Abstract
Background
Neural cell adhesion molecule 1 (NCAM1; CD56) and E-cadherin are both involved in cell-cell adhesion and cell development processes, and their dysregulation is associated with various tumors. We hypothesized that dysregulated NCAM1 could suppress the invasive behavior of ameloblastoma (AB), and its expression was regulated by miR-141-3p.
Material/Methods
Real-time qPCR was performed to examine differences in miR-141-3p expression between AB tissues and normal oral tissues (NOMs). The potential target NCAM1 of miR-141-3p was predicted by bioinformatics analysis, which was validated through dual-luciferase assay. The mRNA and protein levels of NCAM1 were detected by real-time qPCR and Western blot, respectively. Furthermore, the expression and distribution of NCAM1 in AB were investigated through immunohistochemical staining, and immunohistochemical staining of E-cadherin was also performed. After overexpression of NCAM1, the migration of AM-1 cells was examined using wound-healing assay.
Results
Real-time qPCR results confirmed that miR-141-3p was significantly downregulated in AB tissues. According to bioinformatics analysis, NCAM1 was a target of miR-141-3p, which was confirmed by dual luciferase assay. We found that NCAM1 was significantly upregulated in AB tissues at the mRNA and protein levels. Furthermore, NCAM1 and E-cadherin were mainly expressed on the cell membrane of AB. Downregulation of E-cadherin was found in AB tissues. As shown in wound-healing assay results, NCAM1 overexpression significantly inhibited the invasiveness of AM-1 cells.
Conclusions
In this study, highly expressed NCAM1 was found in AB, and it suppressed the migration of AB cells and was regulated by miR-141-3p, suggesting its potential value as a therapeutic target for AB.
MeSH Keywords: Ameloblastoma, Cell Migration Assays, MicroRNAs, Neural Cell Adhesion Molecules
Background
AB is a common benign odontogenic epithelial tumor, characterized by slow growth and local invasiveness [1,2]. According to the latest WHO system, AB is classified into AB, unicystic, and extraosseous/peripheral types [3]. Among them, AB is the most common type, but its etiology and pathogenesis remain to be clarified [4]. Most patients are treated by surgical resection. However, surgical removal often results in facial deformities, and patients undergoing conservative treatment often have relapse [5]. Targeted therapy may be a promising treatment for AB and effective therapeutic targets need to be explored.
Abnormal gene expression can participate in the development of AB. NCAM1 is a cell adhesion family molecule in the immunoglobulin molecule superfamily [6]. It is mainly expressed in the nervous system and is involved in regulating the function of nerve cells and neuronal migration [7]. In human hematopoietic, NCAM1 is expressed in NK cells, rare subsets of T and B lymphocytes, dendritic cells, and neural or mesenchymal stem cells [8–11]. NCAM1 is the best surface antigen for identification of human NK cells [12,13]. Furthermore, NCAM1 is widely expressed during embryogenesis [8]. Recent findings have suggested that abnormally expressed NCAM1 is associated with development of various cancers, such as acute myeloid leukemia [11], small-cell lung cancer [14], and breast cancer [15]. Its dysregulation can be regulated by several factors and has been confirmed as a target of miR-572 in human oligodendroglial cells [16]. Thus, in this study, the expression and function of NCAM1 in AB were analyzed.
MicroRNAs (miRNAs) are endogenous non-coding RNAs about 20–24 nucleotides in size [17]. miRNAs can silence their target genes at post-transcriptional levels [18]. Numerous studies have reported that miRNAs mediate a variety of tumors, including AB [19]. Among them, miR-141-3p can modulate cell migration and invasion in several cancers through silencing downstream targets. For example, low miR-141-3p expression induces prostate cancer bone metastasis through activation of the NF-κB pathway [20]. miR-141-3p suppresses colorectal cancer cell invasion via targeting TRAF5 [21] and can promote invasion and tumorigenesis of cervical cancer cells via inhibiting FOXA2 [22]. Intriguingly, local invasion is one of the main characteristics of AB. E-cadherin, a cell adhesion molecule, is mainly expressed on the membrane of epithelial cells. Increasing evidence shows that abnormal expression of E-cadherin is related to the aggressive behavior of AB [23]. However, the expression and potential molecular mechanisms of miR-141-3p in AB remain to be explored.
In this study, we hypothesized that highly expressed NCAM1 can suppress the migration behavior of AB, and that its expression is regulated by miR-141-3p.
Material and Methods
Tissue specimen collection
From 2014 to 2019, 25 cases of AB and 15 cases of NOM fresh tissues were harvested from the School and Hospital of Stomatology, China Medical University (Shenyang, China). Paraffin-embedded tissue sections and corresponding clinical information were obtained from the Department of Oral Pathology of China Medical University between 2015 and 2016, including 96 cases of AB tissues and 10 cases of paraneoplastic epithelium tissues. In this study, these paraffin-embedded tissue sections were mainly used for immunohistochemistry analysis and we evaluated the association between NCAM1 expression and clinicopathological factors of patients with AB. Our study was approved by the Ethics Committee of the School and Hospital of Stomatology, China Medical University (2016-13) and all participates provided written informed consent. The clinical information of these patients with AB is shown in Table 1. According to the latest WHO system, the tumors were coded based on histopathological presentation.
Table 1.
The clinical feature information of patients with AB.
| Clinicopathological characteristics | Number of case (n=96), % |
|---|---|
| Age (years) | |
| <50 | 70 (72.92) |
| ≥50 | 26 (27.08) |
| Gender | |
| Male | 54 (56.25) |
| Female | 42 (43.75) |
| Location | |
| Maxilla | 18 (18.75) |
| Mandible | 78 (81.25) |
| Pathological type | |
| AB | 96 (100.00) |
| Others | 0 (0.00) |
| Recurrence | |
| Yes | 9 (9.38) |
| No | 87 (90.62) |
Real-time qPCR
Total RNA was extracted from tissues or cells using Trizol (Takara, Japan), which was reverse-transcribed into cDNA. The primers for miR-141-3p, U6, NCAM1, and β-actin are shown in Table 2. The relative expression levels were calculated by the 2−ΔΔCT method. U6 was the internal control of miR-141-3p, and β-actin was the internal control of NCAM1.
Table 2.
The primer sequences for real-time qPCR.
| Target | Primer sequences |
|---|---|
| miR-141-3p | 5′-AGCCGCTAACACTGTCTGGTA-3′ (forward) 5′-CAGAGCAGGGTCCGAGGTA-3′ (reverse) |
| U6 | 5′-ATTGGAACGATACAGAGAAGATT-3′ (forward) 5′-GGGCCATGCTAATCTTCTCTG-3′ (reverse) |
| NCAM1 | 5′-AATTTACCGCGGCAAGAACATC-3′ (forward) 5′-CCTGGCTGGGAACAATATCCAC-3′ (reverse) |
| β-actin | 5′-CTCCATCCTGGCCTCGCTGT-3′ (forward) 5′-GCTGTCACCTTCACCGTTCC-3′ (reverse) |
Western blot analysis
Total protein was extracted from tissues and was measured using a BCA kit. Then, protein samples were transferred onto PVDF membranes. The membranes were blocked for 1 h at 37°C. After that, the membranes were incubated overnight with the primary antibodies anti-NCAM1 (1: 2000; Abcam, UK) and anti-β-actin (1: 1000; Abcam, UK) at 4°C, followed by addition of secondary antibody (1: 5000; Abcam, UK) at room temperature for 1 h. Finally, the protein blots were visualized using an infrared fluorescence scanning imaging system.
Immunohistochemistry and assessment
Tissue sections were dewaxed and dehydrated. Then, 3% hydrogen peroxide was used to eliminate endogenous peroxidase activity. The sections were blocked with 5% goat serum and incubated for approximately 10 min. The sections were incubated overnight with the primary antibodies anti-NCAM1 (1: 200; Abcam, UK) and anti-E-cadherin (1: 200; Abcam, UK) at 4°C, followed by incubation with biotinylated secondary antibody at room temperature for 20 min. Finally, using the DAB kit, the sections were rinsed, counterstained, dehydrated, cleared, and sealed. In the negative control group, PBS was used instead of the primary antibody. The semiquantitative scores were assessed based on staining intensity and the percentage of positive cells. Three fields of vision were selected for scoring at 200 magnification and the scores were averaged. The scoring criteria were: strong positive (4 points), positive (3 points), weak positive (2 points), and negative (1 point).
Dual luciferase reporter assay
The 3′-UTR of NCAM1 containing predicted miR-141-3p binding sites was constructed into the pmirGLO vector (Gene Pharma, China). Then, 293T cells were cultured in 24-well plates and co-transfected with WT (wild-type) or MUT (mutant) and miR-141-3p mimics or miR-NC mimics, respectively. The relative luciferase activities were measured using the Dual Luciferase Reporter Assay System.
Cell culture and transfection
The human ameloblastoma cell line AM-1 was cultured in keratinocyte serum-free medium (KSFM) and incubated at 37°C and 5% CO2. Then, AM-1 cells were seeded into 12-well plates containing 1 ml of KSFM medium per well until the degree of cell fusion reached approximately 80%. For cell transfection, miR-141-3p mimics, NCAM1 overexpression, and their corresponding NCs were transfected into AM-1 cells.
Wound-healing assay
A horizontal line was drawn evenly on the back of the 6-well plates. Transfected cells were then seeded in 6-well plates. Next, the pipette tip was used to scrape the cell-covered surface, making sure that the scrape was perpendicular to the horizontal line. Scratch photos were taken at 0 h and 24 h, and the area and length of each scratch photo were measured using ImageJ software.
Statistical analysis
All statistical data were analyzed using Graphpad Prism 8.0 and SPSS 23.0. Data are expressed as mean±standard deviation (SD) from at least 3 times repeated experiments. The t test was performed to compare differences between the 2 groups. Correlations between NCAM1 and clinicopathological factors were analyzed by one-way chi-square analysis. P<0.05 was considered statistically significant.
Results
miR-141-3p was significantly downregulated in AB tissues
Our previous microarray analysis revealed that miR-141-3p was differentially expressed between AB and NOM tissues. The expression level of miR-141-3p between AB and NOM tissues was verified using real-time qPCR. The results showed that its expression was 12.5 times lower in AB than that in NOM (P<0.001; Figure 1).
Figure 1.
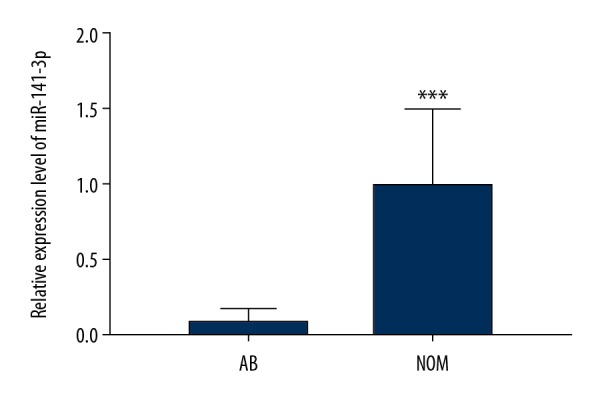
Real-time qPCR results showed that miR-141-3p was significantly downregulated in AB tissues compared to NOM tissues. *** p<0.001.
NCAM1 was significantly upregulated in AB tissues
We assessed and compared the expression levels of NCAM1 between 13 AB tissues and 7 NOM tissues via real-time qPCR. The results suggested that its expression was 4.7 times higher in AB than in NOM at the mRNA level (P<0.05; Figure 2). Moreover, we found that NCAM1 had a higher expression level in AB tissues compared to NOM tissues at the protein level (P<0.05; Figure 3A, 3B).
Figure 2.
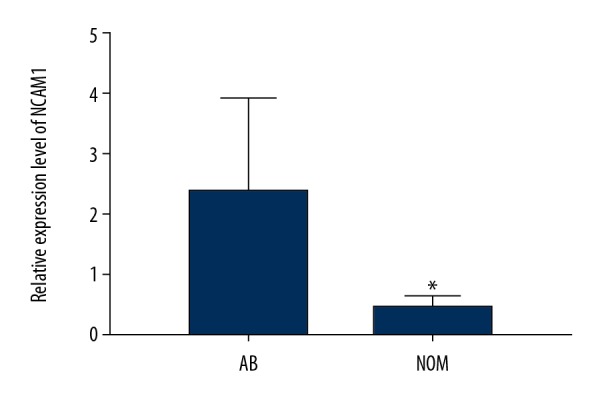
NCAM1 was significantly upregulated in AB tissues compared to NOM tissues according to real-time qPCR results. * p<0.05.
Figure 3.
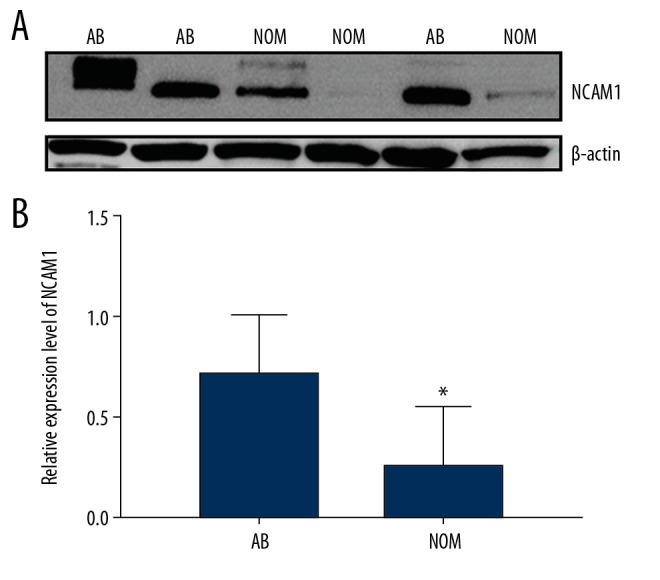
(A, B) Western blot results showed that NCAM1 was significantly upregulated in AB tissues compared to NOM tissues. * p<0.05.
Expression and distribution of NCAM1 and E-cadherin in AB tissues
As shown in immunohistochemistry results, NCAM1 was mainly expressed in the cell membrane of AB. Figure 4A shows ameloblast follicles containing pre-ameloblast-like cells and a central core of loosely arranged stellate reticulum-like cells supported by loose fibrous stroma. NCAM1 was strongly positively expressed in the plasma membrane of ameloblast-like cells and partial stellate reticulum-like cells of follicular AB. However, it was negatively expressed in interstitial collagen fibroblasts. As shown in Figure 4B, tumor epithelial cell proliferation was a network-connected epithelial cord, with central cells surrounded by surrounding cells resembling stellate reticulocytes. NCAM1 was moderately positively expressed in the plasma membrane of peripheral and stellate reticulum in plexiform AB. It is well-established that E-cadherin is associated with tumor invasiveness. Thus, we assessed the expression and distribution of E-cadherin in AB. As shown Figure 4C, we found that E-cadherin was positively expressed in the plasma membrane of the peripheral layer of follicular AB and was weakly positively expressed in the stellate reticulum cells, but it was not expressed in interstitial fibroblasts. In addition, higher NCAM1 expression in AB was found than that in NOM (P<0.001; Table 3). However, the expression of NCAM1 was not significantly correlated with sex, age, recurrence, location, or lesion type of patients with AB (Table 3).
Figure 4.
Immunohistochemistry results showing the expression and distribution of NCAM1 and E-cadherin in AB. Positive NCAM1 expression in follicular AB (A) and plexiform AB (B); (C) Positive E-cadherin expression in follicular AB. Magnification, ×200; scale bar=100 μm. Black arrows represents positive expression and red arrows represents negative expression.
Table 3.
Correlation between NCAM1 expression and clinicopathological factors of patients with AB.
| Clinicopathological factors | Groups | Number of cases | NCAM1 positive expression rate, % | χ2 | P value |
|---|---|---|---|---|---|
| Age | >50 | 26 | 80.77 | 0.07 | 0.93 |
| ≤50 | 70 | 80.00 | |||
| Gender | Male | 54 | 77.78 | 0.46 | 0.50 |
| Female | 42 | 83.33 | |||
| Location | Mandible | 78 | 79.49 | 0.14 | 0.71 |
| Maxillary | 18 | 83.33 | |||
| Type | Solid | 71 | 80.28 | 0.001 | 0.98 |
| Peripheral/desmoplastic | 25 | 80.00 | |||
| Recurrence | Yes | 9 | 77.78 | 0.04 | 0.85 |
| No | 87 | 80.46 | |||
| Total positive rate | AB | 96 | 80.21 | 29.32 | <0.001 |
| NOM | 10 | 0.00 |
NCAM1 was regulated by miR-141-3p
As predicted by bioinformatics analysis results, there was a binding site between miR-141-3p and NCAM1 by the TargetScan database. To validate their relationship, a luciferase reporter plasmid containing NCAM1-WT and NCAM1-MUT of the miR-141-3p binding site was constructed (p<0.05, Figure 5A). NCAM1-WT or NCAM1-MUT luciferase reporter plasmid and miR-141-3p mimics or miR-141-3p NC were co-transfected in 293T cells and assayed for luciferase activity. As displayed in Figure 5B, the relative luciferase activity of NCAM1-WT was significantly decreased, while the relative luciferase activity of the NCAM1-MUT was unchanged. These results suggested that NCAM1 was a target of miR-141-3p.
Figure 5.
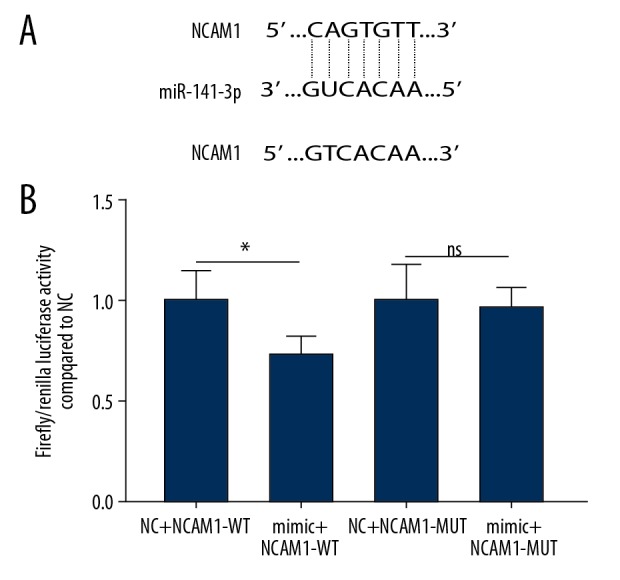
Luciferase reporter assay. Dual luciferase confirmed miR-141-3p and NCAM1 interaction in AM-1 cells (A, B). * p<0.05; ns indicated no statistical significance.
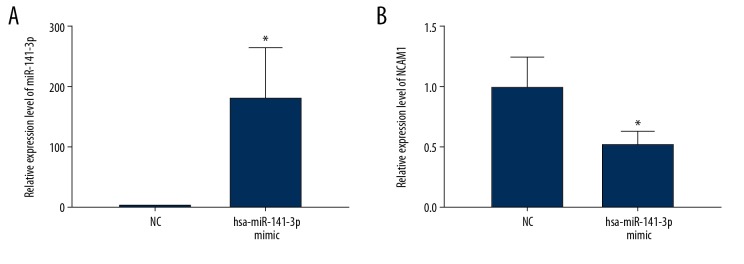
miR-141-3p mimics were used to overexpress miR-141-3p in AM-1 cells. As shown in Figure 6A, miR-141-3p was successfully overexpressed (P<0.05). The expression of NCAM1 was significantly inhibited in AM-1 cells after miR-141-3p overexpression (P<0.05, Figure 6B).
Figure 6.
NCAM1 is regulated by miR-141-3p. (A) Transfection effect of miR-141-3p mimics and inhibitors in AM-1 cell was detected by real-time qPCR. (B) Real-time qPCR showed that NCAM1 was significantly inhibited in AM-1 cells transfected with miR-141-3p mimics. * p<0.05.
NCAM1 inhibits AM-1 cell migration
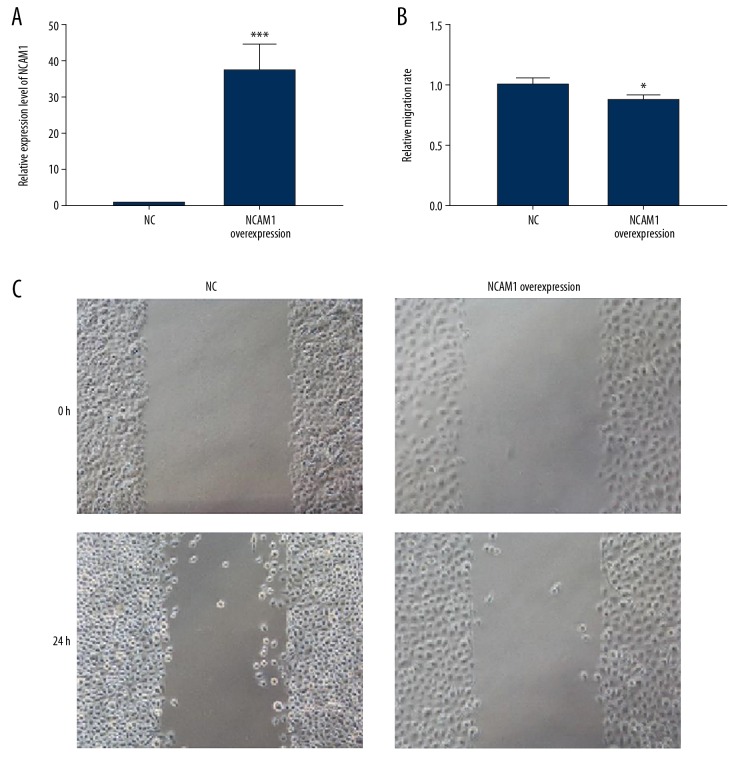
We further observed whether NCAM1 could affect the migration of AB cells. Firstly, RT-qPCR results confirmed that NCAM1 was successfully overexpressed in AM-1 cells (p<0.001, Figure 7A). After that, the cell scratch test was performed. We found that the migration ability of AM-1 cells was significantly inhibited after NCAM1 overexpression (P<0.05, Figure 7B, 7C). These results suggest that NCAM1 inhibits AM-1 cell migration.
Figure 7.
NCAM1 inhibits AM-1 cell migration. (A) Validation of the transfection effects of NCAM1 overexpression in AM-1 cells. (B, C) The inhibitory effect of NCAM1 overexpression on the migration behavior of AM-1 cells, as shown in wound-healing assay results. * p<0.05; *** p<0.001.
Discussion
The etiology and pathogenesis of AB remain to be clarified. In this study, highly expressed NCAM1 was found in AB tissues and it could inhibit migration of AB cells. Furthermore, it was regulated by miR-141-3p in AB cells. Our findings present novel insights into the mechanism of AB.
Tumor formation involves cell adhesions and interaction with surrounding cells and extracellular matrix [24]. Abnormal adhesion may be a determinant of epithelial tumor formation [25,26]. Thus, we observed the effect of adhesion molecule NCAM1 on the biological behavior of AB. The reduction of cell adhesion is closely related to tumor metastasis [27]. For example, the abnormal expression of the cell adhesion molecule CD44 and its variants is related to cell invasion and metastatic potential of various cancers [28–30]. Highly expressed NCAM1 was found in AB tissues at the mRNA and protein levels in our study. Its high expression significantly suppressed the migration and invasion of AB cells. Consistent with previous studies, NCAM1 appears to be involved in the aggressiveness of tumor cells, but not AB. For example, dysregulated NCAM1 is related to the invasiveness ability of human skin squamous cell carcinoma cells [31]. We found that NCAM1 was mainly expressed on the cell membrane of AB. Consistent with Western blot results, immunohistochemistry results confirmed that NCAM1 was highly expressed in AB, but there was not a significant correlation between its expression and clinical features. Our results suggest that NCAM1 is involved in migration and invasion of AB cells.
Studies have found that signaling pathways related to odontogenesis can promote the pathogenesis of AB [2]. Epithelial-mesenchymal transition (EMT) has been reported to activate early dentinogenesis and promote AB development [32]. EMT serves as an early-stage factor for malignant transformation of tumor epithelial cells. Epithelial marker E-cadherin expression loss occurs during EMT, which is closely related to the invasion and metastasis of AB [33]. Consistent with previous studies, we found that E-cadherin was mainly expressed on the cell membrane of AB, indicating the invasive behavior of AB cells [2,33].
In our study, low expression of miR-141-3p was observed in AB tissues. Its low expression has been found in several cancers and is related to cell migration and invasion in colorectal cancer [21], pancreatic cancer [34], cervical cancer [22], and papillary thyroid cancer [35]. Based on the evidence presented above, we speculate that miR-141-3p is involved in migration and invasion of AB cells by targeting NCAM1. Dual luciferase reporter assay confirmed that NCAM1 was a target of miR-141-3p in AM-1 cells. Furthermore, miR-141-3p overexpression significantly inhibited the expression NCAM1 in AM-1 cells. These results suggest that miR-141-3p participates in the development of AB by inhibiting NCAM1.
Conclusions
NCAM1 was significantly overexpressed in AB tissues, and its dysregulation inhibited the invasive behavior of AB. Furthermore, we found that NCAM1 expression was regulated by miR-141-3p, which could be a potential therapeutic target for AB.
Abbreviations
- AB
ameloblastoma
- NCAM1
neural cell adhesion molecule 1
- miRNA
microRNA
- NOM
normal oral mucosa
- EMT
epithelial-mesenchymal transition
Footnotes
Conflicts of interest
None.
Source of support: This work was funded by the National Natural Science Foundation of China (81072197 and 81470758)
References
- 1.Nakano K, Takabatake K, Kawai H, et al. Notch signaling affects oral neoplasm cell differentiation and acquisition of tumor-specific characteristics. Int J Mol Sci. 2019;20(8) doi: 10.3390/ijms20081973. pii: E1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Wang Y, Fan C, et al. Interleukin-8/beta-catenin mediates epithelial-mesenchymal transition in ameloblastoma. Oral Dis. 2019;25(8):1964–71. doi: 10.1111/odi.13173. [DOI] [PubMed] [Google Scholar]
- 3.Wright JM, Vered M. Update from the 4th Edition of the World Health Organization Classification of Head and Neck Tumours: Odontogenic and maxillofacial bone tumors. Head Neck Pathol. 2017;11(1):68–77. doi: 10.1007/s12105-017-0794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada T, Fujitsu K, Ichikawa T, et al. Unicystic ameloblastomatoid cystic craniopharyngioma: pathological discussion and clinical significance of cyst formation in adamantinomatous craniopharyngioma. Pediatr Neurosurg. 2016;51(3):158–63. doi: 10.1159/000442992. [DOI] [PubMed] [Google Scholar]
- 5.Kurppa KJ, Catón J, Morgan PR, et al. High frequency of BRAF V600E mutations in ameloblastoma. J Pathol. 2014;232(5):492–98. doi: 10.1002/path.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siles AM, Martínez-Hernández E, Araque J, et al. Antibodies against cell adhesion molecules and neural structures in paraneoplastic neuropathies. Ann Clin Transl Neurol. 2018;5(5):559–69. doi: 10.1002/acn3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu MJ, Ha JO. CD56 positive diffuse large B-cell lymphoma: A case report and literature review. Int J Clin Exp Pathol. 2013;6(12):3023–25. [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Youssef Y, Robinson C, et al. CD56 expression marks human group 2 innate lymphoid cell divergence from a shared NK cell and group 3 innate lymphoid cell developmental pathway. Immunity. 2018;49(3):464–76.e4. doi: 10.1016/j.immuni.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crossland DL, Denning WL, Ang S, et al. Antitumor activity of CD56-chimeric antigen receptor T cells in neuroblastoma and SCLC models. Oncogene. 2018;37(27):3686–97. doi: 10.1038/s41388-018-0187-2. [DOI] [PubMed] [Google Scholar]
- 10.Huang L, Liu D, Wang N, et al. Integrated genomic analysis identifies deregulated JAK/STAT-MYC-biosynthesis axis in aggressive NK-cell leukemia. Cell Res. 2018;28(2):172–86. doi: 10.1038/cr.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasca D, Szybinski J, Schüler A, et al. NCAM1 (CD56) promotes leukemogenesis and confers drug resistance in AML. Blood. 2019;133(21):2305–19. doi: 10.1182/blood-2018-12-889725. [DOI] [PubMed] [Google Scholar]
- 12.Voigt J, Malone DFG, Dias J, et al. Proteome analysis of human CD56(neg) NK cells reveals a homogeneous phenotype surprisingly similar to CD56(dim) NK cells. Eur J Immunol. 2018;48(9):1456–69. doi: 10.1002/eji.201747450. [DOI] [PubMed] [Google Scholar]
- 13.Wagner JA, Rosario M, Romee R, et al. CD56bright NK cells exhibit potent antitumor responses following IL-15 priming. J Clin Invest. 2017;127(11):4042–58. doi: 10.1172/JCI90387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiskopf K, Jahchan NS, Schnorr PJ, et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest. 2016;126(7):2610–20. doi: 10.1172/JCI81603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghaderi F, Ahmadvand S, Ramezani A, et al. Production and characterization of monoclonal antibody against a triple negative breast cancer cell line. Biochem Biophys Res Commun. 2018;505(1):181–86. doi: 10.1016/j.bbrc.2018.09.087. [DOI] [PubMed] [Google Scholar]
- 16.Mancuso R, Agostini S, Marventano I, et al. NCAM1 is the target of miRNA-572: validation in the human oligodendroglial cell line. Cell Mol Neurobiol. 2018;38(2):431–40. doi: 10.1007/s10571-017-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–33. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Wang G, Qiao X, et al. Downregulated miR-524-5p participates in the tumor microenvironment of ameloblastoma by targeting the Interleukin-33 (IL-33)/suppression of tumorigenicity 2 (ST2) axis. Med Sci Monit. 2020;26:e921863. doi: 10.12659/MSM.921863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S, Wa Q, Pan J, et al. Downregulation of miR-141-3p promotes bone metastasis via activating NF-kappaB signaling in prostate cancer. J Exp Clin Cancer Res. 2017;36(1):173. doi: 10.1186/s13046-017-0645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Z, Li X, Liu S, et al. MiR-141-3p inhibits cell proliferation, migration and invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys Res Commun. 2019;514(3):699–705. doi: 10.1016/j.bbrc.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Li JH, Zhang Z, Du MZ, et al. microRNA-141-3p fosters the growth, invasion, and tumorigenesis of cervical cancer cells by targeting FOXA2. Arch Biochem Biophys. 2018;657:23–30. doi: 10.1016/j.abb.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Kurioka K, Wato M, Iseki T, et al. Differential expression of the epithelial mesenchymal transition factors Snail, Slug, Twist, TGF-beta, and E-cadherin in ameloblastoma. Med Mol Morphol. 2017;50(2):68–75. doi: 10.1007/s00795-016-0149-0. [DOI] [PubMed] [Google Scholar]
- 24.Friedl P, Alexander S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Chu BH, Sen S, et al. Modulating malignant epithelial tumor cell adhesion, migration and mechanics with nanorod surfaces. Biomed Microdevices. 2011;13(1):89–95. doi: 10.1007/s10544-010-9473-7. [DOI] [PubMed] [Google Scholar]
- 26.Sokeland G, Schumacher U. The functional role of integrins during intra- and extravasation within the metastatic cascade. Mol Cancer. 2019;18(1):12. doi: 10.1186/s12943-018-0937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamidi H, Ivaska J. Every step of the way: Integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18(9):533–48. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya R, Mitra T, Ray Chaudhuri S, Roy SS. Mesenchymal splice isoform of CD44 (CD44s) promotes EMT/invasion and imparts stem-like properties to ovarian cancer cells. J Cell Biochem. 2018;119(4):3373–83. doi: 10.1002/jcb.26504. [DOI] [PubMed] [Google Scholar]
- 29.Mueller N, Wicklein D, Eisenwort G, et al. CD44 is a RAS/STAT5-regulated invasion receptor that triggers disease expansion in advanced mastocytosis. Blood. 2018;132(18):1936–50. doi: 10.1182/blood-2018-02-833582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Qin W, Chen Y, et al. Cholesterol inhibits hepatocellular carcinoma invasion and metastasis by promoting CD44 localization in lipid rafts. Cancer Lett. 2018;429:66–77. doi: 10.1016/j.canlet.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 31.Brugiere C, El Bouchtaoui M, Leboeuf C, et al. Perineural invasion in human cutaneous squamous cell carcinoma is linked to neurotrophins, epithelial-mesenchymal transition, and NCAM1. J Invest Dermatol. 2018;138(9):2063–66. doi: 10.1016/j.jid.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 32.Siar CH, Ng KH. Epithelial-to-mesenchymal transition in ameloblastoma: Focus on morphologically evident mesenchymal phenotypic transition. Pathology. 2019;51(5):494–501. doi: 10.1016/j.pathol.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Carreón-Burciaga RG, González-González R, Molina-Frechero N, et al. Differences in E-cadherin and syndecan-1 expression in different types of ameloblastomas. Anal Cell Pathol (Amst) 2018;23:9392632. doi: 10.1155/2018/9392632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui XP, Wang CX, Wang ZY, et al. LncRNA TP73-AS1 sponges miR-141-3p to promote the migration and invasion of pancreatic cancer cells through the up-regulation of BDH2. Biosci Rep. 2019;39(3) doi: 10.1042/BSR20181937. pii: BSR20181937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang M, Huang W, Wu X, et al. MiR-141-3p suppresses tumor growth and metastasis in papillary thyroid cancer via targeting Yin Yang 1. Anat Rec (Hoboken) 2019;302(2):258–68. doi: 10.1002/ar.23940. [DOI] [PubMed] [Google Scholar]