Summary
The order of Bunyavirales includes numerous (re)emerging viruses that collectively have a major impact on human and animal health worldwide. There are no vaccines for human use or antiviral drugs available to prevent or treat infections with any of these viruses. The development of efficacious and safe drugs and vaccines is a pressing matter. Ideally, such antivirals possess pan‐bunyavirus antiviral activity, allowing the containment of every bunya‐related threat. The fact that many bunyaviruses need to be handled in laboratories with biosafety level 3 or 4, the great variety of species and the frequent emergence of novel species complicate such efforts. We here examined the potential druggable targets of bunyaviruses, together with the level of conservation of their biological functions, structure, and genetic similarity by means of heatmap analysis. In the light of this, we revised the available models and tools currently available, pointing out directions for antiviral drug discovery.
Keywords: antiviral drugs; Bunyavirales; endonuclease; nucleoprotein, phylogenetic analysis; viral polymerase
List of abbreviations
- AKAV
Akabane orthobunyaviruses
- ANDV
Andes virus
- BMP
bismonoacylglycero‐phosphate
- BSL
biosafety level
- BUNV
Bunyamwera virus
- CCHFV
Crimean‐Congo hemorrhagic fever virus
- cRNA
complementary RNA
- DC‐SIGN
dendritic cell–specific ICAM‐grabbing nonintegrin
- eIF4F
eukaryotic initiation factor 4F
- EREV
Erve virus
- HAZV
Hazara virus
- HPS
hantavirus pulmonary syndrome
- IFNAR−/−
lack of alpha/beta interferon receptor
- IMPDH
inosine monophosphate dehydrogenase enzyme
- IRF‐3
IFN regulatory factor‐3
- KUPV
Kupe virus
- LACV
La Crosse virus
- LASV
Lassa virus
- LEAV
Leanyer virus
- L‐segment
large segment
- MPRLV
Maporal virus
- M‐segment
middle‐sized segment
- N‐protein
nucleoprotein
- P bodies
processing bodies
- PTV
Punta Toro virus
- PUUV
Puumala virus
- RdRp
RNA‐dependent RNA polymerase
- RVFV
Rift Valley fever virus
- SBV
Schmallenberg virus
- SFTSV
severe fever with thrombocytopenia syndrome virus
- SNV
Sin Nombre virus
- S‐segment
small segment
- UUKV
Uukuniemi virus
- vRNA
viral RNA
1. INTRODUCTION
Currently, two out of eight “prioritized infectious diseases” by the World Health Organization belong to the order of the Bunyavirales (previously the family Bunyaviridae), ie, Crimean‐Congo hemorrhagic fever virus (CCHFV) and Rift Valley fever virus (RVFV).1 The frequent report of novel bunyaviruses, detected in humans, animals, and plants together with the high genetic diversity among its species, led to a reclassification by the International Committee on Taxonomy of Viruses.2 This illustrates the ubiquity, the variety of hosts, and the epidemic potential of viruses in the Bunyavirales order, which is now a collection of nine viral families, comprising 13 genera (Figure S1). The scope of this review includes the four genera that have been associated with human disease, ie, Orthohantaviruses, Orthonairoviruses, Orthobunyaviruses, and Phleboviruses (Figure 1).
Figure 1.
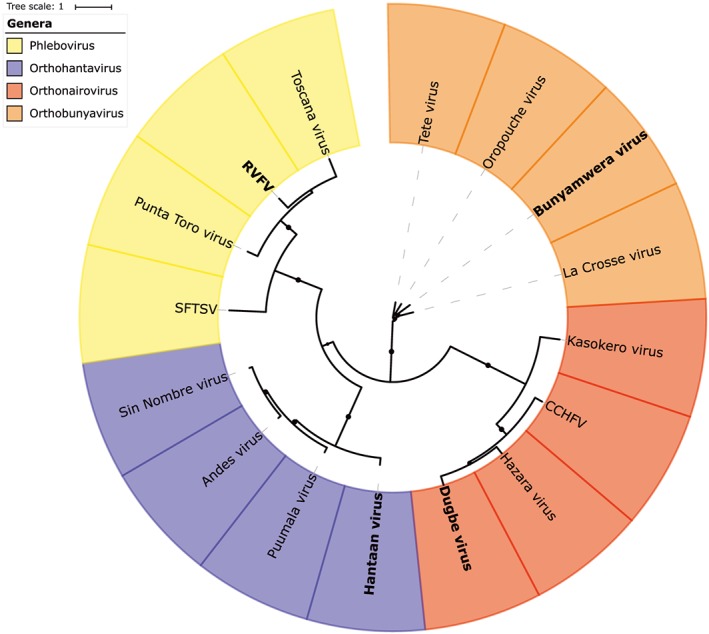
Phylogenetic tree constructed with amino acid sequences of L‐proteins of bunyaviruses from the orthohantaviruses, orthonairoviruses, orthobunyaviruses, and phleboviruses genera. Names in bold indicate the type of species. Crimean‐Congo hemorrhagic fever virus (CCHFV), severe fever with thrombocytopenia syndrome virus (SFTSV), Rift Valley fever virus (RVFV)
Many of these viruses need to be handled in biosafety level (BSL)‐3 or ‐4 laboratories, which complicates the research. Yet, it is important to step up to the challenge since these emerging human viruses are now spread over more than 80 countries in Europe, Asia, Middle East, America, and Africa.3 The great diversity and continuous emergence of new bunyaviral species that cause severe disease make it unfeasible to develop drugs or vaccines for every single virus. Therefore, comprehensive efforts towards the development of antiviral drugs with an extended efficacy against an entire virus family or even virus order need to be made. These offer protection against not only the (re)emerging viruses of today but also from the pandemic threats of tomorrow. To aid in such efforts, we here review the available knowledge of potential druggable targets in the replication cycle of these viruses together with current available tools and models.
2. PARTICLE STRUCTURE AND VIRAL PROTEINS
2.1. Virion and genome
Bunyaviruses have spherical 80‐ to 120‐nm‐sized enveloped virions. Their lipid bilayer envelop is covered with capsomers consisting of transmembrane glycoproteins (Gc and Gn). The genome exists out of negative sense single‐stranded [(−)ss]RNA, which is trisegmented in all human pathogenic genera. These RNA segments, together with oligomers of N‐proteins, form looped RNP. A single L‐protein is bound to each of the RNA segments (Figure 2).
Figure 2.
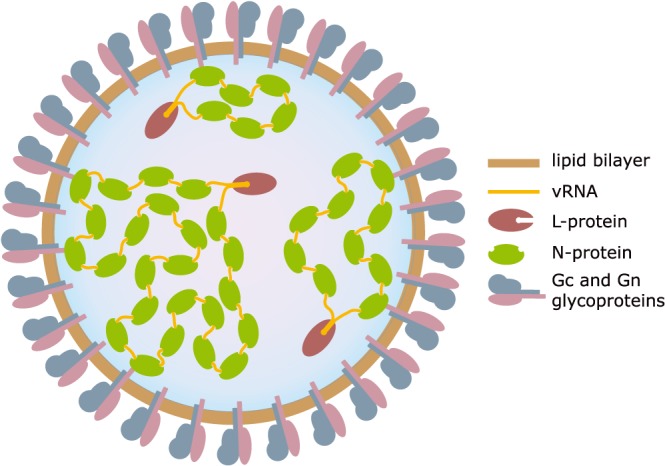
Bunyaviral particle. A schematic illustration of the virion showing the structural proteins (Gn, Gc, L, and N), the vRNA genome (L, M, and S segments), and the RNP complexes
The large segment (L segment) encodes the RNA‐dependent RNA polymerase (RdRp), or L‐protein, which is responsible for the production of complementary RNA (cRNA) and viral RNA (vRNA). The middle‐sized segment (M segment) encodes for the Gc and Gn glycoproteins. Depending on the species, the M segment also codes for the NSm protein. The small segment (S segment) contains the transcript for the nucleoprotein (N‐protein), which, like all above‐mentioned proteins, is encoded in the (−)sense. The S segment further codes for the nonstructural protein NSs in either negative or positive orientation (as in phleboviruses and orthobunyaviruses, respectively) but can also be lacking (as is the case in some orthohantaviruses.4, 5
2.2. L‐protein
The L‐protein is responsible for transcription of the (−)ssRNA and replication of new vRNA. Both these processes occur in the cytosol where newly translated L‐proteins can interact with either additional L‐proteins to facilitate replication or with newly synthesized vRNA to be assembled into new viral particles.5 While the size of the L‐proteins may vary between families, three subdomains, ie, the finger, palm, and thumb can be distinguished in all L‐proteins. The relative orientation of these three subdomains defines the catalytic cavity, which can be attuned to facilitate different stages of replication.6 All L‐proteins recognize highly conserved complementary 3′ and 5′ extremities of the genome segments that either form a double stranded “pan‐handle” bound by the L‐protein or both ends are bound separately to the L‐protein.7, 8 These 5′ and 3′ UTR are acting as promotor and transcription termination signal for the viral polymerase.9, 10 Similar regions were found in influenza viruses, where they also contribute to the activation of several polymerase functions.11 The endonuclease and RdRp domains of the L‐protein are present throughout the bunyaviruses and share functional characteristics and structural similarities with other (−)stranded segmented viruses such as arenaviruses and orthomyxoviruses (Figure 3A).12
Figure 3.
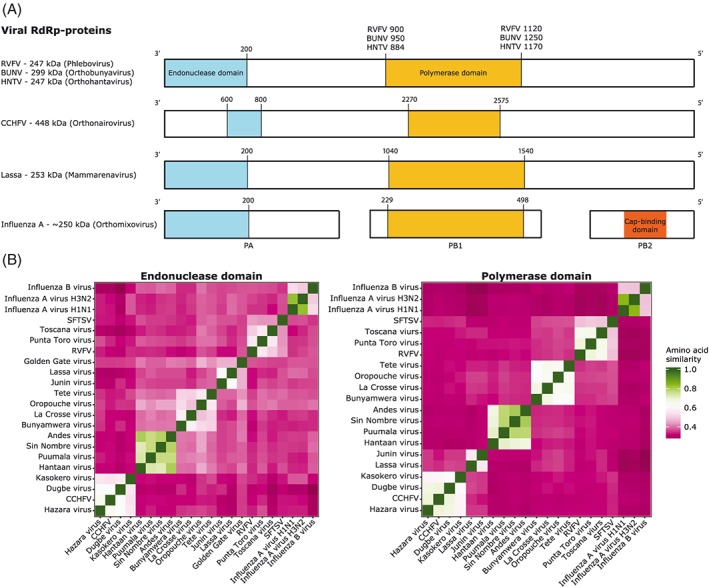
A, Comparison of L‐proteins of segmented (−)ssRNA viruses. The proteins are represented as bars and domains of interest are the endonuclease domain (in blue) and polymerase domain (in yellow). Numbers above the bars indicate the area of the region that has been used in the comparison. B, Heatmaps of the amino acid similarity of endonuclease domains (left) and polymerase domains (right) between (−)ssRNA viruses. Rift Valley fever virus (RVFV), Bunyamwera virus (BUNV), Hantaan virus (HNTV), Crimean‐Congo hemorrhagic fever virus (CCHFV), severe fever with thrombocytopenia syndrome virus (SFTSV)
The endonuclease domain is responsible for cap snatching, which initiates transcription in all orthomyxoviruses, arenaviruses, and bunyaviruses. The endonuclease cleaves the 5′‐end of host mRNAs to acquire primers for viral mRNA transcription. The manganese‐dependent endonuclease domain of bunyaviruses is located at the N‐terminus of the L‐protein, with exception of the nairoviruses where it is located more downstream (Figure 3A).13, 14 The aa similarity of the endonuclease domain of species within the four studied bunyaviral families is 50% to 80% conserved, whereas between families, this is 30% to 45%. The degree of similarity with the arena and influenza domains is 28% to 45% and 22% to 38%, respectively (Figures 3B and S2). However, within the active site of these domains, there are motifs that are highly conserved (H..E/D..PD..E/D).15 Their crucial function and conserved regions makes such domains a suitable target for antiviral endeavors. Successes have already been achieved against the endonuclease of influenza, confirming this enzyme is indeed a druggable target.16 Such compounds include endonuclease inhibitors pimodivir/JNJ872 (formally VX‐787), which is active against a diverse panel of influenza A virus strains and influenza JNJ5806 (formally AL‐794) (J&J/Alios) and S‐033188 (Shionogi).17 This not only shows that the endonuclease is a viable drug target but its conservation among strains and species may also indicate possibilities for the development of pan‐bunya inhibitors. However, it is not self‐evident that such compounds will be equipotent against all bunyaviruses, since subtle differences remain.
A second highly conserved domain among (−)ssRNA viruses is the active RdRp domain in the central region of the L‐proteins of bunyaviruses and arenaviruses and in the PB1 subunit of orthomyxoviruses. While the aa similarity of this region between bunyavirus families is below 50% (Figures 3B and S2), there are highly conserved motifs (preA/F..A..B..C..D..E..H) that extend into arenaviruses and influenza viruses.5 This could make it a target for broad‐acting antivirals. In fact, some polymerase‐targeting broad‐acting antiviral drugs have been reported. Galidesivir (BCX4430, BioCryst Pharmaceuticals) is an adenosine nucleoside RdRp‐inhibitor currently being developed against filoviruses (e.g., Ebola virus and Marburg virus), also exerts activity against coronaviruses, picornaviruses, paramyxoviruses, flaviviruses, orthomyxoviruses, arenaviruses, and bunyaviruses.18 GS‐5734, a monophosphoramidate prodrug of an adenosine analogue, is being developed as anti‐filovirus drug. Its acting range includes also flaviviruses and paramyxoviruses, but not bunyaviruses or arenaviruses.19 Also favipiravir/T‐705 exerts a wide range of anti‐RNA–virus inhibition, including some minor activity against bunyaviruses.20 The antiviral activity of favipiravir is discussed in more detail in Section 4. The similarities between the polymerases of bunyaviruses, arenaviruses, and influenza viruses, ie, their common motifs and similar architecture, could point in the direction of a common ancestor.6 From an antiviral prospective, this could be a very interesting notion, since a single drug might be able to target them all.
2.3. N‐protein
The N‐protein is a variable and multifunctional entity of all bunyaviruses (Figure 4). Their main function, the encapsulation of vRNA and cRNA, is conserved throughout all (–)ssRNA viruses. They do so by forming ribonucleoprotein complexes, thereby protecting the viral genomic material against host defense mechanisms. Structural analysis shows that there is limited overall aa similarity among N‐proteins, both between viral families and genera, but also between bunyaviruses and the other (−)ssRNA viruses.
Figure 4.
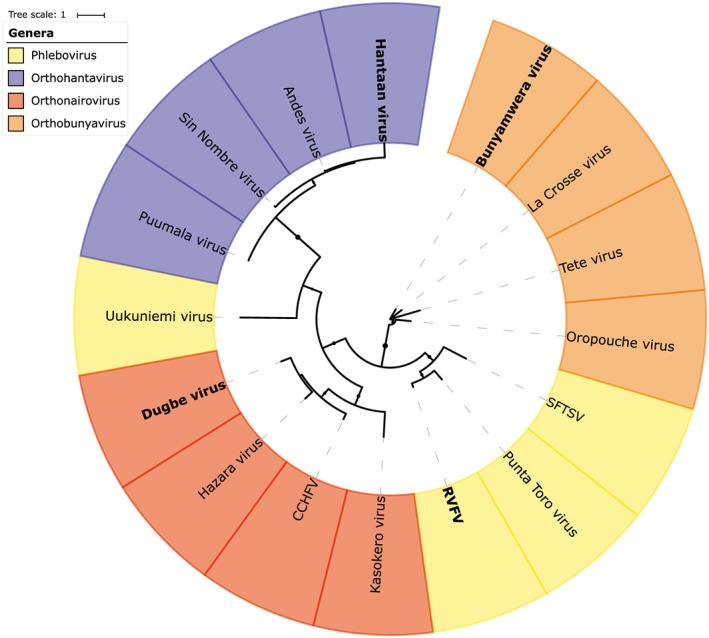
Phylogenetic tree constructed with amino acid sequences of N‐proteins of bunyaviruses from the orthohantaviruses, orthonairoviruses, orthobunyaviruses, and phleboviruses genera. Names in bold indicate the type species. Severe fever with thrombocytopenia syndrome virus (SFTSV), Rift Valley fever virus (RVFV), Crimean‐Congo hemorrhagic fever virus (CCHFV)
The Bunyamwera virus (BUNV) N‐ and C‐terminal aa's protrude outward from the protein core domain and are able to interact with neighboring N‐proteins, causing head‐to‐tail interactions.21 This way, these proteins can form ring‐shaped tetramers. This multimerization might be a flexible feature though, since it was observed that Schmallenberg virus (SBV) N‐proteins form both tetrameric and hexameric multimers.22 N‐N‐protein interactions were also found in BUNV and Leanyer virus (LEAV), suggesting this is at least a genus‐wide conserved feature.23, 24 The structure of LEAV shows two distinct domains together forming the core domain: an N‐lobe and C‐lobe (Figure 5A). Between these two lobes, a positively charged cleft is formed that can hold 11 nt of ssRNA, which together form filamentous RNPs. The ssRNA is bound to the LEAV N‐protein through hydrogen bonds to the backbone and maybe also to the bases. On the surface of the N‐protein, there is a hydrophobic region that is suggested to be involved in N‐N‐protein interactions.24
Figure 5.
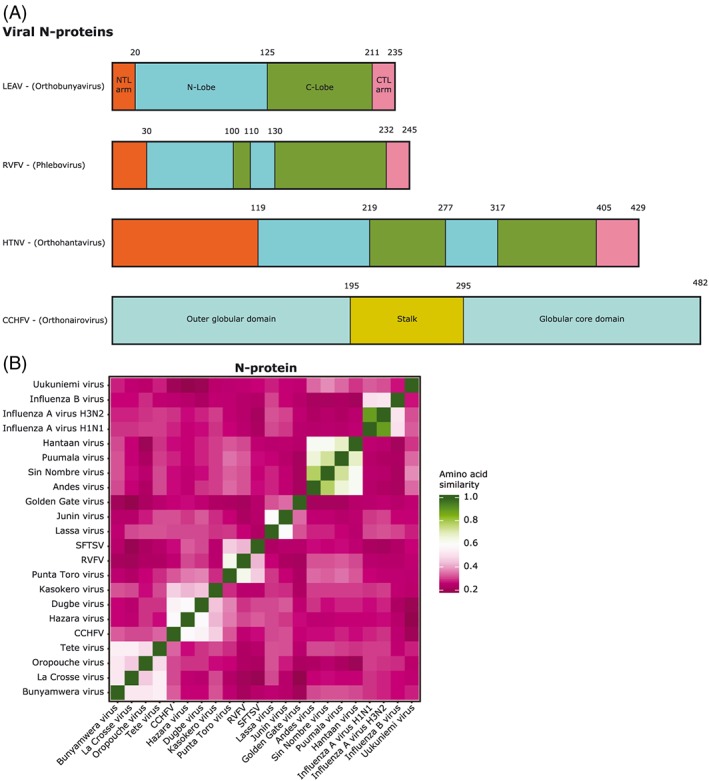
Comparison of N‐proteins of segmented (−)ssRNA viruses. A, Domain distribution comparison between bunyaviruses. The proteins are represented as bars. B, Heatmap of the amino acid similarity of N‐proteins between (−)ssRNA viruses. Leanyer virus (LEAV), Rift Valley fever virus (RVFV), Hantaan virus (HNTV), Crimean‐Congo hemorrhagic fever virus (CCHFV), severe fever with thrombocytopenia syndrome virus (SFTSV)
The typical two lobed structure of N‐proteins was also observed in phleboviruses, but no protruding C‐terminal arm observed. These rather appear to be folded back against the protein.25 Just as in orthobunyaviruses, a strong hydrophobic region was found, which allows interaction with other N‐proteins.26 Even in the sequence dissimilar Uukuniemi virus (UUKV), such N‐N interactions have been observed.27 The hydrophobic region is highly conserved among phleboviruses and is found in a pocket at the N‐lobe surface. This allows the typical ring‐shaped oligomers to form, but these show variability in size and shape. This indicates structural flexibility of N‐protein complexes in combination with RNA. The ring shape was also seen in TOSV N‐protein oligomers, but on introducing RNA to such oligomers, a helical RNP was observed.28 The binding of RNA in this ring‐ or helical‐shaped manner allows it to be concealed. Binding of RNA occurs at the interface of the N‐protein N‐ and C‐lobe where a cleft is formed containing a positively charged patch that allows, in case of RVFV, the binding of six RNA bases. Such a hydrophobic patch is conserved among phleboviruses.25
While the N‐protein of hantaviruses has almost twice the molecular weight as the same protein in orthobunyaviruses and phleboviruses, similar structural properties are observed. There are the N‐ and C‐ terminal arms and subsequent binding pockets that together regulate N‐N‐protein interactions, and there is the hydrophobic cleft at the interface of the N‐ and C‐lobes to accommodate RNA.29 On top of these similarities, there are some additional functionalities attributed to hantaviral N‐proteins. It has been demonstrated that trimers of the protein can substitute the eukaryotic initiation factor 4F (eIF4F) cap‐binding complex, which is responsible for translation initiation. By taking over the helicase function of eIF4A subunits, binding to initiation complexes like eIF4G and binding to 5′ m7G caps just like eIF4E, it makes eIF4F obsolete.30 The N‐protein facilitates the association of ribosomal protein S19 and the viral mRNA 5′ UTR allowing ribosomes to be loaded onto the mRNA.31 Furthermore, N‐proteins are able to interfere with cellular mRNA decay by associating with processing bodies (P bodies), which are hot spots for cap snatching.32 Besides these translational‐enhancing properties, the hantavirus N‐protein has also been found to interfere in host innate immune responses.33
Nairovirus N‐proteins do not share the common N‐ and C‐lobe structure described above. Their N‐proteins have a racket shape that consists of a globular (or head) domain and a stalk domain.34, 35 The globular domain is structurally very similar to the Lassa virus (LASV) N‐protein N‐terminal domain, but despite the similarity, they may not have equivalent functions, since the CCHFV‐ N‐protein lacks the 3′‐5′ exoribonuclease functionality attributed to the LASV N‐protein.35, 36 The stalk domain can alter its relative position, thereby altering the function of the protein. One main function is again the binding of RNA, which seems to drastically modify its structure, and the interaction with the globular domain of neighboring N‐proteins and thereby form circular oligomers.37 Three positively charged regions (two on the globular domain, one on the stalk) are present and may play a role in RNA binding, which is in contrast to other bunyaviral families with their clear RNA‐binding cleft.
However, the RNA‐binding domains overlap with other functional sites; thus, it seems likely that in an RNA‐bound state, the other abilities are inhibited.37 It was shown that CCHFV N‐protein allows cap‐independent translation of viral RNA by interacting with eIF4G, but details about these interactions are lacking.38 The structure of the N‐protein of CCHFV seems to be shared by all nairoviruses, since structures of HAZV, Kupe virus (KUPV), and Erve virus (EREV) showed very similar structures, although functionality might slightly vary.39
Our own aa similarity comparison of the N‐proteins shows there is very limited similarity between viral families; this is substantial within families (Figure 5B) (Supporting Information). Within hantaviruses, the sequence is especially conserved, from 62% up to 78%. We observed that UUKV N‐protein shares lower sequence similarity with other phleboviruses. Lower aa identity of UUKV was reported earlier.26 Whereas the similarity between families is low, their structural and functional comparability seems, with exception of nairoviruses, much higher. The numerous sites for interaction of the N‐protein with other viral proteins, RNA, and host factors, together with their crucial functions in the viral replication cycle, could possibly make good antiviral targets.40 This is supported by findings of small molecule targeting N‐proteins in other (−)ssRNA viruses. Inhibitors against conserved epitopes of the N‐protein of influenza (eg, nucleozin) and RSV (eg, RSV604 and AstraZeneca) have been discovered.41, 42 Antiviral effect against influenza was observed by targeting N‐protein with the nonsteroidal antiinflammatory drug naproxen. It binds the highly conserved RNA binding groove of the N‐protein, demonstrating that this is indeed a druggable target and thus could also be a potential target in bunyaviruses.43
2.4. Glycoproteins
The viral envelope is covered by oligomers of membrane glycoproteins (Gn and Gc), which are derived through endoproteolytic cleavage of the RNA M segment–encoded polyprotein.44 In CCHFV, this polyprotein is cotranslationally cleaved into the PreGn and PreGc precursors, which are then cleaved by SKI‐1 and SKI‐1–like proteases to generate the N‐termini of Gn and Gc, respectively.45 The outward‐directed domains are variable between families and species and are responsible for the attachment to host cell. This variability is reflected in the wide range of host cell receptors that have been described (see Section 3) and could make it difficult to use these domains as a target for broad‐acting antivirals. Moreover, viruses have shown that they can easily mutate their receptor‐binding interface to escape the drug pressure and pay little to no cost in fitness to do so. This would quickly lead to the emergence of drug resistant variants, thus rendering antivirals targeting these domains ineffective. The domain that is directed inwards into the virion is able to interact with the N‐proteins of the RNP or directly with the RNA.46, 47 The disruption of these critical interactions could interfere with the viral replication cycle, making them potentially good antiviral targets.
2.5. Nonstructural proteins
Bunyaviruses use their nonstructural proteins in strategies to interact with apoptotic pathways and to interfere with the host innate immune system. Unlike the structural proteins, these proteins are not conserved throughout the viral order. The nonstructural proteins can be lacking entirely, such is the case in most hantaviruses. The NSs (encoded on the S segment) and NSm (encoded on the M segment) can both be encoded in the (−)sense as in orthobunyaviruses, or the NSs can be encoded in the (+)sense, as is the case for the phenuiviruses.
2.6. NSs protein
The main purpose of the NSs protein is to interfere with the host IFN‐mediated immunity; it therefore is a major virulent factor. The NSs of phleboviruses and orthobunyaviruses is transported to the nucleus of infected cells and prevents the transcription of IFN by suppressing IFN regulatory factor‐3 (IRF‐3). The NSs of RVFV induces a characteristic filament formation in infected cell nuclei. The structure of an RVFV NSs protein was reported, showing a stable core domain that might contain all essential determinants for nuclear translocation and filament formation. While this core domain has only a 15% to 30% sequence identity between phleboviruses, these all have a similar core domain fold, while N‐ and C‐terminal regions are highly variable.48 In the orthobunyaviruses BUNV and La Crosse virus (LACV), IRF‐3 is suppressed through interfering with the assembly and/or leading to degradation of the host RNA polymerase II complex, which has also been seen in RVFV.49, 50 The S segment of some hantaviruses carried by voles and lemmings of the northern hemisphere and American mice and rats have an ORF for an NSs protein, which is not found in the S segments of hantaviruses associated with mice and rats in Europe and Asia. These NSs proteins seem to be able to inhibit the expression of the IFN‐β gene, although in a weaker fashion than BUNV and RVFV.51 The NSs of CCHFV (orthonairovirus) is an inducer of apoptosis through disruption of the mitochondria and activation of caspases.52
2.7. NSm protein
RVFV NSm is thought to interfere with apoptotic executive caspase activity by inhibiting upstream initiator caspases.53 However, for most bunyaviruses, the function of NSm is still uncertain.
The M segment can encode more nonstructural proteins: a mucin‐like domain and GP38.45 RVFV M segment encodes the 14‐kDa cytosolic protein and P78/NSm‐GN. P78 has no association with virulence in mice but appears to play an important role in infectivity of mosquitos.54
While the nonstructural proteins are very variable between, and even within, bunyaviral families, interfering with this viral inhibition of the innate immune response is a proposed antiviral strategy, although examples of such molecules in clinical development are still lacking.55, 56
3. REPLICATION CYCLE
3.1. Viral attachment and entry
The replication cycle of the Bunyavirales is in general very similar among its members (Figure 6). Bunyaviruses may, depending on the family, viral species, or type of host cell, use different cellular targets for attachment and employ different host entry pathways. For the Phenuiviridae family and a variety of bunyaviruses, the most widely described attachment factor is dendritic cell‐specific ICAM‐grabbing nonintegrin (DC‐SIGN). Mutations or knock down of these molecules result in lower infection rates in vitro.57, 58, 59 A second attachment factor in these viruses is L‐SIGN, another human C‐type lectin, which shares a 77% aa sequence homology to DC‐SIGN.60 Even though these factors have not been extensively studied in other bunyaviruses, they are involved in the infection pathway of other viruses such as dengue, Lassa, and hepatitis C virus.61, 62, 63 It was recently shown that heparan sulfate proteoglycan is also an important attachment factor for the SBV and Akabene orthobunyaviruses (AKAV).64 Hantaviruses use integrin as receptor for cell entry: β1 integrin by nonpathogenic hantaviruses and β3 integrin by the pathogenic species in vitro.65
Figure 6.
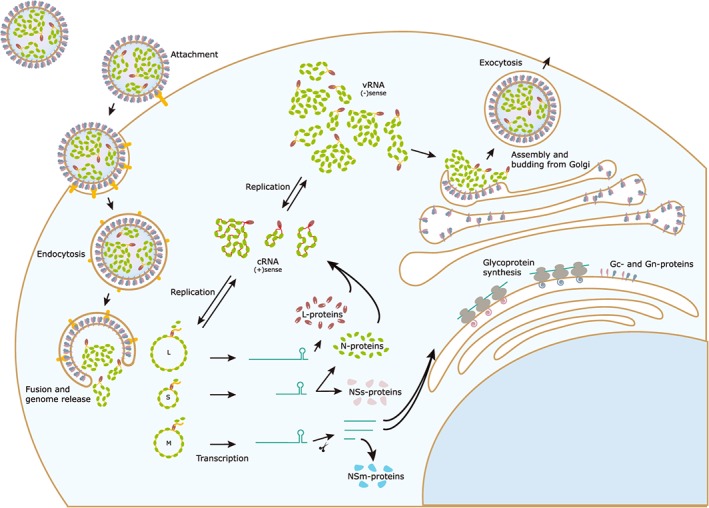
Replication cycle of bunyaviruses. Infection starts with attachment of the virus to the host cell receptors, a process mediated by the viral glycoproteins and endocytosis. Due to a pH drop, the endosomal and viral membrane will fuse, and the genome is released into the cytosol. The viral L‐protein catalyzes primary transcription of viral mRNAs, which will translate into new structural proteins and (depending on the species) nonstructural proteins. The three negative‐sense vRNA segments are converted into positive‐sense cRNA that will serve as template for novel RNPs. These will be transported to the Golgi complex where it will interact with newly translated glycoproteins to commence assembly processes. The new viral particle will bud from the Golgi and will be transported out of the cell
Cell entry varies between bunyaviruses, with clathrin‐mediated, calveolin‐mediated, and independent endocytosis all being described in one or more bunyaviruses, depending also on the host cell type.66, 67 Envelope fusion with the endosomal membrane is thought to be triggered by the acidification of the late endosome, which initiate a conformational change of the viral particle glycoproteins. However, it was recently shown that also potassium channels play a crucial role in this step and thus could be potential druggable targets.58, 68, 69, 70, 71 In UUKV, this is promoted by the presence of the late endosomal phospholipid bis(monoacylglycero)‐phosphate (BMP).72 Essential histidine residues have been identified in the RVFV Gc‐protein by mutational analysis, which serve as sensors for acidification in endosomal lumen and often define the optimal pH value for virus fusion.73, 74 Hantaviruses depend heavily on cholesterol for membrane fusion.75 The Puumala virus (PUUV) Gc has shown to be the membrane fusion effector, and it presents a class II membrane fusion protein fold in its postfusion conformation.76, 77 Such a fold was also observed in RVFV.78
3.2. Transcription and translation of bunyaviral proteins and genome replication
The L‐protein becomes active once it is released in the cytosol and starts by transcribing the three segments of the (−)sense genome. This process is preceded by cap snatching, where a fragment of 12‐ to 18‐nt‐long 5′‐capped primer is stolen from host cell mRNA to initiate the synthesis of viral mRNA's, which will be translated into new viral proteins.79 Translation of the L‐, N‐, and Ns proteins occurs in the cytosol by free ribosomes, while the viral glycoproteins are produced by the ER‐membrane bound ribosomes.80 For the replication of the genome, cRNA, i.e. a (+)sense copy of the genome segments, is created by de novo synthesis, which is suggested to be initiated by an interaction between two L‐proteins.5 If this is indeed the case, targeting this critical interaction could be an efficient way to inhibit bunyavirus replication.
3.3. Assembly and exocytosis
The newly formed glycoproteins instantly form oligomers in the ER membrane. From there, they target to the Golgi to assemble, allowing the new viral particles to bud from the Golgi membrane, which is a unique feature for the bunyaviruses since all other (−)ssRNA viruses bud from the cellular membrane.81, 82 This mechanism was targeted in RVFV by a small molecule, G202‐0362, which seems to inhibit assembly and/or exocytosis by depolymerizing microtubules.83
In the replication complexes, the glycoprotein cytoplasmic tails, which are located on the inside of the virion after budding, are presented outwards towards the cytosol. This could be the bunyavirus alternative to matrix proteins, since the tail domain of the glycoproteins is able to interact with the RNP and initiate assembly of new viral particles.46, 84 The assembly of RVFV is induced by an interaction between the RNP N‐proteins and the viral glycoproteins. This seems to be a nonselective process that results in particles carrying a random amount of genomic segments.85 It is not evident that this mechanism is the same for all bunyaviruses. In CCHFV, the Gn cytosolic tail directly binds to the RNA to initiate assembly.86 This could enable a more specific selection of genomic segments. Besides, it was shown that assembly mechanisms depend not only on the virus species but also on the type of cell that is infected.87
3.4. Available model systems and tools for antiviral research
Since numerous bunyavirus species require BSL‐3 and 4 facilities for handling, only a relatively limited number of research facilities are equipped to study these viruses.
3.5. Biosafety and surrogate models
Working with a surrogate virus may be a good alternative. For several highly pathogenic bunyaviruses, such surrogates are described. The Hazara virus (HAZV) is a CCHFV‐related orthonairovirus that is nonpathogenic to humans and can therefore be used both in in vitro assays to study the effect of antiviral agents and in mouse models at BSL‐2.88 A closer alternative would be the CCHFV AP92 or AP92‐like strains, which have a moderately reduced virulence compared with the wild‐type virus. However, a first lethal case has been reported.89, 90 This, together with the lack of a detailed virulence and safety studies, makes that in most countries this is still classified as a BSL‐4 pathogen. Unlike wild‐type CCHFV, there are no reverse genetics systems available for Hazara or AP92.
For RVFV (a BSL‐3 pathogen), the attenuated MP‐12 strain (BSL‐2) may be a suitable surrogate. MP‐12 is a cell‐culture–attenuated strain derived from the RVFV ZH548 strain. It replicates robustly in vitro and in vivo, making it a suitable research tool (Table 1).91, 92
Table 1.
Overview of viruses from the Bunyavirales order that are important human pathogens or are often used in antiviral research
| Genus | Virus | BSL | Vector Transmission | In Vitro Systems | Reverse Genetics System | Animal Models | Ref |
|---|---|---|---|---|---|---|---|
| Orthohantavirus | Andes virus | 3 | Inhalation aerosolized excrement long‐tailed pigmy rice rat (Oligoryzomys fulvescens) /human‐to human transmission | Vero | Minireplicon system | Syrian hamster | 100, 112, 118, 119, 120 |
| Hantaan virus | 3 | Inhalation aerosolized excrement striped filed mouse (Apodemus agrarius) | Vero | Minireplicon system |
C57BL/6 mice BALB/c mice Syrian hamster |
121, 122, 123 | |
| Puumala virus | 3 | Inhalation aerosolized excrement bank vole (Myodes glareolus) | Vero | ‐ | Syrian hamster | 124, 125 | |
| Sin Nombre virus | 3 | Inhalation aerosolized excrement deer mouse (Peromyscus maniculatus) | Vero, HUVEC | ‐ | Immunosuppressed Syrian hamsters | 120, 126, 127 | |
| Orthonairovirus | CCHFV | 4 | Tick (Hyalomma spp.) /contact with infectious blood or body fluids | Vero, SW‐13, Huh‐7 |
Minireplicon system VLP system |
129S6/SvEv STAT‐1−/− mice IFNAR−/− mice |
97, 104, 128, 129 |
| Dugbe virus | 3 | Tick (Amblyomma variegatum) | BHK | ‐ | 129S6/SvEv mice | 130 | |
| Hazara virus | 2 | Ticks (Ixodes) | Vero, A549, SW‐13 | ‐ | IFNAR−/− mice | 88, 131 | |
| Phlebovirus | RVFV | 3 | Mosquito (mainly Aedes and Culex) /contact with infectious blood or body fluids | Vero, A549, Hek293 |
Minireplicon system VLP system Rescue from plasmid |
IFNAR−/− mice Syrian hamsters |
91, 92, 98, 132, 133, 134 |
| Punta Toro virus | 2 | Sandfly (Phlebotominae) | Vero, LLC‐MK2, Hek293 | ‐ |
C57BL/6 mice Syrian hamsters |
135, 136, 137 | |
| SFTSV | 3 | Tick (Haemaphysalis longicornis) /contact with infectious blood or body fluids | DH82, Vero, Huh‐7 | Minireplicon system |
C57BL/6 mice IFNAR−/− mice |
138, 139, 140, 141 | |
| Toscana virus | 2 | Sandfly (Phlebotominae) | Vero, HeLa, Hek293T | Minireplicon system | BALB/c mice | 142, 143 | |
| Uukuniemi virus | 2 | Tick (Ixodes ricinus) | BHK, A549 |
Minireplicon system Rescue from plasmid |
‐ | 144, 145, 146, 147 | |
| Orthobunyavirus | Bunyamwera virus | 2 | Mosquito (mainly Aedes) | Vero, BHK, A549 | Minireplicon system | Swiss albino suckling mice 129S6/SvEv mice | 148, 149, 150 |
| La Crosse virus | 2 | Mosquito (mainly Aedes) | Vero, RAW264.7 | Minireplicon system | C57BL/6 mice | 151, 152 | |
| Oropouche virus | 3 | Mosquito (mainly Aedes and Culex) | Vero, A549 |
Minireplicon system VLP system Rescue from plasmid |
Swiss albino suckling mice Syrian hamster | 153, 154, 155, 156, 157 |
Abbreviations: BHK, baby hamster kidney; CCHFV, Crimean‐Congo hemorrhagic fever virus; Hek293, human embryonic kidney 293; HUH‐7, human hepatoma‐7; HUVEC, human umbilical vein endothelial cells; IFNAR, interferon α/β receptor; LLC‐MK2, Lilly Laboratories Cell–Monkey Kidney 2; RVFV, Rift Valley fever virus; SFTSV, Severe fever with thrombocytopenia syndrome virus; STAT‐1, signal transducer and activator of transcription 1; VLP, virus‐like particle.
All orthohantaviruses are classified as either BSL‐3 or ‐4 pathogens. The Maporal virus (MPRLV), a New World hantavirus, has been proposed as a potential surrogate, since there is currently no evidence that it can infect humans or cause disease.93
Most orthobunyaviruses are BSL‐2 pathogens, because symptoms in humans are mostly limited to fever, headache, joint and muscle aches, nausea, and rash.94 Many models and tools are available to study these viruses (Table 1).
Not all highly pathogenic bunyaviruses can currently be studied by using surrogates. For example, for severe fever with thrombocytopenia syndrome virus (SFTSV) (BSL‐3), there are currently no attenuated strains or surrogates available.
The majority of bunyaviruses replicate in a wide variety of routinely used cell‐culture systems, allowing to study their replication cycle (and inhibition thereof) in cell‐based assays. The most widely used cell lines are Vero, A549, and BHK‐21 (Table 1).
3.6. Animal infection models
While many bunyaviruses are not (or only low) pathogenic in immunocompetent rodents, RVFV is highly pathogenic in immunocompetent mice and hamsters. The virulent RVFV‐ZH548 strain is pathogenic in wild type mouse models, whereas the attenuated MP‐12 strain only gives similar pathogenicity in mice that lack the alpha/beta interferon receptor (IFNAR−/−) and STAT‐1 KO mice.95, 96 This indicates that the virulent strain possesses viral factors to interfere with IFN‐mediated innate immune response.
IFNAR−/− mice are also highly sensitive to CCHFV, and HAZV in which infection causes acute hepatitis and death of the animals within a few days after infection88 CCHFV infection is robust in STAT‐1 KO mice too.97
An alternative rodent model are Golden Syrian hamsters, which are susceptible to RVFV and Hantaviruses. When devoid of functional STAT‐2, they are also highly susceptible to SFTSV, showing thrombocytopenia, typical marked systemic inflammation, and mortality.98, 99
Hamsters develop a lethal Hantavirus pulmonary syndrome (HPS)‐like disease when infected with ANDV, while infection with the Sin Nombre virus (SNV) results in an asymptomatic infection.100, 101 MPRLV, which does not cause disease in humans, causes partial lethality and HPS‐like disease in these hamsters.102 This makes the Syrian hamster model currently the best Hantavirus infection and HPS‐disease model, making it well suited to study the antiviral efficacy of small molecule inhibitors.103
4. CURRENTLY USED ANTIVIRAL COMPOUNDS IN BUNYAVIRAL INFECTIONS
4.1. Ribavirin
Ribavirin is a purine nucleotide analog approved for the treatment of RSV and Lassa virus and was until 2014 routinely used for the treatment of infections with hepatitis C virus. In vitro antiviral activity against CCHFV has been reported, and the compound was shown to reduce CCHFV viral load and to slow down disease progression in an IFNAR−/− mouse infection model.104 Its antiviral effect derives from multiple mechanisms of action.105 The foremost antiviral mechanism against RNA viruses (described for flaviviruses and paramyxoviruses) is thought to be the inhibition of the inosine monophosphate dehydrogenase enzyme (IMPDH), resulting in the depletion of GTP pools.106
Ribavirin is currently the only drug used in a clinical setting against CCHFV infection in humans. Clinical benefits have been controversial, since randomized placebo–controlled clinical trials are not in line with the Declaration of Helsinki.107 However, available studies indicate that ribavirin is effective during initial viremia when disease is still mild or moderate, but not once patients reach the hemorrhagic stage. Consequently, ribavirin could also be used as a (postexposure) prophylaxis for health care workers at risk of contracting CCHFV.108, 109 The blocking of the viral replication by ribavirin could explain why the drug is beneficial during the viremic phase and not in the later hemorrhagic phase, which is the result of released proinflammatory cytokines and the disruption of coagulation cascades.110 Currently, two phase II trials have been enrolled to get more insight in the safety and efficacy of ribavirin (NCT00992693 & NCT02483260).
Ribavirin has also been used occasionally in the treatment of HPS in humans, but due to the small numbers of studies, no defining conclusions can be drawn.111 However, a significant reduction in mortality in rodents has been observed.112, 113
4.2. Favipiravir
Favipiravir acts as a nucleobase analog that functions in its active form (T‐705‐ribose‐triphosphate) as a substrate by the RdRp and inhibits the RNA polymerase activity.20 It was first described as antiviral drug against influenza, but in vitro and in vivo activity has also been demonstrated against a variety of other RNA viruses, such as arenaviruses, filoviruses, togaviruses, and bunyaviruses. In case of bunyaviruses, its in vitro activity is superior to ribavirin. Its ability to increase survival rates in PTV‐ and RVFV‐infected hamsters and in SFTSV‐ and CCHFV‐infected mice further proof the potency of favipiravir.114
Favipiravir has been approved in Japan for the use in patients infected with novel or reemerging influenza viruses. The drug has also been suggested for the treatment of Ebola infections, and clinical trials have been performed despite concerns of teratogenicity shown in animals.115 These trials showed favipiravir to be well tolerated and decrease the mortality of patients with the low viral load.116 Promising data obtained in preclinical experiments, together with positive results from clinical trials against other RNA viruses resulted in clinical trials with favipiravir against SFTSV (UMIN000022398 & UMIN000029020), leading to a phase three clinical trial (JapicCTI‐183872).117
4.3. Perspectives
The order of the Bunyavirales with its nine viral families, various animal hosts, and wide range of transmission vectors have a major impact and form a severe threat to human health. There are no drugs for the prevention or treatment of bunyavirus infections, and current response to bunya‐related disease is still largely insufficient. Even though bunyaviruses prove a tremendous challenge to the field of antiviral research, it is imperative to step up to the challenge. Antiviral therapy has progressed enormously in the last decades, and history has taught us that it is possible to achieve what was sometimes deemed impossible (eg, one pill a‐day treatment regimen for HIV and a cure for a chronic HCV infection after only 12 wk of treatment). Thus, if sufficient effort is put forward by the scientific community, insight into bunyavirus biology is gained and new small molecule inhibitors will be identified that can potentially be developed as antiviral treatment. This holds truth even against a viral order as diverse as the Bunyavirales. The current paper provides an in‐depth review of the biology of bunyavirus replication, and we expand on how certain viral proteins can be exploited for antiviral drug development, together with the level of conservation of their biological functions, structure, and genetic similarity.
ROLE OF THE FUNDING SOURCE
The Infect‐ERA grant “ESCential” supported this work but had no involvement in any aspect of the content. The corresponding author had full access to all the data and had final responsibility in the decision to submit for publication.
CONFLICT OF INTEREST
The authors have no competing interest.
Supporting information
Figure S1: Phylogenetic tree constructed with amino acid sequences of L‐proteins of bunyaviruses. Names in bold indicate the type species.
Figure S2: Numeric data corresponding to heatmaps. A) Endonuclease domain of RdRp‐proteins. B) Polymerase domain of RdRp‐proteins. C) N‐protein.
Data S1 Supporting information
ACKNOWLEDGEMENTS
This work was kindly supported by the Infect‐ERA grant “ESCential.” We thank Jelle Matthijnssens for his collaboration and Piet Maes for managing our servers.
ter Horst S, Conceição‐Neto N, Neyts J, Rocha‐Pereira J. Structural and functional similarities in bunyaviruses: Perspectives for pan‐bunya antivirals. Rev Med Virol. 2019;29:e2039 10.1002/rmv.2039
REFERENCES
- 1. World Health Organization , www.who.int. 2018 Annual review of diseases prioritized under the Research and Development Blueprint. Geneva, Switzerland: World Health Organization 2018.
- 2. Adams MJ, Lefkowitz EJ, King AMQ, et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch Virol. 2017;162(8):2505‐2538. [DOI] [PubMed] [Google Scholar]
- 3. Berger S. Bunyaviridae infections—misc In: Miscellaneous Bunyaviridae: Global Status. 2017 edition ed. Los Angeles, California: Gideon Informatics, Inc; 2017:7‐11. [Google Scholar]
- 4. Walter CT, Barr JN. Recent advances in the molecular and cellular biology of bunyaviruses. J Gen Virol. 2011;92(Pt 11):2467‐2484. [DOI] [PubMed] [Google Scholar]
- 5. Ferron F, Weber F, de la Torre JC, Reguera J. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 2017;234:118‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerlach P, Malet H, Cusack S, Reguera J. Structural insights into bunyavirus replication and its regulation by the vRNA promoter. Cell. 2015;161(6):1267‐1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barr JN, Wertz GW. Bunyamwera bunyavirus RNA synthesis requires cooperation of 3′‐ and 5′‐terminal sequences. J Virol. 2004;78(3):1129‐1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kolakofsky D. dsRNA‐ended genomes in orthobunyavirus particles and infected cells. Virology. 2016;489:192‐193. [DOI] [PubMed] [Google Scholar]
- 9. Barr JN, Rodgers JW, Wertz GW. The Bunyamwera virus mRNA transcription signal resides within both the 3′ and the 5′ terminal regions and allows ambisense transcription from a model RNA segment. J Virol. 2005;79(19):12602‐12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barr JN, Rodgers JW, Wertz GW. Identification of the Bunyamwera bunyavirus transcription termination signal. J Gen Virol. 2006;87(Pt 1):189‐198. [DOI] [PubMed] [Google Scholar]
- 11. te Velthuis AJ, Turrell L, Vreede FT, Fodor E. Uncoupling of influenza A virus transcription and replication through mutation of the unpaired adenosine in the viral RNA promoter. J Virol. 2013;87(18):10381‐10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das K, Arnold E. Negative‐strand RNA virus L proteins: one machine, many activities. Cell. 2015;162(2):239‐241. [DOI] [PubMed] [Google Scholar]
- 13. Reguera J, Weber F, Cusack S. Bunyaviridae RNA polymerases (L‐protein) have an N‐terminal, influenza‐like endonuclease domain, essential for viral cap‐dependent transcription. PLoS Pathog. 2010;6(9):e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holm T, Kopicki JD, Busch C, et al. Biochemical and structural studies reveal differences and commonalities among cap‐snatching endonucleases from segmented negative‐strand RNA viruses. J Biol Chem. 2018;293(51):19686‐19698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reguera J, Gerlach P, Rosenthal M, et al. Comparative structural and functional analysis of bunyavirus and arenavirus cap‐snatching endonucleases. PLoS Pathog. 2016;12(6):e1005636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Z, Liu T, Zhang J, Zhan P, Liu X. Influenza A virus polymerase: an attractive target for next‐generation anti‐influenza therapeutics. Drug Discov Today. 2018;23(3):503‐518. [DOI] [PubMed] [Google Scholar]
- 17. Koszalka P, Tilmanis D, Hurt AC. Influenza antivirals currently in late‐phase clinical trial. Influenza Other Respi Viruses. 2017;11(3):240‐246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Warren TK, Wells J, Panchal RG, et al. Protection against filovirus diseases by a novel broad‐spectrum nucleoside analogue BCX4430. Nature. 2014;508(7496):402‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lo MK, Jordan R, Arvey A, et al. GS‐5734 and its parent nucleoside analog inhibit filo‐, pneumo‐, and paramyxoviruses. Sci Rep. 2017;7(1):43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. Favipiravir (T‐705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100(2):446‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leonard VH, Kohl A, Osborne JC, McLees A, Elliott RM. Homotypic interaction of Bunyamwera virus nucleocapsid protein. J Virol. 2005;79(20):13166‐13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong H, Li P, Elliott RM, Dong C. Structure of Schmallenberg orthobunyavirus nucleoprotein suggests a novel mechanism of genome encapsidation. J Virol. 2013;87(10):5593‐5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eifan SA, Elliott RM. Mutational analysis of the Bunyamwera orthobunyavirus nucleocapsid protein gene. J Virol. 2009;83(21):11307‐11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niu F, Shaw N, Wang YE, et al. Structure of the Leanyer orthobunyavirus nucleoprotein‐RNA complex reveals unique architecture for RNA encapsidation. Proc Natl Acad Sci U S A. 2013;110(22):9054‐9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferron F, Li Z, Danek EI, et al. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 2011;7(5):e1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raymond DD, Piper ME, Gerrard SR, Smith JL. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci U S A. 2010;107(26):11769‐11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katz A, Freiberg AN, Backstrom V, et al. Oligomerization of Uukuniemi virus nucleocapsid protein. Virol J. 2010;7(1):187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olal D, Dick A, Woods VL Jr, et al. Structural insights into RNA encapsidation and helical assembly of the Toscana virus nucleoprotein. Nucleic Acids Res. 2014;42(9):6025‐6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olal D, Daumke O. Structure of the hantavirus nucleoprotein provides insights into the mechanism of RNA encapsidation. Cell Rep. 2016;14(9):2092‐2099. [DOI] [PubMed] [Google Scholar]
- 30. Mir MA, Panganiban AT. A protein that replaces the entire cellular eIF4F complex. EMBO J. 2008;27(23):3129‐3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haque A, Mir MA. Interaction of hantavirus nucleocapsid protein with ribosomal protein S19. J Virol. 2010;84(23):12450‐12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT. Storage of cellular 5′ mRNA caps in P bodies for viral cap‐snatching. Proc Natl Acad Sci U S A. 2008;105(49):19294‐19299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taylor SL, Krempel RL, Schmaljohn CS. Inhibition of TNF‐α‐induced activation of NF‐κB by hantavirus nucleocapsid proteins. Ann N Y Acad Sci. 2009;1171(Suppl 1):E86‐E93. [DOI] [PubMed] [Google Scholar]
- 34. Carter SD, Surtees R, Walter CT, et al. Structure, function, and evolution of the Crimean‐Congo hemorrhagic fever virus nucleocapsid protein. J Virol. 2012;86(20):10914‐10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo Y, Wang W, Ji W, et al. Crimean‐Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci U S A. 2012;109(13):5046‐5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qi X, Lan S, Wang W, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468(7325):779‐783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X, Li B, Guo Y, et al. Molecular basis for the formation of ribonucleoprotein complex of Crimean‐Congo hemorrhagic fever virus. J Struct Biol. 2016;196(3):455‐465. [DOI] [PubMed] [Google Scholar]
- 38. Jeeva S, Cheng E, Ganaie SS, Mir MA. Crimean‐Congo hemorrhagic fever virus nucleocapsid protein augments mRNA translation. J Virol. 2017;91(15):e00636‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang W, Liu X, Wang X, et al. Structural and functional diversity of nairovirus‐encoded nucleoproteins. J Virol. 2015;89(23):11740‐11749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Levingston Macleod JM, Marmor H, Garcia‐Sastre A, Frias‐Staheli N. Mapping of the interaction domains of the Crimean‐Congo hemorrhagic fever virus nucleocapsid protein. J Gen Virol. 2015;96(Pt 3):524‐537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chapman J, Abbott E, Alber DG, et al. RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob Agents Chemother. 2007;51(9):3346‐3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kao RY, Yang D, Lau LS, et al. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol. 2010;28(6):600‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lejal N, Tarus B, Bouguyon E, et al. Structure‐based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob Agents Chemother. 2013;57(5):2231‐2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guardado‐Calvo P, Rey FA. The envelope proteins of the Bunyavirales . Adv Virus Res. 2017;98:83‐118. [DOI] [PubMed] [Google Scholar]
- 45. Altamura LA, Bertolotti‐Ciarlet A, Teigler J, Paragas J, Schmaljohn CS, Doms RW. Identification of a novel C‐terminal cleavage of Crimean‐Congo hemorrhagic fever virus PreGN that leads to generation of an NSM protein. J Virol. 2007;81(12):6632‐6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strandin T, Hepojoki J, Vaheri A. Cytoplasmic tails of bunyavirus Gn glycoproteins—Could they act as matrix protein surrogates? Virology. 2013;437(2):73‐80. [DOI] [PubMed] [Google Scholar]
- 47. Shi X, Kohl A, Li P, Elliott RM. Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis. J Virol. 2007;81(18):10151‐10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barski M, Brennan B, Miller OK, et al. Rift Valley fever phlebovirus NSs protein core domain structure suggests molecular basis for nuclear filaments. Elife. 2017;6:e29236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leonard VH, Kohl A, Hart TJ, Elliott RM. Interaction of Bunyamwera orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J Virol. 2006;80(19):9667‐9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verbruggen P, Ruf M, Blakqori G, et al. Interferon antagonist NSs of La Crosse virus triggers a DNA damage response‐like degradation of transcribing RNA polymerase II. J Biol Chem. 2011;286(5):3681‐3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jaaskelainen KM, Kaukinen P, Minskaya ES, et al. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon‐beta promoter. J Med Virol. 2007;79(10):1527‐1536. [DOI] [PubMed] [Google Scholar]
- 52. Barnwal B, Karlberg H, Mirazimi A, Tan YJ. The non‐structural protein of Crimean‐Congo hemorrhagic fever virus disrupts the mitochondrial membrane potential and induces apoptosis. J Biol Chem. 2016;291(2):582‐592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Terasaki K, Won S, Makino S. The C‐terminal region of Rift Valley fever virus NSm protein targets the protein to the mitochondrial outer membrane and exerts antiapoptotic function. J Virol. 2013;87(1):676‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kreher F, Tamietti C, Gommet C, et al. The Rift Valley fever accessory proteins NSm and P78/NSm‐GN are distinct determinants of virus propagation in vertebrate and invertebrate hosts. Emerg Microbes Infect. 2014;3(10):e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eifan S, Schnettler E, Dietrich I, Kohl A, Blomström AL. Non‐structural proteins of arthropod‐borne bunyaviruses: roles and functions. Viruses. 2013;5(10):2447‐2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lucas‐Hourani M, Dauzonne D, Jorda P, et al. Inhibition of pyrimidine biosynthesis pathway suppresses viral growth through innate immunity. PLoS Pathog. 2013;9(10):e1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lozach PY, Kühbacher A, Meier R, et al. DC‐SIGN as a receptor for phleboviruses. Cell Host Microbe. 2011;10(1):75‐88. [DOI] [PubMed] [Google Scholar]
- 58. Hofmann H, Li X, Zhang X, et al. Severe fever with thrombocytopenia virus glycoproteins are targeted by neutralizing antibodies and can use DC‐SIGN as a receptor for pH‐dependent entry into human and animal cell lines. J Virol. 2013;87(8):4384‐4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suda Y, Fukushi S, Tani H, et al. Analysis of the entry mechanism of Crimean‐Congo hemorrhagic fever virus, using a vesicular stomatitis virus pseudotyping system. Arch Virol. 2016;161(6):1447‐1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Léger P, Tetard M, Youness B, et al. Differential use of the C‐type lectins L‐SIGN and DC‐SIGN for phlebovirus endocytosis. Traffic. 2016;17(6):639‐656. [DOI] [PubMed] [Google Scholar]
- 61. Goncalves AR, Moraz ML, Pasquato A, Helenius A, Lozach PY, Kunz S. Role of DC‐SIGN in Lassa virus entry into human dendritic cells. J Virol. 2013;87(21):11504‐11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lozach PY, Lortat‐Jacob H, de Lacroix de Lavalette A, et al. DC‐SIGN and L‐SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J Biol Chem. 2003;278(22):20358‐20366. [DOI] [PubMed] [Google Scholar]
- 63. Lozach PY, Burleigh L, Staropoli I, et al. Dendritic cell‐specific intercellular adhesion molecule 3‐grabbing non‐integrin (DC‐SIGN)‐mediated enhancement of dengue virus infection is independent of DC‐SIGN internalization signals. J Biol Chem. 2005;280(25):23698‐23708. [DOI] [PubMed] [Google Scholar]
- 64. Murakami S, Takenaka‐Uema A, Kobayashi T, et al. Heparan sulfate proteoglycan is an important attachment factor for cell entry of Akabane and Schmallenberg viruses. J Virol. 2017;91(15):e00503‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mackow ER, Gavrilovskaya IN. Cellular receptors and hantavirus pathogenesis. Curr Top Microbiol Immunol. 2001;256:91‐115. [DOI] [PubMed] [Google Scholar]
- 66. Garrison AR, Radoshitzky SR, Kota KP, et al. Crimean‐Congo hemorrhagic fever virus utilizes a clathrin‐ and early endosome‐dependent entry pathway. Virology. 2013;444(1‐2):45‐54. [DOI] [PubMed] [Google Scholar]
- 67. Albornoz A, Hoffmann AB, Lozach PY, Tischler ND. Early bunyavirus‐host cell interactions. Viruses. 2016;8(5):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Santos RI, Rodrigues AH, Silva ML, et al. Oropouche virus entry into HeLa cells involves clathrin and requires endosomal acidification. Virus Res. 2008;138(1‐2):139‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Simon M, Johansson C, Mirazimi A. Crimean‐Congo hemorrhagic fever virus entry and replication is clathrin‐, pH‐ and cholesterol‐dependent. J Gen Virol. 2009;90(Pt 1):210‐215. [DOI] [PubMed] [Google Scholar]
- 70. Cifuentes‐Muñoz N, Barriga GP, Valenzuela PD, Tischler ND. Aromatic and polar residues spanning the candidate fusion peptide of the Andes virus Gc protein are essential for membrane fusion and infection. J Gen Virol. 2011;92(Pt 3):552‐563. [DOI] [PubMed] [Google Scholar]
- 71. Hover S, Foster B, Fontana J, et al. Bunyavirus requirement for endosomal K+ reveals new roles of cellular ion channels during infection. PLoS Pathog. 2018;14(1):e1006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bitto D, Halldorsson S, Caputo A, Huiskonen JT. Low pH and anionic lipid‐dependent fusion of Uukuniemi phlebovirus to liposomes. J Biol Chem. 2016;291(12):6412‐6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. The role of histidine residues in low‐pH‐mediated viral membrane fusion. Structure. 2006;14(10):1481‐1487. [DOI] [PubMed] [Google Scholar]
- 74. de Boer SM, Kortekaas J, Spel L, Rottier PJ, Moormann RJ, Bosch BJ. Acid‐activated structural reorganization of the Rift Valley fever virus Gc fusion protein. J Virol. 2012;86(24):13642‐13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kleinfelter LM, Jangra RK, Jae LT, et al. Haploid genetic screen reveals a profound and direct dependence on cholesterol for hantavirus membrane fusion. MBio. 2015;6(4):e00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Willensky S, Bar‐Rogovsky H, Bignon EA, Tischler ND, Modis Y, Dessau M. Crystal structure of glycoprotein C from a hantavirus in the post‐fusion conformation. PLoS Pathog. 2016;12(10):e1005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Barriga GP, Villalón‐Letelier F, Márquez CL, et al. Inhibition of the hantavirus fusion process by predicted domain III and stem peptides from glycoprotein Gc. PLoS Negl Trop Dis. 2016;10(7):e0004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dessau M, Modis Y. Crystal structure of glycoprotein C from Rift Valley fever virus. Proc Natl Acad Sci U S A. 2013;110(5):1696‐1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Barr JN. Bunyavirus mRNA synthesis is coupled to translation to prevent premature transcription termination. RNA. 2007;13(5):731‐736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gerrard SR, Nichol ST. Synthesis, proteolytic processing and complex formation of N‐terminally nested precursor proteins of the Rift Valley fever virus glycoproteins. Virology. 2007;357(2):124‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fontana J, López‐Montero N, Elliott RM, Fernández JJ, Risco C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell Microbiol. 2008;10(10):2012‐2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liljeroos L, Butcher SJ. Matrix proteins as centralized organizers of negative‐sense RNA virions. Front Biosci (Landmark Ed). 2013;18:696‐715. [DOI] [PubMed] [Google Scholar]
- 83. Mudhasani R, Kota KP, Retterer C, Tran JP, Whitehouse CA, Bavari S. High content image‐based screening of a protease inhibitor library reveals compounds broadly active against Rift Valley fever virus and other highly pathogenic RNA viruses. PLoS Negl Trop Dis. 2014;8(8):e3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Overby AK, Pettersson RF, Neve EP. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J Virol. 2007;81(7):3198‐3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wichgers Schreur PJ, Kortekaas J. Single‐molecule FISH reveals non‐selective packaging of Rift Valley fever virus genome segments. PLoS Pathog. 2016;12(8):e1005800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Estrada DF, De Guzman RN. Structural characterization of the Crimean‐Congo hemorrhagic fever virus Gn tail provides insight into virus assembly. J Biol Chem. 2011;286(24):21678‐21686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Salanueva IJ, Novoa RR, Cabezas P, et al. Polymorphism and structural maturation of bunyamwera virus in Golgi and post‐Golgi compartments. J Virol. 2003;77(2):1368‐1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dowall SD, Findlay‐Wilson S, Rayner E, et al. Hazara virus infection is lethal for adult type I interferon receptor‐knockout mice and may act as a surrogate for infection with the human‐pathogenic Crimean‐Congo hemorrhagic fever virus. J Gen Virol. 2012;93(Pt 3):560‐564. [DOI] [PubMed] [Google Scholar]
- 89. Papa A, Chaligiannis I, Kontana N, et al. A novel AP92‐like Crimean‐Congo hemorrhagic fever virus strain, Greece. Ticks Tick Borne Dis. 2014;5(5):590‐593. [DOI] [PubMed] [Google Scholar]
- 90. Salehi‐Vaziri M, Baniasadi V, Jalali T, et al. The first fatal case of Crimean‐Congo hemorrhagic fever caused by the AP92‐like strain of the Crimean‐Congo hemorrhagic fever virus. Jpn J Infect Dis. 2016;69(4):344‐346. [DOI] [PubMed] [Google Scholar]
- 91. Gray KK, Worthy MN, Juelich TL, et al. Chemotactic and inflammatory responses in the liver and brain are associated with pathogenesis of Rift Valley fever virus infection in the mouse. PLoS Negl Trop Dis. 2012;6(2):e1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Scharton D, Bailey KW, Vest Z, et al. Favipiravir (T‐705) protects against peracute Rift Valley fever virus infection and reduces delayed‐onset neurologic disease observed with ribavirin treatment. Antiviral Res. 2014;104:84‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Buys KK, Jung KH, Smee DF, Furuta Y, Gowen BB. Maporal virus as a surrogate for pathogenic New World hantaviruses and its inhibition by favipiravir. Antivir Chem Chemother. 2011;21(5):193‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Elliott RM. Orthobunyaviruses: recent genetic and structural insights. Nat Rev Microbiol. 2014;12(10):673‐685. [DOI] [PubMed] [Google Scholar]
- 95. Vialat P, Billecocq A, Kohl A, Bouloy M. The S segment of rift valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J Virol. 2000;74(3):1538‐1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lang Y, Henningson J, Jasperson D, et al. Mouse model for the Rift Valley fever virus MP12 strain infection. Vet Microbiol. 2016;195:70‐77. [DOI] [PubMed] [Google Scholar]
- 97. Bente DA, Alimonti JB, Shieh WJ, et al. Pathogenesis and immune response of Crimean‐Congo hemorrhagic fever virus in a STAT‐1 knockout mouse model. J Virol. 2010;84(21):11089‐11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Scharton D, Van Wettere AJ, Bailey KW, et al. Rift Valley fever virus infection in golden Syrian hamsters. PLoS One. 2015;10(1):e0116722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gowen BB, Westover JB, Miao J, et al. Modeling severe fever with thrombocytopenia syndrome virus infection in golden Syrian hamsters: importance of STAT2 in preventing disease and effective treatment with favipiravir. J Virol. 2017;91(3):e01942‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hooper JW, Larsen T, Custer DM, Schmaljohn CS. A lethal disease model for hantavirus pulmonary syndrome. Virology. 2001;289(1):6‐14. [DOI] [PubMed] [Google Scholar]
- 101. Wahl‐Jensen V, Chapman J, Asher L, et al. Temporal analysis of Andes virus and Sin Nombre virus infections of Syrian hamsters. J Virol. 2007;81(14):7449‐7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Milazzo ML, Eyzaguirre EJ, Molina CP, Fulhorst CF. Maporal viral infection in the Syrian golden hamster: a model of hantavirus pulmonary syndrome. J Infect Dis. 2002;186(10):1390‐1395. [DOI] [PubMed] [Google Scholar]
- 103. Golden JW, Hammerbeck CD, Mucker EM, Brocato RL. Animal models for the study of rodent‐borne hemorrhagic fever viruses: arenaviruses and hantaviruses. Biomed Res Int. 2015;2015:793257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Oestereich L, Rieger T, Neumann M, et al. Evaluation of antiviral efficacy of ribavirin, arbidol, and T‐705 (favipiravir) in a mouse model for Crimean‐Congo hemorrhagic fever. PLoS Negl Trop Dis. 2014;8(5):e2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Paeshuyse J, Dallmeier K, Neyts J. Ribavirin for the treatment of chronic hepatitis C virus infection: a review of the proposed mechanisms of action. Curr Opin Virol. 2011;1(6):590‐598. [DOI] [PubMed] [Google Scholar]
- 106. Leyssen P, De Clercq E, Neyts J. The anti‐yellow fever virus activity of ribavirin is independent of error‐prone replication. Mol Pharmacol. 2006;69(4):1461‐1467. [DOI] [PubMed] [Google Scholar]
- 107. Arda B, Aciduman A, Johnston JC. A randomized controlled trial of ribavirin in Crimean Congo haemorrhagic fever: ethical considerations. J Med Ethics. 2012;38(2):117‐120. [DOI] [PubMed] [Google Scholar]
- 108. Dokuzoguz B, Celikbas AK, Gok SE, Baykam N, Eroglu MN, Ergonul O. Severity scoring index for Crimean‐Congo hemorrhagic fever and the impact of ribavirin and corticosteroids on fatality. Clin Infect Dis. 2013;57(9):1270‐1274. [DOI] [PubMed] [Google Scholar]
- 109. Ozbey SB, Kader C, Erbay A, Ergonul O. Early use of ribavirin is beneficial in Crimean‐Congo hemorrhagic fever. Vector Borne Zoonotic Dis. 2014;14(4):300‐302. [DOI] [PubMed] [Google Scholar]
- 110. Ergonul O. Treatment of Crimean‐Congo hemorrhagic fever. Antiviral Res. 2008;78(1):125‐131. [DOI] [PubMed] [Google Scholar]
- 111. Moreli ML, Marques‐Silva AC, Pimentel VA, da Costa VG. Effectiveness of the ribavirin in treatment of hantavirus infections in the Americas and Eurasia: a meta‐analysis. Virusdisease. 2014;25(3):385‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Safronetz D, Haddock E, Feldmann F, Ebihara H, Feldmann H. In vitro and in vivo activity of ribavirin against Andes virus infection. PLoS One. 2011;6(8):e23560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ogg M, Jonsson CB, Camp JV, Hooper JW. Ribavirin protects Syrian hamsters against lethal hantavirus pulmonary syndrome—after intranasal exposure to Andes virus. Viruses. 2013;5(11):2704‐2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Furuta Y, Komeno T, Nakamura T. Favipiravir (T‐705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(7):449‐463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Nagata T, Lefor AK, Hasegawa M, Ishii M. Favipiravir: a new medication for the Ebola virus disease pandemic. Disaster Med Public Health Prep. 2015;9(1):79‐81. [DOI] [PubMed] [Google Scholar]
- 116. Sissoko D, Laouenan C, Folkesson E, et al. Experimental treatment with favipiravir for Ebola virus disease (the JIKI Trial): a historically controlled, single‐arm proof‐of‐concept trial in Guinea. PLoS Med. 2016;13(3):e1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tani H, Komeno T, Fukuma A, et al. Therapeutic effects of favipiravir against severe fever with thrombocytopenia syndrome virus infection in a lethal mouse model: Dose‐efficacy studies upon oral administration. PLoS One. 2018;13(10):e0206416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Martinez VP, Bellomo C, San Juan J, et al. Person‐to‐person transmission of Andes virus. Emerg Infect Dis. 2005;11(12):1848‐1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Brown KS, Ebihara H, Feldmann H. Development of a minigenome system for Andes virus, a New World hantavirus. Arch Virol. 2012;157(11):2227‐2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Safronetz D, Falzarano D, Scott DP, Furuta Y, Feldmann H, Gowen BB. Antiviral efficacy of favipiravir against two prominent etiological agents of hantavirus pulmonary syndrome. Antimicrob Agents Chemother. 2013;57(10):4673‐4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wei F, Li JL, Ling JX, et al. Establishment of SYBR green‐based qPCR assay for rapid evaluation and quantification for anti‐Hantaan virus compounds in vitro and in suckling mice. Virus Genes. 2013;46(1):54‐62. [DOI] [PubMed] [Google Scholar]
- 122. Flick K, Hooper JW, Schmaljohn CS, Pettersson RF, Feldmann H, Flick R. Rescue of Hantaan virus minigenomes. Virology. 2003;306(2):219‐224. [DOI] [PubMed] [Google Scholar]
- 123. Wichmann D, Grone HJ, Frese M, et al. Hantaan virus infection causes an acute neurological disease that is fatal in adult laboratory mice. J Virol. 2002;76(17):8890‐8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Stoltz M, Sundstrom KB, Hidmark A, et al. A model system for in vitro studies of bank vole borne viruses. PLoS One. 2011;6(12):e28992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Seto T, Tkachenko EA, Morozov VG, et al. An efficient in vivo method for the isolation of Puumala virus in Syrian hamsters and the characterization of the isolates from Russia. J Virol Methods. 2011;173(1):17‐23. [DOI] [PubMed] [Google Scholar]
- 126. Prescott J, Ye C, Sen G, Hjelle B. Induction of innate immune response genes by Sin Nombre hantavirus does not require viral replication. J Virol. 2005;79(24):15007‐15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Brocato RL, Hammerbeck CD, Bell TM, Wells JB, Queen LA, Hooper JW. A lethal disease model for hantavirus pulmonary syndrome in immunosuppressed Syrian hamsters infected with Sin Nombre virus. J Virol. 2014;88(2):811‐819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Flick R, Flick K, Feldmann H, Elgh F. Reverse genetics for Crimean‐Congo hemorrhagic fever virus. J Virol. 2003;77(10):5997‐6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Devignot S, Bergeron E, Nichol S, Mirazimi A, Weber F. A virus‐like particle system identifies the endonuclease domain of Crimean‐Congo hemorrhagic fever virus. J Virol. 2015;89(11):5957‐5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Boyd A, Fazakerley JK, Bridgen A. Pathogenesis of Dugbe virus infection in wild‐type and interferon‐deficient mice. J Gen Virol. 2006;87(Pt 7):2005‐2009. [DOI] [PubMed] [Google Scholar]
- 131. Flusin O, Vigne S, Peyrefitte CN, Bouloy M, Crance JM, Iseni F. Inhibition of Hazara nairovirus replication by small interfering RNAs and their combination with ribavirin. Virol J. 2011;8:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ikegami T, Peters CJ, Makino S. Rift valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J Virol. 2005;79(9):5606‐5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Habjan M, Penski N, Spiegel M, Weber F. T7 RNA polymerase‐dependent and ‐independent systems for cDNA‐based rescue of Rift Valley fever virus. J Gen Virol. 2008;89(Pt 9):2157‐2166. [DOI] [PubMed] [Google Scholar]
- 134. Reed C, Lin K, Wilhelmsen C, et al. Aerosol exposure to Rift Valley fever virus causes earlier and more severe neuropathology in the murine model, which has important implications for therapeutic development. PLoS Negl Trop Dis. 2013;7(4):e2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Gowen BB, Wong MH, Jung KH, et al. In vitro and in vivo activities of T‐705 against arenavirus and bunyavirus infections. Antimicrob Agents Chemother. 2007;51(9):3168‐3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Fisher AF, Tesh RB, Tonry J, Guzman H, Liu D, Xiao SY. Induction of severe disease in hamsters by two sandfly fever group viruses, Punta toro and Gabek Forest (Phlebovirus, Bunyaviridae), similar to that caused by Rift Valley fever virus. Am J Trop Med Hyg. 2003;69(3):269‐276. [PubMed] [Google Scholar]
- 137. Mendenhall M, Wong MH, Skirpstunas R, Morrey JD, Gowen BB. Punta Toro virus (Bunyaviridae, Phlebovirus) infection in mice: strain differences in pathogenesis and host interferon response. Virology. 2009;395(1):143‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Shimojima M, Fukushi S, Tani H, et al. Effects of ribavirin on severe fever with thrombocytopenia syndrome virus in vitro. Jpn J Infect Dis. 2014;67(6):423‐427. [DOI] [PubMed] [Google Scholar]
- 139. Brennan B, Li P, Zhang S, et al. Reverse genetics system for severe fever with thrombocytopenia syndrome virus. J Virol. 2015;89(6):3026‐3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Tani H, Fukuma A, Fukushi S, et al. Efficacy of T‐705 (Favipiravir) in the treatment of infections with lethal severe fever with thrombocytopenia syndrome virus. mSphere. 2016;1(1):e00061‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Jin C, Liang M, Ning J, et al. Pathogenesis of emerging severe fever with thrombocytopenia syndrome virus in C57/BL6 mouse model. Proc Natl Acad Sci U S A. 2012;109(25):10053‐10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Brisbarre NM, Plumet S, de Micco P, Leparc‐Goffart I, Emonet SF. Toscana virus inhibits the interferon beta response in cell cultures. Virology. 2013;442(2):189‐194. [DOI] [PubMed] [Google Scholar]
- 143. Gori Savellini G, Di Genova G, Terrosi C, et al. Immunization with Toscana virus N‐Gc proteins protects mice against virus challenge. Virology. 2008;375(2):521‐528. [DOI] [PubMed] [Google Scholar]
- 144. Rezelj VV, Overby AK, Elliott RM. Generation of mutant Uukuniemi viruses lacking the nonstructural protein NSs by reverse genetics indicates that NSs is a weak interferon antagonist. J Virol. 2015;89(9):4849‐4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Mazelier M, Rouxel RN, Zumstein M, Mancini R, Bell‐Sakyi L, Lozach PY. Uukuniemi virus as a tick‐borne virus model. J Virol. 2016;90(15):6784‐6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Overby AK, Popov V, Neve EP, Pettersson RF. Generation and analysis of infectious virus‐like particles of Uukuniemi virus (Bunyaviridae): a useful system for studying bunyaviral packaging and budding. J Virol. 2006;80(21):10428‐10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Flick R, Pettersson RF. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I‐catalyzed expression of chimeric viral RNAs. J Virol. 2001;75(4):1643‐1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Odhiambo C, Venter M, Limbaso K, Swanepoel R, Sang R. Genome sequence analysis of in vitro and in vivo phenotypes of Bunyamwera and Ngari virus isolates from northern Kenya. PLoS One. 2014;9(8):e105446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Weber F, Dunn EF, Bridgen A, Elliott RM. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology. 2001;281(1):67‐74. [DOI] [PubMed] [Google Scholar]
- 150. Lowen AC, Noonan C, McLees A, Elliott RM. Efficient bunyavirus rescue from cloned cDNA. Virology. 2004;330(2):493‐500. [DOI] [PubMed] [Google Scholar]
- 151. Borucki MK, Kempf BJ, Blitvich BJ, Blair CD, Beaty BJ. La Crosse virus: replication in vertebrate and invertebrate hosts. Microbes Infect. 2002;4(3):341‐350. [DOI] [PubMed] [Google Scholar]
- 152. Blakqori G, Weber F. Efficient cDNA‐based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J Virol. 2005;79(16):10420‐10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Livonesi MC, De Sousa RL, Badra SJ, Figueiredo LT. In vitro and in vivo studies of ribavirin action on Brazilian orthobunyavirus. Am J Trop Med Hyg. 2006;75(5):1011‐1016. [PubMed] [Google Scholar]
- 154. Acrani GO, Tilston‐Lunel NL, Spiegel M, et al. Establishment of a minigenome system for Oropouche virus reveals the S genome segment to be significantly longer than reported previously. J Gen Virol. 2015;96(Pt 3):513‐523. [DOI] [PubMed] [Google Scholar]
- 155. Tilston‐Lunel NL, Acrani GO, Randall RE, Elliott RM. Generation of recombinant Oropouche viruses lacking the nonstructural protein NSm or NSs. J Virol. 2015;90(5):2616‐2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Rodrigues AH, Santos RI, Arisi GM, et al. Oropouche virus experimental infection in the golden hamster (Mesocrisetus auratus). Virus Res. 2011;155(1):35‐41. [DOI] [PubMed] [Google Scholar]
- 157. Santos RI, Almeida MF, Paula FE, et al. Experimental infection of suckling mice by subcutaneous inoculation with Oropouche virus. Virus Res. 2012;170(1‐2):25‐33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Phylogenetic tree constructed with amino acid sequences of L‐proteins of bunyaviruses. Names in bold indicate the type species.
Figure S2: Numeric data corresponding to heatmaps. A) Endonuclease domain of RdRp‐proteins. B) Polymerase domain of RdRp‐proteins. C) N‐protein.
Data S1 Supporting information


