Summary
Studies have shown that the predictive value of “clinical diagnoses” of influenza and other respiratory viral infections is low, especially in children. In routine care, pediatricians often resort to clinical diagnoses, even in the absence of robust evidence‐based criteria.
We used a dual approach to identify clinical characteristics that may help to differentiate infections with common pathogens including influenza, respiratory syncytial virus, adenovirus, metapneumovirus, rhinovirus, bocavirus‐1, coronaviruses, or parainfluenza virus: (a) systematic review and meta‐analysis of 47 clinical studies published in Medline (June 1996 to March 2017, PROSPERO registration number: CRD42017059557) comprising 49 858 individuals and (b) data‐driven analysis of an inception cohort of 6073 children with ILI (aged 0‐18 years, 56% male, December 2009 to March 2015) examined at the point of care in addition to blinded PCR testing. We determined pooled odds ratios for the literature analysis and compared these to odds ratios based on the clinical cohort dataset.
This combined analysis suggested significant associations between influenza and fever or headache, as well as between respiratory syncytial virus infection and cough, dyspnea, and wheezing. Similarly, literature and cohort data agreed on significant associations between HMPV infection and cough, as well as adenovirus infection and fever. Importantly, none of the abovementioned features were unique to any particular pathogen but were also observed in association with other respiratory viruses.
In summary, our “real‐world” dataset confirmed published literature trends, but no individual feature allows any particular type of viral infection to be ruled in or ruled out. For the time being, laboratory confirmation remains essential. More research is needed to develop scientifically validated decision models to inform best practice guidelines and targeted diagnostic algorithms.
Keywords: children, clinical symptoms, respiratory viruses
List of Abbreviations
- Ab
antibody
- AE
asthma exacerbation
- altered/LOC
altered or loss of consciousness
- anorexia/DF
anorexia/difficulty feeding
- ARI
acute respiratory infection
- BALF
bronchoalveolar lavage fluids
- BCL
bronchiolitis
- CAP
community‐acquired pneumonia
- CC
case‐control
- CF
cystic fibrosis
- CI
confidence interval
- COH
(inception) cohort dataset
- CS
cross‐sectional
- DB
difficulty breathing
- DFA
direct immunofluorescence assay
- EIA
enzyme immunoassay
- EIFA
enzyme immunofluorescence assay
- ETA
endotracheal aspirates
- Flu
influenza
- FS
febrile seizure
- FRI
febrile respiratory illness
- HAdV
human adenovirus
- HBoV‐1
human bocavirus type 1
- HCoV
human coronavirus
- HHP‐6
human herpesvirus 6
- HMPV
human metapneumovirus
- HPIV
human parainfluenza virus
- HRV
human rhinovirus
- IFA
(indirect) immunofluorescence assay
- ILI
influenza‐like illness
- LIT
literature review dataset
- LRTI
lower respiratory tract infections
- NOS
number of studies
- NPA
nasopharyngeal aspirate
- NPS
nasopharyngeal swab
- NS
nasal swabs/secretions
- NW
nasal washing
- OP
observational prospective
- OPS
oropharyngeal swabs
- OR
observational retrospective
- PC
prospective cohort
- PNA
pneumonia
- (p)OR
(pooled) odds ratio
- PPV
positive predictive value
- PROSPERO
International Prospective Register of Systematic Reviews
- PS
pharyngeal swabs
- QI
quality improvement
- RKI
Robert Koch Institute
- RS
respiratory samples
- SARI
severe acute respiratory infection
- RT
rapid test,
- RTI
respiratory tract infection
- TA
tracheal aspirates
- TS
throat swabs
- URTI
upper respiratory tract infections
- WHO
World Health Organization
1. INTRODUCTION
Influenza and acute respiratory infections (ARI) are major contributors to disease burden in the pediatric age group1, 2, 3, 4 with highest mortality rates in resource‐limited settings.5, 6
It has been shown that the positive predictive value of a “clinical” influenza diagnosis in children is as low as 32%.7 In children in particular, influenza symptoms are often nonspecific, making it difficult to distinguish influenza infection from infection because of other respiratory viruses.8 The ability to make accurate “clinical diagnoses” is further hampered by the frequent succession of different respiratorys infection during the winter months.7
For pediatricians in acute care settings, however, it may not always be possible to perform virus diagnostics. Even if diagnostic tests are widely available, presumptive clinical diagnoses will still be influencing clinical decision‐making, such as the use of diagnostics, antivirals, and antibiotics. Clinical bias in the use of diagnostic testing may thus impair epidemiological surveillance and disease burden estimates.9
To address this question further, we explored which clinical features, according to the published literature, may be associated with ARI due to influenza, respiratory syncytial virus (RSV), human adenovirus (HAdV), human rhinovirus (HRV), human metapneumovirus (HMPV), human bocavirus‐1 (HBoV‐1), human parainfluenza virus (HPIV), and human coronavirus (HCoV). We then addressed the same question through analysis of a “real‐world” dataset based on a prospective surveillance of 6073 children aged 0 to 18 years, where detailed clinical presentations and virus diagnoses were assessed and documented in all patients, independent from routine care.10
The objectives of this analysis are as follows:
To identify clinical features linked to specific respiratory viral infection in pediatric clinical trials and observational studies published in Medline
To explore the same question in a real‐world dataset, derived from a pediatric inception cohort.
2. METHODS
2.1. Systematic literature review and meta‐analysis
We searched the English language literature published in Medline (PubMed) from January 01, 1996 to March 21, 2017.
The search protocol was registered in the International Prospective Register of Systematic Reviews (PROSPERO) No. CRD42017059557. The literature search was conducted on March 21, 2017. The search terms are listed in the following online document: https://www.crd.york.ac.uk/PROSPEROFILES/59557_PROTOCOL_20180419.pdf
The publications identified by the initial search were included only if (a) study participants were children 0 to 18 years of age; (b) studies were randomized/nonrandomized clinical trials, observational studies, and/or epidemiological reports; (c) ≥1 association between patients with confirmed viral infection and a clinical symptom had been reported; and if (d) a control group with patients testing negatively for the respective viruses was included.
We excluded animal or in vitro studies, adult studies, case series, studies lacking information on clinical features and outcomes, studies lacking virological data, studies lacking same virus‐negative control groups, studies where data could not be reliably categorized and extracted, overlapping studies addressing chronic conditions or other nonrespiratory infection as well as meta‐analyses, review papers, and conference papers.
In the first round of review, the following data were extracted independently: (1) study location (country), (2) study design, (3) age range, (4) cohort size/number of subjects, (5) sampling and laboratory method, and (6) presenting symptoms including respiratory and extrarespiratory symptoms. Full‐text publications were accessed for a second round of review. XM and BR independently reviewed studies against the predefined inclusion and exclusion criteria, and any eligible discrepancy was resolved by discussion among the reviewer team (3 researchers). The clinical symptoms were grouped into the following 19 distinct symptom categories: altered or loss of consciousness (altered/LOC), anorexia/difficulty feeding, apnea, conjunctivitis, cough, hypoxia, diarrhea, dyspnea, fever, headache, malaise, myalgia, rash, rhinitis, seizures, sore throat, signs of upper respiratory tract infection, vomiting, and wheezing/bronchoconstriction/signs of lower respiratory tract infection (henceforth labeled “wheezing”).
2.2. Inception cohort analysis
The literature review was compared to a well‐described clinical inception cohort11, 12, 13, 14, 15: From December 2009 to April 2015, a specifically trained quality improvement (QI) team performed predefined clinical assessments of 6073 influenza‐like illness (ILI) patients aged 0 to 18 years at the point of care.11, 12, 13, 14, 15 Influenza‐like illness case criteria were defined as evidence of fever with a body temperature ≥38°C and ≥1 respiratory symptom (including cough, rhinitis/coryza, red/sore throat, ear ache, dyspnea, tachypnea, labored breathing, wheezing) or a documented clinician diagnosis of ILI. Clinical assessments were as described previously.10 Nasopharyngeal swabs were collected in universal transport medium (Copan™, Copan Diagnostics, Murrieta, CA) and investigated at the National Reference Centre for Influenza at the Robert Koch Institute, Berlin, for 8 respiratory viruses. The QI program was approved by the institutional review board (Charité EA 24/008/10). Informed consent procedures were waived for the purpose of enhanced quality of care and infection control.10, 11, 12, 13, 14, 15
Nucleic acid was extracted by MagNA Pure 96 DNA and Viral NA Small Volume Kit (Roche Deutschland Holding GmbH, Mannheim, Germany), MagAttract Viral RNA M48 Kit (Qiagen, Hilden, Germany), or RTP DNA/RNA Virus Mini Kit (Invitek, Germany) according to the manufacturer's instructions using a specimen volume of 200, 300, and 400 μL, respectively. Twenty‐five microliters of extracted RNA were subjected to cDNA synthesis applying 200 U M‐MLV Reverse Transcriptase (Invitrogen, USA) in a total reaction volume of 40 μL.
Specimens were analyzed for influenza A and B, RSV, HMPV, HAdV, and HRV by real‐time PCR as published previously.10, 11, 16, 17, 18, 19 Investigation of HCoV (NL63, 229E, OC43, and HKU1), HPIV1‐4, and HBoV‐1 was performed in a total reaction volume of 15 μL containing 1× PCR buffer, 4 mM MgCl2, 0.2 mM dNTP with dUTP, 40 ng/μL BSA, 0.3 U Platinum Taq Polymerase primers and probes (as specified in Supporting Information 1) and 5 μL of cDNA (or nucleic acid for HBoV‐1). Amplification was carried out at 95°C for 300 seconds, followed by 45 cycles at 95°C for 15 seconds and 60°C for 30 seconds.
In summary, the QI program used an unbiased approach where all 19 predefined clinical features were assessed at the point of care, and all 8 viruses were tested in all ILI patients.
2.3. Comparative analysis
The comparative statistical analysis was performed using R with the Metaphor Package software.20 Clinical features associated with viral pathogens were determined independently using pooled odds ratios (pOR), with 95% confidence intervals (CI) for the literature review dataset (LIT) and odds ratio (OR) for the real‐world (inception cohort) dataset (COH). We used 2 × 2 contingency tables to analyze the association between a virus‐positive (versus virus‐negative) case and an individual clinical feature (present/absent) in the literature and inception cohort, respectively. If lower/upper limits of 95% CI were within 1 decimal point of 1.0, we did not consider these OR as significant.
Random effect models for meta‐analysis were applied.20 Heterogeneity testing was done using I 2 statistics. I 2 values <25% were considered low, 25% to 75% as moderate, and values >75% indicated high levels of heterogeneity.21 Publication bias was assessed using funnel plots. A symmetrical plot indicates a lack of publication bias.22 For each used OR calculation, we estimated the exact CI using the mid‐p method.23
3. RESULTS
3.1. Characteristics of the literature review
The initial Medline search yielded 1861 potentially relevant publications. After manual screening of all titles and abstracts, 666 publications were relevant to the topic. Of these, 47 eligible publications were included into the final analysis according to the predefined inclusion and exclusion criteria (Figure 1).
Figure 1.
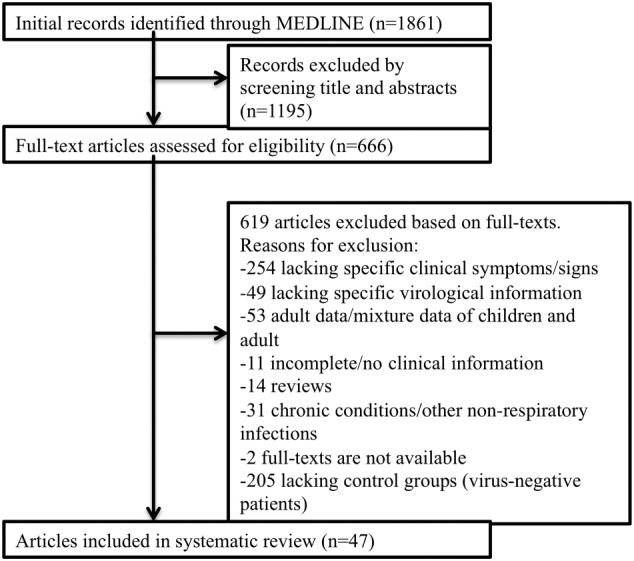
Flow chart describing the systematic literature search and selection of eligible publications
The 47 eligible publications are listed in Table 1.24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70 One quarter of these studies included children aged 0 to 5 years, and 6.4% and 12.8% of studies recruited children aged 0 to 3 years and aged 0 to 2 years, respectively. The systematic literature review yielded 9960 individual cases of laboratory‐confirmed ARI and 39 898 cases with negative test results for the same virus, respectively. The virology methods varied considerably: PCR was used in the majority of studies (88.9%), followed by enzyme‐linked immunoassays (14.9%), direct/indirect immunofluorescence (21.3%), and culture methods (6.4%). The geographic representation was rather broad, with 34.0% of study subjects stemming from the World Health Organization (WHO) region of the Americas, followed by the European Region (21.3%) and Western Pacific Region (21.3%). Fewer studies represented the WHO African Region (8.5%), the Eastern Mediterranean Region (8.5%), and the Southeast Asian Region (6.4%).
Table 1.
Characteristics of 47 eligible published studies
| Author, Year, Reference | Country | Design | Size | Age | Sample Type | Type of RTI | Lab Method | Pathogen |
|---|---|---|---|---|---|---|---|---|
| Ahn 201424 | Korea | OP | 1528 | ≤18 years | NPA | ARI | PCR | HBoV‐1 |
| Akhras 201025 | USA | OR | 256 | <18 years | NPS | ARI | DFA, culture, PCR | RSV, HMPV |
| Ali 201026 | Jordan | OP | 728 | <5 years | NS, TS | ARI | PCR | HMPV |
| Annamalay 201627 | Mozambique | OP | 277 | ≤10 years | NPA | RTI | PCR | HRV |
| Bhandary 201628 | India | CS | 100 | ≤5 years | NPA | RTI | DFA | RSV |
| Broor 201429 | USA | OP | 245 | <5 years | NS, TS | ARI | PCR | Flu A/B, RSV |
| Bryant 201030 | Australia | OP | 446 | ≤16 years | NPA, NS, TS | ILI | DFA, PCR | Flu A |
| Carballal 200231 | Argentina | OR | 168 | <2 years | NPA | Acute LRTI | IFA, culture | HAdV |
| Chang 201232 | USA | OP and CC | 5066 | ≤18 years | NS | ILI | PCR | Flu A |
| Chano 200533 | Canada | CC | 1132 | ≤18 years | NPA, BAL, ETA | RTI | DFA, culture, EIA, PCR | HMPV |
| Chen 201034 | China | OP | 6296 | ≤18 years | NPA | Acute LRTI | PCR | RSV, HMPV |
| Cuevas 200335 | Brazil | OP | 111 | <3 years | NS | Acute LRTI | PCR | RSV, HMPV |
| Esposito 201636 | Italy | OP | 307 | ≤18 years | NS | RTI | PCR | HAdV |
| Fairchok 201037 | USA | Cohort | 318 | ≤30 months | NS | RTI | PCR | Flu A |
| Farng 200238 | China | OR | 48 | ≤18 years | TS, serum | PNA | IFA | HAdV |
| Fischer Langley 201339 | Guatemala | OP | 2413 | <5 years | NPS, OPS | ARI | PCR | RSV |
| Flores 200440 | Portugal | OP | 225 | <3 years | NS | Acute BCL | PCR | RSV |
| Giamberardin 201641 | Brazil | CS | 250 | 24‐59 months | NS, OPS | RTI, asthma | PCR | Flu A/B, HRV, HAdV, HPIV, HCoV |
| Halasa 201542 | Jordan | OP | 3173 | <2 years | NS, TS | RTI, others | PCR | RSV |
| Hite 200743 | UK | CC | 411 | ≤18 years | NS | ILI | RT, culture | Flu A/B |
| Hombrouck 201244 | Belgium | OP | 139 | <5 years | NPS, TS | ILI | PCR | Flu A, RSV, HRV, HMPV, HPIV |
| Hsieh 201445 | China | OP | 1062 | ≤18 years | Serum | Flu season | Ab | Flu A |
| Huai 201746 | China | OP | 14479 | <15 years | NS | SARI | PCR | Flu A/B |
| Jevsnik 201247 | Slovenia | OP | 741 | <6 years | NPS, TS, TA, BAL, sputum | ARI | PCR | HCoV |
| Jin 201048 | China | OP | 645 | <16 years | NPA | ARI | PCR | HCoV |
| Khamis 2012 49 | Oman | OP | 259 | ≤5 years | RS | RTI | PCR | RSV |
| Kuo 201150 | China | CC | 308 | ≤18 years | NPS, TS | ILI | RT, PCR | Flu A |
| Lamarao 201251 | Brazil | CS | 1214 | ≤18 years | NPS | CAP | DFA, PCR | RSV |
| Landa‐Cardena 201252 | Mexico | CS | 124 | <6 years | NS | RTI | PCR | HRV |
| Leung 200953 | China | OP and OR | 1981 | <18 years | NPA | ARI | IFA, PCR | HCoV |
| Martin 201554 | Canada | OP | 219 | ≤2 years | Oral fluid | HHP‐6 history | PCR | HBoV‐1 |
| Moreno‐Valencia 201555 | Mexico | OP | 432 | <12 years | NPS | ARI | PCR | Flu A, RSV, HRV, HMPV, HPIV, HAdV |
| Nitsch‐Osuch 201356 | Poland | OP | 59 | ≤59 months | NS, PS | ILI | RT, PCR | Flu A/B |
| Nokes 200957 | Kenya | OP | 6026 | 1 day‐59 months | NS | PNA | DFA | RSV |
| Nyawanda 201658 | Kenya | OP | 4714 | <5 years | NPS, OPS | ARI | PCR | RSV |
| Pecchini 200859 | Brazil | OP | 455 | <5 years | NPS | Acute LRTI | IFA | RSV |
| Pierangeli 201260 | Italy | OP | 231 | ≤16 years | PS, nasal washing | ILI | PCR | Flu A, RSV, HRV |
| Ramagopal 201661 | India | OP | 80 | 1 month‐3 years | NPS | BCL | PCR | RSV |
| Saha 201062 | India | OP | 69 | 10 months‐12 years | NS, TS | Acute FRI | PCR | Flu A |
| Schuster 201563 | Jordan | OP | 3175 | <2 years | NS, TS | TRI, AE, CF, FS | PCR | HMPV |
| Smit 201264 | Netherlands | OP | 423 | ≤17 years | OPS, NW | ILI | PCR | Flu A |
| Smuts 201165 | South Africa | OP | 220 | 2 months‐5 years | NS | Cough, DB, wheezing | PCR | HRV |
| Tresoldi 201166 | Brazil | Cohort | 61 | ≤18 years | NPS, PS | ILI | PCR | Flu A |
| von Linstow 200467 | Denmark | OP | 374 | ≤2 years | NPS | TRI | IFA, EIFA, PCR | RSV, HMPV |
| Weigl 200368 | Germany | CC | 1316 | ≤2 years | NPA | LRTI | PCR | RSV |
| Yan 201769 | China | OP | 387 | 8 days‐15 years | NPA | Acute LRTI | PCR | RSV, HMPV |
| Zimmerman 201470 | USA | CC | 662 | <2 years | NS, OPS | URTI | PCR | Flu A/B, RSV, HRV, HMPV, HCoV |
Ab, antibody; AE, asthma exacerbation; ARI, acute respiratory infection; BALF, bronchoalveolar lavage fluids; BCL, bronchiolitis; CAP, community‐acquired pneumonia; CC, case‐control; CF, cystic fibrosis; CS, cross‐sectional; DB, difficulty breathing; DFA, direct immunofluorescence assay; EIA, enzyme immunoassay; EIFA, enzyme immunofluorescence assay; ETA, endotracheal aspirates; Flu, influenza; FS, febrile seizure; FRI, febrile respiratory illness; HHP‐6, human herpesvirus 6; IFA, (indirect) immunofluorescence assay; ILI, influenza‐like illness; LRTI, lower respiratory tract infection; NPA, nasopharyngeal aspirate; NPS, nasopharyngeal swab; NS, nasal swabs/secretions; NW, nasal washing; OP, observational prospective; OPS, oropharyngeal swabs; OR, observational retrospective; PC, prospective cohort; PNA, pneumonia; PS, pharyngeal swabs; RS, respiratory samples; SARI, severe acute respiratory infection; RT, rapid test; RTI, respiratory tract infection; TA, tracheal aspirates; TS, throat swabs; URTI, upper respiratory tract infection.
3.2. Associations between clinical features and viral infections based on literature review (LIT)
3.2.1. Fever and wheezing in influenza and RSV infections
In clinical practice, fever is often considered a hallmark of influenza disease, whereas wheezing is viewed as “typical” for RSV infections. Therefore, we studied these clinical associations in detail in the published literature (Figures 2 and 3). Of note, the pooled sample sizes in studies of influenza and RSV were highest (N = 24 661 and N = 29 426, respectively). The funnel and forest plots for other significant associations discussed below are provided in the Supporting Information 2 and 3.
Figure 2.
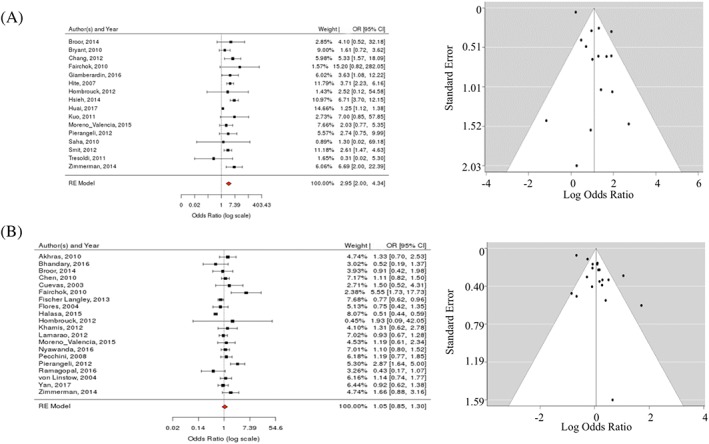
Relationship between fever and A, influenza versus B, RSV: LIT forest and funnel plots
Figure 3.
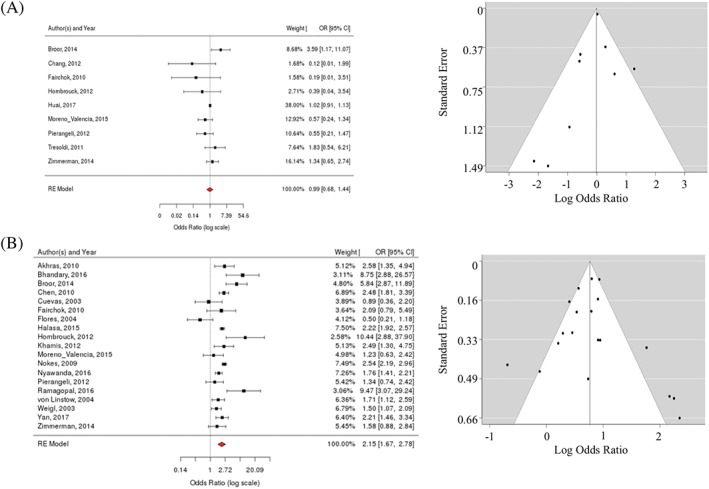
Relationship between wheezing and A, influenza versus B, RSV: LIT forest and funnel plots
Indeed, fever was the single most highly associated feature with regards to influenza infection (pOR = 3.0; 95% CI [2.0, 4.3]; I 2 = 66%) (Figures 2A and 4A). As evident from detailed literature analysis, most studies agreed on a positive correlation, with 1 exception.66 No evidence of publication bias was observed. There was no significant association in the meta‐analysis between fever and RSV (pOR 1.1; 95% CI [0.9, 1.3]; I 2 = 76%), albeit individual studies suggested (positive or negative) associations (Figure 2B).
Figure 4.
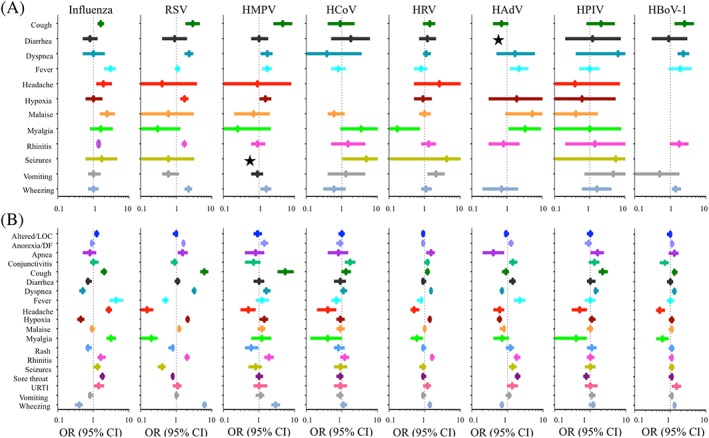
Summary of statistically significant (P < .05) features identified in A, the LIT dataset and in B, the COH dataset. ★ 95% CI exceeding scale: for seizure/HMPV, OR (95% CI) =16.6 (0.6, 438.1) and for or diarrhea/HAdV OR (95% CI) OR = 14.4 (2.5, 82.1)
The meta‐analysis (Figure 3B) also yielded a significant association between wheezing and RSV infection (pOR = 2.2; 95% CI [1.7, 2.8]; I 2 = 86%). In‐depth analysis of individual studies showed positive associations for most RSV studies, with 2 exceptions.35, 40 There was some publication bias in the RSV studies. Of note, wheezing was not significantly associated with influenza (pOR = 1.0; 95% CI [0.7, 1.4]; I 2 = 35%) (Figure 3A).
3.2.2. Clinical associations across all types of respiratory infection
Associations with fever or wheezing however are neither unique to influenza nor to RSV. When we studied the meta‐analyses across all 8 types of respiratory viral infection, multiple overlapping associations were easily identified for different types of respiratory viral infections (Figure 4A). In fact, most associations were shared across multiple types of viral infection, and no clinical feature stood out as unique to any specific type of infection.
In addition to influenza, fever was also significantly associated with HMPV (pOR = 1.7; 95% CI [1.2, 2.3]; I 2 = 45%) and HAdV infections (pOR = 2.2; 95% CI [1.2, 4.1]; I 2 = 11%). Additional features significantly associated with influenza infection included malaise (pOR = 2.4; 95% CI [1.5, 4.0]; I 2 = 48%), headache (pOR = 1.9; 95% CI [1.2, 3.3]; I 2 = 76%), cough (pOR = 1.6; 95% CI [1.3, 2.0]; I 2 = 19%), and rhinitis (pOR = 1.4; 95% CI [1.3, 1.6]; I 2 = 0%).
Wheezing was not only linked to RSV infections but also to HMPV (pOR = 1.6; 95% CI [1.1, 2.2]; I 2 = 54%) and HBoV‐1 infections (pOR = 1.4; 95% CI [1.1, 2.0]; I 2 = 0%). The strongest association with RSV infection in the literature was seen with cough and dyspnea (pORcough = 2.9; 95% CI [1.8, 4.6]; I 2 = 77% and pORdyspnea = 2.3; 95% CI [1.7, 3.0]; I 2 = 84%). Cough and dyspnea were also shared with HMPV infection (pORcough = 4.6; 95% CI [2.5, 8.6]; I 2 = 18% and pORdyspnea = 1.7; 95% CI [1.1, 2.4]; I 2 = 39%), and cough was also linked to influenza and HBoV‐1 infections.
3.3. New associations revealed in the inception cohort (COH)
The same clinical features were now tested in the clinical cohort (Figure 4B). The most striking difference was that associations in the COH dataset yielded narrower CI compared to the LIT dataset.
Several new (positive and negative) associations were revealed in the COH dataset that were not previously observed in the meta‐analysis. For example, influenza was positively linked to myalgia (OR 3.1; 95% CI [2.3, 4.3]) and sore throat (OR = 1.8; 95% CI [1.5, 2.1]). Wheezing (OR = 0.4; 95% CI [0.3, 0.5]) as well as hypoxia (OR = 0.4; 95% CI [0.4, 0.6]), dyspnea (OR = 0.5; 95% CI [0.4, 0.6]), rash, and diarrhea (both OR = 0.7; 95% CI [0.6, 0.9]) were negatively linked to influenza in the COH dataset.
With respect to RSV, anorexia/difficulty feeding and apnea were positively linked to that pathogen (OR = 1.6; 95% CI [1.4, 1.8] and OR = 1.5; 95% CI [1.1, 2.1] respectively). Additional negative associations were also revealed for RSV, namely fever (OR = 0.5; 95% CI [0.4, 0.6]), headache and myalgia (OR = 0.2; 95% CI [0.1, 0.2] and OR = 0.2; 95% CI [0.1, 0.3] respectively), seizures (OR = 0.4; 95% CI [0.3, 0.5]), rash, and sore throat (OR = 0.8; 95% CI [0.6, 0.9] and OR = 0.8; 95% CI [0.7, 0.9], respectively).
Of note, headache was negatively associated with all respiratory viral infections, except for influenza.
3.4. Agreements and disagreements between literature and cohort data (LIT/COH)
Figure 5 provides a “matrix view” of clinical features in relation to the 8 different types of viral infection, in direct comparison between LIT and COH data. This synopsis confirms that in most instances, COH data agreed with published literature trends, usually with higher OR and higher confidence levels (ie, narrower 95% CI) despite a smaller sample size. Exceptions were malaise/influenza, fever/HMPV, vomiting/HRV, and malaise/HAdV, which could not be confirmed in the COH data.
Figure 5.
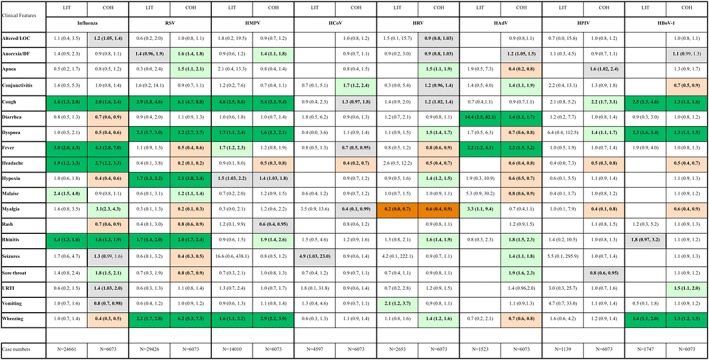
Comparison between literature review (LIT; pOR) and cohort data (COH; OR): Dark green color: positive agreement with statistically significant positive associations in both LIT and COH datasets. Dark red color: negative agreement with statistically significant negative associations in LIT and COH. Light green color: significant positive association in either LIT or COH, but not the other; light red color: significant negative association in either LIT or COH, but not the other; gray color: borderline‐significant associations (ie, CI values close to 1). N: number of study subjects with diagnostic testing and clinical data
The matrix view in Figure 5 also provides an overview of new negative (red) as well as positive (green) associations in the COH column/analysis that were not evident in the LIT column/analysis. Individual clinical features were not distinctive but shared among multiple types of viral infection, even if OR were not always significant.
The synopsis of meta‐analysis and inception cohort suggests that fever is significantly associated with influenza and HAdV infections, but again, the presence of fever does not rule out any of the other types of viral infection. HAdV infections were also linked to diarrhea. Cough is most likely present in influenza, RSV, HMPV, and HBOV‐1 infections but also observed in other respiratory infections. Wheezing was most prominent in RSV, HMPV, and HBOV‐1 infections. Wheezing was less likely to be seen in influenza and HAdV infections in the cohort, whereas the literature review revealed inconclusive results in this regard. HCoV infections showed no agreement bewteen LIT and COH datasets with no data available in the literature on a number of nonrespiratory symptoms. The only negative association confirmed in both LIT and COH datasets was between malaise and HRV infections.
4. DISCUSSION
We are presenting the first systematic literature review and meta‐analysis coupled with the analysis of a matching inception cohort, addressing the question whether respiratory viral infections in children can be differentiated based on individual clinical features. The prospective cohort confirmed several trends that were also identified in the meta‐analysis of the published literature. But the cohort dataset also established new significant associations between individual clinical features and viral infections.
Our matching analysis showed that systemic signs and symptoms such as fever and headache are more common in influenza and HAdV infections, whereas RSV, HMPV, and HBoV‐1 are more likely to manifest with respiratory symptoms such as wheezing, cough, and dyspnea. This trend was observed in the meta‐analysis and confirmed in the inception cohort. The approach of linking individual clinical features to specific viral infections however revealed a major diagnostic challenge to the clinician: None of the associations identified with either method were unique to any specific type of respiratory viral infection, but were shared across several types of infection. Individual clinical signs or symptoms can therefore not be used to reliably rule‐in or rule‐out any specific type of respiratory viral infection. For the time being, physicians need to be aware that clinical diagnoses are insufficiently sensitive, and laboratory testing will remain inevitable.
The results in our matched analysis are broadly in line with literature reviews by Ebell et al71 confirming the suspected association between fever and influenza. The findings by Ebell et al71 however are limited by the fact that viruses other than influenza (such as adenoviruses for example) were not studied. Thornton et al,72 the second literature review in this area, found that wheezing is associated with RSV infections, but this study again lacked associations with other respiratory viral pathogens tested in this study (such as HMPV and HBoV‐1). The latter also differed from the current study in that it was restricted to children with acute cough. Compared to previous reviews, our meta‐analysis included more recent literature sources and lager sample sizes (49 858 versus 6790 and 15 069, respectively) and a slightly greater number of countries (24 versus 4 and 20, respectively). This current meta‐analysis is also the only one linking to a prospective dataset.
Overall, literature data on RSV and influenza (and to a lesser degree, HMPV) seemed more readily available than literature on other respiratory viruses, which remain understudied in children. With the advent of new vaccines and antivirals against various respiratory viruses however, it will soon become critical to distinguish respiratory viral infections and to study the associated disease burden, including in the acute care setting.
The individual studies in our meta‐analysis showed high levels of heterogeneity, especially with regards to inclusion criteria and/or cutoff criteria for specific symptoms such as fever and hypoxia. For instance, 7 studies used a hypoxia definition of oxygen saturation <90% while 2 studies used higher thresholds of <92%29 and <95%60. Similarly, 11 studies defined fever as body temperature ≥38°C, 6 studies used cutoff values at 37.5°C,27,35,37,40,52,65 and 1 study 38.2°C73, while 22 studies did not define fever at all. Various clinical data collection methods, including phone interviews24, 29, 37, 39, 41 questionnaires,26, 27, 36, 40, 45, 49, 52, 58, 68 and surveys,70 were used to obtain information on clinical features; clinical data collection for these studies may thus have been subject to recall bias, interviewer bias, or misclassification bias. Additional issues may arise in the design of control groups in observational and cohort studies33, 68, 70 Among 6 case‐control studies, merely 2 used age‐matched or sex‐matched control groups43, 73 and 1 was randomized.50 Publication bias was also of concern in literature studies as funnel plots indicate that negative associations may have been missed in the published literature.
By contrast, the design of the inception cohort limited the risk of bias and heterogeneity through standardized clinical assessments in a predefined group of patients followed by independent laboratory and data analysis. In inception cohorts, the same data are solicited from all patients, and the same definitions/cutoff criteria for symptoms such as fever and hypoxia are used consistently. Standardized clinical and laboratory data collection in the QI program included predefined positive and negative findings, yielding a complete dataset for the analysis of positive and negative associations. Commonly referenced symptoms such as headache and myalgia may be underreported in infants and toddlers compared to older children. Headache and myalgia can only be elicited when age‐appropriate examination techniques are applied. To avoid observer bias in the inception cohort, a trained QI team elicited these symptoms accurately in all patients, regardless of age.
In clinical routine care in most settings, it will not be feasible to obtain virus diagnostics on the 8 most common respiratory viral pathogens in all patients with ILI, as was the case in this inception cohort. However, it will be important for clinicians to be conscious of awareness bias. One of the greatest challenges may lie in preconceived notions of “typical clinical presentations” in children with specific viral infections. The literature analysis may have revealed some of these inherent biases.
Clinical judgment is not only influenced by the lecture of journal articles: One may speculate that textbook knowledge (as acquired during medical school and residency training) also influences clinical decision‐making. The most commonly referenced pediatric textbook (Nelson's textbook of pediatrics), for example, states that influenza infection is dominated by systemic symptoms such as fever, myalgia, chills, headache, malaise, and anorexia.74 It also suggests that the onset of RSV infections is associated with rhinorrhea, cough, and wheezing, sometimes concomitant with a low‐grade fever.75 For these 2 diseases however, there are no literature references provided, nor specific guidance on the differential diagnosis in clinical practice. Nelson's textbook also states that symptoms of HAdV infection may be difficult to distinguish from similar illnesses caused by other pathogens, such as RSV, HPMV, or HRV.76 However, it does not mention the difficulty in differentiating influenza infections from HAdV and other respiratory viral infections in children, as identified in the current work.
It may be of interest that the mass media have recently picked up on the fact that “not all that looks like the flu may indeed be influenza”: Media reports by CNN, NBC News, and New York Daily News during the recent flu season emphasized that adenovirus infections may mimic symptoms otherwise attributed to influenza.77, 78, 79 This recent media attention underlines the importance of health messages to the general public, for example to avoid the false impression that “flu vaccines don't work.”
The current work has several strengths and limitations. The systematic review was restricted to publications in English and available in PubMed. Articles published elsewhere may have been missed. PubMed and the English language were chosen as they represent the most commonly accessed publications by clinicians.
A total of 205 literature studies had to be excluded because of lack of a “virus‐negative” control groups (Figure 1). In the inception cohort, each patient was simultaneously tested (+/−) for the same viral pathogens using highly sensitive and specific RT‐PCR assays at the National Reference Centre for Influenza at the Robert‐Koch Institute. Even though sample sizes were usually smaller in the inception cohort, prospective data collection resulted in higher confidence levels, because of a comprehensive dataset with predefined variables (8 viruses/19 clinical features) determined in all subjects.
The inception cohort dataset was derived from a single center and 6073 subjects, limiting the generalizability of these findings to other settings with different patient populations or health care systems. The literature meta‐analysis was more global in reach. Overall, cohort sizes were higher in the literature meta‐analysis focusing on influenza (N = 24661), RSV (N = 29462), and HMPV (N = 14010).
As mentioned above, the available literature data on HRV, HAdV, HPIV, and HBoV‐1 infections in children are relatively sparse, with sample sizes ranging from 1139 for HPIV to 2653 for HRV indicating selection bias. Published articles often focused on no more than 1 to 3 viral pathogens at a time and optional reporting of symptoms. The literature review is also limited by the inconsistency of laboratory methods used to detect respiratory viruses: Each published study used slightly different laboratory methods. In the inception cohort, a trained QI team obtained clinical specimens and delivered these to 1 National Reference Center.
Lastly, the effect of antiviral treatment or vaccination status on clinical features was not assessed in this analysis. Among the 6073 patients included in the inception cohort, only 3.3% received antivirals and influenza vaccination rates were 8.2% (data not shown). Future prospective studies or QI programs in different settings (for example in countries with universal influenza vaccination and treatment recommendations) may allow for the analysis of medical interventions.80
5. CONCLUSIONS
We showed that point‐of‐care clinical assessments via mobile application represent a powerful mechanism to identify “typical clinical features” likely to be associated with a specific viral infection.
Many clinical features are shared across different types of respiratory viral infection. This means that even though significant associations between individual clinical features and viral infections have been identified, clinical symptoms alone cannot be used to predict specific respiratory viral infections in a particular patient. Clinicians should be aware that clinical features alone will not “rule‐in” or “rule‐out” any specific type of viral infection. Diagnostic testing for respiratory viruses will remain the cornerstone of accurate diagnoses. Testing should be encouraged to prevent unnecessary prescriptions of antivirals in “similar‐looking” noninfluenza cases, where neuraminidase inhibitors would be ineffective.81, 82
Methodologically, prospective data collection may be more effective in identifying clinical associations than large‐scale meta‐analyses of the medical literature. While some trends in the literature are confirmed, additional features were identified through the inception cohort. In the future, complex decision models considering combinations of symptoms rather than individual features may be more useful to inform best practice. Machine‐learning algorithms may show the way toward “smart” decision software and the targeted use of diagnostics and antivirals.
FUNDING INFORMATION
Robert Koch Institute supported the laboratory work. The Vienna Vaccine Safety Initiative provided the clinical data collection methodology and the design of the QI program and literature review. XM was financially supported by a full tuition scholarship by the Chinese Scholarship Council (201508080019). He received startup funding by the Capital Institute of Pediatrics, Beijing, China.
AUTHOR CONTRIBUTIONS
XM wrote the initial draft of the manuscript and contributed significantly to the laboratory data collection. BR designed the QI program and the literature review and supervised the writing of the manuscript. BS supervised the laboratory work and contributed significantly to the writing of the manuscript. TC conducted the statistical analysis and contributed to the writing of the manuscript. JR supported the laboratory work and contributed significantly to the writing of the manuscript. MA as a key member of the QI staff contributed to the clinical data collection and to the writing of the manuscript. All authors have seen and approved the final version of the manuscript.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare with regards to the work presented.
Supporting information
Supporting Information 1: Primer and Probe sequences for HCoV, HPIV and HBoV‐1
Supporting Information 2: Forest plots and funnel plots for significant associations in the LIT dataset
Supporting Information 3: Heterogeneity analysis for significant associations in the LIT dataset
ACKNOWLEDGEMENTS
The authors kindly thank the members of the QI team, namely Katharina Karsch, Franziska Tief, Susann Muehlhans, Patrick Obermeier, Xi Chen, and Lea Seeber for their contribution to the clinical dataset. The authors also express their gratitude to the laboratory team at the Robert Koch Institute including Barbara Biere, Eleni Adamou, as well as the colleagues at the National Reference Centre for their helpful comments and review of the manuscript.
Ma X, Conrad T, Alchikh M, Reiche J, Schweiger B, Rath B. Can we distinguish respiratory viral infections based on clinical features? A prospective pediatric cohort compared to systematic literature review. Rev Med Virol. 2018;28:e1997 10.1002/rmv.1997
REFERENCES
- 1. Rudan I, Chan KY, Zhang JS, et al. Causes of deaths in children younger than 5 years in China in 2008. Lancet. 2010;375(9720):1083‐1089. [DOI] [PubMed] [Google Scholar]
- 2. Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of child mortality in 2000‐13, with projections to inform post‐2015 priorities: An updated systematic analysis. Lancet. 2015;385(9966):430‐440. [DOI] [PubMed] [Google Scholar]
- 3. Tang JW, Lam TT, Zaraket H, et al. Global epidemiology of non‐influenza RNA respiratory viruses: Data gaps and a growing need for surveillance. Lancet Infect Dis. 2017;17(10):e320‐e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi T, McAllister DA, O'Brien KL, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. Lancet. 2017;390(10098):946‐958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walker CL, Rudan I, Liu L, et al. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381(9875):1405‐1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thompson WW, Shay DK, Weintraub E, et al. Influenza‐associated hospitalizations in the United States. JAMA. 2004;292(11):1333‐1340. [DOI] [PubMed] [Google Scholar]
- 7. Peltola V, Reunanen T, Ziegler T, Silvennoinen H, Heikkinen T. Accuracy of clinical diagnosis of influenza in outpatient children. Clin Infect Dis. 2005;41(8):1198‐1200. [DOI] [PubMed] [Google Scholar]
- 8. Monto AS, Gravenstein S, Elliott M, Colopy M, Schweinle J. Clinical signs and symptoms predicting influenza infection. Arch Intern Med. 2000;160(21):3243‐3247. [DOI] [PubMed] [Google Scholar]
- 9. Jefferson T, Smith S, Demicheli V, Harnden A, Rivetti A, Di Pietrantonj C. Assessment of the efficacy and effectiveness of influenza vaccines in healthy children: Systematic review. Lancet. 2005;365(9461):773‐780. [DOI] [PubMed] [Google Scholar]
- 10. Rath B, Conrad T, Myles P, et al. Influenza and other respiratory viruses: Standardizing disease severity in surveillance and clinical trials. Expert Rev Anti Infect Ther. 2017;15(6):545‐568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tuttle R, Weick A, Schwarz WS, et al. Evaluation of novel second‐generation RSV and influenza rapid tests at the point of care. Diagn Microbiol Infect Dis. 2015;81(3):171‐176. [DOI] [PubMed] [Google Scholar]
- 12. Chen X, Pouran Yousef K, Duwe S, et al. Quantitative influenza follow‐up testing (QIFT)—a novel biomarker for the monitoring of disease activity at the point‐of‐care. PLoS One. 2014;9(3):e92500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rath B, Tief F, Obermeier P, et al. Early detection of influenza A and B infection in infants and children using conventional and fluorescence‐based rapid testing. J Clin Virol. 2012;55(4):329‐333. [DOI] [PubMed] [Google Scholar]
- 14. Rath B, von Kleist M, Tief F, et al. Virus load kinetics and resistance development during oseltamivir treatment in infants and children infected with influenza A (H1N1) 2009 and influenza B viruses. Pediatr Infect Dis J. 2012;31(9):899‐905. [DOI] [PubMed] [Google Scholar]
- 15. Tief F, Hoppe C, Seeber L, et al. An inception cohort study assessing the role of pneumococcal and other bacterial pathogens in children with influenza and ILI and a clinical decision model for stringent antibiotic use. Antivir Ther. 2016;21(5):413‐424. [DOI] [PubMed] [Google Scholar]
- 16. Schulze M, Nitsche A, Schweiger B, Biere B. Diagnostic approach for the differentiation of the pandemic influenza a (H1N1) v virus from recent human influenza viruses by real‐time PCR. PLoS One. 2010;5(4):e9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Biere B, Bauer B, Schweiger B. Differentiation of influenza B virus lineages Yamagata and Victoria by real‐time PCR. J Clin Microbiol. 2010;48(4):1425‐1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chmielewicz B, Nitsche A, Schweiger B, Ellerbrok H. Development of a PCR‐based assay for detection, quantification, and genotyping of human adenoviruses. Clin Chem. 2005;51(8):1365‐1373. [DOI] [PubMed] [Google Scholar]
- 19. Reiche J, Jacobsen S, Neubauer K, et al. Human metapneumovirus: Insights from a ten‐year molecular and epidemiological analysis in Germany. PLoS One. 2014;9(2):e88342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Viechtbauer W. Conducting meta‐analyses in R with the metafor package. J Statistic Softw. 2010;36:1‐48. [Google Scholar]
- 21. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta‐analyses. BMJ. 2003;327(7414):557‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sterne JA, Egger M, Smith GD. Systematic reviews in health care: Investigating and dealing with publication and other biases in meta‐analysis. BMJ. 2001;323(7304):101‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kenneth J, Rothman SG. Modern epidemiology. 2nd ed. Philadelphia, United States: Lippincott‐Raven; 1998. [Google Scholar]
- 24. Ahn JG, Choi SY, Kim DS, Kim KH. Human bocavirus isolated from children with acute respiratory tract infections in Korea, 2010‐2011. J Med Virol. 2014;86(12):2011‐2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Akhras N, Weinberg JB, Newton D. Human metapneumovirus and respiratory syncytial virus: Subtle differences but comparable severity. Infect Dis Rep. 2010;2:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ali SA, Williams JV, Chen Q, et al. Human metapneumovirus in hospitalized children in Amman, Jordan. J Med Virol. 2010;82(6):1012‐1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Annamalay AA, Lanaspa M, Khoo SK, et al. Rhinovirus species and clinical features in children hospitalised with pneumonia from Mozambique. Trop Med Int Health. 2016;21(9):1171‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhandary R, Boloor R. Detection of respiratory syncytial virus using direct fluorescent antibody assay in paediatric patients with acute respiratory tract infection. J Clin Diagn Res. 2016;10. Dc10‐12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Broor S, Dawood FS, Pandey BG, et al. Rates of respiratory virus‐associated hospitalization in children aged <5 years in rural northern India. J Infect. 2014;68(3):281‐289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bryant PA, Tebruegge M, Papadakis G, et al. Clinical and microbiologic features associated with novel swine‐origin influenza A pandemic 2009 (H1N1) virus in children: A prospective cohort study. Pediatr Infect Dis J. 2010;29(8):694‐698. [DOI] [PubMed] [Google Scholar]
- 31. Carballal G, Videla C, Misirlian A, Requeijo PV, Aguilar MC. Adenovirus type 7 associated with severe and fatal acute lower respiratory infections in Argentine children. BMC Pediatr. 2002;2(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang ML, Jordan‐Villegas A, Evans A, et al. Respiratory viruses identified in an urban children's hospital emergency department during the 2009 influenza A (H1N1) pandemic. Pediatr Emerg Care. 2012;28(10):990‐997. [DOI] [PubMed] [Google Scholar]
- 33. Chano F, Rousseau C, Laferriere C, Couillard M, Charest H. Epidemiological survey of human metapneumovirus infection in a large pediatric tertiary care center. J Clin Microbiol. 2005;43(11):5520‐5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen X, Zhang ZY, Zhao Y, Liu EM, Zhao XD. Acute lower respiratory tract infections by human metapneumovirus in children in Southwest China: A 2‐year study. Pediatr Pulmonol. 2010;45(8):824‐831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cuevas LE, Nasser AM, Dove W, Gurgel RQ, Greensill J, Hart CA. Human metapneumovirus and respiratory syncytial virus, Brazil. Emerg Infect Dis. 2003;9(12):1626‐1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Esposito S, Zampiero A, Bianchini S, et al. Epidemiology and clinical characteristics of respiratory infections due to adenovirus in children living in Milan, Italy, during 2013 and 2014. PLoS One. 2016;11: e0152375(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fairchok MP, Martin ET, Chambers S, et al. Epidemiology of viral respiratory tract infections in a prospective cohort of infants and toddlers attending daycare. J Clin Virol. 2010;49(1):16‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Farng KT, Wu KG, Lee YS, Lin YH, Hwang BT. Comparison of clinical characteristics of adenovirus and non‐adenovirus pneumonia in children. J Microbiol Immunol Infect. 2002;35:37‐41. [PubMed] [Google Scholar]
- 39. Fischer Langley G, McCracken J, Arvelo W, et al. The epidemiology and clinical characteristics of young children hospitalized with respiratory syncytial virus infections in Guatemala (2007‐2010). Pediatr Infect Dis J. 2013;32(6):629‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flores P, Rebelo‐de‐Andrade H, Goncalves P, et al. Bronchiolitis caused by respiratory syncytial virus in an area of portugal: Epidemiology, clinical features, and risk factors. Eur J Clin Microbiol Infect Dis. 2004;23(1):39‐45. [DOI] [PubMed] [Google Scholar]
- 41. Giamberardin HI, Homsani S, Bricks LF, et al. Clinical and epidemiological features of respiratory virus infections in preschool children over two consecutive influenza seasons in southern Brazil. J Med Virol. 2016;88(8):1325‐1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Halasa N, Williams J, Faouri S, et al. Natural history and epidemiology of respiratory syncytial virus infection in the Middle East: Hospital surveillance for children under age two in Jordan. Vaccine. 2015;33(47):6479‐6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hite LK, Glezen WP, Demmler GJ, Munoz FM. Medically attended pediatric influenza during the resurgence of the Victoria lineage of influenza B virus. Int J Infect Dis. 2007;11(1):40‐47. [DOI] [PubMed] [Google Scholar]
- 44. Hombrouck A, Sabbe M, Van Casteren V, et al. Viral aetiology of influenza‐like illness in Belgium during the influenza A (H1N1) 2009 pandemic. Eur J Clin Microbiol Infect Dis. 2012;31(6):999‐1007. [DOI] [PubMed] [Google Scholar]
- 45. Hsieh YH, Tsai CA, Lin CY, et al. Asymptomatic ratio for seasonal H1N1 influenza infection among schoolchildren in Taiwan. BMC Infect Dis. 2014;14(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huai Y, Guan X, Liu S, et al. Clinical characteristics and factors associated with severe acute respiratory infection and influenza among children in Jingzhou, China. Influenza Other Respi Viruses. 2017;11(2):148‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jevsnik M, Ursic T, Zigon N, Lusa L, Krivec U, Petrovec M. Coronavirus infections in hospitalized pediatric patients with acute respiratory tract disease. BMC Infect Dis. 2012;12(1):365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jin Y, Song JR, Xie ZP, et al. Prevalence and clinical characteristics of human CoV‐HKU1 in children with acute respiratory tract infections in China. J Clin Virol. 2010;49(2):126‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khamis FA, Al‐Kobaisi MF, Al‐Areimi WS, Al‐Kindi H, Al‐Zakwani I. Epidemiology of respiratory virus infections among infants and young children admitted to hospital in Oman. J Med Virol. 2012;84(8):1323‐1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kuo CY, Huang YC, Huang CG, Tsao KC, Lin TY. Symptomatic predictors for 2009 influenza A virus (H1N1) infection with an emphasis for patients with a negative rapid diagnostic test. PLoS One. 2011;6(12):e28102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lamarao LM, Ramos FL, Mello WA, et al. Prevalence and clinical features of respiratory syncytial virus in children hospitalized for community‐acquired pneumonia in northern Brazil. BMC Infect Dis. 2012;12(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Landa‐Cardena A, Morales‐Romero J, Garcia‐Roman R, et al. Clinical characteristics and genetic variability of human rhinovirus in Mexico. Viruses. 2012;4(2):200‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leung TF, Li CY, Lam WY, et al. Epidemiology and clinical presentations of human coronavirus NL63 infections in Hong Kong children. J Clin Microbiol. 2009;47(11):3486‐3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martin ET, Kuypers J, McRoberts JP, Englund JA, Zerr DM. Human Bocavirus 1 primary infection and shedding in infants. J Infect Dis. 2015;212(4):516‐524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moreno‐Valencia Y, Hernandez‐Hernandez VA, Romero‐Espinoza JA, et al. Detection and characterization of respiratory viruses causing acute respiratory illness and asthma exacerbation in children during three different season (2011‐2014) in Mexico City. Influenza Other Respi Viruses. 2015;9(6):287‐292. 10.1111/irv.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nitsch‐Osuch A, Wozniak‐Kosek A, Korzeniewski K, Zycinska K, Wardyn K, Brydak LB. Clinical features and outcomes of influenza A and B infections in children. Adv Exp Med Biol. 2013;788:89‐96. [DOI] [PubMed] [Google Scholar]
- 57. Nokes DJ, Ngama M, Bett A, et al. Incidence and severity of respiratory syncytial virus pneumonia in rural Kenyan children identified through hospital surveillance. Clin Infect Dis. 2009;49(9):1341‐1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nyawanda BO, Mott JA, Njuguna HN, et al. Evaluation of case definitions to detect respiratory syncytial virus infection in hospitalized children below 5 years in rural western Kenya, 2009‐2013. BMC Infect Dis. 2016;16(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pecchini R, Berezin EN, Felicio MC, et al. Incidence and clinical characteristics of the infection by the respiratory syncytial virus in children admitted in Santa Casa de Sao Paulo Hospital. Braz J Infect. 2008;12(6):476‐479. [DOI] [PubMed] [Google Scholar]
- 60. Pierangeli A, Scagnolari C, Selvaggi C, et al. Virological and clinical characterization of respiratory infections in children attending an emergency department during the first autumn‐winter circulation of pandemic A (H1N1) 2009 influenza virus. Clin Microbiol Infect. 2012;18(4):366‐373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ramagopal G, Brow E, Mannu A, Vasudevan J, Umadevi L. Demographic, clinical and hematological profile of children with bronchiolitis: A comparative study between respiratory synctial virus [RSV] and [non RSV] groups. J Clin Diagn Res 2016; 10: Sc05‐08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Saha A, Jha N, Dubey NK, Gupta VK, Kalaivani M. Swine‐origin influenza A (H1N1) in Indian children. Ann Trop Paediatr. 2010;30(1):51‐55. [DOI] [PubMed] [Google Scholar]
- 63. Schuster JE, Khuri‐Bulos N, Faouri S, et al. Human Metapneumovirus infection in Jordanian children: Epidemiology and risk factors for severe disease. Pediatr Infect Dis J. 2015;34(12):1335‐1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smit PM, Bongers KM, Kuiper RJ, von Rosenstiel IA, Smits PH, Brandjes DP. Characterization of 2009 H1N1 pandemic influenza in a population of Dutch children with influenza‐like signs and symptoms. Acta Paediatr. 2012;101(1):67‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smuts HE, Workman LJ, Zar HJ. Human rhinovirus infection in young African children with acute wheezing. BMC Infect Dis. 2011;11(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tresoldi AT, Pereira RM, Fraga AM, et al. Clinical features and outcome of children and adolescents hospitalized with influenza A (H1N1) virus infection compared with flu‐like symptoms and negative rapid tests for influenza A (H1N1) admitted in the same period of time. J Trop Pediatr. 2011;57(6):481‐483. [DOI] [PubMed] [Google Scholar]
- 67. von Linstow ML, Larsen HH, Eugen‐Olsen J, et al. Human metapneumovirus and respiratory syncytial virus in hospitalized Danish children with acute respiratory tract infection. Scand J Infect Dis. 2004;36(8):578‐584. [DOI] [PubMed] [Google Scholar]
- 68. Weigl JA, Puppe W, Schmitt HJ. Can respiratory syncytial virus etiology be diagnosed clinically? A hospital‐based case‐control study in children under two years of age. Eur J Epidemiol. 2003;18(5):431‐439. [DOI] [PubMed] [Google Scholar]
- 69. Yan XL, Li YN, Tang YJ, et al. Clinical characteristics and viral load of respiratory syncytial virus and human metapneumovirus in children hospitaled for acute lower respiratory tract infection. J Med Virol. 2017;89(4):589‐597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zimmerman RK, Rinaldo CR, Nowalk MP, et al. Influenza and other respiratory virus infections in outpatients with medically attended acute respiratory infection during the 2011‐12 influenza season. Influenza Other Respi Viruses. 2014;8(4):397‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ebell MH, White LL, Casault T. A systematic review of the history and physical examination to diagnose influenza. J Am Board Fam Pract. 2004;17(1):1‐5. [DOI] [PubMed] [Google Scholar]
- 72. Thornton HV, Blair PS, Lovering AM, Muir P, Hay AD. Clinical presentation and microbiological diagnosis in paediatric respiratory tract infection: A systematic review. Br J Gen Pract. 2015;65(631):E69‐E81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Macaubas C, Nguyen KD, Peck A, et al. Alternative activation in systemic juvenile idiopathic arthritis monocytes. Clin Immunol. 2012;142(3):362‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kliegman RMSB, St. Geme J, Schor NF, Behrman RE. Nelson Textbook of Pediatrics In: Influenza Viruses. 19th ed. Philadelphia, United States: Elsevier‐Health Sciences Division; 2011:1121‐1125. [Google Scholar]
- 75. Kliegman RMSB, St. Geme J, Schor NF, Behrman RE. Nelson Textbook of Pediatrics In: Respiratory Syncytial Viruses. 19th ed. Philadelphia, United States: Elsevier‐Health Sciences Division; 2011:1126‐1129. [Google Scholar]
- 76. Kliegman RMSB, St. Geme J, Schor NF, Behrman RE. Nelson Textbook of Pediatrics In: Adenoviruses. 19th ed. Philadelphia, United States: Elsevier‐Health Sciences Division; 2011:1131‐1132. [Google Scholar]
- 77. Scutti S. CNN . If it's not the flu, you might be sick because of this virus. https://edition.cnn.com/2018/01/30/health/adenoviruses-mimic-flu/index.html. Accessed January 30, 2018.
- 78. Fox M. NBC News . Virus looks like flu, acts like flu, but it's not influenza. https://www.aol.com/article/news/2018/01/29/virus-looks-like-flu-acts-like-flu-but-its-not-influenza/23346369/. Accessed January 29, 2018.
- 79. Scotti A. New York daily news . It Feels like the Flu but isn't: Meet the Adenovirus http://www.nydailynews.com/life-style/feels-flu-isn-meet-adenovirus-article-1.3791501. Accessed January 31, 2018.
- 80. Alchikh M, Hoppe C, Papagrigoriou‐Theorodridou MA, et al. Harmonizing disease severity assessments in infants and children: The PEDSIDEA Consortium. In Options for The Control of Influenza IX Conference, Chicago IL, USA; 2016 August 24‐28.
- 81. Kajon AE, Lamson DM, Bair CR, et al. Adenovirus type 4 respiratory infections among civilian adults, Northeastern United States, 2011‐2015(1). Emerg Infect Dis 2018; 24: 201‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lamson DM, Kajon A, Shudt M, Girouard G, St George K. Detection and genetic characterization of adenovirus type 14 strain in students with influenza‐like illness, New York, USA, 2014‐2015. Emerg Infect Dis. 2017;23(7):1194‐1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information 1: Primer and Probe sequences for HCoV, HPIV and HBoV‐1
Supporting Information 2: Forest plots and funnel plots for significant associations in the LIT dataset
Supporting Information 3: Heterogeneity analysis for significant associations in the LIT dataset


