Abstract
Background
The clindamycin-resistant erythromycin-susceptible (CRES) phenotype is rare in Streptococcus agalactiae (group B streptococci). We aimed to determine the molecular characteristics of CRES S. agalactiae using whole genome sequencing (WGS).
Methods
Sixty-six S. agalactiae isolates obtained from blood (N=26), cerebrospinal fluid (N=10), urine (N=17), and vaginal discharge (N=13) between 2010 and 2017 in Korea were subjected to WGS. Based on the WGS data, we analyzed antimicrobial resistance (AMR) determinants, sequence types (STs), capsular polysaccharide (CPS) genotypes, and virulence gene profiles, and constructed a phylogenetic tree. We included the clindamycin-susceptible erythromycin-resistant (CSER) phenotype for comparison.
Results
We identified seven CRES S. agalactiae isolates from urine (N=5) and vaginal discharge (N=2) collected between 2010 and 2011. All CRES isolates harbored AMR determinants of lnu(B), lsa(E), and aac(6′)-aph(2″), revealed ST19 and CPS genotype III, and had a virulence gene profile of rib-lmb-cylE. Phylogenetic tree analysis revealed that all CRES isolates belonged to the same cluster, suggesting a clonal distribution. In contrast, seven CSER isolates showed a diverse distribution and clustered separately from the CRES isolates.
Conclusions
CRES isolates collected between 2010 and 2011 showed a unique cluster with ST19 and CPS genotype III in Korea. This is the first report on WGS-based characteristics of S. agalactiae in Korea.
Keywords: Streptococcus agalactiae, Group B streptococci, Antimicrobial resistance, Whole genome sequencing, Sequence types, Clonal distribution, CRES (clindamycin-resistant erythromycin-susceptible)
INTRODUCTION
Streptococcus agalactiae (group B β-hemolytic streptococci) can cause several invasive infections, including sepsis, infective endocarditis, septic arthritis, and meningitis, especially in neonates and the elderly [1, 2]. S. agalactiae infections are classified as invasive (blood, cerebrospinal fluid, joint fluid, pleural effusion, ascites, and closed pus) or non-invasive (urine and vaginal discharge) [3]. Multilocus sequence typing (MLST) for sequence type (ST) determination has been used to evaluate the clonal distribution or persistence of S. agalactiae from urine and vagina [3].
S. agalactiae possesses numerous virulence factors, including capsular polysaccharide (CPS), alpha and beta antigens of the surface-associated C protein, and the surface protein Rib. CPS is the most important virulence factor and is used for strain typing [4]. Alpha antigen of the surface-associated C protein (encoding gene bca) mediates adherence to the epithelium, whereas beta antigen of the surface-associated C protein (encoding gene bac) is involved in invasiveness and resistance to phagocytic clearance [5, 6]. Protein Rib (encoding gene rib), which exhibits resistance to proteases, confers protective immunity and is detected in most CPS III isolates, which cause severe infections in neonates [5]. lmb and cylE encode human laminin binding protein and beta-hemolysin, respectively.
There are two major phenotypes of macrolide resistance in streptococci: an MLSB phenotype is resistant to macrolides, lincosamides, and streptogramin B, and an M phenotype is resistant to macrolides, but not to lincosamides and streptogramin B [7]. The MLSB phenotype in streptococci can result from induced/constitutive expression of the antimicrobial resistance (AMR) determinants, erm(A) and erm(B), whereas the M phenotype can be caused by the mef(A) determinant [7]. In addition, an L phenotype exists, which is resistant to lincosamides, but not to macrolides [8]. The clindamycin-resistant erythromycin-susceptible (CRES) phenotype corresponds to the L phenotype [9].
The CRES phenotype (L phenotype) is rare in S. agalactiae. It has been described in clinical isolates from Korea and is caused by antimicrobial modification mediated by lnu(B) [10]. Members of the lnu gene family encode nucleotidyl transferase enzymes that catalyze the adenylation of lincomycin and clindamycin. The CRES phenotype is also mediated by two genes of the lsa gene family, namely lsa(C) and lsa(E), which encode ATP-binding proteins that have been classified as class 2 ATP-binding cassette transporters (antibiotics efflux pumps) [11]. While the overall frequency of this phenotype in S. agalactiae was quite low (65/21,186=0.31%), it had increased in 2014–2015 in the USA [11].
We aimed to determine the genetic characteristics of CRES S. agalactiae in Korea based on whole genome sequencing (WGS). To the best of our knowledge, this is the first report on WGS-based characteristics of S. agalactiae in Korea.
MATERIALS AND METHODS
Study design
S. agalactiae isolates collected between 2010 and 2017 were randomly selected from the repository at Gyeongsang National University Hospital (GNUH) in Gyeongnam Province, Korea. We included a total of 66 isolates: invasive isolates from blood (N=26) and cerebrospinal fluid (CSF) (N=10) as well as non-invasive isolates from urine (N=17) and vaginal discharge (N=13); repeated isolates from the same patients were excluded. Bacterial identification was conducted using a Vitek-2 automated identification system (bioMerieux Inc., Marcy l’Etoile, France). All isolates were stored at −70°C to −80°C before being processed for further evaluation.
Patients’ sex and age were obtained from the electronic medical records. In total, 66 patients with a median age of 50.5 years (range, 0–86 years), including 12 children, 54 adults, and 32 males (48.5%), were enrolled. The study protocol was approved by the Institutional Review Board of GNUH (approval number: GNUH 2016-03-010). Informed consent was waived because of the retrospective nature of the study.
Antimicrobial susceptibility testing (AST)
AST was conducted using 11 antimicrobial agents, including β-lactam, tetracycline, macrolide/lincosamide (ML), and fluoroquinolone, to evaluate AMR levels by the broth microdilution method using a Vitek-2 System and an ST-01 test kit (bioMerieux Inc.). CRES S. agalactiae was defined as having a minimum inhibitory concentration of >1 μg/mL for clindamycin and of <0.25 μg/mL for erythromycin. Seven isolates showed the CRES phenotype. We also included seven isolates showing the clindamycin-susceptible erythromycin-resistant (CSER) phenotype for comparison. In addition, we included 13 isolates that were clindamycin-resistant erythromycin-resistant, and 29 isolates that were clindamycin-susceptible erythromycin-susceptible. The seven CRES isolates were recovered from urine (N=5) and vaginal discharge (N=2) during a limited period (March 2010 to August 2011).
Bacterial identification and AST had been performed previously by routine microbiological procedures, whereas WGS and bioinformatics analysis had been conducted for this study.
WGS
S. agalactiae isolates were grown at 35°C in Todd-Hewitt broth (Becton Dickinson, Sparks, MD, USA) for 16–18 hours. Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) after pretreatment with lysozyme (Thermo Fisher Scientific, Waltham, MA, USA) and proteinase K (Qiagen) [12]. S. agalactiae isolates were identified by 16S rRNA gene sequencing with amplifying/sequencing primer set (27F: AGAGTTTGATCMTGGCTCAG and 1485R: TACGGTTACCTTGTTACGAC) developed in-house using an ABI 3730 DNA sequencing instrument (Applied Biosystems, Foster City, CA, USA). The sequencing library was prepared using a TruSeq DNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) for the Illumina MiSeq system. Draft genome sequences of the isolates were determined based on 300-bp paired-end reads. Illumina sequencing data were assembled using SPAdes 3.13.0 (Algorithmic Biology Lab, St. Petersburg Academic University of the Russian Academy of Sciences). For gene-finding and functional annotation, we used the whole genome analysis pipeline of ChunLab (Seoul, Korea). Protein-coding DNA sequences were predicted using Prodigal 2.6.2 (https://github.com/hyattpd/Prodigal) [13].
AMR genotyping
AMR genotyping was conducted based on the contig sequences obtained using ResFinder version 3.2 (https://cge.cbs.dtu.dk/services/ResFinder/) managed by the Center for Genomic Epidemiology [14]. This tool can detect genes conferring resistance to β-lactams, macrolide, lincosamide, tetracycline, quinolone, oxazolidinone, sulfonamide/trimethoprim, glycopeptide, aminoglycoside, phenicol, fosfomycin, nitroimidazole, rifampicin, fusidic acid, and colistin. AMR genotypes were determined based on an identity threshold >90% and a minimum length of 60% as compared with the reference sequence in the database.
MLST
MLST (allelic profile: adhP–pheS–atr–glnA–sdhA–glcK–tkt) was conducted based on the contig sequences using the MLST server (https://cge.cbs.dtu.dk/services/MLST/) managed by the Center for Genomic Epidemiology [15]. The STs were grouped into clonal complexes (CCs), whereby related STs were classified as single-locus variants differing in only one housekeeping gene. An expansion of the goeBURST algorithm implemented in PHYLOViZ (http://www.phyloviz.net/) was used to produce a minimum-spanning tree representing possible relationships among the STs [16].
CPS genotyping
CPS genotyping and detection of the S. agalactiae-specific dltS gene were conducted based on the contig sequences by PCR simulation [17] in the online application, Serial Cloner (http://serialbasics.free.fr/Serial_Cloner.html). The CPS genotypes included Ia, Ib, II, III, IV, V, VI, VII, and VIII.
Virulence gene profiling
The presence of five virulence genes (bca-rib-bac-lmb-cylE) was determined based on the contig sequences by PCR simulation in Serial Cloner [18–20]. Sequence identity of the virulence genes in all simulation PCR-positive strains was confirmed using the basic local alignment search tool (BLAST) (http://blast.ddbj.nig.ac.jp/blastn?lang=ja).
Phylogenetic tree analysis
A phylogenetic tree was constructed using Orthologous Average Nucleotide Identity Tool, which measures similarity among multiple genome sequences based on the OrthoANI algorithm and BLAST calculations, on EZBioCloud (https://www.ezbiocloud.net/tools/orthoani) [21].
Statistical analysis
We used Fisher’s exact probability tests (two-sided) to determine significant differences between CRES and CSER isolates using SPSS Statistics version 22.0 (IBM Corp., Armonk, NY, USA). P<0.05 indicated statistical significance.
RESULTS
We deposited the draft genome sequences of the 66 S. agalactiae isolates into the National Center for Biotechnology Information (NCBI) database (Table 1). The WGS of the S. agalactiae isolate NCTC8181 (accession number UAVB00000000) obtained from environmental milk was used as a reference genome. The number of contigs ranged from eight (for isolate GCH73) to 90 (for isolate GCH63).
Table 1.
Specimen, date of collection, and accession numbers of draft genome sequences of 66 isolates of Streptococcus agalactiae
| Strain | Specimen | Date of specimen collection (yr/month) | Sex | Age (yr) | Accession numbers |
|---|---|---|---|---|---|
| GCH2 | Blood | 2016/03 | M | 75 | WHUI00000000 |
| GCH4 | Blood | 2016/05 | F | 83 | WHUJ00000000 |
| GCH5 | Blood | 2016/06 | F | 78 | WHUK00000000 |
| GCH7 | Blood | 2016/07 | M | 74 | WHUL00000000 |
| GCH8 | Blood | 2016/07 | M | 79 | WACQ00000000 |
| GCH9 | Blood | 2016/07 | M | 0 | VYJX00000000 |
| GCH10 | Blood | 2016/08 | F | 72 | VYJI00000000 |
| GCH11 | Blood | 2016/08 | M | 40 | VYJJ00000000 |
| GCH13 | Blood | 2016/10 | M | 64 | VYJK00000000 |
| GCH14 | Blood | 2016/12 | M | 67 | VYJL00000000 |
| GCH15 | Blood | 2016/12 | M | 76 | VYJM00000000 |
| GCH16 | Blood | 2017/01 | M | 43 | VYJN00000000 |
| GCH18 | Blood | 2017/02 | M | 77 | VYJO00000000 |
| GCH19 | Blood | 2017/02 | M | 60 | VYJP00000000 |
| GCH21 | Blood | 2017/03 | F | 85 | VYJQ00000000 |
| GCH22 | Blood | 2017/05 | M | 46 | VYJR00000000 |
| GCH25 | Blood | 2017/06 | M | 64 | VYJS00000000 |
| GCH26 | Blood | 2017/07 | M | 86 | VYJT00000000 |
| GCH28 | Blood | 2017/08 | M | 54 | VYJU00000000 |
| GCH29 | Blood | 2017/10 | M | 0 | VYJV00000000 |
| GCH30 | Blood | 2017/10 | F | 74 | VYJW00000000 |
| GCH32 | Blood | 2017/12 | M | 51 | VYQL00000000 |
| GCH33 | Blood | 2016/08 | F | 56 | VYQM00000000 |
| GCH34 | Blood | 2016/11 | F | 73 | VYQN00000000 |
| GCH35 | Blood | 2017/05 | M | 84 | VYQO00000000 |
| GCH36 | Blood | 2017/07 | F | 73 | VYQP00000000 |
| GCH37 | CSF | 2014/07 | M | 51 | VYQQ00000000 |
| GCH38 | CSF | 2014/10 | M | 0 | VYQR00000000 |
| GCH39 | CSF | 2014/12 | M | 0 | VYQS00000000 |
| GCH40 | CSF | 2015/08 | F | 0 | VYQT00000000 |
| GCH41 | CSF | 2015/08 | F | 0 | VYQU00000000 |
| GCH42 | CSF | 2016/06 | M | 50 | VYQV00000000 |
| GCH43 | Urine | 2017/02 | M | 32 | VYQW00000000 |
| GCH44 | Urine | 2017/02 | F | 56 | VYQX00000000 |
| GCH45 | Urine | 2017/05 | F | 84 | VYQY00000000 |
| GCH46 | Urine | 2017/07 | M | 61 | VYQZ00000000 |
| GCH47 | Urine | 2017/07 | F | 34 | VYRA00000000 |
| GCH48 | Urine | 2017/08 | F | 73 | VYRB00000000 |
| GCH49 | Urine | 2017/08 | M | 65 | VYRC00000000 |
| GCH50 | Urine | 2017/11 | F | 13 | VYRD00000000 |
| GCH51 | Urine | 2017/12 | M | 53 | VYRE00000000 |
| GCH53 | Vaginal discharge | 2016/04 | F | 34 | VYRF00000000 |
| GCH54 | Vaginal discharge | 2016/05 | F | 33 | VYRG00000000 |
| GCH55 | Vaginal discharge | 2016/10 | F | 33 | VYRH00000000 |
| GCH56 | Vaginal discharge | 2017/01 | F | 22 | VYRI00000000 |
| GCH57 | Vaginal discharge | 2017/01 | F | 33 | VYRJ00000000 |
| GCH58 | Vaginal discharge | 2017/02 | F | 37 | VYRK00000000 |
| GCH59 | Vaginal discharge | 2017/04 | F | 39 | VYRL00000000 |
| GCH60 | Vaginal discharge | 2017/07 | F | 33 | VYRM00000000 |
| GCH61 | Urine | 2010/03 | M | 51 | VYRN00000000 |
| GCH62 | Vaginal discharge | 2010/03 | F | 38 | VYRO00000000 |
| GCH63 | Urine | 2010/04 | M | 44 | VYRP00000000 |
| GCH64 | Urine | 2010/10 | F | 83 | VYRQ00000000 |
| GCH65 | Urine | 2010/12 | F | 49 | VYRR00000000 |
| GCH66 | Vaginal discharge | 2010/12 | F | 42 | VYRS00000000 |
| GCH67 | Urine | 2011/08 | F | 55 | VYRT00000000 |
| GCH68 | CSF | 2011/04 | M | 0 | VYRU00000000 |
| GCH70 | CSF | 2012/01 | F | 0 | VYRW00000000 |
| GCH71 | CSF | 2012/03 | M | 0 | VYRX00000000 |
| GCH72 | CSF | 2012/08 | F | 0 | VYRY00000000 |
| GCH73 | Vaginal discharge | 2014/07 | F | 33 | WHUM00000000 |
| GCH74 | Vaginal discharge | 2014/08 | F | 40 | WHUN00000000 |
| GCH75 | Vaginal discharge | 2016/02 | F | 25 | WHUO00000000 |
| GCH76 | Urine | 2015/11 | F | 79 | WHUP00000000 |
| GCH77 | Urine | 2016/02 | M | 60 | WHUQ00000000 |
| GCH78 | Urine | 2014/10 | M | 0 | WHUR00000000 |
Abbreviations: CSF, cerebrospinal fluid; M, male; F, female.
The goeBURST diagram is shown in Fig. 1. All seven CRES isolates belonged to ST19 (CC19), suggesting a clonal distribution of the CRES isolates (Table 2), whereas the seven CSER isolates belonged to ST19 (CC19) (N=3), ST2/ST1 (CC2) (N=2), or ST23 (N=2).
Fig. 1.
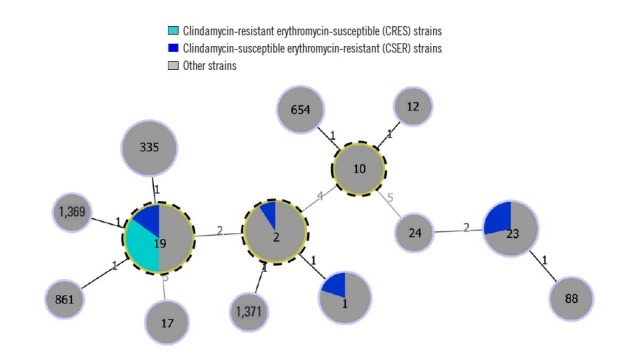
goeBURST diagram of the relationships among STs of 66 S. agalactiae isolates. The numbers in the circles indicate the STs, and the numbers near the lines indicate the number of differing alleles between the two connected STs. Putative CCs are identified by an outer dotted frame and correspond to the STs with the highest number of single locus variants.
Abbreviations: CC, clonal complex; ST, sequence type.
Table 2.
Phenotypic and genotypic features of S. agalactiae for AMR, CPS, ST, and virulence
| Isolate | Antimicrobial susceptibility pattern | Macrolide/lincosamide resistance gene | Tetracycline resistance gene | Aminoglycoside resistance gene | ST | CPS genotype | Virulence gene profile |
|---|---|---|---|---|---|---|---|
| GCH2 | CRER | erm(B), lnu(B), lsa(E), mre(A) | tet(O) | ant(6)-Ia, ant(6)-Ia, aph(3′)-III | 12 | Ib | bca-bac*-lmb-cylE |
| GCH4 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH5 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH7 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH8 | CSES | mre(A) | 654 | Ib | bca-bac*-lmb-cylE | ||
| GCH9 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH10 | CIES | mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH11 | CSES | mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH13 | CSES | mre(A) | 1,371 | VIII | rib-lmb-cylE | ||
| GCH14 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH15 | CSES | mre(A) | 2 | VIII | rib-cylE | ||
| GCH16 | CSES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH18 | CSES | lsa(C), mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH19 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH21 | CREI | lsa(C), mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH22 | CSES | mre(A) | 88 | II | lmb-cylE | ||
| GCH25 | CSES | mre(A) | 1 | VI | bca-lmb-cylE | ||
| GCH26 | CSES | mre(A) | tet(O) | 2 | VIII | rib-lmb-cylE | |
| GCH28 | CSES | mre(A) | tet(M) | 19 | III | rib-lmb-cylE | |
| GCH29 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH30 | CRER | erm(B), mre(A) | tet(M) | 1 | V | lmb-cylE | |
| GCH32 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH33 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH34 | CSES | mre(A) | tet(M) | 24 | Ia | bca-lmb-cylE | |
| GCH35 | CSES | mre(A) | 10 | Ib | bca-bac*-lmb-cylE | ||
| GCH36 | CSES | mre(A) | 10 | Ib | bca-bac*-lmb-cylE | ||
| GCH37 | NA | mre(A) | 88 | Ia | lmb-cylE | ||
| GCH38 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 1,369 | III | bca-lmb-cylE |
| GCH39 | CSES | mre(A) | tet(M) | 17 | III | rib-cylE | |
| GCH40 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH41 | CRER | erm(B), mre(A) | tet(O) | ant(6)-Ia, ant(6)-Ia, aph(3′)-III | 17 | III | rib-cylE |
| GCH42 | CSES | mre(A) | 2 | VIII | rib-cylE | ||
| GCH43 | CREI | lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH44 | CSEI | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH45 | CSER | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH46 | CSES | mre(A) | 654 | Ib | bca-bac*-lmb-cylE | ||
| GCH47 | CRER | erm(A), lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | V | cylE |
| GCH48 | CRER | erm(A), mre(A) | tet(M) | 1 | V | lmb-cylE | |
| GCH49 | CRER | lsa(C), mef(A), mre(A), msr(D) | tet(O) | 861 | III | rib-lmb-cylE | |
| GCH50 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH51 | CRER | erm(B), mre(A) | 10 | Ib | bca-bac*-lmb-cylE | ||
| GCH53 | CSES | mre(A) | tet(M) | 1 | II | lmb-cylE | |
| GCH54 | CSES | mre(A) | 10 | Ib | bca-bac*-lmb-cylE | ||
| GCH55 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH56 | CSES | mre(A) | 2 | VIII | rib-lmb-cylE | ||
| GCH57 | CRER | mre(A) | 654 | Ib | bca-bac*-lmb-cylE | ||
| GCH58 | CREI | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH59 | CRER | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH60 | CSES | mre(A) | tet(M) | 19 | III | rib-lmb-cylE | |
| GCH61 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH62 | CRES | lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH63 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, ant(6)-Ia, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH64 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH65 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH66 | CRES | lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH67 | CRES | erm(B)*, lnu(B), lsa(E), mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH68 | CSES | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH70 | CSES | erm(A), mre(A) | tet(M) | 335 | III | rib-lmb-cylE | |
| GCH71 | CSES | mre(A) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH72 | CSES | erm(B)*, mre(A) | tet(M) | aac(6′)-aph(2″), ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
| GCH73 | CSER | mre(A) | 1 | VI | bca-lmb-cylE | ||
| GCH74 | CSER | mre(A) | tet(M) | 19 | III | rib-lmb-cylE | |
| GCH75 | CSER | mef(A), mre(A), msr(D) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH76 | CSER | erm(A), mre(A) | tet(M) | 19 | V | lmb-cylE | |
| GCH77 | CSER | mef(A), mre(A), msr(D) | tet(M) | 23 | Ia | lmb-cylE | |
| GCH78 | CSER | erm(B)*, mef(A), mre(A), msr(D) | tet(M) | ant(6)-Ia*, aph(3′)-III | 19 | III | rib-lmb-cylE |
Identical nucleotide length<100% of the reference sequence in the database.
Abbreviations: AMR, antimicrobial resistance; CPS, capsular polysaccharide; CRER, clindamycin-resistant erythromycin-resistant; CSES, clindamycin-susceptible erythromycin-susceptible; CIES, clindamycin-intermediate erythromycin-susceptible; CREI, clindamycin-resistant erythromycin-intermediate; CSEI, clindamycin-susceptible erythromycin-intermediate; CSER, clindamycin-susceptible erythromycin-resistant; CRES, clindamycin-resistant erythromycin-susceptible; ST, sequence type.
All CRES isolates showed the lnu(B)-lsa(E) ML resistance genotype (Table 2). However, none of the CSER isolates possessed the lnu(B)-lsa(E) genotype (P=0.001). All CRES isolates showed the aac(6′)-aph(2″) aminoglycoside resistance genotype, whereas none of the CSER isolates did. All isolates did not show AMR genes for β-lactams, quinolone, oxazolidinone, sulfonamide/trimethoprim, glycopeptide, phenicol (except for cat[pC194] in isolate GCH2), fosfomycin, nitroimidazole, rifampicin, fusidic acid, and colistin.
All isolates possessed the dltS gene specific to S. agalactiae, validating species identification. All CRES isolates were CPS III, whereas the CSER isolates possessed diverse CPS types (Table 2).
All CRES isolates exhibited the rib-lmb-cylE profile (Table 2). The CSER isolates had a diverse virulence gene profile. There was a significant difference in the frequency of lnu(B)-lsa(E) or lsa(C) between invasive (N=6/36, 16.7%) and non-invasive (N=13/30, 43.3%, P=0.017) isolates. There was no difference in the frequency of lnu(B)-lsa(E) or lsa(C) between urine (N=9/17, 52.9%) and vaginal discharge (N=4/13, 30.8%, P=0.200).
The phylogenetic tree revealed that all CRES isolates belonged to the same group, whereas CSER isolates belonged to diverse groups, corroborating the clonal distribution of the CRES isolates (Fig. 2). The group distribution on the tree was in accordance with the CPS genotype distribution (e.g., Ia, Ib, III, V, and VIII).
Fig. 2.

Phylogenetic tree of 66 S. agalactiae isolates. The phylogenetic tree was constructed using OAT, based on the OrthoANI algorithm, with S. agalactiae strain NCTC8181 (accession number UAVB00000000) as a reference. Asterisks (*) indicate CRES isolates, daggers (†) indicate CSER isolates. There was a concordance of the group distribution on the tree with the CPS genotype distribution (Ia, Ib, III, V, and VIII).
Abbreviations: CPS, capsular polysaccharide; CRES, clindamycin-resistant erythromycin-susceptible; CSER, clindamycin-susceptible erythromycin-resistant; CRER, clindamycin-resistant erythromycin-resistant; CSES, clindamycin-susceptible erythromycin-susceptible; CSEI, clindamycin-susceptible erythromycin-intermediate; CREI, clindamycin-resistant erythromycin-intermediate; CIES, clindamycin-intermediate erythromycin-susceptible; OAT, Orthologous Average Nucleotide Identity Tool.
The erm(B) sequence of the CRES isolate GCH61 in our study was compared with the previously registered sequence (738 bp) of S. agalactiae isolate KMP104 (RefSeq accession number DQ355148). This sequence (accession number LC512876) contained C222T (N74N), T224C (I75T), and A299G (N100S) nucleotide (amino acid) substitutions in addition to the insertion of an IS1216E element at nucleotide position 642, which resulted in the deletion of a segment spanning nucleotides 642–738 (97 bp) (Fig. 3).
Fig. 3.

Comparison of sequences of GCH61, NUBL-9601, and KMP104. Sequence of erm(B) from isolate GCH61 (accession number VYRN00000000) with the CRES phenotype contained C222T (N74N), T224C (I75T), and A299G (N100S) nucleotide (amino acid) substitutions, in addition to the insertion of an IS1216E element at nucleotide position 642, which resulted in the deletion of a segment spanning nucleotides 642–738 (97 bp). This sequence was identical to that from the CRES isolate NUBL-9601 (accession number LC430933). The sequence of erm(B) from the isolate KMP104 was used as a reference (RefSeq accession number DQ355148).
Abbreviation: CRES, clindamycin-resistant erythromycin-susceptible.
DISCUSSION
Our study revealed that CRES isolates have unique features compared with CSER isolates, including their AMR genotype [lnu(B)-lsa(E) with aac(6′)-aph(2″)], ST19/CC19, CPS type III, virulence gene profile of rib-lmb-cylE, and in terms of cluster on the phylogenetic tree.
We searched for the presence of CRES S. agalactiae isolates in the Isolates Database on the MLST website (https://pubmlst.org/bigsdb?db=pubmlst_sagalactiae_isolates&page=query). Interestingly, only one CRES isolate was previously recovered from a 61-year-old female patient with bacteremia in Kangwon Province in the north of Korea in 2010 [22]. Another CRES isolate of CPS genotype III was isolated from a patient with bacteremia in Bergen, Norway, in 2010 [23]. Three CRES isolates of serotype III were recovered from clinical specimens in Seoul during 2010–2013 [9, 24]. These findings are in line with our observation that all CRES isolates in Gyeongnam Province in the south of Korea were recovered between March 2010 and August 2011. Thus, the CRES isolates appeared to be epidemic in Korea and other countries during this limited period.
In line with a previous study [11], we found a significant difference in the distribution of AMR genotype of lnu(B)-lsa(E) between the CRES and CSER isolates. For all CRES isolates, the lnu(B) locus was adjacent to the lsa(E) locus within the same contigs, with a short 53-bp distance between these two loci. Therefore, in our study, the lnu(B)-lsa(E) gene combination seems to mainly contribute to the CRES phenotype. Further observation of the dynamic changes in the CRES phenotype and the corresponding gene transfer is needed.
The erm(B) confers constitutive resistance (cMLSB) through a conformational change in 23S ribosomal RNA methyltransferase [8]. Deletion of a segment in erm(B) sequence in CRES suggests loss of function of the erm(B) protein in this isolate, resulting in an erythromycin-susceptible phenotype. Interestingly, three CRES isolates (NUBL-9601, NUBL-9602, and NUBL-9603) isolated at a hospital in Seoul during 2010–2013 had the identical sequences (accession numbers LC430933, LC430934, and LC430935) (3) [9, 10]. Furthermore, we observed the IS1216E insertion in erm(B) (accession numbers LC512881, LC512882, LC512883, LC512885, and LC512886) in five isolates (GCH16, GCH19, GCH38, GCH55, and GCH58, respectively) of this study. Thus, IS1216E seems to be common among CRES isolates in Korea.
Four CRES isolates (GCH63, GCH64, GCH65, and GCH67) in our study possessed truncated variant sequences of erm(B) (624 and 678 bps) due to insertion of a TAA stop codon into the open reading frame (accession numbers LC512877, LC512878, LC512879, and LC512880), suggesting that an immature erm(B) protein leads to the erythromycin-susceptible phenotype. Three other phenotypes (GCH50, GCH72, and GCH78) also had truncated variant sequences (660, 678, and 510 bps, respectively) (accession numbers LC512884, LC512887, and LC512888, respectively) harboring the erythromycin-susceptible phenotype. Therefore, we may explain the mechanism of the presence of erm(B)-gene with the erythromycin-susceptible phenotype in CRES S. agalactiae.
This study has several limitations. First, clinical data, such as antibiotic treatment, outcome, and complications, does not suffice to demonstrate the relationship with the AMR genotype or virulence gene profile. Second, we cannot explain why the clonal outbreak occurred only during a limited period and is absent nowadays. Third, we did not determine translational activities and enzymatic functions of the IS1216E insertion-erm(B) and the truncated variant sequences.
In conclusion, CRES isolates were obtained during a limited period (2010–2011) and showed a genetic cluster having ST19 and CPS III in Korea as revealed by WGS. This rare AMR phenotype should be meticulously monitored, and the therapeutic efficacy of optimal antibiotics should be further evaluated.
ACKNOWLEDGMENTS
The authors thank ChunLab (Seoul, Korea) for technical support in WGS. This publication made use of the Streptococcus agalactiae MLST website (https://pubmlst.org/sagalactiae/) hosted at the University of Oxford (Jolley, Maiden 2010, BMC Bioinformatics, 11:595). The development of this website was funded by the Wellcome Trust. The pathogen resources for this study were provided by Gyeongsang National University Hospital Branch of the National Culture Collection for Pathogens (GNUH-NCCP).
Footnotes
AUTHOR CONTRIBUTIONS
TT and SK contributed to the study concept and design, and prepared and revised the manuscript. D-HL collected the bacterial strains and patient information. TM analyzed bioinformatics data and interpreted the acquired results. SL analyzed WGS data and submitted the draft genome sequences to the NCBI database.
CONFLICTS OF INTEREST
No potential conflicts of interest relevant to this article were reported.
RESEARCH FUNDING
This work was supported by a National Research Foundation of Korea grant funded by the Korea government (No. 2018R1D1A3B07041280). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Takahashi T, Sunaoshi K, Sunakawa K, Fujishima S, Watanabe H, Ubukata K, et al. Clinical aspects of invasive infections with Streptococcus dysgalactiae ssp. equisimilis in Japan: differences with respect to Streptococcus pyogenes and Streptococcus agalactiae infections. Clin Microbiol Infect. 2010;16:1097–103. doi: 10.1111/j.1469-0691.2009.03047.x. [DOI] [PubMed] [Google Scholar]
- 2.Shibayama A, Yoshizaki T, Tamaki M, Goto M, Takahashi T. Pyogenic sternoclavicular arthritis caused by Streptococcus agalactiae in an elderly adult with diabetes mellitus. J Am Geriatr Soc. 2016;64:1376–7. doi: 10.1111/jgs.14169. [DOI] [PubMed] [Google Scholar]
- 3.Choi WH, Park HW, Kim S. Persistence of Group B streptococcus in the urogenital area. Ann Lab Med. 2017;37:454–6. doi: 10.3343/alm.2017.37.5.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emaneini M, Mirsalehian A, Beigvierdi R, Fooladi AA, Asadi F, Jabalameli F, et al. High incidence of macrolide and tetracycline resistance among Streptococcus agalactiae strains isolated from clinical samples in Tehran, Iran. Maedica (Buchar) 2014;9:157–61. [PMC free article] [PubMed] [Google Scholar]
- 5.Lindahl G, Stålhammar-Carlemalm M, Areschoug T. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin Microbiol Rev. 2005;18:102–27. doi: 10.1128/CMR.18.1.102-127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oviedo P, Pegels E, Laczeski M, Quiroga M, Vergara M. Phenotypic and genotypic characterization of Streptococcus agalactiae in pregnant women. First study in a province of Argentina. Braz J Microbiol. 2013;44:253–8. doi: 10.1590/S1517-83822013005000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Lee S, Park H, Kim S. Predominance of emm4 and antibiotic resistance of Streptococcus pyogenes in acute pharyngitis in a southern region of Korea. J Med Microbiol. 2019;68:1053–8. doi: 10.1099/jmm.0.001005. [DOI] [PubMed] [Google Scholar]
- 8.Haenni M, Lupo A, Madec JY. Antimicrobial resistance in Streptococcus spp. Microbiol Spectr. 2018;6 doi: 10.1128/microbiolspec.arba-0008-2017. ARBA-0008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moroi H, Kimura K, Ido A, Banno H, Jin W, Wachino JI, et al. Erythromycin-susceptible but clindamycin-resistant phenotype of clinical ermB-PCR-positive Group B streptococci isolates with IS1216E-inserted ermB. Jpn J Infect Dis. 2019;72:420–2. doi: 10.7883/yoken.JJID.2019.015. [DOI] [PubMed] [Google Scholar]
- 10.Seo YS, Srinivasan U, Oh KY, Shin JH, Chae JD, Kim MY, et al. Changing molecular epidemiology of group B streptococcus in Korea. J Korean Med Sci. 2010;25:817–23. doi: 10.3346/jkms.2010.25.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins PA, Law CS, Metcalf BJ, Chochua S, Jackson DM, Westblade LF, et al. Cross-resistance to lincosamides, streptogramins A and pleuromutilins in Streptococcus agalactiae isolates from the USA. J Antimicrob Chemother. 2017;72:1886–92. doi: 10.1093/jac/dkx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida H, Katayama Y, Fukushima Y, Taniyama D, Murata Y, Mizutani T, et al. Draft genome sequence of Streptococcus canis clinical strain TA4, harboring the M-like protein gene and isolated in Japan from a patient with bacteremia. Genome Announc. 2018;6:e01469–17. doi: 10.1128/genomeA.01469-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–61. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Vaz C. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33:128–9. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 17.Murayama SY, Seki C, Sakata H, Sunaoshi K, Nakayama E, Iwata S, et al. Capsular type and antibiotic resistance in Streptococcus agalactiae isolates from patients, ranging from newborns to the elderly, with invasive infections. Antimicrob Agents Chemother. 2009;53:2650–3. doi: 10.1128/AAC.01716-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning SD, Ki M, Marrs CF, Kugeler KJ, Borchardt SM, Baker CJ, et al. The frequency of genes encoding three putative group B streptococcal virulence factors among invasive and colonizing isolates. BMC Infect Dis. 2006;6:116. doi: 10.1186/1471-2334-6-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Schnitzler N, Lütticken R, et al. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect Immun. 1999;67:871–8. doi: 10.1128/iai.67.2.871-878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergseng H, Bevanger L, Rygg M, Bergh K. Real-time PCR targeting the sip gene for detection of group B Streptococcus colonization in pregnant women at delivery. J Med Microbiol. 2007;56:223–8. doi: 10.1099/jmm.0.46731-0. [DOI] [PubMed] [Google Scholar]
- 21.Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–3. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 22.PubMLST. Isolate information: id 4644 (593049) - Streptococcus agalactiae isolates. https://pubmlst.org/bigsdb?page=info&db=pubmlst_sagalactiae_isolates&id=4644. (Updated on Oct 2017)
- 23.PubMLST. Isolate information: id 5773 (iGBS308) - Streptococcus agalactiae isolates. https://pubmlst.org/bigsdb?page=info&db=pubmlst_sagalactiae_isolates&id=5773. (Updated on Oct 2019)
- 24.Ryu H, Park YJ, Kim YK, Chang J, Yu JK. Dominance of clonal complex 10 among the levofloxacin-resistant Streptococcus agalactiae isolated from bacteremic patients in a Korean hospital. J Infect Chemother. 2014;20:509–11. doi: 10.1016/j.jiac.2014.03.005. [DOI] [PubMed] [Google Scholar]


