Abstract
As the SARS-CoV-2 virus race around the world across the different population, there needs to be a consolidated effort to understand the divergence of demographically distributed strains. The emerging trends in SARS-CoV-2 genome data show specific mutation and genetic diversity, which could provide the basis to develop a cocktail of vaccine and may also be used to develop the region-specific diagnostic tool, thus decreasing the chances of testing failures in fields. Since the transmission of SARS-CoV-2 is subject to the extent of human interaction, the insights from the correlation of genetic diversity with epidemiological parameter would give paramount information to tackle this transmission. Previously, studies have also correlated the epidemiological data with gut microbiome and its role in immunomodulation for maintaining health status, and such information could be generated from recovered individuals from different demographic regions. It will help in designing a probiotic-based diet for modulation of the gut microbiome, and that could be another plausible prophylactic treatment option. The genomics data suggest that a specific variant of SARS-CoV-2 gets enriched with the specific demographic region. Overall, demographic data suggests that host influences mutation and expression of the virus. Hence, the experiences from the clinical intervention for that region should be considered in control and treatment strategies.
Keywords: SARS-CoV-2 genome, Genetic diversity, Phylogeny, Gut microbiome, Drug development
Introduction
The recent outbreak of the novel coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has engulfed nearly all the economies. On January 30, 2020, the WHO declared this outbreak a public health emergency of international concern (PHEIC) and at the time of writing nearly 465,915 cases have been reported across 200 countries [1].
Genetic Divergence of SARS-CoV-2
SARS-CoV-2 belongs to Coronaviridae family and has a peculiar feature of spiked coat glycoproteins, have shown a high mutation rate, mortality rate and fatality rate; which is subject to change in upcoming days [2]. The observed mutation rate, strain variability, genetic selection during its interaction with the host population and transmission across the countries creates a challenging clinical scenario. As the virus race around the world, clues to how it is spreading and evolving can be inferred from analysis of available 1455 genomes (as on date 26.03.2020) on the GISAID (Global Initiative on Sharing All Influenza Data) platform [3]. Phylogenetic inferences substantially suggest that SARS-CoV-2, originating in China, has now evolved into various variants which are specific to regions [3].
Interestingly, it highlights that the strain travelled majorly to the western region and has possibly evolved during endemic transmission. Initially, substantial evidence to state the mutation rate in the novel SARS-CoV-2 from Wuhan city was lacking. However, a recent study from Iceland has highlighted that the virus has acquired more than 40 mutations, and the strains grouped into three different clusters [4]. The initial studies identified two major types of virulent SARS-CoV-2 circulating among the Chinese population and the prevalence of the aggressive form decreased after early January 2020, and the milder type has become common owing to selective pressure due to human intervention [5]. Thus, it is crucial to understand the evolution of SARS-CoV-2 across different temperate countries and populations. Interestingly, the spread of SARS-CoV-2 is limited, so far, in the countries which have a high rate of malaria incidences.
Past one month, mostly all the states of India are gripped by SARS-CoV-2. Understanding how it is spreading within the subcontinent would be a gruesome challenge. The genomic epidemiology of SARS-CoV-2 suggests that there is a molecular evolution of the variants which are supposedly endemic, making this pandemic more challenging to tackle. There is an urgent and prompt need to collect, collate and analyse data of every traveller reaching India from different parts of the world. Moreover, based on SARS-CoV-2 infected/carrier cases so far, the inferences should be drawn to develop the template for the massive epidemiological profiling of SARS-CoV-2 strain across the pan-India. Still, the question remains that once the strains find different host genotype does it remain the same genetically from where it has originated. GISAID analysis suggests that there are variation and influence of host on the primary sequence of the virus. Till date, two genomes available from India showed clustering closer to the strains prevalent in China [6]. However, a unique mutation (A930V (24351C > T)) in the spike surface glycoprotein of Indian SARS-CoV-2 genome which is absent in other strains from Wuhan, Italy, USA, and Nepal is reported [7], highlighting the possible host-dependent modification and the endemic evolution of SARS-CoV-2.
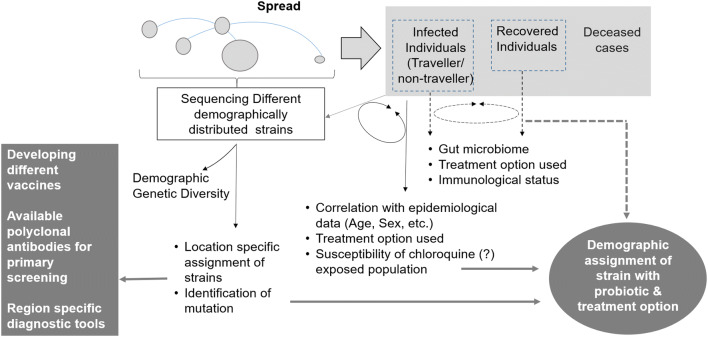
Figure 1 describes an approach to study the transmission of SARS-CoV-2 and its demographic divergence. Identifying the location-specific strain and along with its specific mutation would help in developing a cocktail of vaccine and the region-specific diagnostic tool for covering the demographic genetic diversity of SARS-CoV-2. Linking this data with the extent of human interaction and epidemiological parameter would be important to tackle the mass scale transmission. The gut microbiome has a role in immunomodulation and maintaining health status; hence, a plausible prophylactic treatment option based on a probiotic-based diet could be envisaged through comparative genomics and microbiome studies [8]. The genomics data suggests that a specific variant of a virus gets enriched within the specific demographic region. Hence, the experiences from the clinical intervention for that region should be considered in control and treatment strategies.
Fig. 1.
Key points to answer the transmission, control and cure for SARS-CoV-2 infection
Diagnosis and Epidemiology
Based on the GISAID diversity profile [3], the array of RT-PCR primers, as well as reliable generic sets of primers for primary screening, could be planned. In a country like India, where the virus has been imported from different countries, hence the travel history should be linked with generating complete sequence data for a few random cases. It will not only help in diagnosis but also suggest the possible treatment option to opt for these types of strains. A tight correlation among the pathogenesis of this disease, immunological markers, microbiome dysbiosis, demographic correlation and strain level variations across the demographics should be comprehensively investigated; and should be considered as part of the detailed diagnosis.
Drug Interventions and Pathogenesis of SARS-CoV-2
The data shows that the SARS-CoV-2 strain is clustering and evolving in each demographic region but has a common ancestor. Hence, a drug which delays the progression of infection would be an essential option for treatment for this pandemic while real cure emerges. Recently as a breakthrough treatment of the SARS-CoV-2 infected patients, chloroquine has tested as the potent preventive drug [9]. Chloroquine, an antimalaria agent, has been a widely used drug for decades [10] and has recently shown the apparent efficacy for treating the SARS-COV-2 associated pneumonia in clinical studies [11]. In some cases, other available antiviral drugs also have been tried in combination with chloroquine [12, 13]. With the challenges posed by this virus to address the pathogenesis and to intervene its multiplication cycles in a patient, the alternative medicine [14, 15] could suggest new bio-molecules.
The global data on the SARS-CoV-2 [2] shows that among the closed cases (139,679 cases which had an outcome), nearly 84% have recovered so far. The high titre of antibodies in the plasma from such positive cases could also be an alternate but immediate therapy. There are reports of valproic acid Co-A [16] and Vit B12 [17] in controlling the replication of SARS-CoV-2 via RDRp regulation, and in another study, nine host miRNAs which can potentially target and control SARS-CoV2 genes were identified [7]. Such studies provide insight into host–viral interactions and pathogenesis.
Linking Microbiome, Immune Response and SARS-CoV-2
Gut microbiome profile is linked with food habits, lifestyle and genetic variance of the population [18] and has shown influence on immune response [19]; hence, under the pandemic scenario, this has to be also accounted. Studies have shown for parasitic infection like malaria, feeding the susceptible mice with yoghurt loaded with Lactobacillus and Bifidobacterium reduces the parasite count [20]. The use of probiotics and high fibre diet can also support the immune system against the virus [20, 21]. Similarly, immunity impairment in Tuberculosis (TB) cases was associated with the gut microbiome in Indian subjects [22]. Therefore, the study of the gut microbiome from the SARS-CoV-2 recovered individuals from different demographic regions will suggest possible gut microbiome connections to device the prophylactic measures.
Tackling SARS-CoV-2 Outbreak in India
In light of the distribution of strain variability across the demographic regions, is the virus undergoing a host-dependent mutation and if so the studies on the correlation of SNPs to the strain variability in the diverse population group should be carried out. Additionally, does the divergence of SARS-CoV-2 strains leading to more virulence or it is just adapting to its new host humans? Is there any correlation between the mutation and virulence of the strain? Simultaneously, a comparative analysis could provide a clue to identify the sub-sequences related to the virulence and its evolutionary selection. In conclusion, the key points emerge from diversity and approach described in Fig. 1 that the generic tools and intervention measures for such eventualities need to be envisaged and planned at a global level. Also, the comprehensive investigation targeting gut microbiome and immunomodulation will emerge as a new way of prophylactic treatment which will have demographic relevance.
Acknowledgements
Authors are thankful to the Director, CSIR-National Environmental Engineering Research Institute (CSIR-NEERI) for his support. The manuscript is checked by Knowledge Resource Center, CSIR-NEERI, Nagpur, India and assigned KRC No. CSIR-NEERI/KRC/2020/MARCH/EBGD/2.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coronavirus Disease (COVID-19) Outbreak Situation (2020) WHO. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 2.Coronavirus Cases (2020) Worldometer. https://www.worldometers.info/coronavirus/coronavirus-cases/#daily-cases. Accessed Mar 25, 2020
- 3.Genomic Epidemiology of hCoV-19 (2020). https://www.gisaid.org/epiflu-applications/next-hcov-19-app/. Accessed Mar 25, 2020
- 4.Iceland Scientists Found 40 Mutations of the Coronavirus: Report (2020). https://nypost.com/2020/03/24/iceland-scientists-found-40-mutations-of-the-coronavirus-report-says/. Accessed Mar 25, 2020
- 5.Tang X, Wu C, Li X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020 doi: 10.1093/nsr/nwaa036/5775463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yadav P, Potdar V, Choudhary M, et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020 doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sardar R, Satish D, Birla S, Gupta D. Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host–virus interaction and pathogenesis. bioRxiv. 2020 doi: 10.1101/2020.03.21.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maldonado Galdeano C, Cazorla SI, Lemme Dumit JM, et al. Beneficial effects of probiotic consumption on the immune system. Ann Nutr Metab. 2019;74:115–124. doi: 10.1159/000496426. [DOI] [PubMed] [Google Scholar]
- 9.Colson P, Rolain JM, Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents. 2020;55:105923. doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Bari MAA. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol Res Perspect. 2017;5:e00293. doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao QY, Chen YX, Fang JY. 2019 Novel coronavirus infection and gastrointestinal tract. J Dig Dis. 2020 doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Cao R, Xu M, et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Wang Y, Shao C, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020 doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma H, Patil P, Kolhapure R, Gopalkrishna V. Antiviral activity of the Indian medicinal plant extract, Swertia chirata against herpes simplex viruses: a study by in vitro and molecular approach. Indian J Med Microbiol. 2008;26:322. doi: 10.4103/0255-0857.43561. [DOI] [PubMed] [Google Scholar]
- 16.Patra A, Bhavesh NS. Virtual screening and molecular dynamics simulation suggest Valproic acid Co-A could bind to SARS-CoV2 RNA depended RNA polymerase. OSF. 2020 doi: 10.17605/osf.io/y8uac. [DOI] [Google Scholar]
- 17.Narayanan N, Nair DT (2018) Vitamin B12 may inhibit RNA-dependent-RNA polymerase activity of nsp12 of the COVID-19 virus. Preprint, pp 1–29. 10.20944/preprints201810.0478.v1 [DOI] [PMC free article] [PubMed]
- 18.Purohit HJ. Gut-bioreactor and human health in future. Indian J Microbiol. 2018;58:3–7. doi: 10.1007/s12088-017-0697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schirmer M, Smeekens SP, Vlamakis H, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:1125–1136.e8. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villarino NF, LeCleir GR, Denny JE, et al. Composition of the gut microbiota modulates the severity of malaria. Proc Natl Acad Sci USA. 2016;113:2235–2240. doi: 10.1073/pnas.1504887113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngwa CJ, Pradel G. Coming soon: probiotics-based malaria vaccines. Trends Parasitol. 2015;31:2–4. doi: 10.1016/j.pt.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Maji A, Misra R, Dhakan DB, et al. Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ Microbiol. 2018;20:402–419. doi: 10.1111/1462-2920.14015. [DOI] [PubMed] [Google Scholar]