Summary
MicroRNAs (miRNAs) have emerged as a class of regulatory RNAs in host–pathogen interactions. Aberrant miRNA expression seems to play a central role in the pathology of several respiratory viruses, promoting development and progression of infection. miRNAs may thus serve as therapeutic and prognostic factors for respiratory viral infectious disease caused by a variety of agents. We present a comprehensive review of recent findings related to the role of miRNAs in different respiratory viral infections and discuss possible therapeutic opportunities aiming to attenuate the burden of viral infections. Our review supports the emerging concept that cellular and viral‐encoded miRNAs might be broadly implicated in human respiratory viral infections, with either positive or negative effects on virus life cycle. Copyright © 2016 John Wiley & Sons, Ltd.
Abbreviations
- microRNAs
miRNAs
- nucleotide
nt
- precursor miRNAs
pre‐miRNA
- RNA‐induced silencing complex
RISC
- three prime untranslated region
3ʹ‐UTR
- nuclear factor kappa B
NF‐kB
- interleukin‐1 receptor‐associated kinase
IRAK
- chemokine (C‐C motif) ligand
CCL
- nerve growth factor
NGF
- tropomyosin‐related kinase A
TrkA
- severe acute respiratory syndrome‐coronavirus
SARS‐CoV
- Middle East respiratory syndrome‐coronavirus
MERS‐CoV
- OC43‐coronavirus
OC43‐CoV
- very low‐density lipoprotein receptor
VLDLR
- human metapneumovirus
HMPV
- virus‐associated RNAs
VARNAs
Introduction
MicroRNAs (miRNAs) are small endogenous, non‐coding RNAs, approximately 20–25 nt long. They are RNA‐sequence‐specific post‐transcriptional regulators of gene expression 1. miRNAs are expressed in a wide variety of organisms and originate in the nucleus as primary miRNA transcripts (~1000 nt), which are processed by the dsRNA‐specific endonuclease Drosha into precursor miRNAs (pre‐miRNA). The pre‐miRNA (~70 nt) are transported to the cytoplasm, and further processed by Dicer into mature miRNAs. A single strand of mature miRNA is incorporated into the RNA‐induced silencing complex (RISC), which binds to the three prime untranslated region (3ʹ‐UTR) of target mRNA, and exerts direct effects by blocking the translational process or inducing mRNA degradation, and indirect effects by influencing methylation or targeting of transcriptional factors 2, 3. Over 2000 human miRNAs are currently recognized in the comprehensive miRNA database miRBase 4, and the function of many of these miRNAs in various biological processes including differentiation, proliferation, metabolism, and apoptosis is well established 1. It is estimated that about 60% of human genes may be subjected to miRNA regulation. miRNA systems constitute complex combinatorial networks, where one miRNA may regulate many mRNA, and conversely, one mRNA may be regulated by several miRNAs 5.
Given the breadth of miRNA‐mediated regulation of various biological process and immunity in mammals, the role of miRNAs has recently been highlighted in host–pathogen interactions 6, 7, 8. Host–pathogen interaction is the most important dynamic system in nature, and epigenetic modifications and post‐transcriptional regulation through miRNA systems may provide an accessory source of fast‐acting and readily available phenotypic variation that can be directly carried out by both host and pathogen selection pressures 9. Over the past decade, our knowledge of miRNA processes in various biological systems and host–pathogen interactions has rapidly advanced, but the precise role of miRNAs in the host–pathogen interactions is still unclear 10. A number of studies in recent years report differential expression and biological function of miRNAs in airway cells 11, 12. miRNAs play an important role in physiological and pathological aspects of airway cells including pulmonary development, immune function, fibrosis, and cancer 13, 14. In airway epithelial cells, miRNAs have been shown to affect numerous processes pertaining to respiratory pathogens, such as modulation of innate and adaptive immune responses, cell cycle progression, and apoptosis induction 7, 15.
In this comprehensive review, we discuss recent findings that indicate an important role for miRNAs in various respiratory viral infections. We also discuss the putative significance of these effects on respiratory viral replication, viral cytopathogenicity, and the immune response. Identifying the role of miRNAs in respiratory viral infections may enhance our understanding of the mechanisms of infection and also indicate a potential future for miRNA‐based therapies.
MicroRNA and Virus Interaction
Viruses are obligate intracellular infectious agents that use the host cellular machinery to ensure their own fitness and survival. The success of viruses principally depends on their capability to efficiently use the host machinery to take advantage of basic biological processes 16, 17, 18. miRNA systems have several features that make them ideal tools for virus propagation. They are potent post‐transcriptional gene expression regulators. They are both small and non‐antigenic and can modulate expression of several critical cellular pathways 19, 20, 21. miRNA systems modulate viral replication and pathogenesis in several ways: (i) Host cell miRNAs can positively or negatively affect viral replication and pathogenesis as a result of their biological functions. (ii) Some viruses also encode miRNAs. Current evidence indicates that viral‐encoded miRNAs target several cellular genes involved in cell proliferation and survival, stress responses, and anti‐viral response. (iii) Virus‐encoded miRNAs may also regulate viral gene expression 21, 22, 23, 24, 25, 26, 27. Thus, host‐encoded miRNAs, virus‐encoded miRNAs, and miRNA targets together form a novel regulatory system (miRNA system) between the host and the virus, which contributes to the outcome of infection 19, 28.
Host cellular miRNA expression is profoundly altered following viral infection (Figure 1), which affects the viral life cycle including viral replication, immune responses, and infection outcome 22. On the one hand, these changes could represent a host defense against infection and might therefore act to inhibit virus replication. On the other hand, changes in cellular miRNA may be induced by viruses to prepare a suitable environment for productive viral infection and/or latency 21. Host cellular miRNAs may have direct effects on viral replication, through positive or negative interactions with viral genomes or other viral factors 29. An miRNA system may also contribute to anti‐viral host defense 30. Evidence suggests that miRNA can positively or negatively regulate innate and adaptive immune responses 7. Furthermore, host cell miRNAs have potential to regulate virus tissue tropisms 22. Thus, miRNAs are utilized by viruses to invade host cells, replicate in host cell, evade host immune response, and establish and maintain virus latency 19, 24.
Figure 1.
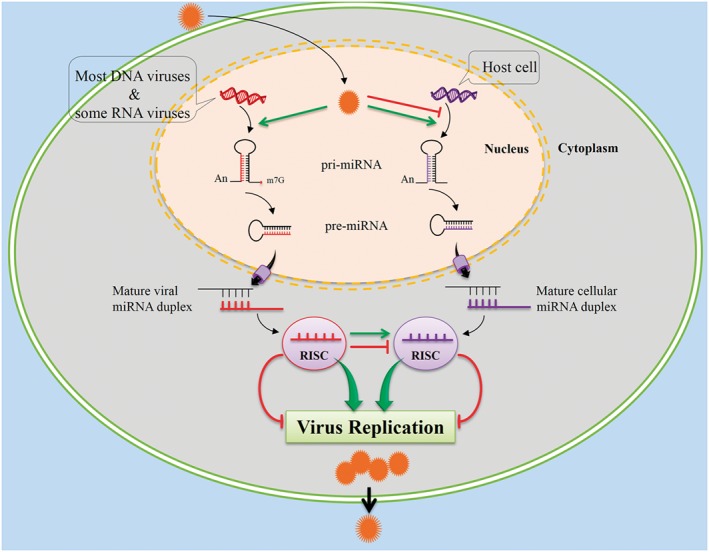
Following viral infection, host cells alter their microRNAs (miRNAs) expression as a defense against infection, while viruses can circumvent host defense and promote their own propagation by affecting host cellular miRNAs expression or by expressing their own miRNAs
There are several reports demonstrating that some viruses take advantage of cellular miRNAs by enhancing the expression of specific cellular miRNAs. This can enhance virus replication, apparently by down‐regulating specific cellular mRNA targets with anti‐viral potential 31, 32, 33. Alternatively, viruses may use the miRNA system to limit their own replication in infected cells, allowing evasion of the host immune response, survival of infected cells, establishment of viral latency, and increased spread to other individuals in a population 34. Furthermore, other studies indicate that viral gene products inhibit cellular miRNA expression 35. Thus, viruses can induce certain cellular miRNAs that affect the virus life cycle positively and inhibit those that affect the virus life cycle negatively 29. Interestingly, some viruses can propagate despite the presence of host cell‐encoded inhibitory miRNA. Viruses may avoid inhibition by cellular miRNAs by several methods including: (i) blocking cellular miRNA biogenesis; (ii) inhibiting cellular miRNA function; or (iii) evolving 3ʹ‐UTR sequences that miRNAs are unable to bind to because they are not complementary to miRNA, are too short, or have complex secondary structures that could restrict binding by RISCs 36. However, some reports demonstrated that specific cellular miRNAs can negatively inhibit virus replication 37, 38, 39.
There are several ways in which the association of cellular and viral‐encoded miRNAs with pathology and their targets can be identified 35. Computational analyses (in silico) for predicting miRNAs and their targets are applied by most studies as the first step of a survey. However, computer‐based predictions of miRNA‐target interactions may or may not exist in reality and should be verified by in vitro and/or in vivo investigations, often involving addition and removal of miRNAs from a system. In recent years, rapid advances in next generation sequencing have been successfully incorporated to analyze miRNAs and their targets 40. In addition, deep sequencing of small RNAs isolated from virus‐infected cells may provide valuable information 41.
MicroRNAs and Respiratory Viruses
Respiratory viruses are the most common global health problem with morbidity and mortality worldwide 42. Respiratory viral infections are responsible for an enormous economic burden, precipitating considerable absence from school and work, large numbers of visits to clinicians, and also represent a major cause of exacerbations of chronic respiratory disease such as asthma and chronic obstructive pulmonary disease 43. Viruses most commonly associated with respiratory infections are orthomyxoviruses, adenoviruses, paramyxoviruses, coronaviruses, picornaviruses, human bocavirus, and human herpesviruses 44. The availability of effective vaccines against respiratory viral infections is limited, and other than the anti‐influenza medications oseltamivir and zanamivir, no clinical anti‐viral treatments for common respiratory viruses are available 45, 46. Novel anti‐viral therapeutic approaches to prevent and treat respiratory viral infection are needed according to the WHO initiative Battle against Respiratory Viruses 47.
In recent years, significant progress has been made in understanding the molecular mechanisms underlying respiratory virus infection and host interaction. Identification and characterization of the miRNA expression profile following respiratory viral infection and its implication in viral infection is an important tool for understanding host–virus interaction, mechanisms of infection, and also therapy strategy development. Here, we summarize the literature data on such host‐respiratory virus implications in humans and discuss how these implications can be used as research tools or targets in the development of novel anti‐viral therapeutics (Table 1).
Table 1.
miRNAs effects in respiratory viral infection
| Virus | miRNAs | Effects | Reference |
|---|---|---|---|
| Influenza | miR‐30 down‐regulation and miR‐223 up‐regulation | Regulate apoptosis | 48 |
| miR‐29 up‐regulation | Regulates apoptosis | 49 | |
| miR‐4276 induction | Inhibits COX6C and caspase‐9 | 50 | |
| miR‐29 up‐regulation | Protects A20 mRNA | 32 | |
| miR‐146a induction | Regulates immune response | 51 | |
| miR‐15b‐3p, miR‐24‐2‐5p, miR‐331‐3p, miR‐124‐3p, and miR‐337‐5p up‐regulation | Regulate anti‐viral response | 52 | |
| miR‐7, miR‐132, miR‐146a, miR‐187, miR‐200c, and miR‐1275 expression | Regulate anti‐viral response | 53 | |
| miR‐106b, miR‐124, and miR‐1254 expression | Regulate human protease genes | 54 | |
| miR‐24 down‐regulation | Up‐regulates furin mRNA | 55 | |
| miR‐21 expression | Inhibits proliferation‐suppressing factors | 56 | |
| miR‐30 family down‐regulation | Contribute to higher proliferation | 57 | |
| miR‐141 up‐regulation | Suppress expression of TGF‐β | 58 | |
| miR‐200a and miR‐223 expression | Regulate neutrophil and IFN‐I response | 59 | |
| miR‐576‐3p down‐regulation | Regulates virus entry | 60 | |
| miR‐323, miR‐491, and miR‐654 expression | Inhibit H1N1 influenza replication | 37 | |
| let‐7c expression | Inhibits M1 protein | 39 | |
| RSV | let‐7f expression | Contributes to delayed viral clearance | 31 |
| let‐7i and miR‐30b inhibition | Enhance viral replication | 61 | |
| miR‐221 silencing | Enhance NGF and TrkA expression | 62 | |
| miR‐125a down‐regulation | Contributes to the virus immune evasion | 63 | |
| Coronavirus | miR‐17, miR‐574‐5p, and miR‐214 up‐regulation | Contribute to virus evade immune elimination | 64 |
| miR‐9 expression | Potentiates NF‐kB activation | 65 | |
| Rhinovirus | miR‐128 and miR‐155 expression | Contribute to the anti‐viral activity against rhinovirus‐1B | 38 |
| miR‐23b expression | Inhibits infections of minor group rhinoviruses | 66 | |
| HMPV | miR‐30a and miR‐16 inhibition | Regulate host cellular response to HMPV virus infection | 67 |
| Adenovirus | miVARNAs expression | Targets cellular and viral genes | 68 |
| miVARNAs expression | Inhibits human pre‐miRNA | 69 | |
| miVARNAs expression | Down‐regulates the TIA‐1 expression | 70 | |
| miVARNAs expression | Down‐regulates the HDGF expression | 71 | |
| miR‐214 expression | Inhibits virus replication | 72 | |
| miR‐466 expression | Down‐regulates the level of CAR protein | 73 | |
| miR‐1, miR‐34, miR‐22, miR‐365, miR‐29, miR‐145, and let‐7 expression | Target Rb‐dependent cell cycle and DNA replication mRNAs | 74 | |
| HCMV | miR‐US4‐1 expression | Inhibits CD8 + T cell response | 75 |
| miR‐UL112 expression | Attenuates NK cell activity | 76 | |
| miR‐UL148D expression | Targets the human chemokine CCL5 | 77 | |
| miR‐UL112‐3p expression | Targets TLR2 and following signaling | 78 | |
| miR‐UL112‐1 expression | Down‐regulates IL‐32 expression | 79 | |
| miR‐US25‐1‐5p expression | Inhibits viral replication | 80 | |
| miR‐US25‐2 expression | Reduces viral replication | 81 | |
| miR‐200 family members expression | Targets the UL122 expression | 82 | |
| miR‐US33 expression | Down‐regulates virus replication | 83 | |
| miR‐UL112‐1 expression | Decreases genomic viral DNA levels | 84 | |
| miR‐UL112‐1 expression | Down‐regulates cellular BclAF1 | 85 | |
| miR‐UL112‐1, miR‐US5‐1, and miR‐US5‐2 expression | Target multiple components of the host secretory pathway | 86 | |
| miR‐UL70‐3p and miR‐UL148D expression | Target the pro‐apoptotic genes | 87 | |
| HHV‐6 | miR‐U86 expression | Targets the HHV‐6A IE gene U86, thereby regulates virus lytic replication | 88 |
miRNAs, microRNAs; TGF‐β, transforming growth factor‐β; COX6C, cytochrome c oxidase VIC; NGF, nerve growth factor; TrkA, tropomyosin‐related kinase A; NF‐kB, nuclear factor kappa B; HMPV, human metapneumovirus; HCMV, human cytomegalovirus; CAR, Coxsackie virus and adenovirus receptor; HDGF, hepatoma‐derived growth factor.
RNA Viruses
Unlike DNA viruses, RNA viruses usually do not encode their own miRNA, and the reasons behind this discrepancy are debated theoretically 89. The majority of RNA viruses replicate in the cytoplasm where they cannot access the nuclear enzyme Drosha, which is required for miRNA processing. Those RNA viruses, which do have access to the nucleus (e.g. influenza and HIV‐1), may avoid encoding their own miRNAs because excision of a primary miRNA from RNA virus genome would induce cleavage and destruction of viral genome 20, 27. In addition, viruses that undergo short lytic replication cycles are less likely to encode miRNAs 22.
Influenza virus
Influenza virus is a common respiratory pathogen that primarily infects airway epithelial cells and leads to clinical outcomes ranging from mild upper respiratory infection to severe pneumonia 90. The host cellular response, specifically miRNA dysregulation, is likely to play a critical role in influenza infection outcome 53, 58. Recent studies show distinct miRNA expression profiles in ill patients with influenza A (H1N1), that is, down‐regulation of miR‐29a, miR‐29c, let‐7g, miR‐146b‐5p, miR‐150, miR‐342‐3p, miR‐769‐5p, miR‐30b, miR‐31, miR‐361‐3p, miR‐362‐3p, miR‐342, miR‐155, miR‐210, and miR‐192. These miRNAs are involved in the regulation of important biological pathways during virus infection, such as mitogen‐activated protein kinase, epidermal growth factor receptor, and toll‐like receptor signaling pathways 91. Additional studies detected high expression of miR‐299‐5p and miR‐335 in influenza patients 91, 92. In contrast, miR‐765, miR‐34b, miR‐519e, miR‐18a, miR‐628‐3p, miR‐185, miR‐576‐3p, miR‐519d, miR‐28‐5p, miR‐26a, miR‐1285, miR‐665, and miR‐30a were down‐regulated in H1N1 patients, and interestingly, miR‐576‐3p could affect viral entry into cells by regulating AP1G1 expression 60. Furthermore, miR‐17, miR‐20a, miR‐106a, and miR‐376c were significantly elevated in H7N9 patients 93.
The virulence of influenza virus may be mediated in part by host cellular miRNAs via dysregulation of pathways critical for anti‐viral immune responses 48. Influenza infection up‐regulates miR‐29 expression, which is involved in regulation of both innate and adaptive immune responses through protection of A20 mRNA 32. miR‐29 acts as an RNA decoy to prevent HuR (human antigen R) from binding to the A20 3ʹ‐UTR and recruiting the RISC 94. A20 is a deubiquitinating enzyme known to play an important role in terminating the anti‐viral immune response by inhibiting nuclear factor kappa B (NF‐kB) and interferon regulatory factor pathways 95. Influenza infection of A549 cells induces expression of miR‐146a, also a negative regulator of NF‐kB 51. Interestingly, one study showed that the zoonotic respiratory hendra virus induces miR‐146a, which promotes viral replication by targeting ring finger protein 11 33. In a study by Huang, et al., up‐regulation of several miRNAs including miR‐15b‐3p, miR‐24‐2‐5p, miR‐331‐3p, miR‐124‐3p, and miR‐337‐5p was demonstrated following H1N1 infection. These miRNAs participate in toll‐like receptor and RIG‐I‐like receptor signaling pathways, and also regulate IL‐1β and TNF receptor‐associated factor 3 52. Furthermore, miR‐7, miR‐132, miR‐146a, miR‐187, miR‐200c, and miR‐1275 accumulate in human lung cell lines in response to infection with two influenza A virus strains, A/Udorn/72 and A/WSN/33, causing down‐regulation of anti‐viral proteins such as interleukin‐1 receptor‐associated kinase (IRAK1) and mitogen‐activated protein kinase 3 53.
Virulence of highly pathogenic influenza viruses may be mediated in part by host cellular miRNAs. For example, the highly pathogenic H5N1 virus induces miR‐141 shortly after infection, which suppresses the expression of transforming growth factor‐β in lung epithelial cells 58. Without sufficient transforming growth factor‐β, the pro‐inflammatory response might not be tightly controlled in cases of highly pathogenic H5N1 infection 96. The 1918 pandemic influenza virus induces a distinct miRNA expression profile in mice compared with non‐lethal influenza A/Texas/36/91, including down‐regulation of miR‐200a and up‐regulation of miR‐223; miR‐223 is a negative modulator of neutrophil activation, and miR‐200a has a role in the type I IFN response 59.
Apoptosis is characteristic of influenza virus infection, and the mechanisms underlying this have advanced understanding of influenza virus replication 49, 50. According to Othumpangat, et al., in the first hours after influenza infection, down‐regulation of miR‐4276 increases cytochrome c oxidase VIC expression, inhibiting viral replication by inducing the apoptotic protein caspase‐9. However, after 6 to 9 h, this effect is completely reversed, thereby prolonging cell survival. This may suggest that influenza virus is able to induce miR‐4276 and inhibit cytochrome c oxidase VIC and caspase‐9 expression, thus promoting viral replication 50. Recent studies have revealed that miR‐29 family members are up‐regulated during influenza infection, especially miR‐29c, which targets the anti‐apoptotic factor B‐cell lymphoma 2‐like 2 contributing to virus‐mediated apoptosis 49. Furthermore, down‐regulation of miR‐30 family members and up‐regulation of miR‐223 during influenza infection lead to increased apoptosis 48.
The expression of host genes required for influenza virus replication can be regulated by multiple cellular miRNAs, for example, miR‐106b, miR‐124, and miR‐1254 regulate human protease genes (ADAMTS7, CPE, DPP3, MST1, and PRSS12) that are essential for influenza replication 54. In addition, down‐regulation of miR‐24 with a concomitant up‐regulation of furin mRNA has been demonstrated during the influenza H5N1 infection in A549 cells. miR‐24 regulates furin‐mediated activation of influenza hemagglutinin precursor and subsequent production of fusion‐competent virions in the host secretory pathway 55.
Some miRNAs play important roles in priming airway cells for repair and regeneration following influenza infection 97. Elevated expression of miR‐21 throughout repair and regeneration corresponds with increased cell proliferation in repairing lungs, because miR‐21 targets proliferation‐suppressing factors 56. The miR‐30 family was significantly down‐regulated during repair consistent with increased expression of its main target p53, which promotes proliferation in recovering lung tissues 57.
While influenza virus can clearly take advantage of cellular miRNAs via their promotion or inhibition, it is also revealed that certain cellular miRNAs can inhibit replication of influenza viruses in infected cells 37, 39. miR‐323, miR‐491, and miR‐654 inhibit replication of the H1N1 influenza A virus in MDCK cells by targeting the same conserved region in the influenza PB1 gene 37. Furthermore, let‐7c inhibits M1 protein expression of the H1N1 influenza A virus in A549 cells 39.
Overall, these results suggest that influenza respiratory infection induces or inhibits expression of certain miRNAs in airway cells that favor viral replication, pathogenesis, and also suppress anti‐viral responses (Figure 2). Thus, cellular miRNAs associated with immune response, apoptosis, and protease genes could be the best candidates for development of miRNA‐based therapies for influenza disease. However, caution must be taken due to the immunopathogenic character of influenza infection.
Figure 2.
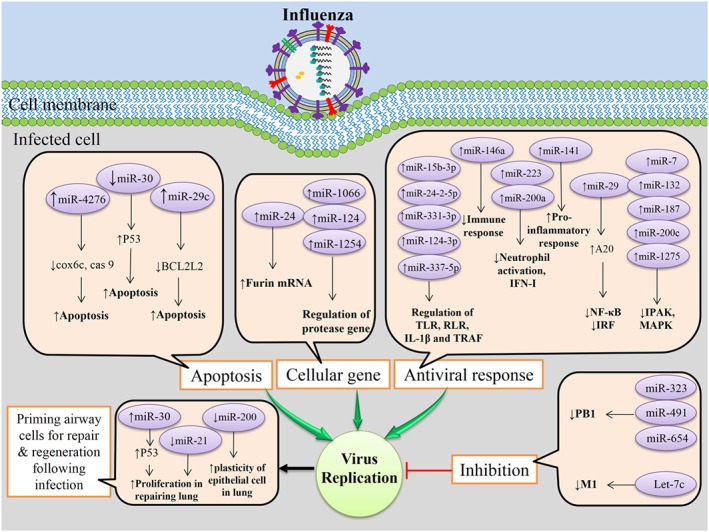
Influenza infection of airway epithelial cells induces or inhibits certain cellular microRNAs (miRNAs) expression in favor of viral replication, pathogenesis, and also suppress anti‐viral responses. However, certain cellular miRNAs can inhibit replication of influenza in infected cells, and certain miRNAs play important roles in priming airway cells for repair and regeneration following influenza infection
Respiratory syncytial virus
RSV is a leading cause of viral respiratory tract disease among infants and young children 98, 99. Worldwide, 33.8 million episodes of RSV‐associated acute lower respiratory tract infections are estimated to occur in children <5 years of age 100. In developed countries, the RSV‐associated mortality rates are reported to be approximately three deaths per 100 000 in children younger than 1 year 101, 102, 103, 104.
RSV infection is a common example of viruses that modulate host miRNA expression to influence the outcome of the anti‐viral host response and viral replication 7, 15, 105. Distinct immune‐associated miRNA expression profiles have been detected in the nasal epithelium of RSV‐positive infants; down‐regulation of miR‐34b, miR‐34c, miR‐125b, miR‐29c, miR125a, miR‐429, and miR‐27b, and up‐regulation of miR‐155, miR‐31, miR‐203a, miR‐16, and let‐7d were detected in these patients. In addition, miR‐125a and miR‐429 were down‐regulated in mild disease, but not in severe disease, and the lack of down‐regulation in severe disease may rationalize the observed differences in disease manifestations following RSV infection 63. miR‐125a regulates the expression of NF‐kB by suppressing the inhibitor protein A20, and chemokine (C‐C motif) ligand (CCL5), an important cytokine in both innate and adaptive immune systems 106.
RSV infection of A549 cells induced let‐7f, miR‐24, miR‐337‐3p, miR‐26b, and miR‐520a‐5p and repressed miR‐198 and miR‐59 expression. Let‐7f expression was RSV G protein dependent, and its expression likely contributes to delayed viral clearance by targeting CCL7 and suppressor of cytokine signaling 3, which are involved in anti‐viral response 31. In another study, let‐7b, let‐7c, let‐7i, and miR‐30b were up‐regulated on RSV infection of monocyte derived dendritic cells and human bronchial epithelial cells, and associated with IFN‐β and/or NF‐kB activation. Interestingly, RSV nonstructural proteins NS1 and NS2 antagonized the up‐regulation of let‐7i and miR‐30b, a process that may favor viral replication 61. The miRNAs described in these studies have a number of experimentally confirmed targets that are associated with RSV replication and pathology. For example, an experimentally confirmed target of the let‐7 family is IL‐13, which appears to enhance the severity of disease 107.
RSV infection modifies the expression of critical neurotrophic factors and receptors such as nerve growth factor (NGF), and its cognate high‐affinity receptor tropomyosin‐related kinase A (TrkA), which prevents apoptosis by increasing expression of the anti‐apoptotic Bcl‐2 family members 108. In human bronchial epithelial cells, high levels of intracellular miR‐221 reduced NGF and TrkA expression, which favor the apoptotic death of infected cells, and attenuate virus infection. RSV infection reduces miR‐221 expression, thus interfering with the apoptotic death of infected cells by increasing NGF and TrkA expression and ultimately promoting viral replication 62.
Overall, these findings suggest that following RSV respiratory infection, an altered expression profile of distinct immune‐associated miRNAs occurs in the airway cells that inhibit viral replication and preserve the airway epithelial barrier. However, the virus concurrently induces or inhibits the expression of other miRNAs that favor viral replication (Figure 3). These conflicting miRNA effects during RSV infection may provide treatment options in susceptible individuals. However, attempts to modulate RSV pathology in clinical practice should be made with caution as RSV immunopathogenesis is complicated and an early RSV vaccine candidate caused serious adverse events during natural RSV infection 109.
Figure 3.
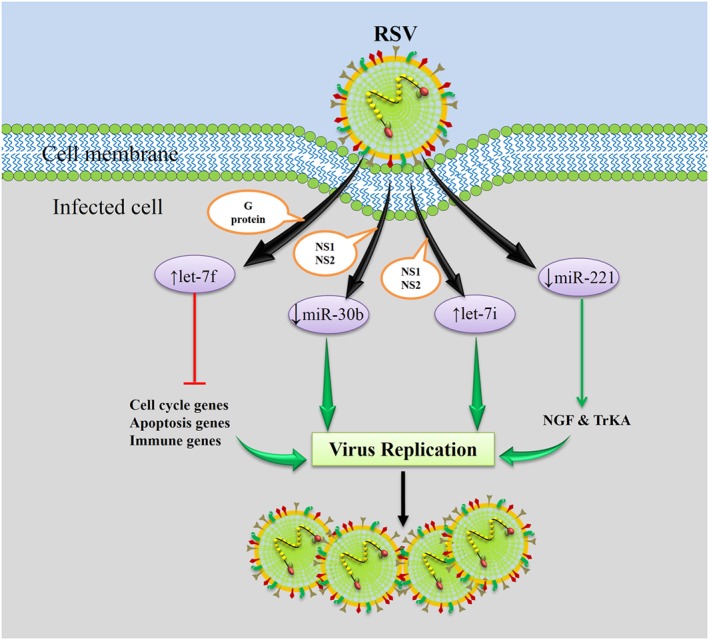
Following RSV respiratory infection, an altered expression profile of certain cellular miRNAs, specifically immune‐associated miRNAs, occurs in order to inhibit viral replication and preserve the airway epithelial barrier; meanwhile, the virus induces or inhibits the expression of other miRNAs that favor viral replication. The RSV G protein enhances let‐7f, the RSV NS1/NS2 proteins decrease miR‐30b and enhance let‐7i, and RSV infection decrease miR‐221, which is an advantage for the virus
Coronaviruses
Coronaviruses can cause a wide spectrum of respiratory infections ranging from mild, upper respiratory tract infections to severe and life‐threatening lower respiratory tract infections 110. There are no in vivo studies regarding the role of miRNAs in coronaviruses infection, but the OC43 virus has been investigated in vitro, and severe acute respiratory syndrome‐coronavirus (SARS‐CoV) and Middle East respiratory syndrome‐coronavirus (MERS‐CoV) were analyzed by in silico methods (Figure 4).
Figure 4.
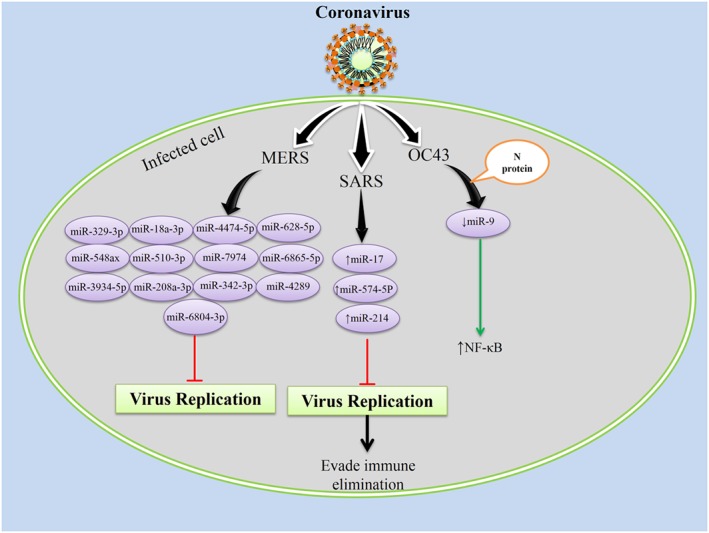
Coronaviruses interact with the host cell at the onset of infection and induces several changes in host cellular microRNAs (miRNAs) expression profile to their own advantage; severe acute respiratory syndrome‐coronavirus (SARS‐CoV) uses cellular miRNAs machinery to evade immune elimination 64; in Middle East respiratory syndrome‐coronavirus (MERS‐CoV), host cells miRNAs would be an anti‐viral therapeutic agent 111, and the N protein of OC43‐coronavirus (OC43‐CoV) causes potentiation of nuclear factor kappa B (NF‐kB) activation via binding to its negative regulator miR‐9 65
The coronavirus OC43 contributes to the common cold worldwide 112. Coronavirus N protein is essential for replication and binds to genomic RNA to form a helical capsid. OC43 N protein potentiates NF‐kB activation by binding to its negative regulator miR‐9. It is not clear whether NF‐kB activation is directly beneficial to viral replication, or whether this is an incidental effect that limits viral virulence. Compared with more pathogenic coronaviruses, reduced OC43 virulence with limited symptoms may promote contact between infected and non‐infected individuals and thus promote spread of the virus within a population 65. This novel mechanism of miRNA‐binding to promote gene activity may provide insight into the mechanisms by which successful RNA viruses avoid the host immune system or cause pathology.
Severe acute respiratory syndrome‐coronavirus is a novel coronavirus that threatened to cause a global pandemic of the severe acute respiratory syndrome in 2002–2003 113. An in silico analysis of miRNA interactions with SARS‐CoV mRNA suggested that the virus might suppress its own replication early during infection by up‐regulation of miR‐17, miR‐574‐5p, and miR‐214. These host miRNAs target all four virulent viral proteins, spike (S), nucleocapsid (N), matrix (M), and envelope (E) 64. Suppression of viral replication may aid evasion of immune surveillance until successful infection of other cells. These results demonstrate how SARS‐CoV might alter host miRNA expression profile to its own advantage.
Middle East respiratory syndrome, caused by a novel human coronavirus MERS‐CoV, has emerged recently 114. An in silico analysis identified miR‐628‐5p, miR‐6804‐3p, miR‐4289, miR‐208a‐3p, miR‐510‐3p, miR‐18a‐3p, miR‐329‐3p, miR‐548ax, miR‐3934‐5p, miR‐4474‐5p, miR‐7974, miR‐6865‐5p, and miR‐342‐3p as having significant sequence similarity to hairpin structures in the MERS‐CoV genome, and they may thus down‐regulate viral gene expression to inhibit viral replication 111. This knowledge may help us to better understand host–virus interactions with the intention to develop new anti‐viral therapies against MERS‐CoV, a highly lethal respiratory disease.
Rhinoviruses
Rhinoviruses are members of the Picornaviridae family. Rhinoviruses cause respiratory infection in humans with severity ranging from the common cold to viral bronchiolitis, and exacerbations of asthma and chronic obstructive pulmonary disease 115. Bondanese, et al. showed that cellular miRNAs miR‐128 and miR‐155 with putative sites in the rhinovirus‐1B coding region can inhibit virus replication. miR‐128 inhibition seemed to increase viral replication by inducing apoptosis. The detection of miR‐155‐mediated anti‐viral activity in bronchial epithelial cells is very relevant because this miRNA has a central role in innate and adaptive immunity. As an example, miR‐155 has been shown to target suppressor of cytokine signaling 1, an inhibitor of type I IFN signaling 38.
A minor group of rhinoviruses including subtypes 1A, 1B, 2, 23, 25, 29, 30, 31, 44, 47, 49, and 62 commonly utilize the very low‐density lipoprotein receptor (VLDLR) for entry into host cells, and cause disease more often than the major group. Recent evidence published by Ouda, et al., showed that down‐regulation of VLDLR by miR‐23b is of significance for host defense against the minor group of rhinoviruses. miR‐23b was induced by RIG‐I‐like receptor signaling resulting in suppression of respiratory infections caused by minor group viruses, specifically rhinovirus‐1B through down‐regulation of its receptor VLDLR 66. In conclusion, these results suggest that miRNAs play an important role in human anti‐viral responses against rhinovirus infection (Figure 5).
Figure 5.
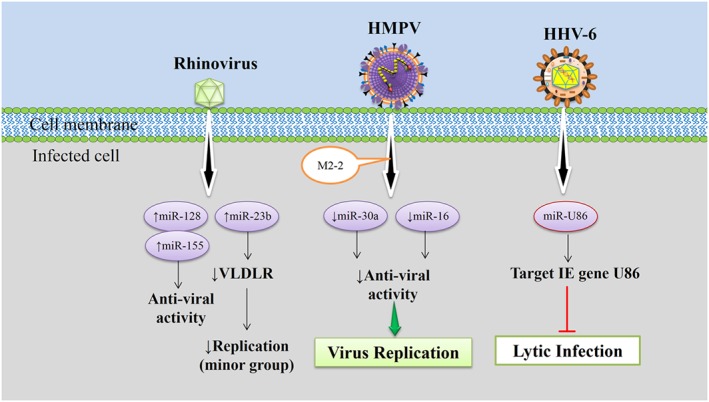
In rhinoviruses infection, cellular microRNAs play anti‐viral responses against viruses 38, human metapneumovirus (HMPV) M2‐2 regulates the host cell microRNAs response to infection 67, and HHV‐6A miR‐U86 targets the HHV‐6A IE gene U86, thereby regulating virus lytic replication 116.
Human metapneumovirus
Human metapneumovirus causes acute respiratory disease in infants, the elderly, and immunocompromised individuals ranging from mild upper respiratory illness to more serious lower respiratory illness 117. Limited literature is available regarding the role of miRNAs in human metapneumovirus (HMPV) infection. Deng, et al. reported that host airway epithelial cells alter their miRNA expression profile upon HMPV infection as a defense mechanism against the virus. The HMPV M2‐2 protein acted as a key viral protein that regulated host cell miRNA expression, specifically antagonizing miR‐30a and miR‐16 (Figure 5). Interestingly, M2‐2‐mediated miR‐16 suppression was interferon dependent, whereas suppression of miR‐30a was interferon independent 67.
The aforementioned data suggest a new way in which HMPV regulates the host cell response to infection. There are currently no licensed therapeutics or vaccines against HMPV. These and future studies may help the development of effective miRNA‐based therapies.
DNA Viruses
Many DNA viruses encode their own miRNAs, because they generally replicate in the nucleus and have access to the canonical miRNA pathway (except poxviruses) 27. Most DNA viruses establish long‐term latent or persistent infections and take advantage of virus‐encoded and host cell miRNAs 22. Because of the fact that viral miRNAs, unlike viral proteins, are non‐immunogenic, viruses have developed their own miRNAs in order to escape and suppress both host innate and adaptive immune responses 118.
Adenoviruses
Adenoviruses cause mild to serious respiratory tract infections in many age groups 119. Adenovirus infection has a great impact on cellular miRNA expression profiles 120, 121. A total of 44 miRNAs demonstrated high expression and 36 miRNAs low expression following adenovirus type 3 infection in human laryngeal epithelial cells 120. A temporal study demonstrated dramatic changes in cellular miRNA expression patterns during the course of adenovirus type 2 infection in lung fibroblast cells; up‐regulation of miR‐22, miR‐320, let‐7, miR‐181b, miR‐155, miR‐125, miR‐27, and miR‐191 and down‐regulation of miR‐21, miR‐31, let‐7 family, miR‐30 family, and miR‐23/27 cluster was detected. These miRNAs have been associated with host immune evasion and inflammatory responses, as well as in virus entry, replication, and propagation 121.
Adenoviruses encode a set of highly abundant miRNAs that are generated by Dicer‐mediated cleavage of the larger non‐coding virus‐associated RNAs (VARNAs) I and II. VARNAs are dsRNA molecules similar in structure to cellular pre‐miRNAs. They are transported by exportin 5 into the cytoplasm, and processed to functional viral miRNAs (miVARNAs) 122. miVARNAs actively target the expression of cellular genes involved in cell proliferation, DNA repair, or RNA regulation 68. VARNAs are expressed at very high levels in adenovirus‐infected cells and potently inhibit human pre‐miRNA via inhibition of nuclear export of pre‐miRNA, competition for exportin 5 to facilitate their transportation, and inhibition of Dicer activity by direct binding to Dicer 69, 123. Adenovirus miVARNAs target cellular and viral genes that are important for the virus cell cycle. Hepatoma‐derived growth factor inhibits adenovirus growth. However, the expression level of hepatoma‐derived growth factor significantly decreased in response to miVARNAs under replication‐deficient conditions, and this suppression was also observed during the early phase of viral infection under replication‐competent conditions 71. Adenovirus miVARNAs also target cellular genes involved in cell growth, gene expression and DNA repair. The TIA‐1 (cytotoxic granule‐associated RNA binding protein) is down‐regulated at mRNA and protein levels in infected cells expressing functional miVARNAs and in transfected cells 70.
Conversely, cellular miRNAs may play a role in anti‐adenovirus replication by regulating virus gene expression. It was shown that cellular miR‐214 inhibits adenovirus replication by regulating the translation of viral E1A protein, which is key to the activation of other adenovirus genes, while inhibition of miR‐214 increases the productive efficiency of the virus 72. Lam, et al. showed that cellular miR‐466 can effectively down‐regulate human Coxsackie virus and adenovirus receptor protein expression 73. Furthermore, a subset of cellular miRNAs including miR‐1, miR‐34, miR‐22, miR‐365, miR‐29, miR‐145, and let‐7 was shown to coordinately target retinoblastoma‐dependent cell cycle and DNA replication mRNAs to restrict proliferation 74.
Taken together, these results suggest that miVARNA‐mediated silencing can represent a novel mechanism used by adenoviruses to control cellular or viral gene expression, and are potential therapeutic targets. The actions of cellular miRNAs may also be exploited to combat adenovirus infection (Figure 6).
Figure 6.
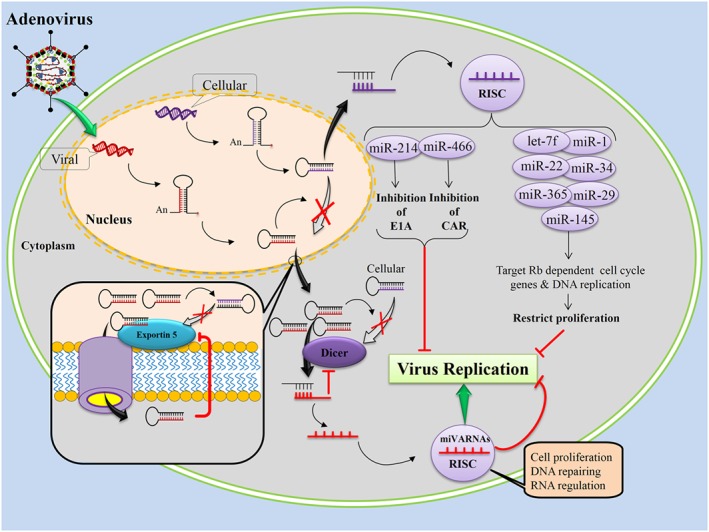
Adenovirus encodes viral miRNAs (miVARNAs) that potently inhibit human pre‐microRNA (miRNA) via inhibition the nuclear export of pre‐miRNA, competition for the exportin 5, and inhibition of Dicer activity by direct binding of Dicer. The miVARNAs are able to target cellular and viral genes that are important for virus cell cycle. Adenovirus miVARNAs target cellular genes involved in cell proliferation, DNA repairing, and RNA regulation. However, cellular miRNAs may play a role in anti‐adenovirus replication by regulating virus gene expression.
Human cytomegalovirus
Human cytomegalovirus (HCMV), a DNA virus, infects a broad range of human cell types and disrupts cellular processes through a variety of mechanisms. For example, HCMV uses several of its own encoded proteins to disrupt the MHC class I pathway 124 and the fine balance between a beneficial and a destructive immune response 82, and uses several of its own encoded miRNAs to disrupt a variety of cellular pathways such as TLR2/IRAK1/NF‐kB signaling 125. This virus therefore induces a complex and diverse pathogenesis, and is an opportunistic pathogen causing lung infection in immunocompromised individuals 126. Host cell miRNA expression levels may determine the cellular site of HCMV infection. As an example, host miR‐200 family members target the HCMV protein UL112 resulting in repression of this viral protein, and cells permissive for lytic HCMV replication demonstrate low levels of these miRNAs 127. However, HCMV can also selectively alter the expression of some cellular miRNAs to help its own replication 128. For example, significant up‐regulation of miR‐96, miR‐182, and miR‐183 have been observed following infection 77. A study by Fu, et al., indicated that expression of host miRNAs may be affected by latent HCMV; at least 49 miRNAs were differentially expressed; 39 were up‐regulated and 10 were down‐regulated accordingly 76. In addition, HCMV encodes its own miRNAs that target both viral and cellular genes in order to regulate viral replication, viral latency, cell survival, and anti‐viral immunity (Figure 7) 79.
Figure 7.
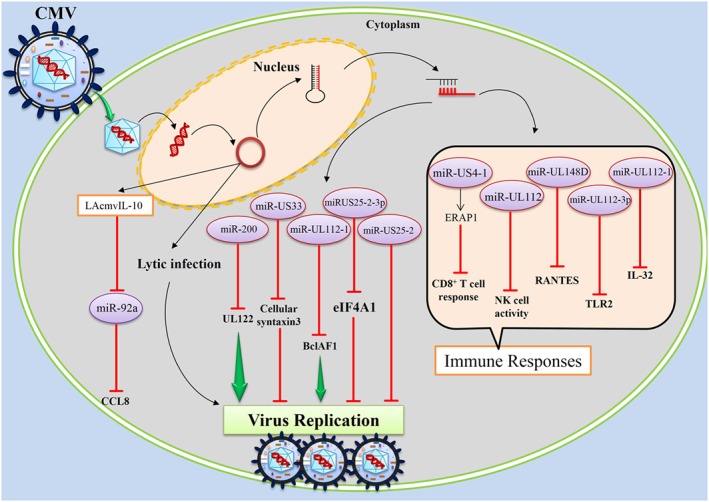
Human cytomegalovirus, a DNA virus, encodes its own microRNAs, and human cytomegalovirus microRNAs target both viral and cellular genes in order to; first regulation of viral replication, second regulation of viral latency infection, and third regulation of cellular anti‐viral immunity.
Human cytomegalovirus has miRNAs that help escape and suppress both host innate and adaptive immune responses 118. HCMV‐encoded miR‐UL148D modulates host immune response by directly targeting the mRNA of human chemokine CCL5 78. HCMV miR‐UL112 attenuates NK cell activity by inhibition of type I IFN secretion 75, down‐regulation of IL‐32 expression 129, and TLR2 targeting, causing significant modulation of the downstream signaling pathway (TLR2/IRAK1/NF‐kB) 125. HCMV may evade CD8+ T‐cells by altering MHC class 1 antigen expression; HCMV miR‐US4‐1 targets the endoplasmic reticulum aminopeptidase 1, a key step in the MHC class I antigen‐processing pathway 80. Furthermore, HCMV expresses miR‐US25‐2 and, in addition, increases cellular miR‐17p expression, both of which target tissue inhibitor of metalloproteinase 3. Reduced tissue inhibitor of metalloproteinase 3 expression following HCMV infection reduces signaling via the MHC class I‐like ligand MICA 81.
Some HCMV‐encoded miRNAs suppress virus replication and lytic infection, which could help the virus to establish or maintain latent infection. It has been reported that HCMV miR‐US25‐1‐5p was highly expressed during lytic and latent infections, and inhibited viral replication 83; HCMV miR‐US25‐2 reduces viral replication by targeting the RNA helicase eIF4A1, which is a requisite for translation of viral mRNA 130, and HCMV miR‐US33 negatively influences virus replication, possibly by suppression of the HCMV gene US29 84 or cellular syntaxin 3 expression 131. Premature expression of HCMV miR‐UL112‐1 during infection resulted in a significant decrease in genomic viral DNA levels, suggesting a functional role for miR‐UL112‐1 in regulating the expression of genes involved in viral replication 85. Finally, HCMV encodes latency‐associated CMV‐IL‐10, a homologue for cellular IL‐10 associated with latent infection. Latency‐associated CMV‐IL‐10 has been shown to suppress miR‐92a, resulting in up‐regulation of its target CCL8. The mechanisms for both miR‐92a suppression and how CCL8 up‐regulation might promote latent infection are unclear, but seem to be associated with increased immune‐regulatory cellular IL‐10. These results provide insight into how HCMV can alter host gene expression 86.
Human cytomegalovirus‐encoded miRNAs do not only suppress virus lytic replication but can also enhance virus replication. For example, HCMV‐restricting cellular BclAF1 is down‐regulated late in infection by HCMV‐encoded miR‐UL112‐1 to promote virus production 87. Furthermore, multiple HCMV‐encoded miRNAs coordinately regulate reorganization of the secretory pathways responsible for controlling cytokine secretion and facilitate formation of the viral assembly compartment for efficient infectious virus production. In this aspect, HCMV‐encoded miRNAs such as miR‐UL112‐1, miR‐US5‐1, and miR‐US5‐2 target multiple components of the host secretory pathways, including VAMP3, RAB5C, RAB11A, SNAP23, and CDC42 132. Additionally, HCMV employs its miRNA repertoire to counter cellular apoptosis and autophagy, particularly the mitochondrial‐dependent intrinsic pathway of apoptosis. The pro‐apoptotic genes MOAP1, PHAP, and ERN1 are identified as potential targets for miR‐UL70‐3p and miR‐UL148D, respectively 133. Finally, a viral intergenetic non‐coding RNA element, composed of highly conserved sequences throughout HCMV clinical strains, selectively degrades the cellular miR‐17 family members of the miR‐17‐92 cluster and accelerates virus production 134.
Overall, these results suggest that identification and characterization of the HCMV‐encoded miRNAs that are expressed during lytic and latent infection are crucial to understanding their roles in HCMV persistence, pathogenesis, and disease. Knowledge of host and viral miRNAs expressed during HCMV infection can thus provide a precise insight into viral pathogenesis and may help researchers to develop new therapeutic approaches.
Human herpesvirus 6
HHV‐6, a DNA virus in the betaherpesvirus subfamily, is associated with several human diseases. Complications of acute respiratory tract infection such as pneumonia and sinusitis in young children are associated with HHV‐6 as is limbic encephalitis following hematopoietic stem cell transplantation. In addition, HHV‐6 salivary gland replication and subsequent secretion in saliva is the epidemiologically proven source of transmission 88, 135. As discussed previously, herpesvirus‐derived miRNAs play considerable roles in modulating both cellular and viral gene expression, thereby facilitating a suitable environment for productive viral infection and/or latency. Like other human herpesviruses, HHV‐6 encodes its own miRNAs, promoting efficient viral infection 116, 136. An miRNA encoded by HHV‐6A (miR‐U86) targets the HHV‐6A IE gene U86, thereby regulating lytic replication, as revealed by growth analyses of mutant viruses (Figure 5) 116. However, HHV‐6B encodes at least four pre‐miRNAs at two positions within the genome in an antisense orientation related to predicted HHV‐6B‐specific genes 136. These data suggest that HHV‐6, like other herpesviruses, encodes its own miRNAs, but the precise function of these miRNAs in HHV6B requires further investigation.
Conclusions
This comprehensive review attempts to highlight the role of miRNAs in replication as well as pathogenesis of respiratory viral infections. miRNAs modulate a variety of cellular processes by regulating multiple targets, promoting or inhibiting the development of viral infection 29. Increasing evidence regarding disrupted miRNA expression and function following viral infection makes them promising targets for therapeutic interventions 137. The development of miRNA‐based therapy for respiratory infection is less advanced compared with other viral infections, such as hepatitis C. Improved knowledge on the cross‐talk between host cells and viruses should increase our understanding of the molecular basis for viral pathogenesis and may enable us to develop better therapeutic strategies 23.
Therapeutic modulation of miRNAs can be achieved through miRNA inhibitors to disrupt miRNA function or miRNA mimics to increase miRNA function 138. The application of miRNA‐based therapies is in its beginning, and important difficulties remain. A significant barrier to miRNA‐based therapy is the development of essential pharmaceutical strategies for targeted delivery to specific sites with minimum toxicity 139, 140. In support of this, novel nanotechnologies and delivery methods are under development for efficient and effective delivery 140, 141.
Alongside the critical role of miRNAs in the regulation of viral respiratory infection and their potential to be targeted by new therapeutics, caution must be taken because excessive inhibition or overexpression of miRNAs might predispose patients to cellular abnormalities, impaired immunity, or even cancer. The relevance of miRNAs in viral infection has been proven broadly; however, the exact role of each miRNA on viral pathogenesis remains to be determined, and future studies are warranted. Enhancing the knowledge on miRNAs may open opportunities to use them in clinical practice in order to develop more accurate and powerful diagnostic and therapeutic strategies.
Acknowledgements
This work was supported by Iran University of Medical Sciences Grant (28601).
Tahamtan, A. , Inchley, C. S. , Marzban, M. , Tavakoli‐Yaraki, M. , Teymoori‐Rad, M. , Nakstad, B. , and Salimi, V. (2016) The role of microRNAs in respiratory viral infection: friend or foe?. Rev. Med. Virol., 26: 389–407. doi: 10.1002/rmv.1894.
References
- 1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. DOI: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics 2010; 11: 597–610. DOI: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3. Fabbri M, Garzon R, Cimmino A, et al. MicroRNA‐29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences USA 2007; 104: 15805–10. DOI: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Griffiths‐Jones S, Grocock RJ, Van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research 2006; 34: D140–D4. DOI: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hammond SM. An overview of microRNAs. Advance Drug Delivery Reviews 2015; 87: 3–14. DOI: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Connell RM, Rao DS, Chaudhuri AA, et al. Physiological and pathological roles for microRNAs in the immune system. Nature Reviews Immunology 2010; 10: 111–22. DOI: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 7. Globinska A, Pawelczyk M, Kowalski ML. MicroRNAs and the immune response to respiratory virus infections. Expert Review of Clinical Immunology 2014; 10: 963–71. DOI: 10.1586/1744666X.2014.913482. [DOI] [PubMed] [Google Scholar]
- 8. Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiological Reviews 2011; 91: 827–87. DOI: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 9. Gomez‐Diaz E, Jorda M, Peinado MA, et al. Epigenetics of host‐pathogen interactions: the road ahead and the road behind. PLoS Pathogens 2012; 8e1003007: . DOI: 10.1371/journal.ppat.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oglesby IK, Bray IM, Chotirmall SH, et al. miR‐126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. Journal of Immunology 2010; 184: 1702–9. DOI: 10.4049/jimmunol.0902669. [DOI] [PubMed] [Google Scholar]
- 11. Hassan T, McKiernan PJ, McElvaney NG, et al. Therapeutic modulation of miRNA for the treatment of proinflammatory lung diseases. Expert Review of Anti‐Infective Therapy 2012; 10: 359–68. DOI: 10.1586/eri.11.175. [DOI] [PubMed] [Google Scholar]
- 12. Foster PS, Plank M, Collison A, et al. The emerging role of microRNAs in regulating immune and inflammatory responses in the lung. Immunological Reviews 2013; 253: 198–215. DOI: 10.1111/imr.12058. [DOI] [PubMed] [Google Scholar]
- 13. Angulo M, Lecuona E, Sznajder JI. Role of MicroRNAs in lung disease. Archivos de Bronconeumología 2012; 48: 325–30. DOI: 10.1016/j.arbr.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sittka A, Schmeck B. MicroRNAs in the lung. Advances in Experimental Medicine and Biology 2013; 774: 121–34. DOI: 10.1007/978-94-007-5590-1_7. [DOI] [PubMed] [Google Scholar]
- 15. Rossi GA, Silvestri M, Colin AA. Respiratory syncytial virus infection of airway cells: role of microRNAs. Pediatric Pulmonology 2015; 10: 359–68. DOI: 10.1002/ppul.23193. [DOI] [PubMed] [Google Scholar]
- 16. DiMaio D. Viruses, masters at downsizing. Cell Host & Microbe 2012; 11: 560–1. DOI: 10.1016/j.chom.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 17. Guo YE, Steitz JA. Virus meets host microRNA: the destroyer, the booster, the hijacker. Molecular and Cellular Biology 2014; 34: 3780–7. DOI: 10.1128/MCB.00871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scaria V, Hariharan M, Maiti S, et al. Host‐virus interaction: a new role for microRNAs. Retrovirology 2006; 3: 68 DOI: 10.1186/1742-4690-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haasnoot J, Berkhout B. RNAi and cellular miRNAs in infections by mammalian viruses. Methods in Molecular Biology 2011; 721: 23–41. DOI: 10.1007/978-1-61779-037-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cullen BR. Five questions about viruses and microRNAs. PLoS Pathogens 2010; 6e1000787: . DOI: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes and Development 2009; 23: 1151–64. DOI: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cullen BR. Viruses and microRNAs. Nature Genetics 2006; 38: S25–S30. DOI: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 23. Dykxhoorn DM. MicroRNAs in viral replication and pathogenesis. DNA and Cell Biology 2007; 26: 239–49. DOI: 10.1089/dna.2006.0559. [DOI] [PubMed] [Google Scholar]
- 24. Liu D‐G. MicroRNAs in human virus genomes: helping hands for viral Infection. MicroRNA 2014; 3: 75–85. DOI: 10.2174/2211536603666140825193447. [DOI] [PubMed] [Google Scholar]
- 25. Grundhoff A, Sullivan CS. Virus‐encoded microRNAs. Virology 2011; 411: 325–43. DOI: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annual Review of Microbiology 2010; 64: 123 DOI: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kincaid RP, Sullivan CS. Virus‐encoded microRNAs: an overview and a look to the future. PLoS Pathogens 2012; 8e1003018: . DOI: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scaria V, Hariharan M, Pillai B, et al. Host–virus genome interactions: macro roles for microRNAs. Cellular Microbiology 2007; 9: 2784–94. DOI: 10.1111/j.1462-5822.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 29. Gottwein E. Roles of microRNAs in the life cycles of mammalian viruses. Current Topics in Microbiology and Immunology 2013; 371: 201–27. DOI: 10.1007/978-3-642-37765-5_8. [DOI] [PubMed] [Google Scholar]
- 30. Cullen BR. MicroRNAs as mediators of viral evasion of the immune system. Nature Immunology 2013; 14: 205–10. DOI: 10.1038/ni.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bakre A, Mitchell P, Coleman JK, et al. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. The Journal of General Virology 2012; 93: 2346–56. DOI: 10.1099/vir.0.044255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X, Dong C, Sun X, et al. Induction of the cellular miR‐29c by influenza virus inhibits the innate immune response through protection of A20 mRNA. Biochemical and Biophysical Research Communications 2014; 450: 755–61. DOI: 10.1016/j.bbrc.2014.06.059. [DOI] [PubMed] [Google Scholar]
- 33. Stewart CR, Marsh GA, Jenkins KA, et al. Promotion of Hendra virus replication by microRNA 146a. Journal of Virology 2013; 87: 3782–91. DOI: 10.1128/JVI.01342-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Müller S, Imler J‐L. Dicing with viruses: microRNAs as antiviral factors. Immunity 2007; 27: 1–3. DOI: 10.1016/j.immuni.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 35. Sullivan CS, Ganem D. MicroRNAs and viral infection. Molecular Cell 2005; 20: 3–7. DOI: 10.1016/j.molcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 36. Cullen BR. How do viruses avoid inhibition by endogenous cellular microRNAs? PLoS Pathogens 2013; 9e1003694: . DOI: 10.1371/journal.ppat.1003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song L, Liu H, Gao S, et al. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. Journal of Virology 2010; 84: 8849–60. DOI: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bondanese VP, Francisco‐Garcia A, Bedke N, et al. Identification of host miRNAs that may limit human rhinovirus replication. World Journal of Biological Chemistry 2014; 5: 437 DOI: 10.4331/wjbc.v5.i4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma YJ, Yang J, Fan XL, et al. Cellular microRNA let‐7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. Journal of Cellular and Molecular Medicine 2012; 16: 2539–46. DOI: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cristiano F, Veltri P. Methods and Techniques for miRNA data analysis. Methods in Molecular Biology 2016; 1375: 11–23. DOI: 10.1007/7651_2015_238. [DOI] [PubMed] [Google Scholar]
- 41. Svoboda P. A toolbox for miRNA analysis. FEBS Letters 2015; 589: 1694–701. DOI: 10.1016/j.febslet.2015.04.054. [DOI] [PubMed] [Google Scholar]
- 42. Salimi V, Tavakoli‐Yaraki M, Yavarian J, et al. Prevalence of human respiratory syncytial virus circulating in Iran. Journal of Infection and Public Health 2015; S1876‐0341: 00102‐1. DOI: 10.1016/j.jiph.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 43. Kurai D, Saraya T, Ishii H, et al. Virus‐induced exacerbations in asthma and COPD. Frontiers in Microbiology 2013; 4: 293 DOI: 10.3389/fmicb.2013.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. del Valle MJ, Cornejo‐Tapia A, Weilg P, et al. Incidence of respiratory viruses in Peruvian children with acute respiratory infections. Journal of Medical Virology 2015; 87: 917–24. DOI: 10.1002/jmv.24159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tahamtan A, Tavakoli‐Yaraki M, Rygiel TP, et al. Effects of cannabinoids and their receptors on viral infections. Journal of Medical Virology 2016; 88: 1–12. DOI: 10.1002/jmv.24292. [DOI] [PubMed] [Google Scholar]
- 46. Salimi V, Hennus MP, Mokhtari‐Azad T, et al. Opioid receptors control viral replication in the airways. Critical Care Medicine 2013; 41: 205–14. DOI: 10.1097/CCM.0b013e31826767a8. [DOI] [PubMed] [Google Scholar]
- 47. Legand A, Briand S, Shindo N, et al. Addressing the public health burden of respiratory viruses: the Battle against Respiratory Viruses (BRaVe) Initiative. Future Virology 2013; 8: 953–68. DOI: 10.2217/fvl.13.85. [DOI] [Google Scholar]
- 48. Li Y, Li J, Belisle S, et al. Differential microRNA expression and virulence of avian, 1918 reassortant, and reconstructed 1918 influenza A viruses. Virology 2011; 421: 105–13. DOI: 10.1016/j.virol.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guan Z, Shi N, Song Y, et al. Induction of the cellular microRNA‐29c by influenza virus contributes to virus‐mediated apoptosis through repression of antiapoptotic factors BCL2L2. Biochemical and Biophysical Research Communications 2012; 425: 662–7. DOI: 10.1016/j.bbrc.2012.07.114. [DOI] [PubMed] [Google Scholar]
- 50. Othumpangat S, Noti JD, Beezhold DH. Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276. Virology 2014; 468: 256–64. DOI: 10.1016/j.virol.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Terrier O, Textoris J, Carron C, et al. Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR‐146a. The Journal of General Virology 2013; 94: 985–95. DOI: 10.1099/vir.0.049528-0. [DOI] [PubMed] [Google Scholar]
- 52. Huang L, Ma J, Sun Y, et al. Altered splenic miRNA expression profile in H1N1 swine influenza. Archives of Virology 2015; 160: 979–85. DOI: 10.1007/s00705-015-2351-0. [DOI] [PubMed] [Google Scholar]
- 53. Buggele WA, Johnson KE, Horvath CM. Influenza A virus infection of human respiratory cells induces primary microRNA expression. Journal of Biological Chemistry 2012; 287: 31027–40. DOI: 10.1074/jbc.M112.387670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meliopoulos VA, Andersen LE, Brooks P, et al. MicroRNA regulation of human protease genes essential for influenza virus replication. PLoS One 2012; 7e37169: . DOI: 10.1371/journal.pone.0037169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loveday E‐K, Diederich S, Pasick J, et al. Human microRNA‐24 modulates highly pathogenic avian‐origin H5N1 influenza A virus infection in A549 cells by targeting secretory pathway furin. The Journal of General Virology 2015; 96: 30–9. DOI: 10.1099/vir.0.068585-0. [DOI] [PubMed] [Google Scholar]
- 56. Zhong Z, Dong Z, Yang L, et al. miR‐21 induces cell cycle at S phase and modulates cell proliferation by down‐regulating hMSH2 in lung cancer. Journal of Cancer Research and Clinical Oncology 2012; 138: 1781–8. DOI: 10.1007/s00432-012-1287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li J, Donath S, Li Y, et al. miR‐30 regulates mitochondrial fission through targeting p53 and the dynamin‐related protein‐1 pathway. PLoS Genetics 2010; 6e1000795: . DOI: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lam W‐Y, Yeung AC‐M, Ngai KL‐K, et al. Effect of avian influenza A H5N1 infection on the expression of microRNA‐141 in human respiratory epithelial cells. BMC Microbiology 2013; 13: 104 DOI: 10.1186/1471-2180-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li Y, Chan EY, Li J, et al. MicroRNA expression and virulence in pandemic influenza virus‐infected mice. Journal of Virology 2010; 84: 3023–32. DOI: 10.1128/JVI.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tambyah PA, Sepramaniam S, Mohamed Ali J, et al. microRNAs in circulation are altered in response to influenza A virus infection in humans. PLoS One 2013; 8e76811: . DOI: 10.1371/journal.pone.0076811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thornburg NJ, Hayward SL, Crowe JE. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF‐kB. MBio 2012; 3: e00220–12. DOI: 10.1128/mBio.00220-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Othumpangat S, Walton C, Piedimonte G. MicroRNA‐221 modulates RSV replication in human bronchial epithelium by targeting NGF expression. PLoS One 2012; 7e30030: . DOI: 10.1371/journal.pone.0030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Inchley CS, Sonerud T, Fjærli HO, et al. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infectious Diseases 2015; 15: 150 DOI: 10.1186/s12879-015-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS One 2009; 4e7837: . DOI: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lai FW, Stephenson KB, Mahony J, et al. Human Coronavirus OC43 Nucleocapsid Protein Binds MicroRNA 9 and Potentiates NF‐kB Activation. Journal of Virology 2014; 88: 54–65. DOI: 10.1128/JVI.02678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ouda R, Onomoto K, Takahasi K, et al. Retinoic acid‐inducible gene I‐inducible miR‐23b inhibits infections by minor group rhinoviruses through down‐regulation of the very low density lipoprotein receptor. Journal of Biological Chemistry 2011; 286: 26210–9. DOI: 10.1074/jbc.M111.229856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deng J, Ptashkin RN, Wang Q, et al. Human metapneumovirus infection induces significant changes in small noncoding RNA expression in airway epithelial cells. Molecular Therapy Nucleic Acids 2014; 3e163: . DOI: 10.1038/mtna.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Carnero E, Sutherland JD, Fortes P. Adenovirus and miRNAs. BBA‐Gene Regulatory Mechanisms 1809; 2011: 660–7. DOI: 10.1016/j.bbagrm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bellutti F, Kauer M, Kneidinger D, et al. Identification of RISC‐associated adenoviral microRNAs, a subset of their direct targets, and global changes in the targetome upon lytic adenovirus 5 infection. Journal of Virology 2015; 89: 1608–27. DOI: 10.1128/JVI.02336-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Aparicio O, Carnero E, Abad X, et al. Adenovirus VA RNA‐derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Research 2010; 38: 750–63. DOI: 10.1093/nar/gkp1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kondo S, Yoshida K, Suzuki M, et al. Adenovirus‐encoding virus‐associated RNAs suppress HDGF gene expression to support efficient viral replication. PLoS One 2014; 9e108627: . DOI: 10.1371/journal.pone.0108627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yanagawa‐Matsuda A, Kitamura T, Higashino F, et al. E1A expression might be controlled by miR‐214 in cells with low adenovirus productivity. Virus Research 2012; 170: 85–90. DOI: 10.1016/j.virusres.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 73. Lam W, Cheung AC, Tung CK, et al. miR‐466 is putative negative regulator of Coxsackie virus and adenovirus receptor. FEBS Letters 2015; 589: 246–54. DOI: 10.1016/j.febslet.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 74. Marzi MJ, Puggioni EM, Dall'Olio V, et al. Differentiation‐associated microRNAs antagonize the Rb–E2F pathway to restrict proliferation. Journal of Cell Biology 2012; 199: 77–95. DOI: 10.1083/jcb.201206033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huang Y, Chen D, He J, et al. Hcmv‐miR‐UL112 attenuates NK cell activity by inhibition type I interferon secretion. Immunology Letters 2015; 163: 151–6. DOI: 10.1016/j.imlet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 76. Fu M, Gao Y, Zhou Q, et al. Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA. Gene 2014; 536: 272–8. DOI: 10.1016/j.gene.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 77. Stark TJ, Arnold JD, Spector DH, et al. High‐resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. Journal of Virology 2012; 86: 226–35. DOI: 10.1128/JVI.05903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kim S, Lee S, Shin J, et al. Human cytomegalovirus microRNA miR‐US4‐1 inhibits CD8+ T cell responses by targeting the aminopeptidase ERAP1. Nature Immunology 2011; 12: 984–91. DOI: 10.1038/ni.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hook L, Hancock M, Landais I, et al. Cytomegalovirus microRNAs. Current Opinion Virology 2014; 7: 40–6. DOI: 10.1016/j.coviro.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Huang Y, Qi Y, Ma Y, et al. The expression of interleukin‐32 is activated by human cytomegalovirus infection and down regulated by HCMV‐miR‐UL112‐1. Virology Journal 2013; 10: 51 DOI: 10.1186/1743-422X-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Esteso G, Luzón E, Sarmiento E, et al. Altered microRNA expression after Infection with human cytomegalovirus leads to TIMP3 downregulation and Increased shedding of metalloprotease substrates, including MICA. Journal of Immunology 2014; 193: 1344–52. DOI: 10.4049/jimmunol.1303441. [DOI] [PubMed] [Google Scholar]
- 82. Craigen JL, Yong KL, Jordan NJ, et al. Human cytomegalovirus infection up‐regulates interleukin‐8 gene expression and stimulates neutrophil transendothelial migration. Immunology 1997; 92: 138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jiang S, Qi Y, He R, et al. Human cytomegalovirus microRNA miR‐US25‐1‐5p inhibits viral replication by targeting multiple cellular genes during infection. Gene 2015; 570: 108–14. DOI: 10.1016/j.gene.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 84. Shen Z‐Z, Pan X, Miao L‐F, et al. Comprehensive analysis of human cytomegalovirus microRNA expression during lytic and quiescent infection. PLoS One 2014; 9e88531: . DOI: 10.1371/journal.pone.0088531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Grey F, Meyers H, White EA, et al. A human cytomegalovirus‐encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathogens 2007; 3e163: . DOI: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Poole E, Avdic S, Hodkinson J, et al. Latency‐associated viral interleukin‐10 (IL‐10) encoded by human cytomegalovirus modulates cellular IL‐10 and CCL8 secretion during latent infection through changes in the cellular microRNA hsa‐miR‐92a. Journal of Virology 2014; 88: 13947–55. DOI: 10.1128/JVI.02424-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lee SH, Kalejta RF, Kerry J, et al. BclAF1 restriction factor is neutralized by proteasomal degradation and microRNA repression during human cytomegalovirus infection. Proceedings of the National Academy of Sciences USA 2012; 109: 9575–80. DOI: 10.1073/pnas.1207496109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gambarino S, Mantovani S, Astegiano S, et al. Lower respiratory tract viral infections in hospitalized adult patients. Minerva Medica 2009; 100: 349–55. [PubMed] [Google Scholar]
- 89. Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes and Development 2011; 25: 1881–94. DOI: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Short KR, Kroeze EJV, Fouchier RA, et al. Pathogenesis of influenza‐induced acute respiratory distress syndrome. Lancet Infectious Diseases 2014; 14: 57–69. DOI: 10.1016/S1473-3099(13)70286-X. [DOI] [PubMed] [Google Scholar]
- 91. Song H, Wang Q, Guo Y, et al. Microarray analysis of microRNA expression in peripheral blood mononuclear cells of critically ill patients with influenza A (H1N1). BMC Infectious Diseases 2013; 13: 257 DOI: 10.1186/1471-2334-13-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kumar M, Ahmad T, Sharma A, et al. Let‐7 microRNA–mediated regulation of IL‐13 and allergic airway inflammation. Journal of Allergy and Clinical Immunology 2011; 128: 1077–85. e10. DOI: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 93. Zhu Z, Qi Y, Ge A, et al. Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza A (H7N9) virus infection in humans. Viruses 2014; 6: 1525–39. DOI: 10.3390/v6041525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Balkhi MY, Iwenofu OH, Bakkar N, et al. miR‐29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Science Signaling 2013; 6: ra63. DOI: 10.1126/scisignal.2004177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Maelfait J, Roose K, Bogaert P, et al. A20 (Tnfaip3) deficiency in myeloid cells protects against influenza A virus infection. PLoS Pathogens 2012; 8e1002570‐e: . DOI: 10.1371/journal.ppat.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Paglinawan R, Malipiero U, Schlapbach R, et al. TGFβ directs gene expression of activated microglia to an anti‐inflammatory phenotype strongly focusing on chemokine genes and cell migratory genes. Glia 2003; 44: 219–31. DOI: 10.1002/glia.10286. [DOI] [PubMed] [Google Scholar]
- 97. Tan KS, Choi H, Jiang X, et al. Micro‐RNAs in regenerating lungs: an integrative systems biology analysis of murine influenza pneumonia. BMC Genomics 2014; 15: 587 DOI: 10.1186/1471-2164-15-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Birkhaug IM, Inchley CS, Aamodt G, et al. Infectious burden of respiratory syncytial virus in relation to time of birth modifies the risk of lower respiratory tract infection in infancy: the Norwegian Mother and Child Cohort. Pediatric Infectious Disease Journal 2013; 32: e235–41. DOI: 10.1097/INF.0b013e31828ab9ff. [DOI] [PubMed] [Google Scholar]
- 99. Stoppelenburg AJ, Salimi V, Hennus M, et al. Local IL‐17A potentiates early neutrophil recruitment to the respiratory tract during severe RSV infection. PLoS One 2013; 8: e78461. DOI: 10.1371/journal.pone.0078461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nair H, Nokes DJ, Gessner BD, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta‐analysis. Lancet 2010; 375: 1545–55. DOI: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Byington CL, Wilkes J, Korgenski K, et al. Respiratory syncytial virus–associated mortality in hospitalized infants and young children. Pediatrics 2015; 135: e24–e31. DOI: 10.1542/peds.2014-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hardelid P, Pebody R, Andrews N. Mortality caused by influenza and respiratory syncytial virus by age group in England and Wales 1999–2010. Influenza and Other Respiratory Viruses 2013; 7: 35–45. DOI: 10.1111/j.1750-2659.2012.00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Robinson RF. Impact of respiratory syncytial virus in the United States. American Journal of Health‐System Pharmacy 2008; 65: S3–S6. DOI: 10.2146/ajhp080438. [DOI] [PubMed] [Google Scholar]
- 104. Tempia S, Walaza S, Viboud C, et al. Mortality associated with seasonal and pandemic influenza and respiratory syncytial virus among children <5 years of age in a high HIV prevalence setting‐South Africa, 1998–2009. Clinical Infectious Diseases 2014; 58: 1241–9. DOI: 10.1093/cid/ciu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Inchley CS, Sonerud T, Fjaerli HO, et al. Reduced Dicer expression in the cord blood of infants admitted with severe respiratory syncytial virus disease. BMC Infectious Diseases 2011; 11: 59 DOI: 10.1186/1471-2334-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Graff JW, Dickson AM, Clay G, et al. Identifying functional microRNAs in macrophages with polarized phenotypes. Journal of Biological Chemistry 2012; 287: 21816–25. DOI: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Polikepahad S, Knight JM, Naghavi AO, et al. Proinflammatory role for let‐7 microRNAS in experimental asthma. Journal of Biological Chemistry 2010; 285: 30139–49. DOI: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Othumpangat S, Gibson LF, Samsell L, et al. NGF is an essential survival factor for bronchial epithelial cells during respiratory syncytial virus infection. PLoS One 2009; 4e6444: . DOI: 10.1371/journal.pone.0006444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kim HW, Canchola JG, Brandt CD, et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. American Journal of Epidemiology 1969; 89: 422–34. [DOI] [PubMed] [Google Scholar]
- 110. Gralinski LE, Baric RS. Molecular pathology of emerging coronavirus infections. American Journal of Epidemiology 2015; 235: 185–95. DOI: 10.1002/path.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hasan MM, Akter R, Ullah MS, et al. A computational approach for predicting role of human microRNAs in MERS‐CoV genome. Advances Bioinformatics 2014; 2014: 967946 DOI: 10.1155/2014/967946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ren L, Zhang Y, Li J, et al. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Science Reports 2015; 5: 11451 DOI: 10.1038/srep11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. New England Journal of Medicine 2003; 348: 1953–66. DOI: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 114. Banik G, Khandaker G, Rashid H. Middle East respiratory syndrome coronavirus “MERS‐CoV”: current knowledge gaps. Paediatric Respiratory Reviews 2015; 16: 197–202. DOI: 10.1016/j.prrv.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rossi GA, Colin AA. Infantile respiratory syncytial virus and human rhinovirus infections: respective role in inception and persistence of wheezing. European Respiratory Journal 2015; 45: 774–89. DOI: 10.1183/09031936.00062714. [DOI] [PubMed] [Google Scholar]
- 116. Nukui M, Mori Y, Murphy EA. A human herpesvirus 6A‐encoded microRNA: role in viral lytic replication. Journal of Virology 2015; 89: 2615–27. DOI: 10.1128/JVI.02007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Van den Hoogen BG, de Jong JC, Groen J, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nature Medicine 2001; 7: 719–24. DOI: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kim S, Seo D, Kim D, et al. Temporal landscape of microRNA‐mediated host‐virus crosstalk during productive human cytomegalovirus infection. Cell Host & Microbe 2015; 17: 838–51. DOI: 10.1016/j.chom.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 119. Lynch JP, Fishbein M, Echavarria M. Adenovirus. Seminars in Respiratory and Critical Care Medicine 2011; 32: 494–511. DOI: 10.1055/s-0031-1283287. [DOI] [PubMed] [Google Scholar]
- 120. Qi Y, Tu J, Cui L, et al. High‐throughput sequencing of microRNAs in adenovirus type 3 infected human laryngeal epithelial cells. Journal of Biomedicine and Biotechnology 2010; 2010915980: . DOI: 10.1155/2010/915980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhao H, Chen M, Tellgren‐Roth C, et al. Fluctuating expression of microRNAs in adenovirus infected cells. Virology 2015; 478: 99–111. DOI: 10.1016/j.virol.2015.01.033. [DOI] [PubMed] [Google Scholar]
- 122. Aparicio O, Razquin N, Zaratiegui M, et al. Adenovirus virus‐associated RNA is processed to functional interfering RNAs involved in virus production. Journal of Virology 2006; 80: 1376–84. DOI: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. Journal of Virology 2004; 78: 12868–76. DOI: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Trgovcich J, Cebulla C, Zimmerman P, et al. Human cytomegalovirus protein pp 71 disrupts major histocompatibility complex class I cell surface expression. Journal of Virology 2006; 80: 951–63. DOI: 10.1128/JVI.80.2.951-963.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Landais I, Pelton C, Streblow D, et al. Human cytomegalovirus miR‐UL112‐3p targets TLR2 and modulates the TLR2/IRAK1/NFkB signaling pathway. PLoS Pathogen 2015; 11e1004881: . DOI: 10.1371/journal.ppat.1004881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Shteinberg M, Shaked‐Mishan P, Kinarti A, et al. Asymptomatic carriage of Pneumocystis jirovecii and cytomegalovirus in lungs of immunocompetent patients. Lung 2014; 192: 875–9. DOI: 10.1007/s00408-014-9644-z. [DOI] [PubMed] [Google Scholar]
- 127. O'Connor CM, Vanicek J, Murphy EA. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. Journal of Virology 2014; 88: 5524–32. DOI: 10.1128/JVI.00481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wang FZ, Weber F, Croce C, et al. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. Journal of Virology 2008; 82: 9065–74. DOI: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kim Y, Lee S, Kim S, et al. Human cytomegalovirus clinical strain‐specific microRNA miR‐UL148D targets the human chemokine RANTES during infection. PLoS Pathogens 2012; 8e1002577: . DOI: 10.1371/journal.ppat.1002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Qi M, Qi Y, Ma Y, et al. Over‐expression of human cytomegalovirus miR‐US25‐2‐3p downregulates eIF4A1 and inhibits HCMV replication. FEBS Letters 2013; 587: 2266–71. DOI: 10.1016/j.febslet.2013.05.057. [DOI] [PubMed] [Google Scholar]
- 131. Guo X, Qi Y, Huang Y, et al. Human cytomegalovirus miR‐US33‐5p inhibits viral DNA synthesis and viral replication by down‐regulating expression of the host Syntaxin3. FEBS Letters 2015; 589: 440–6. DOI: 10.1016/j.febslet.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 132. Hook LM, Grey F, Grabski R, et al. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host & Microbe 2014; 15: 363–73. DOI: 10.1016/j.chom.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Babu SG, Pandeya A, Verma N, et al. Role of HCMV miR‐UL70‐3p and miR‐UL148D in overcoming the cellular apoptosis. Molecular and Cellular Biochemistry 2014; 393: 89–98. DOI: 10.1007/s11010-014-2049-8. [DOI] [PubMed] [Google Scholar]
- 134. Lee S, Song J, Kim S, et al. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host & Microbe 2013; 13: 678–90. DOI: 10.1016/j.chom.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 135. Cone RW, Huang M‐L, Hackman RC, et al. Coinfection with human herpesvirus 6 variants A and B in lung tissue. Journal of Clinical Microbiology 1996; 34: 877–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Tuddenham L, Jung JS, Chane‐Woon‐Ming B, et al. Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B. Journal of Virology 2012; 86: 1638–49. DOI: 10.1128/JVI.05911-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Grassmann R, Jeang K‐T. The roles of microRNAs in mammalian virus infection. BBA‐Gene Regulatory Mechanisms 2008; 1779: 706–11. DOI: 10.1016/j.bbagrm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Esau CC, Monia BP. Therapeutic potential for microRNAs. Advance Drug Delivery Reviews 2007; 59: 101–14. DOI: 10.1016/j.addr.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 139. Soifer HS, Rossi JJ, Sætrom P. MicroRNAs in disease and potential therapeutic applications. Molecular Therapy 2007; 15: 2070–9. DOI: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 140. Patton JS, Byron PR. Inhaling medicines: delivering drugs to the body through the lungs. Nature Reviews Drug Discovery 2007; 6: 67–74. DOI: 10.1038/nrd2153. [DOI] [PubMed] [Google Scholar]
- 141. Tahamtan A, Tabarraei A, Moradi A, et al. Chitosan nanoparticles as a potential nonviral gene delivery for HPV‐16 E7 into mammalian cells. Artificial Cells, Nanomedicine and Biotechnology 2015; 43: 366–72. DOI: 10.3109/21691401.2014.893522. [DOI] [PubMed] [Google Scholar]


