SUMMARY
Most coronaviruses cause respiratory or intestinal infections in their animal or human host. Hence, their interaction with polarized epithelial cells plays a critical role in the onset and outcome of infection. In this paper, we review the knowledge regarding the entry and release of coronaviruses, with particular emphasis on the severe acute respiratory syndrome and Middle East respiratory syndrome coronaviruses. As these viruses approach the epithelial surfaces from the apical side, it is not surprising that coronavirus cell receptors are exposed primarily on the apical domain of polarized epithelial cells. With respect to release, all possibilities appear to occur. Thus, most coronaviruses exit through the apical surface, several through the basolateral one, although the Middle East respiratory syndrome coronavirus appears to use both sides. These observations help us understand the local or systematic spread of the infection within its host as well as the spread of the virus within the host population. Copyright © 2014 John Wiley & Sons, Ltd.
Abbreviations used
- ACE2
angiotensin‐converting enzyme II
- BCoV
bovine coronavirus
- Calu‐3
a human lung cancer cell line
- CCoV
canine coronavirus
- CFR
case fatality rate
- CoVs
coronaviruses
- E
envelope protein
- fAPN
feline aminopeptidase‐N
- FCoV
feline coronavirus
- HCoVs
human coronaviruses
- HE
haemagglutinin esterase
- IBV
avian coronavirus infectious bronchitis virus
- IEC
intestinal epithelial cells
- LLC‐PK1
LLC‐pig kidney 1
- M
membrane protein
- MERS‐CoV
Middle East respiratory syndrome virus
- MHV
murine hepatitis virus
- N
nucleocapsid protein
- pAPN
porcine aminopeptidase‐N
- PEDV
porcine epidemic diarrhea virus
- PRCoV
porcine respiratory coronavirus
- S
spike protein
- SARS‐CoV
severe acute respiratory syndrome‐associated coronavirus
- TCoV
turkey coronavirus
- TGEV
transmissible gastroenteritis
INTRODUCTION
Coronaviruses (CoVs) comprise a large family of enveloped, positive‐stranded RNA viruses that infect a broad range of animal hosts as well as humans. These viruses can cause a wide variety of diseases, in particular respiratory and enteric, but also including hepatic, renal and neuronal infection 1, 2. CoVs are divided into three genera, namely, Alphacoronavirus, Betacoronavirus and Gammacoronavirus, as well as a tentative new genus, the Deltacoronavirus 3. Well‐known representatives are porcine transmissible gastroenteritis virus (TGEV), porcine respiratory CoV (PRCoV) and porcine epidemic diarrhea virus (PEDV); canine CoV (CCoV), feline CoV (FCoV), bovine CoV (BCoV), human CoVs (HCoVs) including HCoV‐229E, HCoV‐OC43, HCoV‐NL63 and HCoV‐HKU1, severe acute respiratory syndrome‐associated CoV (SARS‐CoV) and Middle East respiratory syndrome virus (MERS‐CoV); murine hepatitis virus (MHV); and the avian CoV infectious bronchitis virus (IBV) and turkey CoV (TCoV) 4.
The CoV genome typically encodes four structural proteins: the spike protein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N); some CoVs additionally have a haemagglutinin‐esterase (HE) protein 5, 6, 7. The S glycoprotein of CoV is the dominant surface protein and is responsible for virus attachment and membrane fusion 8, 9, 10, 11.
Epithelia are formed of cells that line the cavities in the body and also cover flat surfaces. Epithelial cells cover the inner and outer linings of body cavities and act as a primary barrier to infection by microorganisms, entering their host via body cavities, such as the respiratory or intestinal tract 12. Primary replication of CoVs is often confined to respiratory or gastrointestinal tract epithelial cells 13. Epithelial cells are functionally polarized; their surface exhibits two distinguishable regions, which are called apical domain and basolateral domain 14. The apical membrane faces the luminal (external) compartment and contains proteins that determine the cells' primary functions such as secretion and absorption, whereas the basolateral domain faces the systemic (internal) compartment, that is, tissues and blood 15, 16.
As CoVs generally spread through the fecal–oral or respiratory route, polarized epithelial cells constitute their first natural barrier. Hence, their interaction with these cells determines for a major part the outcome of the infection 17. In this paper, we review the entry and release of several CoVs in polarized epithelial cells as this information will contribute to our understanding of the pathogenesis of these viruses.
CORONAVIRUS ENTRY INTO POLARIZED EPITHELIAL CELLS
The entry of several CoVs in polarized epithelial cells has been investigated for several decades. As early as 1994, Rossen and colleagues analyzed the entry of TGEV in filter‐grown polarized LLC‐pig kidney 1 (LLC‐PK1) cells, a line of pig kidney epithelial cells, using radioactive labeling, immunoprecipitation and electron microscopy. The results showed that TGEV infection was restricted to the apical plasma membrane 18. In 2001, Rossen et al. similarly analyzed the entry of FCoV and CCoV in polarized epithelial LLC‐PK1 cells expressing the recombinant feline aminopeptidase‐N (fAPN), which acts as a receptor for these viruses, and compared it with TGEV. The results showed that FCoV and CCoV, like TGEV, establish their infection into polarized epithelial cells specifically by entry through the apical membrane 19. The same pattern has subsequently also been found with a large number of other CoVs including HCoV‐229E, HCoV‐OC43, HCoV‐NL63, HCoV‐HKU1, BCoV, SARS‐CoV and MHV 4, 17, 18, 20, 21, 22, 23, 24, 25, 26, 27, 28. Recently, Pratelli reported a study with CCoV in filter‐grown polarized epithelia primary dog kidney cells (A72 cells) and epithelia feline kidney cells (CrFK cells) showing productive infection both through the apical and basolateral cell membrane 29.
It is clear that the S glycoprotein of CoV mediates viral attachment to the cellular receptor and subsequent entry into cells 10, 11. Thus, exploration of the distribution of cellular receptor in polarized epithelial in relation to CoV entry is important for understanding virus invasion. It is well known that porcine aminopeptidase‐N (pAPN), the cellular receptor of TGEV, is primarily expressed on the apical side in polarized epithelial cells 18, 30. Rossen et al. chose LLC‐PK1 cells, which are derived from the proximal tubule of porcine kidney, as their in vitro model for determining the association of TGEV entry with localization of pAPN 18. Furthermore, the authors performed a confocal laser scanning microscope method of filter‐grown LLC‐PK1 cells, which allows optical sections to be cut either in the horizontal plane (XY section) or in the vertical plane (XZ section) to determine the plasma membrane distribution of the pAPN receptor. The results showed that when the LLC‐PK1 cells were not fully polarized, pAPN was expressed on both apical and basolateral sides; in contrast, expression of the pAPN was limited to the apical side in fully polarized LLC‐PK1 monolayers 18, consistent with TGEV entry into these cells to be mediated by pAPN. Similarly, also the entry of HCoV‐229E and FCoV through the apical domain of polarized epithelial cells correlated with the apical expression of their cognate aminopeptidase‐N (APN) receptor 19, 20, 24, 31, 32, 33, 34. Besides, the apical distribution of angiotensin‐converting enzyme II (ACE2; HCoV‐NL63 receptor) mediated the apical entry of HCoV‐NL63 24, 26. For their study of polarized MHV entry, Rossen et al. used porcine LLC‐PK1 cells stably expressing the carcinoembryonic antigen receptor for MHV (MHVR) and confirmed that the apical entry of the virus could be explained by the specific apical expression of its receptor 23, 35. As far as the entry of SARS‐CoV is concerned, Tseng et al. first reported the apical entry of this virus in polarized Calu‐3 (a human lung cancer cell line) cells in 2005 using immunofluorescence staining, confocal microscopy and transmission electron microscopy 17. In the same year, Jia et al. investigated interactions between SARS‐CoV and human airway epithelia using native tissue and a primary culture model of polarized, well‐differentiated tracheal and bronchial epithelia 21. They also confirmed SARS‐CoV receptor, ACE2, to be expressed in greater abundance on the apical surface of the polarized cells 21, 36, 37. Furthermore, Ren et al. analyzed the entry of vesicular stomatitis virus (VSV) pseudotypes bearing SARS‐CoV S protein in polarized Vero (a line of monkey kidney cell line), Calu‐3 and Caco‐2 cells (a human colon cancer cell line) using confocal immunofluorescence and surface biotinylation 12. Their results indicated that SARS‐CoV S mediated apical entry into polarized epithelial cells. Furthermore, the authors used human respiratory tissues in an immunohistochemical assay. They found a strong expression of ACE2 on the epithelium of almost all tracheal glands, and no ACE2 was detected in the lower bronchi of any of the tissues. These results clearly showed ACE2 to be present on the epithelia of certain parts of the respiratory tract and to mediate SARS‐CoV entry 12, 38. The findings indicate the importance of CoV receptors in the context of entry of these viruses into polarized epithelial cells. It can thus be concluded that CoVs bind to particular host cell molecules and that their specific entry route is mediated by the distribution of these molecules on polarized epithelial cells. The most likely mechanism for the sorting of intracellular budding viruses to the apical or basolateral plasma membrane side involves their interaction with a membrane cellular receptor, which is polarized and targeted to a specific destination. To some extent, it explains the entry of CoV 29. In our recent study, we infected polarized Vero cells and intestinal epithelial cells (IEC) with PEDV and revealed by using immunofluorescence assays the apical entry of PEDV into both these cell types (unpublished data). Currently available reports have claimed that pAPN might serve as a functional receptor for PEDV 39, 40, 41. Interestingly, Vero cells are of monkey rather than pig origin and do not express pAPN; therefore, the apical entry of PEDV is independent of the pAPN molecule. Identification of the actual cellular receptor of PEDV is important for understanding the interaction between PEDV and its receptor. Whether there are additional other mechanisms or factors contributing to the entry of CoVs into polarized cells remains unclear.
CORONAVIRUS RELEASE
Coronaviruses are assembled in their host cells from the structural proteins and genome RNA by budding into the endoplasmic reticulum and early Golgi membranes, after which virions are transported through the Golgi complex and secreted out of the cell 42. Such processes are complex, and as a consequence, understanding the release of CoVs from polarized epithelial cells is more complicated than their entry. It has been reported that TGEV was preferentially released from the apical plasma membrane. In this study, the amounts of infectious TGEV particles released into the apical and basolateral media of infected LLC‐PK1 cells were determined by plaque assay. The results showed that 30‐fold more pfu had accumulated in the apical medium than in the basolateral medium 18. The same pattern was also found upon infections of polarized epithelial cells with HCoV‐229E, HCoV‐OC43, HCoV‐NL63, HCoV‐HKU1, SARS‐CoV and BCoV by virus titration or real‐time PCR 12, 17, 20, 21, 23, 24, 25, 26, 27, 28, 43. In 1997, Lin et al. demonstrated that BCoV isolated from enteric (enteric BCoV, EBCoV) and respiratory (respiratory BCoV, RBCoV) tract infections was released through the apical surfaces of the polarized HRT‐18G cells, an epithelioid human rectal tumor cell line 28. Later, in 2000, the same was shown by Wang et al. for HCoV‐229E infection of polarized airway epithelia 20, whereas the apical release of HCoV‐OC43 from polarized primary epithelial cells was reported by Dijkman et al. in 2013 24. In contrast to these examples of apical release, however, CCoV, FCoV and MHV were found to be released from the opposite side 12, 17, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 43. Thus, Rossen et al. studied FCoV and CCoV release from polarized porcine epithelial LLC‐PK1 cells stably expressing the recombinant fAPN and observed the progeny viruses to accumulate preferentially in the basolateral medium of the epithelial cells 19. To investigate whether the differential release of different CoVs is determined by the cells rather than the viruses, these same authors also analyzed the release of TGEV and MHV from the same polarized cells. Using the porcine LLC‐PK1 expressing the MHV receptor to make them susceptible to this murine virus, they confirmed by virus titration the apical release of TGEV, whereas infectious MHV was predominantly released into the basolateral fluid 18, 22. For PEDV, an important pathogen circulating in Asia, but which recently also emerged in Europe 44, 45, we investigated the release in polarized VERO and IEC cells and found the virus to be secreted apically using virus titration and real‐time PCR (unpublished data). Taken together, the observations demonstrate that there are two patterns for CoV release from the polarized epithelial cells. As epithelial cells are the initial target cells for most CoV infections, the pivotal role in the pathogenesis of viral infections is obvious. The virus specifically released from the apical surface is targeted to the lumen; hence, the resulting infection is more likely to be restricted to the epithelial surface. In contrast, basolateral release should provide access to the blood and lymph vessels, resulting in a systemic infection 23, 46. Clarification of the different release routines of CoVs may provide important insight into the mechanisms of transmission and pathogenesis and will facilitate the design of effective antiviral strategies to control CoV infection.
POLARIZED INFECTIONS BY SEVERE HUMAN RESPIRATORY SYNDROME CORONAVIRUSES IN EPITHELIAL CELLS
As HCoV infections are significant threat to public health, a thorough understanding of their infections in polarized cells is important. Particularly, in comparison with the usually milder HCoV‐229E, HCoV‐OC43 and HCoV‐NL63 viruses, the infections of humans by SARS‐CoV and MERS‐CoV are very severe because of their high mortality rates.
Infection of SARS‐CoV in polarized epithelia has been examined by several investigators 12, 17, 21, 43. Tseng et al. infected an established cell line of human bronchial epithelial origin, Calu‐3, which is a relevant cell culture model for SARS‐CoV infection despite its pulmonary adenocarcinoma origin 17. The functional receptor for SARS‐CoV, ACE2, is preferentially expressed on the apical surface of these cells 12, 17, 21, 43. Although the authors, as mentioned earlier, demonstrated that SARS‐CoV enters these cells through the apical domain of polarized Calu‐3 cells, they also found the release to be almost exclusively through this domain. Because SARS‐CoV naturally enters its host through the mucosa of the respiratory tract and the eyes, Jia et al. described the entry and release of the virus in polarized human airway epithelia. Their results were consistent with those of Tseng et al. 17, 21. They used native lung tissue and a model of well‐differentiated cultures of primary human airway epithelia and showed ACE2 receptor expression to increase and appear more abundantly on the apical side with the differentiation state of epithelia. SARS‐CoV preferentially exited via the apical surface of the well‐differentiated cells 10, 21, 47.
In September 2012, the MERS‐CoV drew attention as a new cause of severe respiratory illness in humans in the Middle East 48, 49, 50. Patients with confirmed MERS‐CoV infection presented with a spectrum of disease symptoms ranging from mild influenza‐like illness to severe pneumonia accompanied by respiratory and renal failure and resulting in death; the case fatality rate (CFR) presently stands at 45%, in contrast to the 7% CFR of SARS 51. For MERS‐CoV, no animal reservoir or intermediate host(s) has been definitely implicated in transmission. Limited human‐to‐human transmission has occurred within several clusters of cases in many countries 52. MERS‐CoV infects primary human bronchial epithelial Calu‐3 cells and primary human kidney cells, and CD26 (also known as dipeptidyl peptidase 4, DPP4) was identified as the cellular receptor for MERS‐CoV 53. Tao et al. indicated that MERS‐CoV could not infect the cell lines without CD26 expression including ACE2‐expressing A549 cells, embryonic kidney 293 cells and ACE2‐expressing 293 cells and infected the primary human bronchial epithelial Calu‐3 cells, which are target cell lines for MERS‐CoV and SARS with the distribution of CD26 being indiscriminately expressed on the entire membrane of cells by sequential images caught by z‐scanning and ACE2 being distinctly expressed on the apical side. Then, these two HCoVs infected both sides of the polarized epithelial Calu‐3 cells. SARS‐CoV infected and released almost exclusively through the apical side, whereas MERS‐CoV was indeed capable of doing so through either side and released through both routes. It should be noted that there was a nearly 100‐fold‐lower titer released from the apical side when infection was carried out from the basolateral rather than apical routes 54. Because of the bilateral entry and release, MERS‐CoV caused the lateral spread, human‐to‐human transmission and vertical transmission, whereas viral particles can be detected in serum and plasma 49, 50.
So far, two main patterns of entry into and release from polarized epithelial cells apply to most CoVs. One is apical entry and apical release, and the other is apical entry and basolateral release. The apical release allows a rapid lateral spread over the respiratory or intestinal epithelium; virus is deposited from infected cells into the lung or gut lumen followed by efficient infection of new nearby target cells. A schematic drawing of the process is shown in Figure 1A. SARS‐CoV, for instance, mainly infects through the respiratory tract 12, 17, 43. The new progeny virions are released from the apical domain of polarized epithelial cells and subsequently infect adjacent epithelial cells. However, these virions do not easily cross the epithelial cell layer and infect tissues; hence, the infection is maintained at the surface of the epithelial cells and causes a massive lateral spread. This condition enables efficient horizontal transmission by respiratory secretions, body fluids or body contact and leads to a widespread dissemination into the population 55, 56. In contrast, basolateral release supports the vertical transmission from the infected epithelia to blood and lymph vessels, consequently promoting the establishment of a systemic infection 57, 58, and a proposed schematic drawing is shown in Figure 1B. Virions reach underlying cells and tissues, pass into the bloodstream and are transported around the body, circulating through body fluids. MHV, CCoV and FCoV, which were found to exhibit this pattern of virus release from polarized epithelial cells, cause a systemic infection 19, 22, 23. The current knowledge regarding the entry and release of several CoVs is summarized in Table 1.
Figure 1.
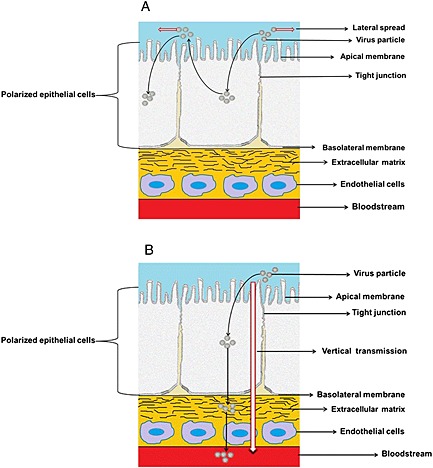
Two patterns for coronavirus entry and release from polarized epithelial cells. (A) Entry and release through the apical membrane facilitates lateral spread. (B) Entry at the apical membrane and release through the basolateral membrane facilitates vertical transmission
Table 1.
Entry and release of selected coronaviruses from polarized epithelial cells
| Coronavirus | Entry | Release | Reference |
|---|---|---|---|
| TGEV | Apical | Apical | [18] |
| PEDV | Apical | Apical | Unpublished data |
| PRCoV | ? | ? | |
| CCoV | Apical | Basolateral | [19] |
| Apical/basolateral | Apical/basolateral | [29] | |
| FCoV | Apical | Basolateral | [19] |
| HCoV‐229E | Apical | Apical | [20] |
| HCoV‐OC43 | Apical | Apical | [24] |
| HCoV‐NL63 | Apical | Apical | [26] |
| HCoV‐HKU1 | Apical | Apical | [25] |
| MHV | Apical | Basolateral | [22] |
| SARS‐CoV | Apical | Apical | [17] |
| [21] | |||
| [12] | |||
| [43] | |||
| [53] | |||
| BCoV | Apical | Apical | [27] |
| [28] | |||
| IBV | ? | ? | |
| MERS‐CoV | Apical/basolateral | Apical/basolateral | [54] |
BCoV, bovine coronavirus; CCoV, canine coronavirus; FCoV, feline coronavirus; HCoV, human coronavirus; IBV, avian coronavirus infectious bronchitis virus; MERS‐CoV, Middle East respiratory syndrome virus; MHV, murine hepatitis virus; PEDV, porcine epidemic diarrhea virus; PRCoV, porcine respiratory coronavirus; SARS‐CoV, severe acute respiratory syndrome‐associated coronavirus; TGEV, transmissible gastroenteritis virus.
CONCLUSIONS
Studying the infection of polarized epithelial cells by CoVs is important for understanding the molecular basis of the pathogenesis of these viruses. The polarized distribution of cellular receptors for CoVs determines the entry domain of most CoVs; their polarized release helps explain their pathogenesis.
Multiple factors should be considered in the analysis of the polarized entry and release of CoVs. First, the characteristics of CoVs have an impact on polarized entry, especially on target cell tropisms. For instance, HCoV‐229E has a preference to infect nonciliated cells, unlike HCoV‐NL63, HCoV‐HKU1 and HCoV‐OC43 24. Experimental polarized cell lines might not reflect the real infection in hosts. Second, laboratory‐adapted strains were commonly used in most of the studies; nonetheless, the wild strains might differ in certain infection properties when characterizing the polarized entry or release from the epithelial cells. Finally, as recombination plays a key role in the evolution of the CoVs 59, 60, 61, especially of HCoVs, most of their target cells were the same, potentially facilitating recombination. Whether it makes a contribution to the polarized entry and release should be investigated in further work.
The entry of several CoVs in polarized epithelial cells is still unclear, as illustrated by PRCoV, IBV and PEDV; for the latter two, this is due particularly to the uncertainty of their cellular receptors. Further elucidation of CoV entry of polarized cells and identification of molecules that are involved in this process should be realized in the future to understand CoV invasion of their target cells in detail. This holds even more for understanding the release of CoVs, as little is still known about the mechanisms and pathways that direct these viruses to specific membrane destinations for their targeted removal from infected cells.
CONFLICT OF INTEREST
The authors have no competing interest
ACKNOWLEDGEMENTS
We thank Drs. Susan Weiss and Joshua M. Thornbrough, Department of Microbiology, Perelman School of Medicine, University of Pennsylvania, and Dr. Peter JM Rottier, Virology Division, Utrecht University, for critical reading and discussion of this manuscript. This work is supported by the National Natural Science Foundation of China (31340003 and 31372438).
Cong Y. and Ren X. (2014), Coronavirus entry and release in polarized epithelial cells: a review, Rev. Med. Virol., 24: 308–315. DOI: 10.1002/rmv.1792
REFERENCES
- 1. Masters PS. The molecular biology of coronaviruses. Advances in Virus Research 2006; 66: 193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Enjuanes L, Gorbalenya AE, de Groot RJ, et al Nidovirales In Encyclopedia of Virology, 3rd edn, Mahy BWJ, Van Regenmortel MHV. (eds). Elsevier: Oxford, 2008; 419–430. [Google Scholar]
- 3. Adams MJ, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Archives of Virology 2012; 157(7): 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Groot RJ, Baker SC, Baric R, et al Coronaviridae In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses, King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (eds). Elsevier Academic Press: San Diego, 2011; 774–796. [Google Scholar]
- 5. Bost AG, Carnahan RH, Lu XT, Denison MR. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. Journal of Virology 2000; 74(7): 3379–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Müller MA, van der Hoek L, Voss D, et al Human coronavirus NL63 open reading frame 3 encodes a virion‐incorporated N‐glycosylated membrane protein. Virology Journal 2010; 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verheije MH, Hagemeijer MC, Ulasli M, et al The coronavirus nucleocapsid protein is dynamically associated with the replication‐transcription complexes. Journal of Virology 2010; 84(21): 11575–11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang N, Shi X, Jiang L, et al Structure of MERS‐CoV spike receptor‐binding domain complexed with human receptor DPP4. Cell Research 2013; 23(8): 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shim BS, Stadler K, Nguyen HH, et al Sublingual immunization with recombinant adenovirus encoding SARS‐CoV spike protein induces systemic and mucosal immunity without redirection of the virus to the brain. Virology Journal 2012; 9: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Josset L, Menachery VD, Gralinski LE, et al Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio 2013; 4(3): e00165‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012; 4(6): 1011–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ren X, Glende J, Al‐Falah M, et al Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome‐associated coronavirus. Journal of General Virology 2006; 87(6): 1691–1695. [DOI] [PubMed] [Google Scholar]
- 13. Lednicky JA, Waltzek TB, McGeehan E, Loeb JC, Hamilton SB, Luetke MC. Isolation and genetic characterization of human coronavirus NL63 in primary human renal proximal tubular epithelial cells obtained from a commercial supplier, and confirmation of its replication in two different types of human primary kidney cells. Virology Journal 2013; 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ikonen E, Simons K. Protein and lipid sorting from the trans‐Golgi network to the plasma membrane in polarized cells. Seminars in Cell and Developmental Biology 1998; 9(5): 503–509. [DOI] [PubMed] [Google Scholar]
- 15. Catalán M, Niemeyer MI, Cid LP, Sepúlveda FV. Basolateral ClC‐2 chloride channels in surface colon epithelium: regulation by a direct effect of intracellular chloride. Gastroenterology 2004; 126(4): 1104–1114. [DOI] [PubMed] [Google Scholar]
- 16. Yang D, Shcheynikov N, Zeng W, et al IRBIT coordinates epithelial fluid and HCO3‐secretion by stimulating the transporters pNBC1 and CFTR in the murine pancreatic duct. Journal of Clinical Investigation 2009; 119(1): 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tseng CT, Tseng J, Perrone L, Worthy M, Popov V, Peters CJ. Apical entry and release of severe acute respiratory syndrome‐associated coronavirus in polarized Calu‐3 lung epithelial cells. Journal of Virology 2005; 79(15): 9470‐9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rossen JW, Bekker CP, Voorhout WF, Strous GJ, van der Ende A, Rottier PJ. Entry and release of transmissible gastroenteritis coronavirus are restricted to apical surfaces of polarized epithelial cells. Journal of Virology 1994; 68(12): 7966–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rossen JW, Kouame J, Goedheer AJ, Vennema H, Rottier PJ. Feline and canine coronavirus are released from the basolateral side of polarized epithelial LLC‐PK1 cells expressing the recombinant feline aminopeptidase‐N cDNA. Archives of Virology 2001; 146(4): 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang G, Deering C, Macke M, et al Human coronavirus 229E infects polarized airway epithelial from the apical surface. Journal of Virology 2000; 74(19): 9234–9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jia HP, Look DC, Shi L, et al ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. Journal of Virology 2005; 79(23): 14614–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rossen JW, Voorhout WF, Horzinek MC, van der Ende A, Strous GJ, Rottier PJ. MHV‐A59 enters polarized murine epithelial cells through the apical surface but is released basolaterally. Virology 1995; 210(1): 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rossen JW, Bekker CP, Strous GJ, et al A murine and a porcine coronavirus are released from opposite surfaces of the same epithelial cells. Virology 1996; 224(1): 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dijkman R, Jebbink MF, Koekkoek SM, et al Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. Journal of Virology 2013; 87(11): 6081–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pyrc K, Sims AC, Dijkman R, et al Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. Journal of Virology 2010; 84(21): 11255–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Donaldson EF, Yount B, Sims AC, Burkett S, Pickles RJ, Baric RS. Systematic assembly of a full‐length infectious clone of human coronavirus NL63. Journal of Virology 2008; 82(23): 11948–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schultze B, Zimmer G, Herrler G. Virus entry into a polarized epithelial cell line (MDCK): similarities and dissimilarities between influenza C virus and bovine coronavirus. Journal of General Virology 1996; 77 (Pt 10): 2507–2514. [DOI] [PubMed] [Google Scholar]
- 28. Lin X, O'Reilly KL, Storz J. Infection of polarized epithelial cells with enteric and respiratory tract bovine coronaviruses and release of virus progeny. American Journal of Veterinary Research 1997; 58(10): 1120–1124. [PubMed] [Google Scholar]
- 29. Pratelli A. Basic science track. Entry and release of canine coronavirus from polarized epithelial cells. New Microbiologica 2011; 34(1): 25–32. [PubMed] [Google Scholar]
- 30. Delmas B, Gelfi J, L'Haridon R, et al Aminopeptidase N is a major receptor for the entero‐pathogenic coronavirus TGEV. Nature 1992; 357(6377): 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tusell SM, Schittone SA, Holmes KV. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. Journal of Virology 2007; 81(3): 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nomura R, Kiyota A, Suzaki E, et al Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. Journal of Virology 2004; 78(16): 8701–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bertram S, Dijkman R, Habjan M, et al TMPRSS2 activates the human coronavirus 229E for cathepsin‐independent host cell entry and is expressed in viral target cells in the respiratory epithelium. Journal of Virology 2013; 87(11): 6150–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tekes G, Hofmann‐Lehmann R, Bamk‐Wolf B, Maier R, Thiel HJ, Thiel V. Chimeric feline coronavirus that encode type II spike protein on type I genetic background display accelerated viral growth and altered receptor usage. Journal of Virology 2010; 84(3): 1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langereis MA, Zenq Q, Heesters BA, Huizinga EG, de Groot RJ. The murine coronavirus hemagglutinin‐esterase receptor‐binding site: a major shift in ligand specificity through modest changes in architecture. PLoS Pathogen 2012; 8(1): e1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Struck AW, Axmann M, Pfefferle S, Drosten C, Meyer B. A hexapeptide of the receptor‐binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antiviral Research 2012; 94(3): 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hou Y, Peng C, Yu M, et al Angiotensin‐converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS‐CoV entry. Archives of Virology 2010; 155(10): 1563–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Current Opinion in Virology 2012; 2(3): 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li BX, Ge JW, Li YJ. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 2007; 365(1): 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nam E, Lee C. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Veterinary Microbiology 2010; 144(1‐2): 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oh JS, Song DS, Yang JS, et al Effect of soluble porcine aminopeptidase N on antibody production against porcine epidemic diarrhea virus. Journal of Veterinary Science 2004; 5(4): 353–357. [PubMed] [Google Scholar]
- 42. Klumperman J, Locker JK, Meijer A, Horzinek MC, Geuze HJ, Rottier PJ. Coronavirus M protein accumulate in the Golgi complex beyond the site of virion budding. Journal of Virology 1994; 68(10): 6523–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sims AC, Burkett SE, Yount B, Pickles RJ. SARS‐CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Research 2008; 133(1): 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park SJ, Song DS, Park BK. Molecular epidemiology and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field isolates in Korea. Archives of Virology 2013; 158(7): 1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao Y, Kou Q, Ge X, Zhou L, Guo X, Yang H. Phylogenetic analysis of porcine epidemic diarrhea virus field strains prevailing recently in China. Archives of Virology 2013; 158(3): 711–715. [DOI] [PubMed] [Google Scholar]
- 46. Krautkrämer E, Lehmann MJ, Bollinger V, Zeier M. Polar release of pathogenic Old World hantaviruses from renal tubular epithelial cells. Virology Journal 2012; 9: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McBride R, Fielding BC. The role of severe acute respiratory syndrome (SARS)‐coronavirus accessory protein in virus pathogenesis. Viruses 2012; 4(11): 2902–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bermingham A, Chand MA, Brown CS, et al Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveillance 2012; 17(40): 20290. [PubMed] [Google Scholar]
- 49. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine 2012; 367(19): 1814–1820. [DOI] [PubMed] [Google Scholar]
- 50. Lim PL, Lee TH, Rowe EK. Middle East respiratory syndrome coronavirus (MERS CoV): update 2013. Current Infectious Disease Reports 2013; 15(4): 295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peiris JS, Yuen KY, Osterhaus AD, Stöhr K. The severe acute respiratory syndrome. New England Journal of Medicine 2003; 349(25): 2431–2441. [DOI] [PubMed] [Google Scholar]
- 52. Health Protection Agency (HPA) UK Novel Coronavirus Investigation team . Evidence of person‐to‐person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveillance 2013; 18(11): 20427. [DOI] [PubMed] [Google Scholar]
- 53. Lu G, Hu Y, Wang Q, et al Molecular basis of binding between novel human coronavirus MERS‐CoV and its receptor CD26. Nature 2013; 500(7461): 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tao X, Hill TE, Morimoto C, Peters CJ, Ksiazek TG, Tseng CT. Bilateral entry and release of Middle east respiratory syndrome‐coronavirus induces profound apoptosis of human bronchial epithelial cells. Journal of Virology 2013; 87(17): 9953–9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ludlow M, Lemon K, de Vries RD, et al Measles virus infection of epithelial cells in the macaque upper respiratory tract is mediated by subepithelial immune cells. Journal of Virology 2013; 87(7): 4033–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Erbar S, Maisner A. Nipah virus infection and glycoprotein targeting in endothelial cells. Virology Journal 2010; 7: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simmons HE, Dunham JP, Zinn KE, Munkvold GP, Holmes EC, Stephenson AG. Zucchini yellow mosaic virus (ZYMV, Potyvirus): vertical transmission, seed infection and cryptic infections. Virus Research 2013; 176(1–2): 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Piedimonte G, Walton C, Samsell L. Vertical transmission of respiratory syncytial virus modulates pre‐ and postnatal innervation and reactivity of rat airways. PLoS One 2013; 8(4): e61309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pyrc K, Dijkman R, Deng L, et al Mosaic structure of human coronavirus NL63, one thousand years of evolution. Journal of Molecular Biology 2006; 364(5): 964–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lau SK, Lee P, Tsang AK, et al Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. Journal of Virology 2011; 85(21): 11325–11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Woo PC, Lau SK, Yip CC, et al Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. Journal of Virology 2006; 80(14): 7136–7145. [DOI] [PMC free article] [PubMed] [Google Scholar]


