Summary
Lujo virus is a novel Old World arenavirus identified in Southern Africa in 2008 as the cause of a viral hemorrhagic fever (VHF) characterized by nosocomial transmission with a high case fatality rate of 80% (4/5 cases). Whereas this outbreak was limited, the unprecedented Ebola virus disease outbreak in West Africa, and recent Zika virus disease epidemic in the Americas, has brought into acute focus the need for preparedness to respond to rare but potentially highly pathogenic outbreaks of zoonotic or arthropod‐borne viral infections. A key determinant for effective control of a VHF outbreak is the time between primary infection and diagnosis of the index case. Here, we review the Lujo VHF outbreak of 2008 and discuss how preparatory measures with respect to developing diagnostic capacity might be effectively embedded into existing national disease control networks, such as those for human immunodeficiency virus, tuberculosis, and malaria.
Keywords: Arenaviridae, diagnostic capacity, Ebola virus disease, lessons, Lujo virus, Mammarenavirus, preparedness, viral hemorrhagic fever, Zika virus
1. Introduction
There are 4 virus families known to cause viral hemorrhagic fever (VHF) in humans: Arenaviridae, Bunyaviridae, Filoviridae, and Flaviviridae. While all VHFs can involve bleeding, hemorrhage is mostly a less common complication of severe infection. The general clinical picture for severe disease is one of grave multisystem syndrome with damage to the vascular system, and sometimes severe neurological symptoms,1 although many infections may also take a milder course. The natural reservoir hosts of these enveloped RNA viruses include a range of mammalian species, particularly rodents and bats. Most VHF viruses are transmitted to humans via direct contact with host body fluids or excreta, sometimes through an intermediate mammalian host. The Bunyaviridae and Flaviviridae VHF viruses are transmitted by insect vectors (ticks and mosquitoes). The CDC also now list 2 Paramyxoviridae (Hendra virus and Nipah virus) as VHF viruses. While they are not associated with hemorrhage many other aspects of the epidemiology and clinical presentation of these zoonotic viral infections show commonalities with the established VHFs.2
Several outbreaks of VHF in humans are recorded each year globally.3 With the glaring exception of the recent Ebola virus disease (EVD) epidemic in West Africa, VHF outbreaks are typically small, limited to less than 100 patients. The median number of patients for the 17 previous EVD outbreaks is 65.4 Possibly due to the generally limited size of outbreaks, viruses associated with VHF have not been considered a priority for research funding. Consequently, existing diagnostics and therapeutics are limited, as is our understanding of the epidemiology, transmission, and animal reservoirs for some of these viruses. However, the recent EVD epidemic in West Africa has shown that VHF outbreaks can occur where least expected (eg, West Africa, whereas most previous outbreaks were in Central Africa)4, 5 and can rapidly spread out of control. As of February 28, 2016, the recent West African EVD outbreak had infected nearly 29 000 people, with more than 11 000 deaths.6 Fragile and under‐resourced health systems in these countries were sluggish in identifying the disease and were unable to respond rapidly and comprehensively enough to stop the spread of the disease.7 The situation was further compounded by an initially slow and uncoordinated international response that has been widely condemned.8, 9, 10, 11 The unprecedented magnitude of the West African EVD outbreak, along with the significant number of EVD survivors with persistent detectable virus in various body fluids (semen and ocular fluid) after recovering from the disease12, 13 and/or complications14 plus the discovery that large numbers of people with no history of VHF are seropositive for Ebola virus,15, 16 has challenged our previous notions of the acute nature of these viral infections of humans and called to question our previous low‐priority categorization of these infections with respect to research and health program funding. A retrospective study from Sierra Leone documented serological evidence for infection with a range of VHF viruses (including Ebola and Marburg) in 2% to 8% of patients using acute phase sera from Lassa virus negative febrile patients (collected October 2006‐2008), suggesting that there could be Ebola and Marburg infections that are not characterized by rampant human‐to‐human transmission,17 similar to the established endemic nature of viruses like Dengue virus, Lassa virus, Hantavirus,18 and Rift Valley fever virus.19, 20 As of 2016 the EVD epidemic is no longer out of control, but flare‐ups continue: on March 17 Sierra Leone declared an end to a flare‐up that started in January, yet on the very same day, a new patient was confirmed in Guinea leading to 5 deaths as of March 24, 2016, prompting Liberia to close their shared border. This experience emphasizes the need to develop regional and national research networks to better understand the underlying causes of these outbreaks.
Lujo virus (LUJV) was discovered after an outbreak of VHF in Lusaka (Zambia) and Johannesburg (South Africa) in 2008 (Figure 1) and was the first novel VHF‐causing virus to be identified in Africa since the discovery of Ebola virus in 1976.21, 22 Although the LUJV outbreak was limited to just 5 people, mortality was high (80%), with the low threshold of suspicion of VHF among health care workers resulting in diagnostic delay and nosocomial transmission. Here, we review the Lujo VHF outbreak of 2008 in light of the lessons learnt from the recent EVD epidemic in West Africa and the current Zika virus (ZIKV) disease epidemic in the Americas and discuss the possible measures that could be taken by health authorities in Zambia and regionally, to efficiently integrate timely diagnosis of rare zoonotic diseases into existing health care, laboratory infrastructure, and human resource capacity development programmes.
Figure 1.
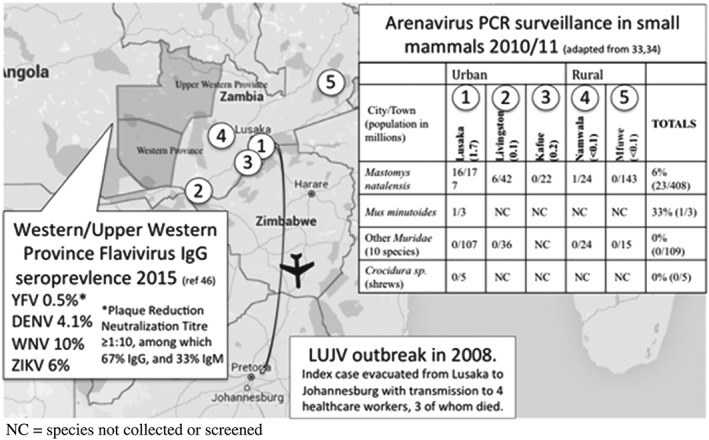
Map illustrating cross border transmission of Lujo virus (LUJV) in 2008, the results of small mammal Arenavirus surveillance in 2010/2011, and Flavivirus seroprevalence studies undertaken in Zambia in 2015. NC denotes that species are not collected or screened. DENV, Dengue virus; PCR, polymerase chain reaction; WNV, West Nile virus; YFV, Yellow fever virus; ZIKV, Zika virus
2. The Lujo Viral Hemorrhagic Fever Outbreak of 2008
In Zambia and South Africa in 2008, a novel arenavirus (LUJV) infected 5 people, killing the index case and 3 health care workers. The index case was a white female aged 36 years who lived on a peri‐urban farm close to Zambia's capital, Lusaka. On September 2, she experienced a sudden onset of severe headache, myalgia, fever, and sore throat and self‐medicated with antipyretics and analgesics.23, 24 On September 4 she travelled by air to South Africa to attend a wedding on September 6, returning to Zambia on September 7 (day 5 of her illness), when she reported diarrhea and vomiting24 (reports diarrhea and vomiting on day 2). Her condition continued to worsen such that on day 7 of her illness she visited a private clinic in Lusaka complaining of severe chest pains, fever, rash, and sore throat for which she was given an assortment of medications (including antiemetic, antipyretic, analgesic, and broad spectrum antibiotics). The next 2 days, her condition rapidly degenerated as she experienced severe myalgias, facial swelling with central nervous system symptoms such as confusion and seizures. She was hospitalized on day 9 and evacuated the following day by air ambulance to a private hospital in Johannesburg, South Africa. On physical examination, the patient exhibited edema of the face and neck, rash, and acute respiratory distress syndrome, but no hemorrhage was observed. Clinical laboratory tests showed that she had elevated liver transaminases, thrombocytopenia, and granulocytosis. The observation of a possible tick bite lead to a tentative diagnosis of Rickettsiosis, and the patient received intravenous cefepime, clarithromycin, and linezolid, along with lactated Ringer's solution and dobutamine.24 Although intensive care treatment was instituted, together with hemodialysis and inotropic and vasopressor therapy, the patient's condition degenerated rapidly with hemodynamic collapse and death on day 13 of her illness. No postmortem was conducted.
Patients 2 to 5 are described in detail elsewhere24 and included 1 paramedic (patient 2) involved in the initial evacuation of the index case. Patient 2 was diagnosed with suspected thrombotic thrombocytopenic purpura, which was then changed to suspected viral haemorrhagic fever a day later, after the epidemiological link with patient 1 was made.24 Patient 3 was an intensive‐care‐unit nurse that cared for the index case, and patient 4 was a cleaner who disinfected the room after the death of the index case. Patients 2 to 4 fell ill 9 to 13 days after probable exposure/contact with the index case, and all resulted in death. All 3 nosocomially transmitted patients were unwell for 10 to 13 days in the community before they were admitted and an epidemiological link with the index case established as well as VHF infection control measures implemented. Patient 3 was initiated on ribavirin on or around the same day that VHF was suspected in patient 2 (September 29 or 30, 2008). Patient 4 fell ill and sought care at her local clinic on September 27, but when seen as an outpatient at her local hospital 6 days later (3 days after the VHF alert and contact tracing commenced), she was initiated on therapy for tuberculosis (TB). She was admitted 2 days later, at which point the contact tracing team made contact with her, and she was referred to the teaching hospital for treatment.
Patient 5 was a 47‐year‐old white female who also worked in the intensive care unit and had contact with patient 2 (but not with the index case), just 2 days before the VHF alert was raised. There were noted lapses in personal protection, but fortunately by the time she fell ill she was known to the contact tracing team, and ribavirin was administered on day 2 of her illness based on suspected VHF. After being given ribavirin, patient 5 became seriously ill needing mechanical ventilation, but gradually recovered and was discharged after 42 days in hospital. She suffered prolonged neurological sequelae for up to 6 months after discharge from hospital.24
The clinical presentation and course of Lujo VHF was quite consistent across all 4 fatal patients, starting with myalgia, headache, and fever, followed by onset of rash and pharyngitis on days 4 and 5. Vomiting and diarrhea were present from days 3 to 7, and then the condition deteriorated with thrombocytopenia and elevated transaminases, severe neurological symptoms, hemodynamic collapse, and death.24 Patient 5 received many of the same treatments as patients 1 to 4, with the key differences that might have contributed to her survival being prompt initiation of treatment with ribavirin, recombinant factor VIIa, N‐acetylcysteine, and atorvastatin.24
3. Old World and New World Arenaviruses
The family Arenaviridae consists of 2 genera, Mammarenavirus and Reptarenavirus, which infect mammals and reptiles, respectively.25 Arenavirus particles are enveloped and spherical in shape and possess a bisegmented single‐stranded ambisense RNA genome comprising a large (L) and small (S) RNA segment, each contained within its own helical nucleocapsid.26 The L segment encodes a viral RNA‐dependent RNA polymerase and a smaller protein termed Z‐protein. The S segment encodes a viral nucleoprotein and viral glycoprotein precursor (Figure 2). Based on antigenic properties, geographical distribution, and phylogenetic analysis; mammalian arenaviruses are divided into 2 distinct groups: New World (NW) arenaviruses (Tacaribe serocomplex) and Old World (OW) arenaviruses (Lassa‐lymphocytic choriomeningitis serocomplex)25 (Figure 2). The NW arenaviruses that are known to infect humans include Junín virus, Guanarito virus, Machupo virus, Sabìa virus, and Chapare virus. Although LUJV is only the third OW arenavirus that is known to be pathogenic in humans, along with Lassa fever virus and lymphocytic choriomeningitis virus (Table 1), studies utilizing modern molecular tools including next generation sequencing technology are rapidly identifying new arenaviruses in rodent hosts.27 Epidemiologically, the assumption is that these viruses are generally well adapted to their rodent hosts, and those that might be pathogenic in humans cause only mild febrile illness, otherwise more arenaviruses would have been previously discovered. NW arenaviruses appear to be more commonly associated with human disease, possibly influenced by the use of different receptors28: OW arenaviruses such as Lassa fever virus use α‐dystroglycan (αDG) as a cellular receptor, which may be highly prevalent in the membranes of monocytes and dendritic cells,29 but the natural ligand of αDG, laminin, does not prevent virus infection in vitro and other candidate receptors (Axl, Tyro3, LSECtin and DC‐SIGN), including some shared with Ebola, have been shown in vitro to facilitate cell entry.30 The primary receptor for NW arenaviruses is transferin receptor 1 (TfR1), which is widely distributed and would facilitate a broad cell tropism,31 and there is in vitro evidence that even a single mutation can confer tropism to human cells.32
Figure 2.
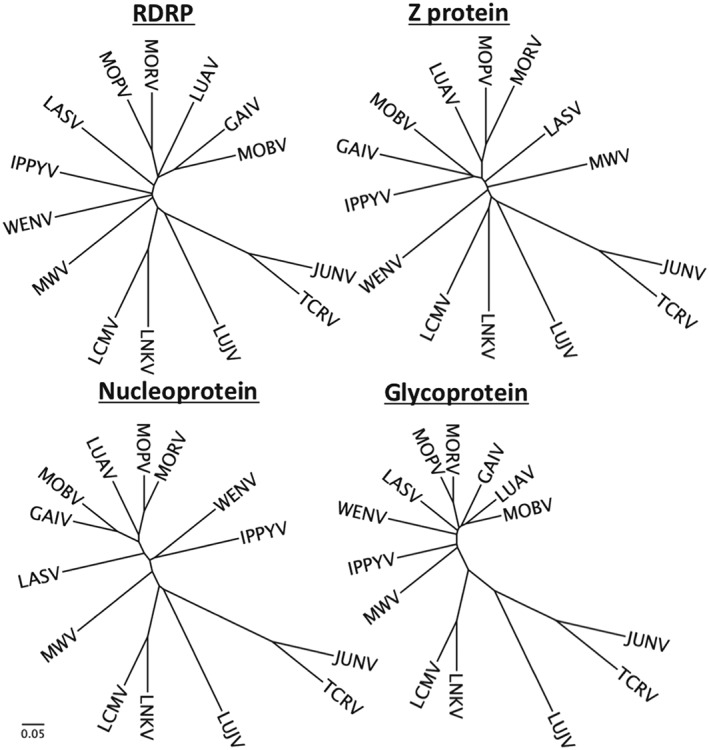
Phylogentic trees of all 4 arenavirus‐encoded proteins for representative Old World viruses, along with Junín virus (JUNV) and Tacaribe virus (TCRV). Phylogenetic trees of amino acid sequences are generated on Clustal Omega using default parameters. NCBI accession numbers and sequence files used are available on request from corresponding author. Scale is equal to substitutions per site. Lassa (strain Josiah), Lymphocytic choriomeningitis virus (LCMV) (strain Armstrong). GAIV, Gairo virus; IPPYV, Ippy virus; LASV, Lassa fever virus; LNKV, Lusaka New‐Kasama virus; LUAV, Lusaka‐Namwala virus; LUJV, Lujo virus; MOBV, Mobala virus; MOPV, Mopeia virus; MORV, Morogoro virus; MWV, Merino walk virus; RDRP, RNA‐dependent RNA polymerase; WENV, Wenzhou virus
Table 1.
Summary of mammalian arenaviruses and their associated epidemiological featuresa
| Virus, Abbreviation and Isolation/Detection Date | Isolated | Lineage/Clade | Natural Host | Geographic Distribution | Disease in Humans |
|---|---|---|---|---|---|
| Old World Arenaviruses | |||||
| Lymphocytic choriomeningitis virus, LCMV, 1933 | Yes | LCM | Mus musculus Linnaeus (house mouse) Apodemus sylvaticus Linnaeus (long‐tailed field mice) | Americas, Europe | Undifferentiated febrile illness, aseptic meningitis; rarely serious. Laboratory infections common, usually mild but 5 fatal cases. |
| Lassa virus, LASV, 1969 | Yes | Lassa | Mastomys sp. (Multimammate rat) | West Africa, imported cases in Europe, Japan, USA | Lassa fever; mild to severe and fatal disease. Laboratory infection common and often severe. |
| Mopeia virus, MOPV, 1977 | Yes | Mopeia | Mastomys natalensis (Multimammate rat) | Mozambique, Zimbabwe | Unknown |
| Mobala virus, MOBV, 1983 | Yes | Mobala | Praomys sp. (soft‐furred mouse) | Central African Republic | Unknown |
| Ippy virus, IPPYV, 1984 | Yes | Lassa | Arvicanthis sp. (unstriped grass rats) Praomys sp. (soft‐furred mouse) | Central African Republic | Unknown |
| Merino Walk, MWV, 1985 | Yes | Merino | Myotomys unisulcatus sp. (Busk Karoo rat) | South Africa | Unknown |
| Menekre, 2005 | No | Mopeia | Hylomyscus sp. (African wood mouse) | Ivory Coast | Unknown |
| Gbagroube, 2005 | No | Lassa | Mus (Nannomys) setulosus (African pigmy mouse) | Ivory Coast | Unknown |
| Morogoro, 2007 | No | Mopeia | Mastomys natalensis (Multimammate rat) | Tanzania | Unknown |
| Kodoko, 2007 | Yes | LCM | Mus (Nannomys) minutoides (savannah pygmy mouse) | Guinea | Unknown |
| Lujo virus, LUJV, 2008 | Yes | Lujo | Unknown | Zambia, South Africa | Fatal hemorrhagic fever |
| Lemniscomys, 2008 | No | Lassa | Lemniscomys rosalia (Single‐striped grass mouse) Mastomys natalensis (Multimammate rat) | Tanzania | Unknown |
| Lunk virus, LNKV, 2008 | No | LCM | Mus minutoides (savannah pygmy mouse) | Tanzania | Unknown |
| Luna virus, LUAV, 2009 | Yes | Lusaka‐Namwala | Mastomys natalensis (Multimammate rat) | Zambia | Unknown |
| Whenzou, 2014 | No | Rattus norvegicus (Brown rat) | China | Unknown | |
| Gairo, 2015 | No | Mobala | Mastomys natalensis (Multimammate rat) | Tanzania | Unknown |
| New World Arenaviruses | |||||
| Tacaribe, 1956 | Yes | B | Originally isolated from Artibeus sp. (bats) but later in vivo experiments on Artibeus jamaciensis suggested they are not the reservoir hosts54 | Trinidad, West Indies | Unknown. One suspected laboratory case that was moderately symptomtic. |
| Junín, 1958 | Yes | B | Calomys musculinus (drylands vesper mouse) | Argentina | Argentinian hemorrhagic fever. Laboratory infection common often severe. |
| Machupo, 1963 | Yes | B | Calomys callosus (large vesper mouse) | Bolivia | Bolivian hemorrhagic fever. Laboratory infection common often severe. |
| Cupixi, 1965 | Yes | B | Oryzomys gaeldi (rice rat) | Brazil | Unknown |
| Amapari,1965 | Yes | B | Neacomys guianae (Guiana Bristly mouse) | Brazil | Unknown |
| Parana, 1970 | Yes | A | Oryzomys buccinatus (Paraguayan Rice Rat) | Paraguay | Unknown |
| Tamiami, 1970 | Yes | A | Sigmodon hispidus (hispid cotton rat) | Florida, USA | Antibodies detected |
| Pichinde, 1971 | Yes | A | Oryzomys albigularis (Tomes's Rice rat) | Colombia | Occasional mild laboratory infection. |
| Latino, 1973 | Yes | C | Calomys callosus (large vesper mouse) | Bolivia | Unknown |
| Flexal, 1977 | Yes | A | Oryzomys spp. (Rice rats) | Brazil | One severe laboratory infection recorded |
| Guanarito, 1989 | Yes | B | Zygodontomys brevicauda (Short‐tailed Cane mouse) | Venezuela | Venezuelan hemorrhagic fever |
| Sabia, 1993 | Yes | B | Unknown | Brazil | Viral hemorrhagic fever, 2 severe laboratory infections recorded. |
| Oliveros, 1996 | Yes | C | Bolomys obscuris (Dark bolo mouse) | Argentina | Unknown |
| Whitewater Arroyo, 1997 | Yes | D | Neotoma spp. (Wood rats) | USA: New Mexico, Oklahoma, Utah, California, Colorado | Unknown |
| Pirital, 1997 | Yes | A | Sigmodon alstoni (Alston's Cotton Rat) | Venezuela | Unknown |
| Pampa, 1997 | Yes | Bolomys sp. | Argentina | Unknown | |
| Bear Canyon, 1998 | Yes | D | Peromyscus californicus (California mouse), Neotoma macrotis (large‐eared woodrat) | USA: California | Unknown |
| Ocozocoautla de Espinosa, 2000 | No | B | Peromyscus mexicanus (Mexican deer mouse) | Mexico | Unknown |
| Allpahuayo, 2001 | Yes | A | Oecomys bicolor, (Bicolored Arboreal Rice Rat) Aecomys paricola | Peru | Unknown |
| Tonto Creek, 2001 | Yes | D | Neotoma albigula (white‐throated woodrat) | USA: Arizona | Unknown |
| Big Brushy Tank, 2002 | Yes | D | Neotoma albigula (white‐throated woodrat) | USA: Arizona | Unknown |
| Real de Catorce, 2005 | No | D | Neotoma leucodon (White‐toothed Woodrat) | Mexico | Unknown |
| Catarina, 2007 | Yes | D | Neotoma micropus (Southern Plains Woodrat) | USA: Texas | Unknown |
| Skinner Tank, 2008 | Yes | D | Neotoma mexicana (Mexican woodrat) | USA: Arizona | Unknown |
| Chapare, 2008 | Yes | B | Unknown | Bolivia | Single fatal hemorrhagic fever case |
| Middle Pease River, 2013 | No | D | Neotoma micropus (southern plains woodrats) | USA: Oklahoma, Texas, New Mexico | Unknown |
| Patawa, 2015 | Yes | A | Oecomys spp. (Arboreal Rice Rat) | French Guiana | Unknown |
| Pinhal, 2015 | No | ? | Calomys tener (Delicate vesper mouse) | Brazil | Unknown |
Adapted from (26, CDC website).
4. Searching for the Lujo Virus Reservoir Host
There have been 2 studies aimed at finding the natural animal host of LUJV and to more broadly investigate the prevalence and molecular epidemiology of arenaviruses in rodents and small mammals in Zambia.33, 34 Combining data from both studies, arenaviruses were identified in kidney tissues by polymerase chain reaction (PCR) in about 6% (23/408) of captured Natal multimammate rodents (Mastomys natalensis) and 33% (1/3) of African Pygmy Mice (Mus minutoides). Among 114 other animals tested (mainly Muridae species) no arenaviruses were detected (Figure 1). Ninety six per cent (23/24) of arenavirus positive rodents were captured in peri‐urban environments close to large human populations (Figure 1). Although the studies did not detect LUJV, 2 other novel arenaviruses were identified: Lusaka‐Namwala Virus,33 a Lassa fever‐like virus; and Lusaka New‐Kasama virus,34 a novel lymphycytic choriomeningitis‐related virus. The capacity of these novel viruses to infect humans is unknown.
5. Phylogenetic Analysis of Lujo virus
For other segmented RNA viruses, most notably influenza virus and SARS‐CoV (severe acute respiratory syndrome coronavirus), reassortment and/or recombination are central to their importance as human pathogens, giving rise to the sudden emergence of novel species of global pandemic potential. There has hence been great concern that arenaviruses, with their established capacity to cause severe disease in humans, and their segmented RNA genomes could also give rise to novel species with pandemic potential. Recombinant mammarenaviruses have been produced in the laboratory for vaccine development purposes,36, 37 and reptarenaviruses are highly recombinant (due to the pet trade and the housing of diverse snake species in close proximity),38 but for wild‐type mammarenaviruses with their segmented genomes and overlapping host species, the evidence for reassorted or recombinant species of either NW or OW mammarenaviruses is weak.39, 40 The variable position of some OW arenaviruses on different branches, depending on which viral protein is analyzed, is suggestive of possible historical recombination events, but the branch lengths (Figure 2) and sequence identities (Table 2) suggest these events have been followed by significant divergence. When analyzing only a tiny fraction of the total number of quasi species in existence, more conserved regions might masquerade as evidence of recombination using some analysis tools.39 As indicated in Table 2, the viral nucleoprotein appears to be more conserved than the other 3 viral proteins.
Table 2.
Identity matrix showing amino acid percentage identity, for all 4 viral proteins, between LUJV and representative OW and NW arenaviruses
| Arenavirus Protein | ||||
|---|---|---|---|---|
| RDRP | Z | NP | GPC | |
| OW Arenviruses | ||||
| WENV | 45 | 43 | 60 | 44 |
| IPPYV | 46 | 42 | 57 | 42 |
| GAIV | 45 | 44 | 59 | 42 |
| MWV | 43 | 43 | 57 | 42 |
| LASV | 43 | 49 | 59 | 42 |
| MOBV | 44 | 46 | 59 | 43 |
| LUAV | 44 | 47 | 60 | 43 |
| MOPV | 45 | 51 | 57 | 43 |
| MORV | 45 | 44 | 57 | 43 |
| LNKV | 43 | 44 | 61 | 44 |
| LCMV | 43 | 46 | 60 | 44 |
| NW Arenavruses | ||||
| TCRV | 36 | 29 | 48 | 39 |
| JUNV | 37 | 30 | 48 | 40 |
Abbreviations: GAIV, Gairo virus; JUNV, Junín virus; LASV, Lassa fever virus; LCMV, Lymphocytic choriomeningitis virus; LNKV, Lusaka New‐Kasama virus; LUAV, Lusaka‐Namwala virus; MOBV, Mobala virus; MOPV, Mopeia virus; MORV, Morogoro virus; MWV, Merino walk virus; NW, New World; OW, Old World; TCRV, Tacaribe virus; WENV, Wenzhou virus.
Phylogenetically LUJV is interesting, because amino acid identities show it is clearly among the OW arenaviruses, phylogenetic trees of amino acid sequences for all 4 viral proteins, consistently suggest that LUJV is the closest OW relative of the NW arenaviruses (Figure 2). All 4 viral proteins are similarly positioned for LUJV with respect to their closest relatives (Lusaka New‐Kasama virus and lymphocytic choriomeningitis virus) (Figure 2) which makes a recent recombinational origin highly unlikely, suggesting LUJV is an established virus in nature but that we simply have not yet identified its reservoir host.
6. Epidemiology of Lujo virus
The index case had regular contact with animals because she kept dogs, cats, and horses at her premises, and the outbreak response team found evidence of rodents, the natural host of all known arenaviruses,23 around the stables. Patient 1 reportedly cut her shin on a broken bottle on August 30, 3 days before she became ill,23 and it is plausible that the wound came into contact with rodent feces/urine, but in Lusaka, whether on peri‐urban farms or in crowded townships, people are in close contact with rodents, and if the natural host is a common rodent species, it begs the question of why LUJV infections are not more common in humans? Taken together with previous surveillance studies that did not detect LUJV in 420 wild‐captured rodents,33, 34 it seems plausible to speculate that a rare and unlikely transmission event led to the infection of a human by LUJV. The environment around the farm would support other small mammal species (rabbits, genets, civets, etc.), but as arenaviruses seem to have coevolved with their rodent hosts, the phylogenetic evidence suggests that the natural host of LUJV should also be a rodent.35
It might be a rare species, or one that is rarely in contact with human settlement, and/or transmission to humans might require a vector such as a tick, which might explain the possible requirement for the presence of other domestic animals such as horses. While the main route of arenavirus transmission is through contact with urine or feces, the Tacaribe virus was purportedly isolated from mosquitoes as well as bats and has recently been detected in ticks.41 The physicians who attended the index case of Lujo VHF in South Africa recorded what they thought could be a potential tick bite on the patient's foot.24 Although this may be coincidental, future surveillance of ticks and mosquitoes for novel RNA viruses is possibly warranted, particularly in light of the recent ZIKV disease outbreak in the Americas42 and a recent next generation sequencing study of mosquitoes in China identified multiple novel flaviviruses.43
7. What Limited the Lujo Viral Hemorrhagic Fever Outbreak?
There are several features of the LUJV outbreak that may have contributed to the limited spread of the virus: the index case was relatively wealthy, living on a peri‐urban farm, and seeking care in a small private hospital. For this reason she had minimal contact with other people while she was ill. Also, human‐to‐human transmission of LUJV appears to occur in the late stages of the infection, maybe during the last 3 days before death,24 a likely smaller window of transmission compared with EVD.44 While the 2008 outbreak did not spread to urban populations, in a possible future scenario, an infected individual could travel to crowded urban centers, dramatically increasing the risk of uncontainable spread. At the private hospital involved in the LUJV outbreak, the level of awareness for possible VHF was low,24 and without intervention this is likely also to be the case at over‐crowded government clinics that serve poor communities in Lusaka. Health‐seeking behavior may involve visiting traditional healers that would also delay diagnosis, as documented in West Africa during the recent EVD epidemic.45 Zambia's high burden of human immunodeficiency virus/TB, malnutrition, and other diseases of poverty could also impact on the size and impact of a future outbreak. Taking all these factors into consideration, it would be dangerously complacent to think that the magnitude and spread of a potential future LUJV outbreak will be similar to that of 2008.
8. Lujo virus Diagnostic Preparedness
The unpredictable nature of VHF outbreaks presents a challenge to poorly resourced health systems across Africa, as to what level of resources we should commit to rare but potentially high‐impact outbreaks. The LUJV outbreak originated in Zambia, a country with no prior recorded VHF outbreak, although there is recent evidence from a flavivirus seroprevalence study undertaken in western and north‐western provinces of low‐level exposure to Yellow fever virus (plaque reduction neutralization titre ≥1:10 0.5% [66.6% IgG + ve. 33.3% IgM + ve]), Dengue virus (4.1% IgG + ve), West Nile virus (10% IgG + ve), and ZIKV (6% IgG + ve).46 A filovirus modeling study based on reservoir host distribution suggests Zambia is very low risk for Ebola, but conversely, is at the center of a putative “Marburg belt,” although there have been no recorded cases of Marburg VHF in Zambia.47 With ever increasing international travel within Africa, and globally, all countries are potentially at risk from human importation of VHF and should have in place some kind of diagnostic capacity, at the very minimum, to provide some kind of diagnostic service until regional/international assistance is mobilized.
For VHF outbreaks in Africa the process of pathogen identification has historically been outsourced to United States and European biosafety level 4 (BSL‐4) laboratories, but the development of rapid molecular diagnostic tests for known VHF pathogens, and the increasing availability of molecular diagnostic platforms on the continent, supported by human immunodeficiency virus and TB diagnostic capacity development initiatives, makes a national or regional primary diagnostic response highly feasible.48 WHO collaborating centers for VHF diagnosis now include 5 African research institutes, in South Africa, Gabon, Kenya, Uganda, and Senegal, but in late 2013, after the first reports of mysterious and sudden deaths in Guinea in December, it took 4 months before Ebola virus was identified on March 22, 2014, in European BSL‐4 laboratories.10 The subsequent international response has been widely criticized as being unacceptably slow,10 with this initial 4‐month window between infection of the index case and identification of the causal agent a key failure that allowed the virus to take hold and spread regionally. A range of factors, both human (population demographics, health seeking behavior, burial practices, government response, etc.) and viral (pathogenicity and transmissibility of the specific virus strain) have probable impact on eventual outbreak size and impact, but molecular confirmation of the presence of a hemorrhagic fever virus is now the seminal event that gives local and international health officials the confirmation they need to mobilize a comprehensive infection control response. Having functional molecular diagnostic capacity nationally or regionally is key to the control of future VHF outbreaks.
The first consideration for laboratory diagnosis of highly pathogenic viruses is biological safety. History has shown that laboratories are high‐risk environments,49 and there needs to be a comprehensive plan and standard operation procedures in place, to ensure worker safety and outbreak prevention. VHF viruses are BSL‐4 pathogens, but due to the cost of construction and maintenance, these facilities are available at just a few centers and are primarily required for infecting cell culture or culturing dangerous pathogens. For diagnosis in the field or at a national reference laboratory, the West African EVD outbreak has led to well‐established protocols for “relatively” safe collection of specimens and specimen handling for molecular diagnosis,50 with emphasis and training on appropriate personal protective equipment and specimen handling techniques. Importantly, these safety measures need to be applied to specimens collected from any contacts of the index case, before the specific etiological agent is confirmed. For known VHF pathogens there are more molecular diagnostic assays becoming available.48 WHO recently approved 6 new rapid diagnostic tests for EVD; 3 real‐time PCR tests, 2 immunochromatographic tests, and 1 multiplex PCR test.51 A modest stock of such diagnostics, including positive and negative controls, reordered on expiry, would cost little and could be embedded into ongoing training and skills development activities. In contrast to the traditional technology of cell culture, molecular techniques do not run the risk of amplifying infectious material.
In Zambia, the University of Zambia School of Veterinary Medicine BSL‐3 laboratory has been nominated by the Zambian Ministry of Health as the national outbreak response diagnostic facility. Diagnosis of suspected patients of VHF is currently performed using conventional real‐time PCR with sets of primers for the detection of Ebola, Marburg, Lujo, and Lassa fever viruses.52 Sanger sequencing facilities are also available but are of limited use for detecting unknown/novel VHF viruses (species or strains) that are not detected by the available assays. Plans are being drawn up to invest in next generation sequencing technology, through the new Illumina MiniSeq and/or Oxford Nanopore minION sequencer, the latter of which has already been used in the field to study the molecular epidemiology of Ebola.53 In the absence of suspected VHF patients, these technologies will be actively used for research projects on other infectious disease priorities, building the human resource capacity to offer rapid pathogen identification services in the event of future VHF or respiratory virus outbreaks.
9. Conclusions
Lujo virus causes severe hemorrhagic fever with highly permissive human‐to‐human transmission and high case fatality. The animal reservoir and mode of transmission to humans are unknown, and the virus is phylogenetically equidistant from other major OW arenaviruses. The limited nature of the LUJV outbreak in 2008 was fortuitous, but the identity, location, and scale of possible future arenavirus or other VHF outbreaks cannot be predicted. For this reason the development of diagnostic capacity across the region is essential to facilitate a rapid and effective response. For known VHF pathogens, national governments should ensure that appropriate and effective means for diagnostic response is embedded within their leading research institutions. For identifying novel VHF pathogens, the required technology is becoming increasingly more available and affordable and could be used for a range of research activities, training, and building up the skills and experience of personnel to respond effectively to novel infectious disease diagnostic challenges.
Abbreviations
- BSL
biosafety level
- EVD
Ebola virus disease
- LUJV
Lujo virus
- NW
New World
- OW
Old World
- PCR
polymerase chain reaction
- TB
tuberculosis
- VHF
viral hemorrhagic fever
- ZIKV
Zika virus
Simulundu, E. , Mweene, A. S. , Changula, K. , Monze, M. , Chizema, E. , Mwaba, P. , Takada, A. , Ippolito, G. , Kasolo, F. , Zumla, A. , and Bates, M. (2016) Lujo viral hemorrhagic fever: considering diagnostic capacity and preparedness in the wake of recent Ebola and Zika virus outbreaks. Rev. Med. Virol., 26: 446–454. doi: 10.1002/rmv.1903.
References
- 1. Wilson MR, Peters CJ. Diseases of the central nervous system caused by lymphocytic choriomeningitis virus and other arenaviruses. Handb Clin Neurol. 2014;123:671–681. doi: 10.1016/B978-0-444-53488-0.00033-X [DOI] [PubMed] [Google Scholar]
- 2. CDC: Viral Hemorrhagic Fevers Fact Sheet. Branch SP , (ed). CDC: Atlanta, 2016.
- 3. Rosenberg R. Detecting the emergence of novel, zoonotic viruses pathogenic to humans. Cell Mol Life Sci. 2015;72:1115–1125. doi: 10.1007/s00018-014-1785-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ebola Outbreaks 2000–2014. . http://www.cdc.gov/vhf/ebola/outbreaks/history/summaries.html [16th December 2015].
- 5. Peterson AT, Bauer JT, Mills JN. Ecologic and geographic distribution of filovirus disease. Emerg Infect Dis. 2004;10:40–47. doi: 10.3201/eid1001.030125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO: Ebola Situation Report . WHO: Geneva, 2016.
- 7. Tomori O. Will Africa's future epidemic ride on forgotten lessons from the Ebola epidemic? BMC Med. 2015;13:116. doi: 10.1186/s12916-015-0359-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Check HE. Ebola failures prompt WHO rethink. Nature. 2015;521:137. doi: 10.1038/521137a [DOI] [PubMed] [Google Scholar]
- 9. Gostin LO, Friedman EA. A retrospective and prospective analysis of the west African Ebola virus disease epidemic: robust national health systems at the foundation and an empowered WHO at the apex. Lancet. 2015;385:1902–1909. doi: 10.1016/S0140-6736(15)60644-4 [DOI] [PubMed] [Google Scholar]
- 10. Ippolito G, Di Caro A. Capobianchi MR. The chronology of the international response to Ebola in western Africa: lights and shadows in a frame of conflicting position and figures. Infect Dis Rep. 2015;7:5957. doi: 10.4081/idr.2015.5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ippolito G, Lanini S, Brouqui P, et al Ebola: missed opportunities for Europe‐Africa research. Lancet Infect Dis. 2015;15:1254–1255. doi: 10.1016/S1473-3099(15)00236-4 [DOI] [PubMed] [Google Scholar]
- 12. Deen GF, Knust B, Broutet N, et al Ebola RNA persistence in Semen of Ebola virus disease survivors—preliminary report. N Engl J Med. 2015. doi: 10.1056/NEJMoa1511410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodriguez LL, De Roo A, Guimard Y, et al Persistence and genetic stability of Ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179 Suppl (1): S170–S176. doi: 10.1086/514291 [DOI] [PubMed] [Google Scholar]
- 14. Gulland A. Thousands of Ebola survivors experience serious medical complications. BMJ. 2015;351:h4336. doi: 10.1136/bmj.h4336 [DOI] [PubMed] [Google Scholar]
- 15. Bellan SE, Pulliam JR, Dushoff J, Meyers LA. Ebola control: effect of asymptomatic infection and acquired immunity. Lancet. 2014;384:1499–1500. doi: 10.1016/S0140-6736(14)61839-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leroy EM, Baize S, Volchkov VE, et al Human asymptomatic Ebola infection and strong inflammatory response. Lancet. 2000;355:2210–2215. [DOI] [PubMed] [Google Scholar]
- 17. Schoepp RJ, Rossi CA, Khan SH, Goba A, Fair JN. Undiagnosed acute viral febrile illnesses, Sierra Leone. Emerg Infect Dis. 2014;20:1176–1182. doi: 10.3201/eid2007.131265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klempa B, Koulemou K, Auste B, et al Seroepidemiological study reveals regional co‐occurrence of Lassa‐ and Hantavirus antibodies in Upper Guinea, West Africa. Trop Med Int Health. 2013;18:366–371. doi: 10.1111/tmi.12045 [DOI] [PubMed] [Google Scholar]
- 19. Heinrich N, Saathoff E, Weller N, et al High seroprevalence of Rift Valley fever and evidence for endemic circulation in Mbeya region, Tanzania, in a cross‐sectional study. PLoS Negl Trop Dis. 2012;6: e1557. doi: 10.1371/journal.pntd.0001557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LaBeaud AD, Muchiri EM, Ndzovu M, et al Interepidemic Rift Valley fever virus seropositivity, northeastern Kenya. Emerg Infect Dis. 2008;14:1240–1246. doi: 10.3201/eid1408.080082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. After Marburg . Ebola. Lancet. 1977;1:581–582. [PubMed] [Google Scholar]
- 22. Ebola haemorrhagic fever in Zaire, 1976. Bull World Health Organ. 1978;56:271–293. [PMC free article] [PubMed] [Google Scholar]
- 23. Paweska JT, Sewlall NH, Ksiazek TG, et al Nosocomial outbreak of novel arenavirus infection, southern Africa. Emerg Infect Dis. 2009;15:1598–1602. doi: 10.3201/eid1510.090211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sewlall NH, Richards G, Duse A, et al Clinical features and patient management of Lujo hemorrhagic fever. PLoS Negl Trop Dis. 2014;8: e3233 doi: 10.1371/journal.pntd.0003233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Radoshitzky SR, Bao Y, Buchmeier MJ, et al Past, present, and future of arenavirus taxonomy. Arch Virol. 2015;160:1851–1874. doi: 10.1007/s00705-015-2418-y [DOI] [PubMed] [Google Scholar]
- 26. Management I: 00.003 . Arenaviridae. In: ICTVdB—The Universal Virus Database, version 3. 2006 ed. Columbia University: New York, USA, 2006.
- 27. Gryseels S, Rieger T, Oestereich L, et al Gairo virus, a novel arenavirus of the widespread Mastomys natalensis: genetically divergent, but ecologically similar to Lassa and Morogoro viruses. Virology. 2015;476:249–256. doi: 10.1016/j.virol.2014.12.011 [DOI] [PubMed] [Google Scholar]
- 28. McLay L, Liang Y, Ly H. Comparative analysis of disease pathogenesis and molecular mechanisms of New World and Old World arenavirus infections. J Gen Virol. 2014;95:1–15. doi: 10.1099/vir.0.057000-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sevilla N, Kunz S, Holz A, et al Immunosuppression and resultant viral persistence by specific viral targeting of dendritic cells. J Exp Med. 2000;192:1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shimojima M, Stroher U, Ebihara H, Feldmann H, Kawaoka Y. Identification of cell surface molecules involved in dystroglycan‐independent Lassa virus cell entry. J Virol. 2012;86:2067–2078. doi: 10.1128/JVI.06451-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Radoshitzky SR, Abraham J, Spiropoulou CF, et al Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abraham J, Kwong JA, Albarino CG, et al Host‐species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic new world clade B arenaviruses. PLoS Pathog. 2009;5:e1000358. doi: 10.1371/journal.ppat.1000358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ishii A, Thomas Y, Moonga L, et al Novel arenavirus, Zambia. Emerg Infect Dis. 2011;17:1921–1924. doi: 10.3201/eid1710.101452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ishii A, Thomas Y, Moonga L, et al Molecular surveillance and phylogenetic analysis of Old World arenaviruses in Zambia. J Gen Virol. 2012;93:2247–2251. doi: 10.1099/vir.0.044099-0 [DOI] [PubMed] [Google Scholar]
- 35. Briese T, Paweska JT, McMullan LK, et al Genetic detection and characterization of Lujo virus, a new hemorrhagic fever‐associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455 doi: 10.1371/journal.ppat.1000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng BY, Ortiz‐Riano E, de la Torre JC, Martinez‐Sobrido L. Generation of recombinant arenavirus for vaccine development in FDA‐approved Vero cells. J Vis Exp. 2013. doi: 10.3791/50662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koma T, Patterson M, Huang C, et al Machupo virus expressing GPC of the candid#1 vaccine strain of junín virus is highly attenuated and immunogenic. J Virol. 2016;90:1290–1297. doi: 10.1128/JVI.02615-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stenglein MD, Jacobson ER, Chang LW, et al Widespread recombination, reassortment, and transmission of unbalanced compound viral genotypes in natural arenavirus infections. PLoS Pathog. 2015;11:e1004900 doi: 10.1371/journal.ppat.1004900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emonet SF, de la Torre JC, Domingo E, Sevilla N. Arenavirus genetic diversity and its biological implications. Infect Genet Evol. 2009;9:417–429. doi: 10.1016/j.meegid.2009.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manning JT, Forrester N, Paessler S. Lassa virus isolates from Mali and the Ivory Coast represent an emerging fifth lineage. Frontiers in Microbiology. 2015;6:1037. doi: 10.3389/fmicb.2015.01037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sayler KA, Barbet AF, Chamberlain C, et al Isolation of Tacaribe virus, a Caribbean arenavirus, from host‐seeking Amblyomma americanum ticks in Florida. PLoS One. 2014;9: doi: 10.1371/journal.pone.0115769e115769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petersen E, Wilson ME, Touch S, et al Rapid spread of Zika virus in the Americas—implications for public health preparedness for mass gatherings at the 2016 Brazil Olympic Games. Int J Infect Dis. 2016;44:11–15. doi: 10.1016/j.ijid.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 43. Shi M, Lin XD, Vasilakis N, et al Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the flaviviridae and related viruses. J Virol. 2015;90:659–669. doi: 10.1128/JVI.02036-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carroll MW, Matthews DA, Hiscox JA, et al Temporal and spatial analysis of the 2014‐2015 Ebola virus outbreak in West Africa. Nature. 2015;524:97–101. doi: 10.1038/nature14594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manguvo A, Mafuvadze B. The impact of traditional and religious practices on the spread of Ebola in West Africa: time for a strategic shift. Pan Afr Med J. 2015;22 Supp (1): 9. doi: 10.11694/pamj.supp.2015.22.1.6190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Babaniyi OA, Mwaba P, Mulenga D, et al Risk assessment for Yellow fever in western and north‐western provinces of Zambia. Journal of Global Infectious Diseases. 2015;7:11–17. doi: 10.4103/0974-777X.150884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peterson AT, Carroll DS, Mills JN, Johnson KM. Potential mammalian filovirus reservoirs. Emerg Infect Dis. 2004;10:2073–2081. doi: 10.3201/eid1012.040346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Racsa LD, Kraft CS, Olinger GG, Hensley LE. Viral hemorrhagic fever diagnostics. Clin Infect Dis. 2016;62:214–219. doi: 10.1093/cid/civ792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martini GA. Marburg virus disease. Postgrad Med J. 1973;49:542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. WHO : How to safely collect blood samples by phlebotomy from patients suspected to be infected with Ebola. WHO: Geneva, 2016.
- 51. WHO : Emergency guidance—selection and use of Ebola in vitro diagnostic (IVD) assays. WHO: Geneva, 2015.
- 52. Ogawa H, Miyamoto H, Ebihara H, et al Detection of all known filovirus species by reverse transcription‐polymerase chain reaction using a primer set specific for the viral nucleoprotein gene. J Virol Methods. 2011;171:310–313. doi: 10.1016/j.jviromet.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quick J, Loman NJ, Duraffour S, et al Real‐time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cogswell‐Hawkinson A, Bowen R, James S, et al Tacaribe virus causes fatal infection of an ostensible reservoir host, the Jamaican fruit bat. J Virol. 2012;86:5791–5799. doi: 10.1128/JVI.00201-12 [DOI] [PMC free article] [PubMed] [Google Scholar]


