Abstract
Rapid, sensitive, selective, convenient, and cost‐effective pathogen diagnosis is important to prevent further spread of pandemic diseases, minimize social and economic losses, and to facilitate right clinical therapy. Over the past few years, various sensor‐based diagnostic systems outperforming conventional pathogenic diagnostic assays have been developed. Among them, colorimetric biosensors detecting target molecules by the naked eye have attracted much attention due to their simplicity, practicality, and cost‐effectiveness. Recently, nanomaterials have been adopted as a versatile signal transduction and amplification tool for rapid and sensitive detection of pathogenic bacteria and viruses. Here, recent trends and advances are reviewed in detecting and diagnosing pathogenic bacteria and viruses using colorimetric biosensors employing various nanomaterials. In addition, it is discussed how nanomaterials and bioreceptors can be better integrated together to develop rapid and sensitive colorimetric detection system in the future.
Keywords: bacteria, colorimetric biosensors, nanomaterials, pathogens, viruses
Colorimetric diagnostic systems for detecting pathogenic bacteria and viruses using various nanomaterials are reviewed. Nanomaterials integrated into colorimetric detection systems allow rapid, sensitive, and convenient detection of pathogens through their unique catalytic activities, aggregation, destabilization, and other characteristics. Advantages and limitations of nanomaterial‐based colorimetric diagnostic systems are discussed together with future directions of research.
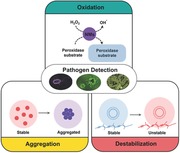
1. Introduction
Infectious diseases caused by pathogenic microorganisms, such as food‐borne (e.g., Clostridium botulinum) and water‐borne (e.g., Vibrio cholera) pathogens and nosocomial pathogens (e.g., methicillin‐resistant Staphylococcus aureus), and viruses (e.g., influenza, Middle East respiratory syndrome coronavirus, and Zika virus) are an increasingly serious threat to public health and global economy.1, 2 According to the World Health Organization (WHO), for example, influenza spreads rapidly and easily to become an epidemic disease, which causes 250 000–500 000 deaths every year around the world. In addition, infectious diseases caused by food‐borne pathogens are an important cause of morbidity and mortality. WHO reported that food‐borne diseases caused 240 000 deaths globally in 2010. In order to prevent spreading of such infectious diseases and better treat patients with the right therapeutics at the right time, development of rapid, accurate, and sensitive methods to detect and identify pathogens at the early stage is important.
Various conventional approaches to detect and identify pathogenic bacteria and viruses have been developed and employed in the field. These approaches include culture and colony‐counting methods, enzyme‐linked immunosorbent assay, and polymerase‐chain‐reaction‐based methods.3, 4, 5, 6, 7 However, these conventional methods are rather time‐consuming, labor‐intensive, and often require special and expensive equipments.8, 9 Due to such limitations, WHO suggests a strong need to develop diagnostic methods that meet so‐called “ASSURED” criteria for diagnostics, meaning affordable, sensitive, specific, user friendly, robust and rapid, equipment free, and deliverable to those who need them.10, 11 To meet “ASSURED” criteria, many different diagnostic methods based on creative systems including optical sensors, electrical sensors, microfluidic devices, DNA microarrays, and NMR sensors have been developed.3, 12, 13, 14, 15, 16, 17, 18, 19 Among them, optical biosensors are preferred because they are easy to implement while meeting “ASSURED” criteria. Various types of optical biosensors including surface plasmon resonance, localized surface plasmon resonance, surface‐enhanced Raman scattering, colorimetric, and photoluminescence methods have been reported.20, 21, 22 Among these, colorimetric biosensors are attractive because the presence of target pathogens can be detected rapidly, easily, selectively, and cost‐effectively by the color change, even by the naked eye.23 However, several limitations, such as low sensitivity, complexity, and the large sample volume required, should be overcome to replace conventional diagnostic methods.
To overcome these limitations of colorimetric detection methods, nanomaterials (NMs), including nanoparticles (NPs), nanorods, nanowires, and other nanostructured materials, have been utilized as a platform in biosensor development due to their excellent chemical, physical, optical, and electronical properties.24, 25, 26 For example, NMs have superior surface‐to‐volume ratio, and some of them exhibit desirable electrical, magnetic, and/or plasmonic characteristics.27 With these unique physical and chemical properties of NMs, various NM‐based biosensors for detecting pathogens have been developed.28, 29 NMs have also been used in signal amplification, labeling, and separation of molecules in colorimetric detection of pathogens.29, 30 Different types of NMs such as gold (Au) NP, copper (Cu) NP, cadmium sulfide quantum dots, and magnetic NMs (MNMs) have been investigated for the development of rapid and sensitive optical sensors.31, 32, 33, 34, 35, 36, 37 In particular, noble metals such as Au and silver (Ag) NMs with unique optical properties are used to induce a distinct color change by aggregation, morphology transition, and chemical reaction on the surface of NMs.38, 39
Here, we review the recent developments made in the field of colorimetric detection of pathogens (Table 1 ). Rather than extensively reviewing the field, which can be found from other review papers, here we present key aspects of colorimetric diagnosis systems employing NMs. Colorimetric detection methods recently developed can be largely grouped into three categories including oxidation by peroxidase‐like NMs, aggregation of NMs, and destabilization of NM structure, which are discussed with examples. Also, future perspectives on developing colorimetric biosensors with enhanced NMs are provided.
Table 1.
Summary of recent studies on colorimetric detection of pathogenic bacteria and viruses using various NMs
| Colorimetric strategy | Target pathogen | Nanomaterials | Functionalized biomolecules | Assay time | Detection limit | Reference |
|---|---|---|---|---|---|---|
| Oxidation by peroxidase‐like NMs | Escherichia coli O157:H7 | Pt‐coated Fe3O4 NPa) | Antibody | 30 min | 10 CFUb) mL−1 | 48 |
| Salmonella typhimurium | Fe3O4 NP | DNA aptamers | 10 min | 7.5 × 105 CFU mL−1 | 49 | |
| Influenza A (H1N1) virus | Au NP | Antibody | 1 min | 10.79 pg mL−1 | 55 | |
| Influenza A (H3N2) virus | Au NP | Antibody | 1 min | 11.62 PFUc) mL−1 | 55 | |
| Influenza A (H3N2) virus | CNTd)‐Au nanohybrid | Antibody | 10 min | 3.4 PFU mL−1 | 56 | |
| Norovirus | Graphene‐Au nanohybrid | Antibody | 10 min | 92.7 pg mL−1 | 57 | |
| Aggregation of NMs | Acetobacter aceti | Ag, Au NP | Citrate ions | 20 min | 0.05 OD600 e) in 200 µL | 74 |
| Bacillus species | Ag, Au NP | Citrate ions | 20 min | 0.05 OD600 in 200 µL | 74 | |
| B. natto | Ag, Au NP | Citrate ions | 20 min | 0.05 OD600 in 200 µL | 74 | |
| B. subtilis | Au NP | Protein | – | 4.5 × 103 CFU mL−1 | 61 | |
| CRPA (carbapenem‐resistant Pseudomonas aeruginosa) | Ag, Au NP | Citrate ions | 20 min | 0.05 OD600 in 200 µL | 74 | |
| E. coli | Au NP | Protein | 10 min | 8 CFUs mL−1 | 65 | |
| E. coli | Ag, Au NP | Citrate ions | 20 min | 0.05 OD600 in 200 µL | 74 | |
| E. coli O157:H7 | Au NP | Antibody | 4 h | 3 ± 1 bacteria mL−1 | 63 | |
| Lactobacillus species | Au NP | Antibody | 1 min | 105 CFU mL−1 | 59 | |
| Listeria monocytogenes | Au NP | Antibody | 4 h | 3 ± 1 bacteria mL−1 | 63 | |
| Mycobacterium tuberculosis | Au NP | DNA | 30 min | 1 pmol of synthetic DNA | 64 | |
| Plasmodium falciparum | Au NP | DNA | 30 min | 1 pmol of synthetic DNA | 64 | |
| Rhodopseudomonas | Ag, Au NP | Citrate ions | 20 min | 0.05 OD600 in 200 µL | 74 | |
| Salmonella aureus | Au NP | Protein | 10 min | 8 CFUs mL−1 for E. coli | 65 | |
| S. typhimurium | Au NP | Antibody | 4 h | 3 ± 1 bacteria mL−1 | 63 | |
| S. typhimurium | Au NP | DNA aptamer | – | 56 CFU mL−1 | 69 | |
| Staphylococcus | Ag, Au NP | Citrate ions | 20 min | 0.05 OD600 in 200 µL | 74 | |
| S. aureus | Au NP | Antibody | 1 min | 120 CFU mL−1 | 59 | |
| Cyprinid herpesvirus‐3 (CyHV‐3) | Au NP | DNA | 20 min | 10 fg CyHV‐3‐Au NP DNA (30 virions) | 67 | |
| Human papillomavirus | Ag NP | PNAf) | – | 1.03 × 10−9 m | 70 | |
| Influenza A (H1N1) virus | Au NP | Antibody | 20 min | 7.6 HAUg) | 60 | |
| Influenza A (H3N2) virus | Au NP | Antibody | 20 min | 7.6 HAU | 60 | |
| Influenza A (H1N1) virus | Glycan‐functionalized Au NP | Antibody | 90 min | 8 HAh) | 62 | |
| Influenza A (H5N1) virus | Glycan‐functionalized Au NP | Antibody | 90 min | 8 HA | 62 | |
| Middle East respiratory syndrome coronavirus | Ag NP | PNA | – | 1.53 × 10−9 m | 70 | |
| Mycobacterium tuberculosis | Ag NP | PNA | – | 1.27 × 10−9 m | 70 | |
| Destabilization of NMs | Salmonella choleraesuis | PDAi) | Lysine | 48 h | 1–10 CFU mL−1 | 77 |
| Influenza A (H1N1) virus | PEP‐PDAj) | Peptide | 5 min | 105 PFU | 76 |
NP: nanoparticle
CFU: colony‐forming units
PFU: plaque‐forming units
CNT: carbon nanotube
OD600: optical density at a wavelength of 600 nm
PNA: peptide nucleic acid
HAU: hemagglutinin units per 50 µL
HA: hemagglutinin
PDA: polydiacetylene
PEP‐PDA: peptide‐functionalized PDA.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2. Colorimetric Detection Based on Oxidation by Peroxidase‐Like NMs
Enzymes have been used as a signal‐transduction tool in colorimetric assays due to their high sensitivity and specificity.40, 41 However, there are several limitations of employing enzymes in biosensor systems, such as high cost, short lifetime, and limited operational conditions.42 As an alternative approach, several NMs possessing enzyme‐like characteristics have been developed as artificial enzymes.43, 44, 45 Currently, Au NP, platinum (Pt) NP, MNMs, and nanohybrids based on carbon nanotubes (CNTs) and graphene oxide are utilized as peroxidase‐like mimics for enhanced colorimetric detection of pathogenic bacteria and viruses (Figure 1 A). These NMs can decompose the O—O bond of H2O2 into an OH• radical with high oxidizing capability.44, 46 The OH• radical generated then can oxidize a peroxidase substrate such as 3,3′,5,5′‐tetramethyl‐benzidine (TMB) and 2,2′‐azino‐bis (3‐ethylbenzothiazoline‐6‐sulfonic acid) (ABTS2−) with H2O2, yielding blue and green colors, respectively, in aqueous solution.
Figure 1.
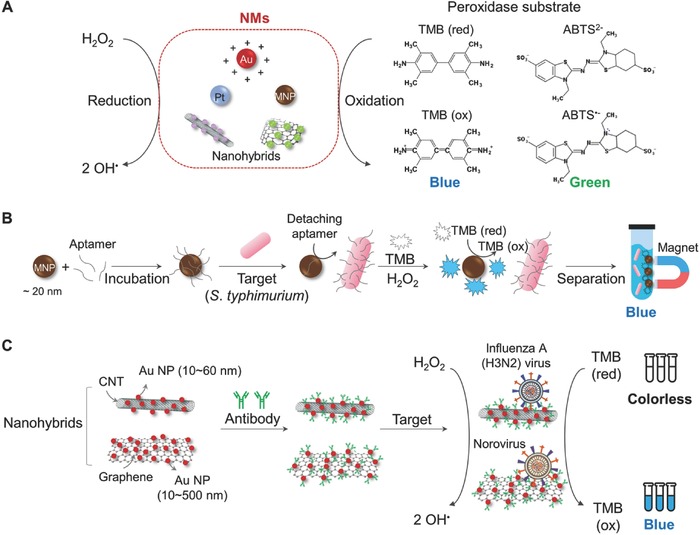
Schematic illustration of colorimetric detection based on oxidation by peroxidase‐like NMs. A) Scheme of oxidizing peroxidase substrates by various peroxidase‐like NMs. B) Schematic to detect S. typhimurium using aptamer‐adsorbed MNPs. C) Detection of influenza (H3N2) virus using a nanohybrid of CNTs and Au NPs, and norovirus‐like particles in human serum using a nanohybrid of graphene and Au NPs. MNP: magnetic nanoparticle; TMB: 3,3′,5,5′‐tetramethyl‐benzidine; ABTS: 2,2′‐azino‐bis(3‐ethylbenzothiazoline‐6‐sulfonic acid); CNT: carbon nanotube. The contents of (B) and (C) are described in refs. 49, 56, 57 and they are redrawn here.
Peroxidase‐like NMs conjugated with various pathogen‐specific bioreceptors, such as antibodies, DNA probes, and DNA aptamers, have been utilized for specific detection of pathogenic bacteria and viruses.47 For example, Pt was functionalized with antibodies to detect Escherichia coli.48 In this assay, the peroxidase activity of Pt induced a color change from colorless (TMB) to blue (oxidized TMB). As another example, MNP (Fe3O4, ≈20 nm) coupled with a Salmonella typhimurium‐specific aptamer through adsorption was utilized to successfully detect S. typhimurium (Figure 1B).49 Here, MNP served as a peroxidase‐like NM converting TMB (colorless) to oxidized TMB (blue).50, 51 Single‐stranded DNA (ssDNA) aptamers with negatively charged phosphate backbone were adsorbed on the surface of positively charged MNP, without any surface modification. Adsorbed aptamers were detached from MNP and bound to the target pathogen, exposing the surface of MNP to H2O2. Exposed MNP then generates the OH• radical, which converts TMB to oxidized TMB, making a colorless‐to‐blue change upon pathogen presence. This method requires several steps: binding of specific ssDNA aptamers desorbed from the MNP–aptamer complex to the corresponding pathogenic cells in the reaction solution, removal of the reaction solution containing pathogen–aptamers with the MNPs being held by a magnet, and then addition of a colorimetric detection mixture (H2O2 and TMB) to induce a color change (Figure 1B). This method is advantageous as only a small amount of the colorimetric detection mixture is needed for a visible and clear color change, and there is no need for surface modification on MNPs for attaching ssDNA aptamers. However, it is compromised by the inconvenience of multiple steps as described above. When the mechanism of peroxidase activity of MNP is better understood through further studies, MNPs as a colorimetric sensing platform will find various applications in diagnostics.
Nanohybrids exhibit synergetic effects on significantly enhancing both catalytic (e.g., peroxidase‐like) activities and detection capabilities of biosensors.52 Since the assay time of enzymatic oxidation methods is highly dependent on catalytic performance, nanohybrids with enhanced catalytic properties of NMs have gained much interest. In particular, CNTs and graphene have received much attention for biosensor applications due to their outstanding electrical properties, based on high surface area and great electron mobility.52 When hybridized with other NMs, the high conductivity of CNTs and graphene significantly enhances catalytic activity.53, 54 Among various NMs, Au NPs are one of the most widely used NMs hybridized with CNTs or graphene due to their unique optical properties and peroxidase‐mimicking activities.55 The use of CNT or graphene hybridized with Au NPs showed further improved colorimetric‐sensing performance. For example, sensitive detection of influenza A (H3N2) virus was possible using CNT–Au NP nanohybrid conjugated with influenza A (H3N2) hemagglutinin (HA) monoclonal antibodies. (Figure 1C).56 The limit of detection (LOD) of the CNT–Au NP nanohybrid system was 3.4 plaque‐forming units (PFU) per mL, which was several hundred times more sensitive than conventional enzyme‐linked immunosorbent assay. In another example, norovirus in human serum could be sensitively detected by enhanced peroxidase‐like activity of a graphene–Au NP nanohybrid (Figure 1C).57 The LOD of the graphene–Au NP nanohybrid system was as low as 92.7 pg mL−1, which was 41 times lower than that could be detected with commercialized norovirus‐detection kits. Most studies employing nanohybrids for colorimetric diagnostics have so far been limited to utilizing Au NPs. Thus, it will be desirable to investigate other NMs having different properties in making nanohybrids with CNT and graphene so that new nanohybrid‐based colorimetric diagnostic systems having improved catalytic activities and consequently enhanced sensing performance can be developed for the efficient detection of even more diverse pathogenic bacteria and viruses.
3. Colorimetric Detection Based on Aggregation of NMs
As another approach to using NMs in colorimetric methods, direct use of NMs has been investigated by taking advantage of their unique optical properties. Electrostatic aggregation of some NMs induces a color change depending on the size of NMs and their degree of aggregation.58 The change of absorbance occurs within the range of visible light so that it can be observed by the naked eye. This visual color change upon the aggregation of NMs provides an easy and practical platform for the colorimetric detection of pathogenic bacteria and viruses (Figure 2 A). Among the various NMs, Au and Ag NMs have been most widely used in aggregation‐based colorimetric assays. Stabilized solutions of Au NPs and Ag NPs exhibit red and yellow colors, respectively, which are changed to blue (or purple) and red, respectively, upon their aggregation. To control the aggregation degree and enhance the sensitivity and specificity of NM binding to the target pathogenic bacteria and viruses, the surface of NMs is modified with biomolecules such as DNA, RNA, antibodies, and proteins, and small molecules (Figure 2A).
Figure 2.
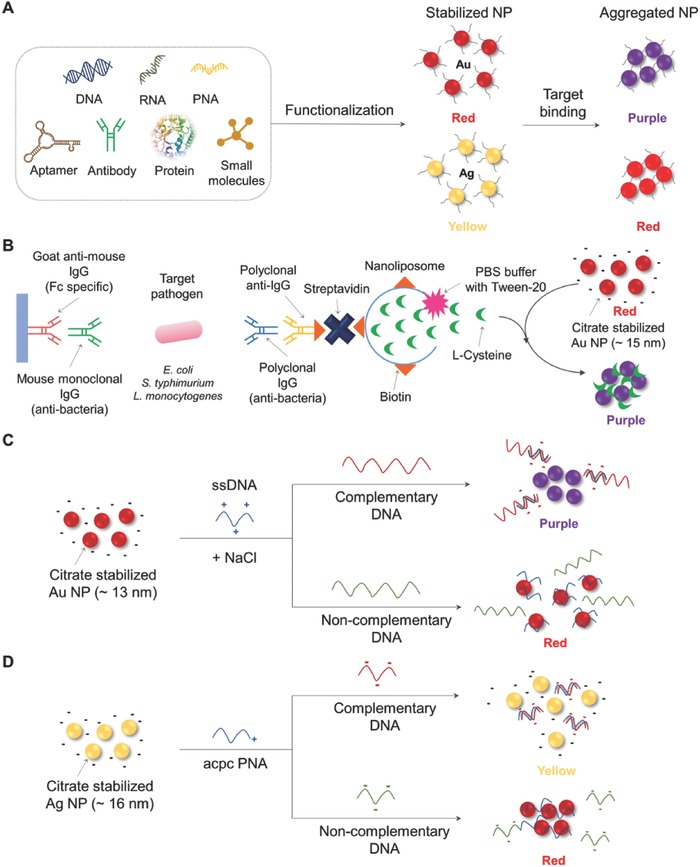
Schematic illustration of colorimetric detection based on aggregation of NMs. A) Scheme of aggregation of NPs with various biomolecular functionalization. B) Signal‐amplified detection of E. coli, S. typhimurium, and L. monocytogenes using cysteine‐containing nanoliposomes (redrawn from ref. 63). C) Schematic to detect the Cy‐HV3 DNA using aggregation of citrate‐stabilized Au NP in the presence of dsDNA (redrawn from ref. 67). D) Schematic to detect Middle East respiratory syndrome coronavirus DNA using redispersion of Ag NP in the presence of dsDNA (redrawn from ref. 70). PNA: peptide nucleic acid; IgG: immunoglobulin G; PBS: phosphate buffer saline; Cy‐HV3: Cyprinid herpesvirus‐3; ssDNA: single‐stranded DNA; acpc PNA: (2S)‐aminocyclopentane‐(1S)‐carboxylic acid pyrrolidinyl PNA.
Interaction of bioreceptor‐modified NMs with target molecules induces aggregation, generating a visible color change. For example, pathogen‐specific antibodies and proteins have been conjugated on the surface of Au NPs.43 An antibody–antigen binding induces aggregation of Au NP around the pathogens, which generates a colorimetric response.43, 59 To conjugate proteins on the surface of Au NPs, surface modification of the Au NPs is performed, for example, with N‐hydroxysuccinimide (NHS), which can interact with the amine groups in antibodies and proteins.59, 60, 61 Also, Au NPs functionalized with a virus‐specific polysaccharide (glycan) have been developed for detecting avian‐adapted influenza (vieH5N1) virus and human‐adapted influenza (shaH1N1) virus.62 However, steric hindrance of bulky bioreceptors limits the amount of bioreceptors (e.g., low bioreceptor concentration) that can be conjugated onto NMs, which hampers sensitive colorimetric detection. To overcome this limitation, several attempts have been made to increase the sensitivity of the colorimetric method. For example, nanoliposomes were used as a signal‐amplification tool to sensitively detect food‐borne pathogens (Figure 2B).63 Since one nanoliposome contains many cysteine residues, which can induce aggregation of Au NPs, even one target pathogen could successfully be detected. Manufacturing the nanoliposome system requires several steps involving binding and washing processes, as shown in Figure 2B; (i) goat anti‐mouse IgG antibodies (Fc specific) are coated on polystyrene microwell plate; (ii) mouse monoclonal antibodies (pathogen specific) are introduced for increasing specificity and reducing cross‐reactivity; (iii) the target pathogens (E. coli O157:H7, S. typhimurium, and Listeria monocytogenes) are allowed to bind to the monoclonal antibodies; (iv) polyclonal antibodies (pathogen specific) are bound to target pathogens in the sandwich mode; and (v) biotinylated IgG molecules are introduced for binding streptavidin, which can bind biotin‐modified nanoliposomes. This multistep process required for diagnostics is a major disadvantage of using this system for a wide range of applications, which urges development of simpler fabrication methods.
The above methods require manufacturing of bioreceptor–NM hybrids, which requires modification of NMs and/or bioreceptors, which is complex and time consuming. Thus, development of a simple fabrication method using unmodified NMs has been pursued for facilitating commercialization of colorimetric pathogenic detection assays.3 For example, thiol–Au bonding is widely adopted as a DNA conjugation technique due to its high bonding strength.64, 65 However, modification of bioreceptors (e.g., DNA and protein) is required. Thus, adsorption of unmodified DNA or peptide nucleic acid (PNA) to unmodified Au NPs has been utilized to detect pathogenic bacteria and viruses.66 For example, DNA of Cyprinid herpesvirus‐3 was successfully detected with citrate‐stabilized Au NPs (Figure 2C).67 Also, dsDNA and ssDNA can be differentiated by colorimetric assay in the presence of salt (e.g., NaCl) due to their different electrostatic properties. While dsDNA forms a stable double‐helix geometry with the negatively charged phosphate backbone, ssDNA with a relatively flexible phosphate backbone exposes its bases to Au NPs.68 Thus, ssDNA can be adsorbed onto the surface of negatively charged Au NPs by van der Waals attractions, resulting in the formation of a colloidal suspension. On the other hand, dsDNA cannot be adsorbed. Thus, in the presence of herpesvirus‐3 DNA, the probe ssDNA binds to form dsDNA, causing aggregation of Au NPs and consequently visible color change from red to purple. A similar strategy was applied to detect S. typhimurium using Au NP adsorbed with ssDNA aptamers.69
In another case, (2S)‐aminocyclopentane‐(1S)‐carboxylic acid pyrrolidinyl peptide nucleic acid (acpc PNA) was adsorbed on the surface of citrate‐stabilized Ag NP to detect Middle East respiratory syndrome coronavirus, Mycobacterium tuberculosis, and human papillomavirus (Figure 2D).70 The acpc PNA probe contains a single positive charge that causes aggregation of negatively charged Ag NPs, resulting in color change from yellow to red. When acpc PNAs are bound to the target DNAs, Ag NPs are stabilized in the solution. This system was integrated to make paper‐based analytical devices, which are widely used for the point‐of‐care platform with simplicity, low‐cost, portability, and disposability.71, 72, 73
Interestingly, detection and differential identification of pathogens including carbapenem‐resistant Pseudomonas aeruginosa, Acetobacter aceti, Rhodopseudomonas, Bacillus, Staphylococcus, and E. coli were also possible using unmodified Ag NP and Au NP without bioreceptors.74 This was based on unique fingerprint UV–vis absorbance caused by interactions between NPs and proteins originating from different pathogens (e.g., surface proteins and excreted proteins). Although the sensitivity and accuracy of detecting diverse pathogens using this method might not be as high as the other methods employing specific bioreceptors, the simpler fabrication offered by this method deserves further studies for improved performance.
4. Colorimetric Detection Based on Destabilization of NM Structure
Methods for colorimetric detection of pathogens based on the change and destabilization of NM structure have also been developed. One of the most popularly used colorimetric biosensing NM structures is based on polydiacetylene (PDA) vesicles with unique chromatic properties.61, 75 Blue‐colored PDA vesicles are synthesized by 1,4‐photopolymerization of self‐assembled diacetylene monomers (e.g., 10,12‐pentacosadyionic acid (PDCA)). For the detection of pathogens, pathogen‐specific antibodies, aptamers, and/or peptides are attached to PDA vesicles as shown in Figure 3 A. Upon binding of corresponding pathogens to the bioreceptor–PDA vesicle, a color change from blue to red occurs (Figure 3A). For example, a peptide‐functionalized PDA (PEP–PDA) vesicle system was developed to detect influenza H1N1 virus by the naked eye (Figure 3B).76 In the presence of the H1N1 virus, the color of the PEP–PDA vesicles changes from blue to red due to the increase of the energy bandgap of PDA. The high affinity of the functionalized peptide sequences to the HA protein of the influenza H1 strain makes the PEP–PDA vesicles highly specific to the H1N1 virus.
Figure 3.
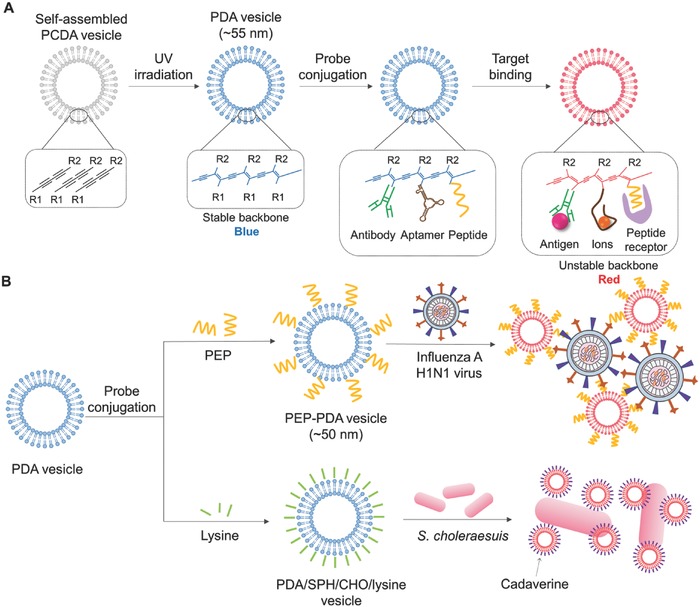
Schematic illustration of colorimetric detection based on destabilization of NM structure. A) Scheme of a color transition of PDA vesicles by their structure change on target binding. B) Scheme of detecting influenza H1N1 virus with PEP–PDA vesicle system (redrawn from ref. 76), and detecting Salmonella choleraesuis with PDA/SPH/CHO/lysine vesicle system (strategy described in ref. 77). PCDA: 10,12‐pentacosadyionic acid; PDA: polydiacetylene; polydiacetylene; PEP: HA1‐specific peptide; SPH: sphingomyelin; CHO: cholesterol.
In another recent study, a PDA/sphingomyelin (SPH)/cholesterol (CHO)/lysine vesicle system was used to detect Salmonella choleraesuis in chicken (Figure 3B).77 To fabricate a blue PDA/SPH/CHO/lysine vesicle, a mixture of a self‐assembled PCDA vesicle, SPH, CHO, and lysine was photopolymerized by UV irradiation (254 nm). SPH and CHO were introduced for improving the colorimetric response of PDA vesicles. Lysine is decarboxylated to cadaverine by pyridoxal phosphate‐containing enzymes in S. choleraesuis. Consequently, cadaverine destabilizes the PDA backbone, resulting in a color change of the PDA/SPH/CHO/lysine vesicles from blue to red. Although highly sensitive detection (e.g., ≈10 cells) using PDA vesicle could be achieved, it required long assay time (e.g., 48 h). Thus, further studies are required to significantly reduce the assay time while maintaining or further improving the sensitivity.
5. Conclusion and Future Perspectives
Here, we have reviewed recent studies on colorimetric diagnostics of pathogenic bacteria and viruses utilizing various NMs with unique optical, electrical, and catalytic properties. As described above, sensitivity and specificity of colorimetric pathogen diagnostics can be enhanced by employing NMs as signal transduction and amplification tools. However, there are still challenges remaining to further enhance performance of NM‐based colorimetric diagnostic systems.
First, various NMs of low cost need to be investigated. Noble metal (Au, Ag, and Pt) NMs with unique optical, electrical, and catalytic properties give significantly enhanced colorimetric sensing performance, but the systems can be too expensive for large‐scale applications in clinical and environmental settings. As not all NMs exhibit the unique optical properties required for colorimetric responses, testing of various NMs for their suitability will be the first step. For example, white or colorless silicon (Si) NPs are not suitable for use in colorimetric detection, although Si is one of the most abundant elements on the Earth. For making colorless Si NPs colorful, doping of various organic dyes on Si NPs has recently been reported and utilized in colorimetric diagnostics.10, 78
Second, optimizing the size and shape of NMs needs to be performed for enhancing the sensitivity and selectivity of colorimetric pathogenic diagnostics. Properties of NMs, such as catalytic properties, are often dependent on the size and shape of NMs.25, 79 However, there have been only few studies on optimizing the size and shape of NMs in colorimetric biosensors.56, 57, 80 In general, decreasing the size of NMs increases the specific surface area of NMs, resulting in enhanced properties, including peroxidase‐like activities.57 Thus, optimization of the size and shape of NMs will lead to the development of an improved system showing a colorimetric sensing performance.
Third, simple methods for fabricating colorimetric diagnostic systems need to be developed for facilitating commercialization. Conjugation/hybridization of target‐specific bioreceptors on the surface of NMs is a necessary step in making a colorimetric diagnostic system. However, methods for modifying NMs and bioreceptors and also conjugation are time consuming and labor intensive.81 Thus, integration of NM synthesis, modification of NMs and bioreceptors, and their conjugation in one‐step or fewer steps than conventional fabricating calorimetric diagnostic methods will significantly reduce the time, effort, and costs required for fabricating the diagnostic system. Modification of NMs with small molecules (e.g., citrate and amino acid residues) during the synthesis of NMs has already been reported.82 Thus, one can naturally come up with an idea of conjugating/hybridizing target‐specific bioreceptors (e.g., DNA, RNA, antibodies, and proteins) during the synthesis of NMs. However, NM synthesis is mostly performed under rather harsh conditions, which can denature or degrade such bioreceptor molecules of interest. Thus, it is of great interest to use biosynthesized NMs, as their biosynthesis steps are compatible with bioreceptor conjugation/hybridization.83 We and others have reported biosynthesis of various metal NPs using various biological systems.84, 85 As such strategies and methods are being continuously developed, it is expected that rapid, sensitive, selective, convenient, and cost‐effective colorimetric detection systems will be developed and more widely applied for the diagnosis of a more diverse range of pathogens and also other agents and molecules of interest.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
Y.C. and J.H.H. contributed equally to this work. The authors thank Seung Min Yoo for her advice in planning the manuscript. This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea (NRF‐2012M1A2A2026556 and NRF‐2012M1A2A2026557).
Biographies
Yoojin Choi is a Ph.D. student in the Department of Chemical and Biomolecular Engineering, KAIST. She is working on biological synthesis of nanomaterials and their applications in various fields. Prior to undertaking her Ph.D. study, she received her M.A. in environmental science and engineering at the Gwangju Institute of Science and Technology (GIST) and B.A. in environmental and chemical engineering at Chonbuk National University in the Republic of Korea.

Ji Hyeon Hwang is an M.S. student in the Department of Chemical and Biomolecular Engineering, KAIST. She is working on biosensors and pathogen diagnostics. Prior to undertaking her M.S. study, she received her B.A. in chemical and biological engineering at Korea University in the Republic of Korea.

Sang Yup Lee is a distinguished professor in the Department of Chemical and Biomolecular Engineering, KAIST. He is currently the Dean of KAIST Institutes, Director of BioProcess Engineering Research Center, and Director of Bioinformatics Research Center. His research interests are metabolic engineering, systems biology and biotechnology, industrial biotechnology, synthetic biology, and nanobiotechnology.

Choi Y., Hwang J. H., Lee S. Y., Small Methods 2018, 2, 1700351 10.1002/smtd.201700351
References
- 1. Global Burden of Foodborne Diseases, WHO, Geneva, Switzerland 2015.
- 2. WHO, Pandemic and Epidemic Diseases http://www.who.int/csr/disease/en/ (accessed: November 2017).
- 3. Yoo S. M., Lee S. Y., Trends Biotechnol. 2016, 34, 7.26506111 [Google Scholar]
- 4. Singh R., Mukherjee M. D., Sumana G., Gupta R. K., Sood S., Malhotra B. D., Sens. Actuators, B 2014, 197, 385. [Google Scholar]
- 5. Lifson M. A., Ozen M. O., Inci F., Wang S., Inan H., Baday M., Henrich T. J., Demirci U., Adv. Drug Delivery Rev. 2016, 103, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bridle H., Balharry D., Gaiser B., Johnston H., Environ. Sci. Technol. 2015, 49, 10762. [DOI] [PubMed] [Google Scholar]
- 7. Vidic J., Manzano M., Chang C.‐M., Jaffrezic‐Renault N., Vet. Res. 2017, 48, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Velusamy V., Arshak K., Korostynska O., Oliwa K., Adley C., Biotechnol. Adv. 2010, 28, 232. [DOI] [PubMed] [Google Scholar]
- 9. Ahmed A., Rushworth J. V., Hirst N. A., Millner P. A., Clin. Microbiol. Rev. 2014, 27, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun Q., Zhao G., Dou W., Anal. Methods 2015, 7, 8647. [Google Scholar]
- 11. Peeling R. W., Holmes K. K., Mabey D., Ronald A., Sex. Transm. Infect. 2006, 82, v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mannoor M. S., Zhang S., Link A. J., McAlpine M. C., Proc. Natl. Acad. Sci. USA 2010, 107, 19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu M., Wang R., Li Y., Talanta 2017, 162, 511. [DOI] [PubMed] [Google Scholar]
- 14. Ahn J.‐H., Choi S.‐J., Im M., Kim S., Kim C.‐H., Kim J.‐Y., Park T. J., Lee S. Y., Choi Y.‐K., Appl. Phys. Lett. 2017, 111, 113701. [Google Scholar]
- 15. Yoo S. M., Baek Y.‐K., Shin S. H. R., Kim J.‐H., Jung H.‐T., Choi Y.‐K., Lee S. Y., J. Nanosci. Nanotechnol. 2016, 16, 6520. [DOI] [PubMed] [Google Scholar]
- 16. Huh Y. S., Park T. J., Lee E. Z., Hong W. H., Lee S. Y., Electrophoresis 2008, 29, 2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoo S. M., Keum K. C., Yoo S. Y., Choi J. Y., Chang K. H., Yoo N. C., Yoo W. M., Kim J. M., Lee D., Lee S. Y., Biotechnol. Bioprocess Eng. 2004, 9, 93. [Google Scholar]
- 18. Zhao Y., Li Y., Jiang K., Wang J., White W. L., Yang S., Lu J., Food Control 2017, 71, 110. [Google Scholar]
- 19. Idil N., Hedström M., Denizli A., Mattiasson B., Biosens. Bioelectron. 2017, 87, 807. [DOI] [PubMed] [Google Scholar]
- 20. Nie S., Emory S. R., Science 1997, 275, 1102. [DOI] [PubMed] [Google Scholar]
- 21. Zheng S., Kim D.‐K., Park T. J., Lee S. J., Lee S. Y., Talanta 2010, 82, 803. [DOI] [PubMed] [Google Scholar]
- 22. Park T. J., Lee S. J., Kim D.‐K., Heo N. S., Park J. Y., Lee S. Y., Talanta 2012, 89, 246. [DOI] [PubMed] [Google Scholar]
- 23. Piriya V. S. A., Joseph P., Daniel S. C. G. K., Lakshmanan S., Kinoshita T., Muthusamy S., Mater. Sci. Eng., C 2017, 78, 1231. [DOI] [PubMed] [Google Scholar]
- 24. Kelly K. L., Coronado E., Zhao L. L., Schatz G. C., J. Phys. Chem. B 2003, 107, 668. [Google Scholar]
- 25. Ray P. C., Chem. Rev. 2010, 110, 5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yin T., Qin W., TrAC, Trends Anal. Chem. 2013, 51, 79. [Google Scholar]
- 27. Fang Y., Ramasamy R., Biosensors 2015, 5, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rowland C. E., Brown C. W., Delehanty J. B., Medintz I. L., Mater. Today 2016, 19, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mustafa F., Hassan R. Y., Andreescu S., Sensors 2017, 17, 2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou C.‐H., Zhao J.‐Y., Pang D.‐W., Zhang Z.‐L., Anal. Chem. 2014, 86, 2752. [DOI] [PubMed] [Google Scholar]
- 31. Wang J., Wu X., Wang C., Rong Z., Ding H., Li H., Li S., Shao N., Dong P., Xiao R., Wang S., ACS Appl. Mater. Interfaces 2016, 8, 19958. [DOI] [PubMed] [Google Scholar]
- 32. Kang T., Yoo S. M., Yoon I., Lee S. Y., Kim B., Nano Lett. 2010, 10, 1189. [DOI] [PubMed] [Google Scholar]
- 33. Cao Y. C., Jin R., Mirkin C. A., Science 2002, 297, 1536. [DOI] [PubMed] [Google Scholar]
- 34. Oh S. Y., Heo N. S., Shukla S., Cho H.‐J., Vilian A. T. E., Kim J., Lee S. Y., Han Y.‐K., Yoo S. M., Huh Y. S., Sci. Rep. 2017, 7, 10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim D.‐K., Yoo S. M., Park T. J., Yoshikawa H., Tamiya E., Park J. Y., Lee S. Y., Anal. Chem. 2011, 83, 6215. [DOI] [PubMed] [Google Scholar]
- 36. Donmez M., Yilmaz M. D., Kilbas B., J. Hazard. Mater. 2017, 324, 593. [DOI] [PubMed] [Google Scholar]
- 37. Mocan T., Matea C. T., Pop T., Mosteanu O., Buzoianu A. D., Puia C., Iancu C., Mocan L., J. Nanobiotechnol. 2017, 15, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Halas N. J., Lal S., Chang W.‐S., Link S., Nordlander P., Chem. Rev. 2011, 111, 3913. [DOI] [PubMed] [Google Scholar]
- 39. Jain P. K., Huang X., El‐Sayed I. H., El‐Sayed M. A., Acc. Chem. Res. 2008, 41, 1578. [DOI] [PubMed] [Google Scholar]
- 40. Kim H.‐S., Oh B.‐K., BioChip J. 2014, 8, 1. [Google Scholar]
- 41. Wang X., Wu L., Ren J., Miyoshi D., Sugimoto N., Qu X., Biosens. Bioelectron. 2011, 26, 4804. [DOI] [PubMed] [Google Scholar]
- 42. Yang H.‐H., Zhang S.‐Q., Chen X.‐L., Zhuang Z.‐X., Xu J.‐G., Wang X.‐R., Anal. Chem. 2004, 76, 1316. [DOI] [PubMed] [Google Scholar]
- 43. Verma M. S., Rogowski J. L., Jones L., Gu F. X., Biotechnol. Adv. 2015, 33, 666. [DOI] [PubMed] [Google Scholar]
- 44. Gao L., Zhuang J., Nie L., Zhang J., Zhang Y., Gu N., Wang T., Feng J., Yang D., Perrett S., Yan X., Nat. Nanotechnol. 2007, 2, 577. [DOI] [PubMed] [Google Scholar]
- 45. Dong Y.‐L., Zhang H.‐G., Rahman Z. U., Su L., Chen X.‐J., Hu J., Chen X.‐G., Nanoscale 2012, 4, 3969. [DOI] [PubMed] [Google Scholar]
- 46. Jv Y., Li B., Cao R., Chem. Commun. 2010, 46, 8017. [DOI] [PubMed] [Google Scholar]
- 47. Chen J., Andler S. M., Goddard J. M., Nugen S. R., Rotello V. M., Chem. Soc. Rev. 2017, 46, 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kwon D., Lee S., Ahn M. M., Kang I. S., Park K.‐H., Jeon S., Anal. Chim. Acta 2015, 883, 61. [DOI] [PubMed] [Google Scholar]
- 49. Park J. Y., Jeong H. Y., Kim M. I., Park T. J., J. Nanomaterials 2015, 2015, 2. [Google Scholar]
- 50. Park K. S., Kim M. I., Cho D.‐Y., Park H. G., Small 2011, 7, 1521. [DOI] [PubMed] [Google Scholar]
- 51. Woo M.‐A., Kim M., Jung J., Park K., Seo T., Park H., Int. J. Mol. Sci. 2013, 14, 9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khalil I., Julkapli M. N., Yehye A. W., Basirun J. W., Bhargava K. S., Materials 2016, 9, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. González V. J., Martín‐Alberca C., Montalvo G., García‐Ruiz C., Baselga J., Terrones M., Martin O., Carbon 2014, 78, 10. [Google Scholar]
- 54. Guo S., Sun S., J. Am. Chem. Soc. 2012, 134, 2492. [DOI] [PubMed] [Google Scholar]
- 55. Ahmed S. R., Kim J., Suzuki T., Lee J., Park E. Y., Biotechnol. Bioeng. 2016, 113, 2298. [DOI] [PubMed] [Google Scholar]
- 56. Ahmed S. R., Kim J., Suzuki T., Lee J., Park E. Y., Biosens. Bioelectron. 2016, 85, 503. [DOI] [PubMed] [Google Scholar]
- 57. Ahmed S. R., Takemeura K., Li T.‐C., Kitamoto N., Tanaka T., Suzuki T., Park E. Y., Biosens. Bioelectron. 2017, 87, 558. [DOI] [PubMed] [Google Scholar]
- 58. Quinten M., Kreibig U., Surf. Sci. 1986, 172, 557. [Google Scholar]
- 59. Verdoodt N., Basso C. R., Rossi B. F., Pedrosa V. A., Food Chem. 2017, 221, 1792. [DOI] [PubMed] [Google Scholar]
- 60. Liu Y., Zhang L., Wei W., Zhao H., Zhou Z., Zhang Y., Liu S., Analyst 2015, 140, 3989. [DOI] [PubMed] [Google Scholar]
- 61. Qiu S., Lin Z., Zhou Y., Wang D., Yuan L., Wei Y., Dai T., Luo L., Chen G., Analyst 2015, 140, 1149. [DOI] [PubMed] [Google Scholar]
- 62. Zheng L., Wei J., Lv X., Bi Y., Wu P., Zhang Z., Wang P., Liu R., Jiang J., Cong H., Liang J., Chen W., Cao H., Liu W., Gao G. F., Du Y., Jiang X., Li X., Biosens. Bioelectron. 2017, 91, 46. [DOI] [PubMed] [Google Scholar]
- 63. Bui M.‐P. N., Ahmed S., Abbas A., Nano Lett. 2015, 15, 6239. [DOI] [PubMed] [Google Scholar]
- 64. Veigas B., Pedrosa P., Carlos F. F., Mancio‐Silva L., Grosso A. R., Fortunato E., Mota M. M., Baptista P. V., J. Nanobiotechnol. 2015, 13, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shafiee H., Asghar W., Inci F., Yuksekkaya M., Jahangir M., Zhang M. H., Durmus N. G., Gurkan U. A., Kuritzkes D. R., Demirci U., Sci. Rep. 2015, 5, 8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li H., Rothberg L. J., J. Am. Chem. Soc. 2004, 126, 10958. [DOI] [PubMed] [Google Scholar]
- 67. Saleh M., El‐Matbouli M., J. Virol. Methods 2015, 217, 50. [DOI] [PubMed] [Google Scholar]
- 68. Bloomfield V., Crothers D. M., Nucleic Acids: Structures, Properties and Functions, University Science Books, Sausalito, CA, USA: 1999. [Google Scholar]
- 69. Ma X., Song L., Zhou N., Xia Y., Wang Z., Int. J. Food Microbiol. 2017, 245, 1. [DOI] [PubMed] [Google Scholar]
- 70. Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C. S., Anal. Chem. 2017, 89, 5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Martinez A. W., Phillips S. T., Butte M. J., Whitesides G. M., Angew. Chem., Int. Ed. 2007, 46, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cate D. M., Adkins J. A., Mettakoonpitak J., Henry C. S., Anal. Chem. 2015, 87, 19. [DOI] [PubMed] [Google Scholar]
- 73. Yetisen A. K., Akram M. S., Lowe C. R., Lab Chip 2013, 13, 2210. [DOI] [PubMed] [Google Scholar]
- 74. Li D., Dong Y., Li B., Wu Y., Wang K., Zhang S., Analyst 2015, 140, 7672. [DOI] [PubMed] [Google Scholar]
- 75. Jung Y. K., Kim T. W., Kim J., Kim J.‐M., Park H. G., Adv. Funct. Mater. 2008, 18, 701. [Google Scholar]
- 76. Song S., Ha K., Guk K., Hwang S.‐G., Choi J. M., Kang T., Bae P., Jung J., Lim E.‐K., RSC Adv. 2016, 6, 48566. [Google Scholar]
- 77. de Oliveira T. V., de N., Soares F. F., de Andrade N. J., Silva D. J., Medeiros E. A. A., Badaró A. T., Food Chem. 2015, 172, 428. [DOI] [PubMed] [Google Scholar]
- 78. Bagwe R. P., Yang C., Hilliard L. R., Tan W., Langmuir 2004, 20, 8336. [DOI] [PubMed] [Google Scholar]
- 79. An K., Somorjai G. A., ChemCatChem 2012, 4, 1512. [Google Scholar]
- 80. Xu S., Ouyang W., Xie P., Lin Y., Qiu B., Lin Z., Chen G., Guo L., Anal. Chem. 2017, 89, 1617. [DOI] [PubMed] [Google Scholar]
- 81. Karakoti A. S., Shukla R., Shanker R., Singh S., Adv. Colloid Interface Sci. 2015, 215, 28. [DOI] [PubMed] [Google Scholar]
- 82. Turkevich J., Stevenson P. C., Hillier J., Discuss. Faraday Soc. 1951, 11, 55. [Google Scholar]
- 83. Park T. J., Lee K. G., Lee S. Y., Appl. Microbiol. Biotechnol. 2016, 100, 521. [DOI] [PubMed] [Google Scholar]
- 84. Lee K. G., Hong J., Wang K. W., Heo N. S., Kim D. H., Lee S. Y., Lee S. J., Park T. J., ACS Nano 2012, 6, 6998. [DOI] [PubMed] [Google Scholar]
- 85. Park T. J., Lee S. Y., Heo N. S., Seo T. S., Angew. Chem., Int. Ed. 2010, 49, 7019. [DOI] [PubMed] [Google Scholar]


