Abstract
Cluster of differentiation 44 (CD44) as a transmembrane glycoprotein is found to be expressed in non-small cell lung cancer (NSCLC), is significantly associated with NSLC progression, metastasis and drug resistance. This study aimed to explore whether CD44 inhibition improves the sensitivity of epidermal growth factor receptor (EGFR) wild-type NSCLC cells to cisplatin and how it affects wild-type EGFR in NSCLC cells. Small interfering RNA was used to knockdown CD44 expression in EGFR wild-type NSCLC cell line H460. Results suggested that CD44 downregulation reduced cell growth, promoted G0/G1 cell cycle arrest and induced cell apoptosis in H460 cells and these effects were evidently enhanced when in combination with cisplatin. Deactivation of EGFR signaling pathway including EGFR phosphorylation and its downstream molecules, targets ERK, AKT1 and SRC which were also observed in CD44-silenced H460 cells with or without EGF stimulation. Furthermore, the CD44 expression level was positively correlated with wild-type EGFR level in human lung adenocarcinoma tissues and CD44 inhibition significantly accelerated the degradation of EGFR, indicating that enhanced sensitivity of H460 cells to cisplatin by downregulation of CD44 might be due to EGFR degradation. This study demonstrated that suppression of CD44 deactivated EGFR signals in NSCLC cells with wild-type EGFR, thereby contributing to the inhibition of cell proliferation and the reinforcement of cisplatin sensitivity. It is suggested that downregulation of CD44 could be a novel potential therapeutic strategy for the treatment of EGFR wild-type NSCLC.
Keywords: non-small cell lung cancer, CD44, cisplatin, EGFR signaling, combination therapy
Introduction
Non-small cell lung cancer (NSCLC) is one of the leading causes of cancer related death in the world, with its 5-year survival rate <20% after diagnosis (1). Although targeted molecular therapy has achieved great success in treatment of NSCLC, it is usually limited to a group of patients harboring drug-sensitive epidermal growth factor receptor (EGFR) mutations (2). Platinum-based chemotherapy remains the main treatment option for NSCLC with wild-type EGFR, but the efficacy is still not satisfactory. Combination therapy has been widely studied and used to increase the efficacy of tyrosine kinase inhibitors (TKIs) or chemotherapeutics on EGFR wild-type lung cancer cells (3-5). Nevertheless, new therapeutic targets are urgently needed in order to improve the therapeutic outcome of the current therapy for these NSCLC patients.
Cluster of differentiation 44 (CD44), a transmembrane glycoprotein, serves as an oncogenic regulator as well as a cancer stem cell marker in numerous kinds of malignancies (6). CD44 is found to be over-expressed in cancer tissues and was significantly associated with progression, migration and multi-drug resistance of various cancers such as colorectal cancer, breast cancer and lung cancer (7,8). Previous studies have shown that the expression of CD44 is correlated with EGFR level in a variety of neoplasms (9-11). It has been indicated that the TKI erlotinib treatment significantly downregulated the CD44 level and inhibited breast cancer cell migration and invasion (9). Moreover, one study also has shown that the EGFR ligand, EGF increased the expression of CD44 as well as the phosphorylation of ERK, STAT3 and AKT in SKBR3 breast cancer cells (12). On the other hand, it was indicated that CD44 is a promoting modulator for EGFR activation. For example, Perez et al (13) showed that CD44 augmented tumorigenesis and progression in head and neck squamous cell carcinoma through interaction with EGFR. This provides direct evidence for the relationship between CD44 and EGFR signaling. Recently, it has been shown that CD44s, a splicing isoform of CD44, could stabilize protein level of receptor tyrosine kinases (RTKs) through interaction with Rab7A and the absence of CD44 facilitated Rab7A-mediated trafficking of EGFR to lysosomes in glioblastoma cells, contributing to EGFR degradation (14). In breast cancer, specific CD44 subtypes are recruited as co-receptors in the EGFR signaling pathway in a ligand-dependent manner and their specificity is determined by the ligand rather than the receptor itself (15). Hyaluronan facilitates transforming growth factor-β1 (TGF-β1)-dependent activation of MAPK/ERK by promoting the interaction between CD44 and EGFR, thereby promoting cellular proliferation of fibroblasts (16). CD44 appears to be both a co-regulator of RTK signaling and a downstream target of EGFR signaling. However, the relationship of CD44 and EGFR or the role of CD44 in modulation of EGFR signaling in NSCLC cells has not been well investigated.
The present study hypothesized that blocking CD44 may result in altered EGFR signaling and increase sensitivity of wild-type EGFR NSCLC cells to chemotherapeutics such as cisplatin. The present study thus focused on wild-type EGFR NSCLC cell line H460 and investigated the role of CD44 in regulation of EGFR signaling as well as its impact on platinum-based chemotherapy. The present study will provide new perspectives for enhancing the efficacy of chemotherapeutics in clinical treatment for EGFR wild-type NSCLC patients.
Materials and methods
Cell culture
Human EGFR wild-type NSCLC cell line H460 was obtained from the School of Biomedical Sciences, Chinese University of Hong Kong. The cell line was cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) and 100 µg/ml streptomycin/100 U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.). All cells were maintained at 37°C under a humidified atmosphere with 5% CO2 in an incubator (SHEL LAB, Inc.). Cells were passaged by trypsin/EDTA (Gibco; Thermo Fisher Scientific, Inc.) and the culture medium was changed every other day.
RNA interference
The expression of CD44 was downregulated using pre-designed target-specific small interfering RNA (siRNAs). Cells were transfected with siRNA by using the jetPRIME reagent (Polyplus-Transfection SA). The control siRNA (sictr, siN05815122147) and CD44 siRNA (siCD44, siG000000960B) were purchased from Guangzhou RiboBio Co., Ltd. The target sequence of CD44 siRNA was 5′-CCG CTT TGC AGG TGT ATT C-3′. By using the Basic Local Alignment Search Tool (BLAST) at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi), the targeted sequence was found to match well and specifically with both standard and variant forms (transcript variant 1-8, x1-x19) of CD44, suggesting that the designed siCD44 can knock down CD44 and its variant forms. The transfection efficiency was evaluated by RT-qPCR and western blot analysis. A total of 50 nM siCD44 or sictr mixed with jetPRIME reagent (Polyplus-Transfection SA) was added to the cells for 24 h, following which the solution was replaced with normal culture medium. Follow-up experiments were conducted 48 h after transfection.
Cell viability
Cells were seeded at a density of 10,00-3,000 cells/well in 96-well plates (Guangzhou Jet Bio-Filtration Co., Ltd.). Then cells were cultured overnight in an incubator at 37°C. Next, cells were cultured with or without cisplatin at different concentrations (2.5, 5, 10 and 20 µM). Cell viability was detected using a Cell Counting Kit (CCK)-8 (Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol whereby the CCK-8 was diluted with RPMI-1640 (1:10). The culture medium was replaced before adding 100 µl diluted solution per well. The plates were incubated for another 3 h in an incubator. The optical density at 450 nm was then measured with a microplate reader (CYTATION 3; Agilent Technologies, Inc.).
Cell cycle analysis
Cells were seeded into 6-well plates at 1.5×105 cells/well. They were later transfected with siRNAs for 24 h and then treated with or without cisplatin (10 µM) for 24 h. The cells were harvest after digestion by 0.25% trypsin, washed with PBS and fixed with cold 70% ethanol for at least 24 h at −20°C. Fixed cells were washed with PBS and then allowed to incubate for 30 min at 37°C prior to analysis with a propidium iodide solution: 50 µg/ml propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.) and 10 mg/ml RNase A (Roche Diagnostics). Fluorescence intensity of PI was measured by flow cytometry (BD FACS Canto™; Becton, Dickinson and Company), which reflects the individual nuclear DNA content. The proportion of G0/G1, S and G2/M cells in cell cycle distribution were analyzed by ModFit LT software (MFLT32; Verity Software House, Inc.). The experiment was repeated three times.
Apoptosis assay
Apoptosis was analyzed with an Annexin V-FITC/PI detection kit (Invitrogen; Thermo Fisher Scientific, Inc.). Transfected cells were collected and were stained with 5 µl Annexin V-FITC and 10 µl PI per sample in the dark for 10 min at room temperature. Finally, cell apoptosis was detected and quantified by a flow cytometer (BD FACS Canto™; Becton, Dickinson and Company). A total of ~10,000 cells were collected for data analysis.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated using Trizol reagent (Thermo Fisher Scientific, Inc.) based on the manufacturer's protocol. cDNA was synthesized from mRNA using a FastKing RT reagent kit (Tiangen, Inc.). The RT reaction was performed at 42°C for 15 min and 95°C for 3 min. QPCR was performed with a SYBR Green Real Time PCR kit (Thermo Fisher Scientific, Inc.) on CFX96 Touch Real Time PCR System (BioRad Laboratories, Inc.) under the following conditions: 95°C for 1 min, then 40 cycles of 95°C for 5 sec and 60°C for 15 sec. The primers used for qPCR were as follows: Human CD44, forward 5′-TGG AGA AAA ATG GTC GCT ACA G-3′, reverse 5′-GGG CAA GGT GCT ATT GAA AGC-3′; human GAPDH, forward 5′-CTG GGC TAC ACT GAG CAC C-3′, reverse 5′-AAG TGG TCG TTG AGG GCA ATG-3′. The formula 2−ΔΔCq was employed to analyze the fold change of mRNA expression levels (17).
Western blotting
Western blotting was used to test the EGFR signaling pathway-associated proteins, as well as the levels of proteins associated with the cell cycle and apoptosis. RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.) containning protease inhibitor PhosSTOP EASYpack (Roche Diagnostics) was used for total protein extraction according to the manufacturer's protocol. Equal amounts of protein samples (25 µg) were separated by 10% SDS-PAGE gel and transferred to a polyvinylidene fluroride hybridization transfer membrane (Immobilon-p; EMD Millipore). After blocking with 5% bovine serum albumin (BSA, Sigma-Aldrich; Merck KGaA) at room temperature for 1 h and the membranes were probed with the corresponding primary antibodies incubated at 4°C overnight. Afterwards, membranes were inclubated with HRP-conjugated secondary antibodies at room temperature for 2 h. Chemiluminescent signals were finally reacted with ECL blotting detection reagents (Clarity; Bio-Rad Laboratories, Inc.) and the signals were detected by the ChemiDoc™ Imaging System (Bio-Rad Laboratories, Inc.). Band density were evaluated by ImageJ 1.48v (National Institutes of Health). Primary antibodies against phosphorylated (p)-EGFR (Tyr1173; cat. no. 4407; 1:1,000), Src (cat. no. 2123; 1:1,000), p-Src (Tyr416; cat. no. 6943; 1:1,000), CD44 (cat. no. 5640s; 1:1,000), ERK (cat. no. 4695; 1:1,000), p-ERK (cat. no. 4370; 1:1,000), CDK2 (cat. no. 2546s; 1:1,000), CDK4 (cat. no. 12790s; 1:1,000), CDK6 (cat. no. 13331s; 1:1,000), Bax (cat. no. 2774; 1:1,000) and Bcl-2 (cat. no. 4223; 1:1,000) was from Cell Signaling Technology, Inc. Primary antibodies against EGFR (cat. no. ET1603-37; 1:1,000), p-EGFR (Y1068; cat. no. ET1612-30; 1:1,000), GAPDH (cat. no. M1310-2; 1:1,000), AKT1/2/3 (cat. no. ET1609-51; 1:1,000), p-AKT1 (Ser473; cat. no. ET1607-73; 1:1,000), as well as the secondary antibodies of goat anti-rabbit IgG HRP (cat. no. HA1006; 1:1,000) and goat anti-mouse IgG HRP (cat. no. HA1001; 1:1,000) were purchased from HUABIO. Anti-c-Myc (cat. no. 469301-22; 1:200) was purchased from Calbiochem, while anti-CyclinD1 (cat. no. SC450; 1:200) was purchased from Santa Cruz Biotechnology, Inc. Cycloheximide was purchased from MedChemExpress.
Immunofluorescence assay
H460 cells were seeded in 8-well chamber (EMD Milipore) at ~1×104 cells/well. Cells were transfected with CD44 siRNA as well as control siRNA with JetPrime. After 48 h incubation, the cells were washed three times with PBS, fixed for 15 min in 4% paraformaldehyde and permeabilized with 0.5% TritonX-100 (Sigma-Aldrich; Merck KGaA) in PBS for 20 min at room temperature. They were later washed three times with PBS and blocked with 5% BSA for 30 min at room temperature. Cells were incubated with rabbit anti-EGFR (cat. no. ET1603-37; 1:400; HUABIO) and mouse anti-CD44 (cat. no. 5640s; 1:800; Cell Signaling Technology, Inc.) for 1 h at room temperature. Subsequently, the slide was washed with PBS three times and incubated with secondary antibody conjugated to the fluorescent dye Alexa Fluor™ 488 goat anti-rabbit (1966932; 1:800; Invitrogen; Thermo Fisher Scientific, Inc.) and Alexa Fluor™ 568 goat anti-mouse (1906484; 1:1,600; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h in dark at room temperature, and mounted with DAPI (Abcam) mounting medium at room temperature for 5 min. Finally, the images were captured using a fluorescent microscope (Nikon Ts2R; Nikon Corporation).
The cancer genome atlas (TCGA) data collection and bioinformatics analysis
The scatter diagram was based on Log2(RSEM+1) analysis using data from 255 lung adenocar-cinoma samples in TCGA (https://portal.gdc.cancer.gov/) and tumor samples (n=224) with EGFR wild-type were selected. The cBioPortal (http://www.cbioportal.org/) for Cancer Genomics provides information on EGFR mutation.
Statistical analysis
Statistical analysis of the data was carried out using GraphPad Prism 7.0 (GraphPad Software, Inc.). The data are expressed as mean ± SD of three independent experiments. Unpaired student t test, or one-way ANOVA with a post hoc Tukey test was used for analysis of difference between two groups or among three or more groups, respectively. The spearman correlation between EGFR and CD44 was analyzed by GraphPad Prism 7.0 (GraphPad Software, Inc.). P<0.05 was considered as significant difference.
Results
Downregulation of CD44 inhibits growth of EGFR wild-type NSCLC cells and the effect is enhanced when combined with cisplatin treatment
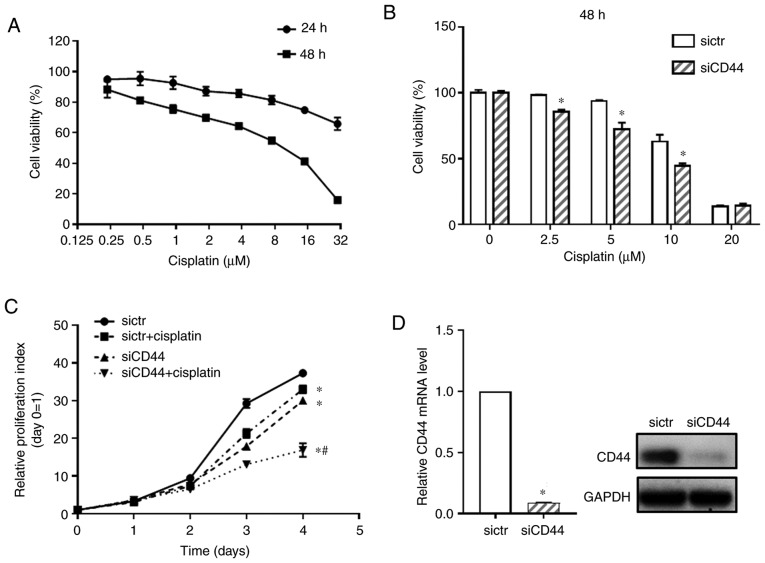
First, the cytotoxicity of cisplatin on EGFR wild-type NSCLC cell H460 was detected (Fig. 1A). Cell viability of H460 cells treated with cisplatin at concentrations from 0.2 to 30 µM were tested by using the CCK-8 assay and IC50 of cisplatin at 48 h was 6.852±0.405 µM. To examine the functional relevance of CD44 expression in cisplatin sensitivity of EGFR wild-type NSCLC cells, the CD44 level in the H460 cell line was downregulated using specific RNA interference. It was confirmed that, compared with blank cells, transfection with sictr did not significantly affect CD44 expression and cell growth of H460 cells (Fig. S1). Thus, the H460 cells transfected with sictr were used for direct comparison with that transfected with siCD44 in the following experiments. As shown in Fig. 1B, after knockdown of CD44, cisplatin significantly inhibited cell growth of H460 at concentration of 2.5, 5 and 10 µM when compared with the sictr group and 10 µM for cisplatin was selected following in vitro studies. The proliferative curve of H460 was further detected after silencing CD44 combined with cisplatin treatment (10 µM). As shown in Fig. 1C, knockdown of CD44 significantly inhibited cell growth of H460. Additionally, the present study found that downregulation of CD44 followed by cisplatin treatment inhibited the proliferation of H460 cell compared with sictr + cisplatin group. Fig. 1D shows that knockdown efficiency of siCD44 in H460 was >90% at 48 h post transfection based on RT-qPCR assessment. Although there was no synergistic effect of siCD44 and cisplatin, their combination was effective in suppressing tumor cells. The results also indicated cisplatin treatment might be more effective and could achieve a better outcome for wild-type EGFR positive NSCLC patients with low CD44 expression.
Figure 1.
Knockdown of CD44 inhibits cell growth of H460 cells and enhances cisplatin sensitivity. (A) Cisplatin cytotoxicity of H460 for 48 h was tested by a CCK-8 assay. (B) sictr and siCD44 were transfected into H460 cells followed by cisplatin treatment at concentrations of 2.5, 5 and 10 µM for 48 h. Cell viability of H460 was detected by CCK-8 assay. (C) After transfection with sictr and siCD44 respectively, H460 cells were treated with or without cisplatin (10 µM) and proliferative curves of H460 cells were assessed at 0, 1, 2, 3 and 4 days post treatment using CCK-8. (D) The knockdown efficiency of CD44 in H460 was evaluated using reverse transcription-quantitative PCR and western blotting. Data are displayed as mean ± SD of 3 independent experiments. *P<0.05 vs. sictr; #P<0.05 vs. sictr + cisplatin. CCK-8, Cell Counting Kit-8; sictr, control siRNA; siCD44, CD44 siRNA.
Downregulation of CD44 promotes G0/G1 cell cycle arrest and apoptosis in H460 cells and the effects are significantly enhanced when in combination with cisplatin treatment
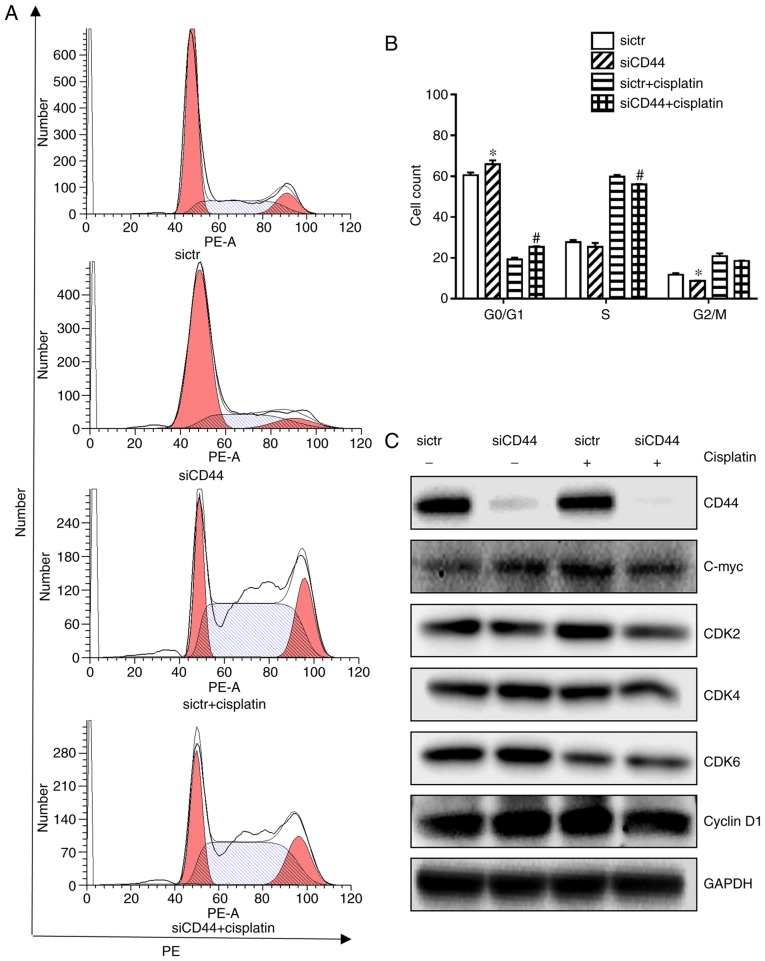
It has been reported that heterodimeric complexes of CDK4/6 with cyclin D and CDK3 with cyclin C regulated cell-cycle transition from G0 to G1 and early phases of G1 through the phosphorylation of retinoblastoma protein (18) while S and G2/M phase control were mediated by CDK2 and CDK1 (19). It was further observed that knockdown of CD44 in H460 cells led to a significant G0/G1 cell cycle arrest compared with the control (Fig. 2A and B), which was associated with downregulation of cell cycle related proteins such as C-myc, CDK2, CDK4, and CDK6 (Fig. 2C). Cisplatin alone induced a slight S phase arrest in H460 cells (Fig. 2B) with decreased expression of CDK6 (Fig. 2C). Its combination with CD44 silencing resulted in a significant increase of G0/G1 populations in H460 cells (Fig. 2B) with a further repression of corresponding proteins (Fig. 2C). The cyclin D1 expression remain unchanged (Fig. 2C), suggesting that neither cisplatin treatment nor siCD44 could influence cyclin D1 expression in the NSCLC cell line H460. The cell cycle arrest is believed to be mainly relying on the downregulation of CDKs as well as other cyclins, such as cyclin C.
Figure 2.
Effects of siCD44, cisplatin or their combination on cell cycle distribution of H460 cells. H460 cells were seeded into 6-well plate at 1.5×105 cells/well and transfected with sictr and siCD44 respectively for 24 h, followed by treatment of cisplatin at concentration of 10 µM for another 24 h. (A) Transfected cells were stained with PI for cell cycle analysis by flow cytometry. Cell distribution was analyzed by MFLT32. (B) Corresponding statistical analysis of cell distribution. (C) The protein expressions of CD44, c-Myc, CDK2, CDK4, CDK6 and cyclin D1 in H460 were analyzed by western blotting, with GAPDH as loading control. Data were presented as mean ± SD of 3 independent experiments. *P<0.05 vs. sictr; #P<0.05 vs. sictr + cisplatin. sictr, control siRNA; siCD44, CD44 siRNA; PI, propidium iodide.
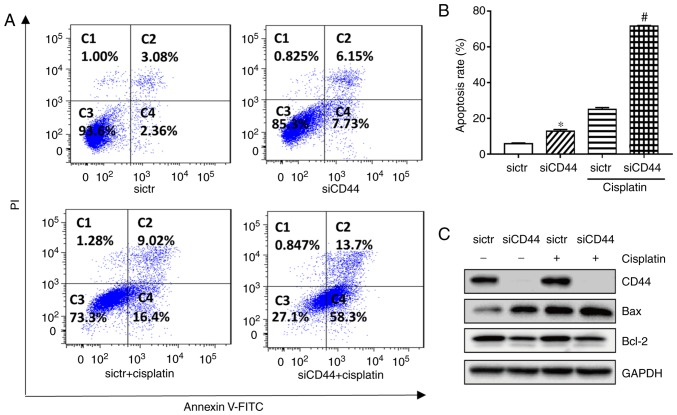
Flow cytometry results indicated that knockdown of CD44 significantly increased the apoptotic population from 5.8 to 13.2% (Fig. 3A and B) in H460 cells. Western-blot analysis showed that expression of the pro-apoptotic protein Bax was increased while that of anti-apoptotic protein Bcl-2 was decreased after knockdown of CD44 in H460 cells (Fig. 3C). The apoptotic rate of cisplatin alone was 25.4% in H460, while its combination with CD44 knockdown resulted in an enhanced anticancer effect. The apoptotic population in the combination group increased to 72.0% in H460, while the corresponding protein Bax and Bcl-2 level was changed (Fig. 3). The results demonstrated that suppression of CD44 promoted cell cycle arrest and cell apoptosis of EGFR wild-type NSCLC cells and further increased the antitumor effect of cisplatin.
Figure 3.
Effects of siCD44, cisplatin or their combination on cell apoptosis of H460 cells. H460 cells were seeded into 6-well plate at 1.5×105 cells/well and transfected with sictr or siCD44 respectively for 24 h, followed by treatment of cisplatin at concentration of 10 µM for another 48 h. Cell apoptosis of H460 was determined with an Annexin V-FITC/PI detection kit by (A) flow cytometry, with (B) the apoptotic rate calculated from early (Annexin V+/PI-) and late (Annexin V+/PI+) apoptotic populations. (C) Protein expression of CD44, BAX and Bcl-2 was analyzed by western blotting, with GAPDH as loading control. Data were presented as mean ± SD of 3 independent experiments. *P<0.05 vs. sictr; #P<0.05 vs. sictr + cisplatin. sictr, control siRNA; siCD44, CD44 siRNA; PI, propidium iodide.
EGFR signaling is attenuated by CD44 silencing in EGFR wild-type NSCLC cells
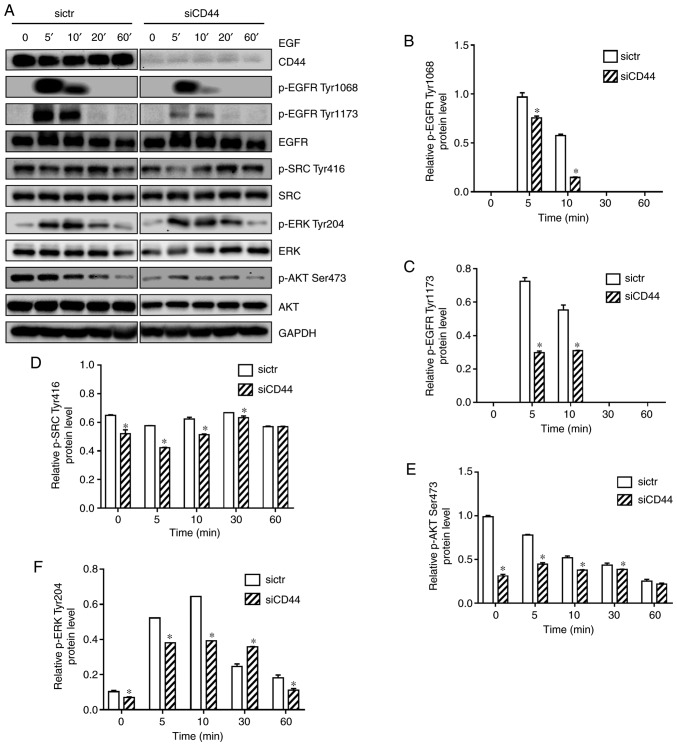
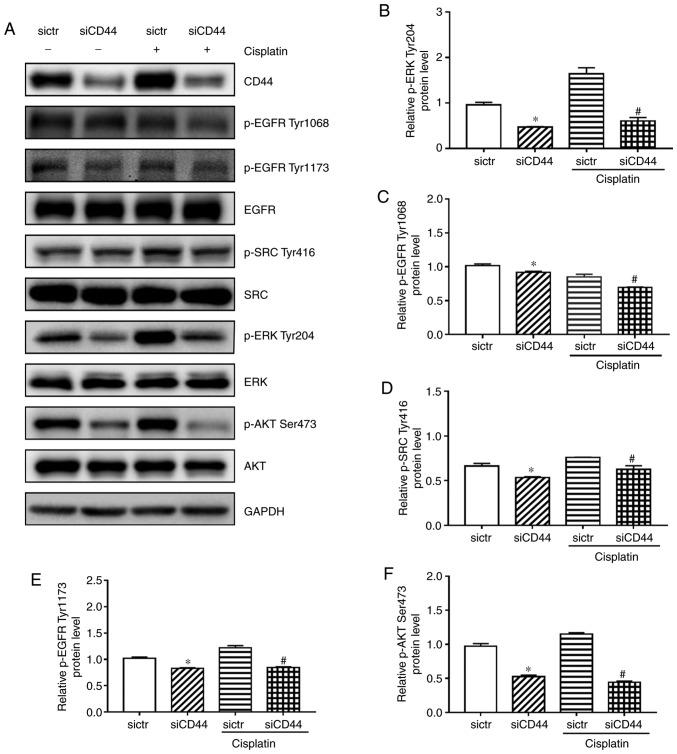
In order to investigate the effect of silencing CD44 on EGFR signaling in H460 cells, western blotting was performed to discover the expression level of EGFR, AKT and ERK as well as their phosphorylated form. EGF was used to stimulate the EGFR signaling pathway in H460 cells. As shown in Fig. 4A, EGF-mediated activation of EGFR in H460 cells was reduced in the siCD44 group, as reflected by protein expression of p-EGFR (Tyr1068 and Tyr1173) within 60 min. Furthermore, protein expression of p-SRC (Tyr416), p-AKT (Ser473), and p-ERK (Tyr204) was decreased, suggesting that the activation of EGFR downstream signaling including SRC, ERK and AKT was attenuated. All experiments were performed in triplicate and the quantification results for the protein band intensity are shown in Fig. 4B-F. The results indicated that knockdown of CD44 attenuated EGF-mediated stimulation of EGFR signaling in H460 cells. It is further confirmed that siCD44 led to direct inhibition of EGFR signaling (Fig. 5A), which might contribute to the increased sensitivity of EGFR wild-type NSCLC cells to chemotherapy.
Figure 4.
Downregulation of CD44 attenuates EGF-mediated activation of EGFR signaling, potentially through enhanced EGFR degradation. (A) Cells were seeded into 6-well plate at 1.5×105 cells/well and transfected with sictr or siCD44 for 24 h. After stimulation by EGF (50 ng/ml) for 0, 5, 10, 20 and 60 min, total protein were collected for western blotting. The protein expression of EGFR, p-EGFR Tyr1068, p-EGFR Tyr1173, SRC, p-SRC Tyr416, AKT, p-AKT Ser473, ERK and p-ERK Tyr204 were assessed. Accordingly, the relative expressions of (B) p-EGFR Tyr1068, (C) p-EGFR Tyr1173, (D) p-SRC Tyr416, (E) p-AKT Ser473, and (F) p-ERK Tyr204 to GAPDH were calculated by ImageJ. Data were presented as mean ± SD of 3 independent experiments. *P<0.05 vs. sictr. sictr, control siRNA; siCD44, CD44 siRNA; p, phosphorylated; EGFR, epidermal growth factor receptor.
Figure 5.
Influences of siCD44, cisplatin or their combination on EGFR signaling in H460 cells. H460 cells were seeded into 6-well plate at 1.5×105 cells/well and transfected with sictr or siCD44 for 24 h. (A) The protein levels of (B) p-ERK Tyr204 (C) p-EGFR Tyr1068, (D) p-SRC Tyr416, (E) p-EGFR Tyr1173, (F) p-AKT Ser473, were assessed by western blotting, with their relative expression to GAPDH calculated using ImageJ. Data are presented as mean ± SD of 3 independent experiments. *P<0.05 vs. sictr; #P<0.05 vs. sictr + cisplatin. sictr, control siRNA; siCD44, CD44 siRNA; p, phosphorylated; EGFR, epidermal growth factor receptor.
CD44 inhibition enhances cisplatin sensitivity in H460 cells due to possible deactivation of EGFR signaling
The current study demonstrated that EGFR signaling pathway was attenuated after silencing of CD44 in H460 cells (Fig. 4), which could be the reason for siCD44-induced higher efficacy of cisplatin. In order to confirm that, the EGFR signaling activation including the phosphorylation of EGFR, SRC, AKT and ERK was observed after knockdown of CD44 as well as in combination with cisplatin. While cisplatin alone had a limited effect on EGFR signaling, as revealed by the unchanged protein expression levels of p-EGFR (Tyr1068 and Tyr1173), p-SRC (Tyr416), p-AKT (Ser473), and p-ERK (Tyr204), cisplatin plus CD44 knockdown led to significantly repressed phosphorylation of these proteins (Fig. 5). These results confirmed that suppression of EGFR signaling by siCD44 contributed to the increased efficacy of cisplatin on EGFR wild-type NSCLC cells.
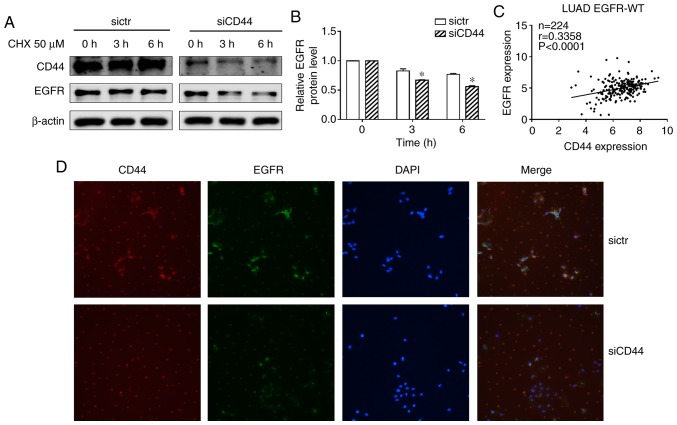
Downregulation of CD44 enhances EGFR degradation in H460 and EGFR level is positively correlated with CD44 expression in EGFR wild-type NSCLC tissues
A previous study suggested that CD44 may interfere with EGFR degradation (14). In the present experiments, a protein synthesis inhibitor cycloheximide (CHX) was used to block protein synthesis in NSCLC cells. The EGFR level decreased after treatment with CHX in the siCD44 group suggesting that CD44 stabilized the EGFR level while CD44 silencing accelerated EGFR degradation (Fig. 6A and B). This indicated that suppression of CD44 expression contributed to a reduction in EGFR activation via increased degradation of EGFR. An immunofluorescence (IF) assay was also performed to stain CD44 and EGFR to confirm this result. As shown in Fig. 6D, after knockdown of CD44, EGFR IF staining was significantly decreased, which suggested that siCD44 promoted EGFR degradation, thereby resulting in deactivation of EGFR downstream signaling. On the other hand, in order to elucidate the correlation between CD44 and EGFR in lung cancer, the CD44 and EGFR expression data of lung adenocarcinoma patient tissues with wild-type EGFR was obtained from TCGA database. The correlation between CD44 and EGFR mRNA level was analyzed and the result showed that CD44 was positively associated with EGFR level in EGFR wild-type lung cancer tissues (Fig. 6C). This further confirmed the findings and pointed to the importance of screening CD44 expression in lung cancer tissue to achieve a better outcome for lung cancer patients undergoing chemotherapy.
Figure 6.
Downregulation of CD44 enhances EGFR degradation in H460 and EGFR level is positively correlated with CD44 expression in EGFR wild-type lung cancer tissues. (A) H460 cells were transfected with sictr or siCD44 for 48 h, followed by treatment with CHX for 0, 3 and 6 h. Protein expression of CD44 and EGFR were analyzed by western-blotting. (B) Their relative expression to GAPDH was calculated by Image J. (C) The correlation between CD44 and EGFR in EGFR wild-type lung adenocarcinoma tissues (n=224). (D) Immunofluorescence staining of CD44 and EGFR (magnification, ×400) after knockdown of CD44 as well as vehicle control in H460 cells. Data were presented as mean ± SD. *P<0.05 vs. sictr. sictr, control siRNA; siCD44, CD44 siRNA; p, phosphorylated; EGFR, epidermal growth factor receptor; CHX, cycloheximide.
Discussion
EGFR mutations or hyperactivation is common among lung cancer patients. While EGFR TKIs achieved a favorable outcome in EGFR-mutant patients and were recommended as first-line treatment for patients with positive EGFR mutation (20), chemotherapeutics including cisplatin, carboplatin, etoposide and docetaxel were used as the first line of treatment for EGFR-wild-type patients (21). However, the outcome of chemotherapy in EGFR wild-type NSCLC tumors remains poor (22). Cisplatin, a commonly used chemotherapeutic, is widely used for treatment of NSCLC patients, while both its cytotoxicity and effective dose are relatively high (23). It is important to find novel therapeutic strategies to increase the efficacy of cisplatin while reducing its side effects. The current study pointed to the functional role of CD44 in regulating wild-type EGFR activation, which provided a way to increase the effectiveness of cisplatin in treatment of EGFR wild-type NSCLC cells.
CD44 is commonly considered to be a cancer stem cell marker and it is found overexpressed in lung cancer tissues especially in lung cancer stem cells. High expression of CD44 in lung cancer tissue was significantly correlated with poor prognosis of NSCLC patients (24). A previous finding showed that the absence of CD44 in lung cancer cells could result in suppression of cancer stem cell properties including downregulation of tumor occurrence and reversing multi-drug resistance (25). Knockdown of CD44 might benefit numerous chemotherapeutic treatments in wild-type EGFR NSCLC. The present results showed that suppression of CD44 expression increased the sensitivity of H460 to cisplatin. Additionally, cell cycle arrest and cell apoptosis were induced by knockdown of CD44 and the effects were enhanced when in combination with cisplatin. These results suggested EGFR wild-type NSCLC patients with low CD44 expression might achieve a favorable outcome from chemotherapeutic treatment.
Previous studies have indicated that expression of CD44 and EGFR was significantly correlated in a variety of cancer types; both were overexpressed in cancer tissues (14,24,26,27), and they can be considered as prognostic factors for cancer patients. It was also reported that a CD44 isoform could attenuate Rab7-induced EGFR degradation by lysosomes in glioblastoma (14), which provides a novel mechanism for CD44 activated the EGFR signaling pathway. The current study further investigated the related mechanism and found that EGF-mediated activation of EGFR signaling was restrained by CD44 silencing. In addition, knockdown of CD44 augments EGFR degradation in CHX in the treatment group, indicating the mechanism underlying the relationship between CD44 and EGFR was consistent with previous findings (14). The modulation of EGFR expression as well as its downstream signaling pathway by siCD44 was further supported by the results of the IF assay and bioinformatics data analysis. The clinical sample data from TCGA database indicated the positive association between CD44 and EGFR mRNA level in EGFR wild-type lung cancer tissue samples. CD44 might be involved in EGFR degradation through interaction with wild-type EGFR in NSCLC cells, thereby affecting its downstream AKT and ERK. Downregulation of CD44 may benefit lung cancer patients undergoing chemotherapy such as cisplatin treatment.
The current results demonstrated that CD44 inhibition enhanced cisplatin sensitivity in H460 cells, which might be due to the downregulating of EGFR signaling activation. Knockdown of CD44 decreased the EGFR protein level and phosphorylation and affected the stimulation of the down-stream signaling, thereby enhancing sensitivity of lung cancer cells to cisplatin. The present results suggested that down-regulation of CD44 could be considered as a useful strategy for cisplatin treatment in wild-type EGFR NSCLC and the expression level of CD44 in NSCLC tissue might be viewed as an effective prognostic factor for NSCLC patients.
Supplementary Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81803237 and 81602578) and the Joint Funds of the Southwest Medical University & Luzhou, China (grant no. 2018LZXNYD-ZK34).
Availability of data and materials
The analyzed data sets generated during the present study are available from corresponding author on reasonable request.
Authors' contributions
ML, XW and DY designed and conceived the study. JY, YZhang, HZ and YW performed the experiments. JY, XW and YZhao analyzed the data. JL, ZX, JS, YZhao, ZZ, FD, PJK, CHC, LL and CH provided the resources and experiments for the study. XW wrote the first draft. ML, HZ and XW reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Noone AM, Howlader N, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al., editors. SEER cancer statistics review, 1975-2015. National Cancer Institute; Bethesda, MD: https://seer.cancer.gov/csr/1975_2015/, based on November 2017, SEER data submission, posted to the SEER web site. Accessed September 10, 2018. [Google Scholar]
- 2.Jiang W, Cai G, Hu PC, Wang Y. Personalized medicine in non-small cell lung cancer: A review from a pharmacogenomics perspective. Acta Pharm Sin B. 2018;8:530–538. doi: 10.1016/j.apsb.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang JC, Ren YG, Zhao J, Long F, Chen JY, Jiang Z. Shikonin enhances sensitization of gefitinib against wild-type EGFR non-small cell lung cancer via inhibition PKM2/stat3/cyclinD1 signal pathway. Life Sci. 2018;204:71–77. doi: 10.1016/j.lfs.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T, Koizumi T, Agatsuma T, Yasuo M, Tsushima K, Kubo K, Eda S, Kuraishi H, Koyama S, Hachiya T, Ohura N. A phase II trial of erlotinib in patients with EGFR wild-type advanced non-small-cell lung cancer. Cancer Chemother Pharmacol. 2012;69:1241–1246. doi: 10.1007/s00280-012-1831-0. [DOI] [PubMed] [Google Scholar]
- 5.Su SF, Li M, Geng YC, Yang WG, Ma Z, Li QS, Hu YX, Ou Yang WW, Liu LF, Lu B. Randomized phase II study of pemetrexed-cisplatin or docetaxel-cisplatin plus thoracic intensity-modulated radiation therapy in patients with stage IV lung adenocarcinoma. Am J Cancer Res. 2019;9:1235–1245. [PMC free article] [PubMed] [Google Scholar]
- 6.Toole BP, Slomiany MG. Hyaluronan: A constitutive regulator of chemoresistance and malignancy in cancer cells. Semin Cancer Biol. 2008;18:244–250. doi: 10.1016/j.semcancer.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Zhao S, Karnad A, Freeman JW. The biology and role of CD44 in cancer progression: Therapeutic implications. J Hematol Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park NR, Cha JH, Jang JW, Bae SH, Jang B, Kim JH, Hur W, Choi JY, Yoon SK. Synergistic effects of CD44 and TGF-β1 through AKT/GSK-3β/β-catenin signaling during epithelial-mesenchymal transition in liver cancer cells. Biochem Biophys Res Commun. 2016;477:568–574. doi: 10.1016/j.bbrc.2016.06.077. [DOI] [PubMed] [Google Scholar]
- 9.Xu H, Wu K, Tian Y, Liu Q, Han N, Yuan X, Zhang L, Wu GS, Wu K. CD44 correlates with clinicopathological characteristics and is upregulated by EGFR in breast cancer. Int J Oncol. 2016;49:1343–1350. doi: 10.3892/ijo.2016.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grass GD, Tolliver LB, Bratoeva M, Toole BP. CD147, CD44, and the epidermal growth factor receptor (EGFR) signaling pathway cooperate to regulate breast epithelial cell invasiveness. J Biol Chem. 2013;288:26089–26104. doi: 10.1074/jbc.M113.497685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wobus M, Rangwala R, Sheyn I, Hennigan R, Coila B, Lower EE, Yassin RS, Sherman LS. CD44 associates with EGFR and erbB2 in metastasizing mammary carcinoma cells. Appl Immunohistochem Mol Morphol. 2002;10:34–39. doi: 10.1097/00129039-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Kil WH, Lee J, Oh SJ, Han J, Jeon M, Jung T, Lee SK, Bae SY, Lee HC, et al. Zerumbone suppresses EGF-induced CD44 expression through the inhibition of STAT3 in breast cancer cells. Oncol Rep. 2014;32:2666–2672. doi: 10.3892/or.2014.3514. [DOI] [PubMed] [Google Scholar]
- 13.Perez A, Neskey DM, Wen J, Pereira L, Reategui EP, Goodwin WJ, Carraway KL, Franzmann EJ. CD44 interacts with EGFR and promotes head and neck squamous cell carcinoma initiation and progression. Oral Oncol. 2013;49:306–313. doi: 10.1016/j.oraloncology.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Zhang H, Liu S, Kim CK, Xu Y, Hurley LA, Nishikawa R, Nagane M, Hu B, Stegh AH, et al. Internalized CD44s splice isoform attenuates EGFR degradation by targeting Rab7A. Proc Natl Acad Sci USA. 2017;114:8366–8371. doi: 10.1073/pnas.1701289114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morath I, Jung C, Lévêque R, Linfeng C, Toillon RA, Warth A, Orian-Rousseau V. Differential recruitment of CD44 isoforms by ErbB ligands reveals an involvement of CD44 in breast cancer. Oncogene. 2018;37:1472–1484. doi: 10.1038/s41388-017-0030-1. [DOI] [PubMed] [Google Scholar]
- 16.Meran S, Luo DD, Simpson R, Martin J, Wells A, Steadman R, Phillips AO. Hyaluronan facilitates transforming growth factor-β1-dependent proliferation via CD44 and epidermal growth factor receptor interaction. J Biol Chem. 2011;286:17618–17630. doi: 10.1074/jbc.M111.226563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Chohan TA, Qayyum A, Rehman K, Tariq M, Akash MSH. An insight into the emerging role of cyclin-dependent kinase inhibitors as potential therapeutic agents for the treatment of advanced cancers. Biomed Pharmacother. 2018;107:1326–1341. doi: 10.1016/j.biopha.2018.08.116. [DOI] [PubMed] [Google Scholar]
- 19.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018;17:38. doi: 10.1186/s12943-018-0777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park K, Vansteenkiste J, Lee KH, Pentheroudakis G, Zhou C, Prabhash K, Seto T, Voon PJ, Tan DSW, Yang JCH, et al. Pan-Asian adapted ESMO clinical practice guidelines for the management of patients with locally-advanced unresectable non-small-cell lung cancer: A KSMO-ESMO initiative endorsed by CSCO, ISMPO, JSMO, MOS, SSO and TOS. Ann Oncol. 2020;31:191–201. doi: 10.1016/j.annonc.2019.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Su C, Zhou F, Shen J, Zhao J, O'Brien M. Treatment of elderly patients or patients who are performance status 2 (PS2) with advanced non-small cell lung cancer without epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) translocations-Still a daily challenge. Eur J Cancer. 2017;83:266–278. doi: 10.1016/j.ejca.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Huang TH, Wu TH, Guo YH, Li TL, Chan YL, Wu CJ. The concurrent treatment of Scutellaria baicalensis Georgi enhances the therapeutic efficacy of cisplatin but also attenuates chemotherapy-induced cachexia and acute kidney injury. J Ethnopharmacol. 2019;243:112075. doi: 10.1016/j.jep.2019.112075. [DOI] [PubMed] [Google Scholar]
- 24.Roudi R, Madjd Z, Korourian A, Mehrazma M, Molanae S, Sabet MN, Shariftabrizi A. Clinical significance of putative cancer stem cell marker CD44 in different histological subtypes of lung cancer. Cancer Biomark. 2014;14:457–467. doi: 10.3233/CBM-140424. [DOI] [PubMed] [Google Scholar]
- 25.Quan YH, Lim JY, Choi BH, Choi Y, Choi YH, Park JH, Kim HK. Self-targeted knockdown of CD44 improves cisplatin sensitivity of chemoresistant non-small cell lung cancer cells. Cancer Chemother Pharmacol. 2019;83:399–410. doi: 10.1007/s00280-018-3737-y. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Z, Shao N, Weng H, Li W, Zhang J, Zhang L, Yang L, Ye S. Correlation between epidermal growth factor receptor and tumor stem cell markers CD44/CD24 and their relationship with prognosis in breast invasive ductal carcinoma. Med Oncol. 2015;32:275. doi: 10.1007/s12032-014-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rho JH, Ladd JJ, Li CI, Potter JD, Zhang Y, Shelley D, Shibata D, Coppola D, Yamada H, Toyoda H, et al. Protein and glycomic plasma markers for early detection of adenoma and colon cancer. Gut. 2018;67:473–484. doi: 10.1136/gutjnl-2016-312794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analyzed data sets generated during the present study are available from corresponding author on reasonable request.