- 86. Ko A, Lee EW, Yeh JY, Yang MR, Oh W, Moon JS, Song J. MKRN1 induces degradation of West Nile virus capsid protein by functioning as an E3 ligase. Journal of Virology 2010; 84(1): 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Oh W, Yang MR, Lee EW, Park KM, Pyo S, Yang JS, Lee HW, Song J. Jab1 mediates cytoplasmic localization and degradation of West Nile virus capsid protein. Journal of Biological Chemistry 2006; 281(40): 30166–30174. [DOI] [PubMed] [Google Scholar]
- 88. Yang JS, Ramanathan MP, Muthumani K, Choo AY, Jin SH, Yu QC, Hwang DS, Choo DK, Lee MD, Dang K, Tang W, Kim JJ, Weiner DB. Induction of inflammation by West Nile virus capsid through the caspase‐9 apoptotic pathway. Emerging Infectious Diseases 2002; 8(12): 1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yang MR, Lee SR, Oh W, Lee EW, Yeh JY, Nah JJ, Joo YS, Shin J, Lee HW, Pyo S, Song J. West Nile virus capsid protein induces p53‐mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cellular Microbiology 2008; 10(1): 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Alvarez E, Dediego ML, Nieto‐Torres JL, Jimenez‐Guardeno JM, Marcos‐Villar L, Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non‐structural protein 3 and is ubiquitinated. Virology 2010; 402(2): 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dunigan DD, Dietzgen RG, Schoelz JE, Zaitlin M. Tobacco mosaic virus particles contain ubiquitinated coat protein subunits. Virology 1988; 165(1): 310–312. [DOI] [PubMed] [Google Scholar]
- 92. Fan Z, Zhuo Y, Tan X, Zhou Z, Yuan J, Qiang B, Yan J, Peng X, Gao GF. SARS‐CoV nucleocapsid protein binds to hUbc9, a ubiquitin conjugating enzyme of the sumoylation system. Journal of Medical Virology 2006; 78(11): 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li FQ, Xiao H, Tam JP, Liu DX. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Letters 2005; 579(11): 2387–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li Q, Xiao H, Tam JP, Liu DX. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus by interaction with Ubc9. Advances in Experimental Medicine and Biology 2006; 581: 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gladue DP, Holinka LG, Fernandez‐Sainz IJ, Prarat MV, O'donell V, Vepkhvadze N, Lu Z, Rogers K, Risatti GR, Borca MV. Effects of the interactions of classical swine fever virus Core protein with proteins of the SUMOylation pathway on virulence in swine. Virology 2010; 407(1): 129–136. [DOI] [PubMed] [Google Scholar]
- 96. Chiu MW, Shih HM, Yang TH, Yang YL. The type 2 dengue virus envelope protein interacts with small ubiquitin‐like modifier‐1 (SUMO‐1) conjugating enzyme 9 (Ubc9). Journal of Biomedical Science 2007; 14(3): 429–444. [DOI] [PubMed] [Google Scholar]
- 97. Watanabe Y, Emori Y, Ooshika I, Meshi T, Ohno T, Okada Y. Synthesis of TMV‐specific RNAs and proteins at the early stage of infection in tobacco protoplasts: transient expression of the 30K protein and its mRNA. Virology 1984; 133(1): 18–24. [DOI] [PubMed] [Google Scholar]
- 98. Reichel C, Beachy RN. Degradation of tobacco mosaic virus movement protein by the 26S proteasome. Journal of Virology 2000; 74(7): 3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. Cdc34p ubiquitin‐conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. Journal of Virology 2008; 82(14): 6911–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Barajas D, Nagy PD. Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology 2010; 397(2): 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]


