Summary
Objective Surveillance programs and research for acute respiratory infections in remote Aboriginal communities are complicated by difficulties in the storage and transport of frozen samples to urban laboratories for testing. This study assessed the sensitivity of a simple method for transporting respiratory samples from a remote setting for viral PCR compared with frozen specimens.
Methods We sampled every individual who presented to a remote Aboriginal community clinic in a non‐epidemic respiratory season. Two anterior nasal swabs were collected from each participant. The left nare specimen was mailed to the laboratory via routine postal services. The right nare specimen was transported frozen. Testing for 16 viruses was undertaken using real‐time multiplex PCR.
Results A total of 140 participants were enrolled who contributed 150 study visits. Respiratory illnesses accounted for 10% of the reasons for presentation. Sixty‐one viruses were identified in 50 (33.3%) presentations for 40 (28.6%) individuals; bocavirus and rhinovirus were the most common viruses identified (14.0% and 12.6% of episodes respectively). The sensitivity for any virus detected in mailed specimens was 67.2% (95%CI 55.4, 78.9) compared to 65.6% (95%CI 53.7, 77.5) for frozen specimens.
Conclusion The mailing of unfrozen nasal specimens from remote communities does not compromise the viability of the specimen for viral studies.
Keywords: respiratory viruses, specimen transport, remote area, respiratory infection
Application réussie d’une méthode simple de transport d’échantillons pour la surveillance des virus respiratoires dans les communautés indigènes éloignées en Australie
Objectif: Les programmes de surveillance et de recherche pour les infections respiratoires aiguës dans les collectivités aborigènes éloignées sont compliqués par les difficultés pour le stockage et le transport des échantillons congelés vers les laboratoires urbains pour être testés. Cette étude a évalué la sensibilité d’une méthode simple pour le transport des échantillons respiratoires d’un endroit éloigné pour subir une PCR virale par rapport à celle des échantillons congelés.
Méthodes: Des échantillons ont été prélevés chez chaque individu qui s’est présentéà une clinique de la communauté aborigène éloignée au cours d’une saison non épidémique pour les infections respiratoires. Deux écouvillons nasaux antérieurs ont été recueillis par participant. Le spécimen nasal gauche a été posté au laboratoire par l’intermédiaire des services postaux de routine. Le spécimen nasal droit a été transporté congelé. Les échantillons ont été testés pour 15 virus à l’aide de la PCR multiplex en temps réel.
Résultats: 140 participants qui ont contribuéà 150 visites ont été inscrits dans l’étude. Les maladies respiratoires représentaient 10% des raisons de visite. 61 virus ont été identifiés chez 50 (33,3%) des visites de 40 (28,6%) individus; bocavirus et rhinovirus étaient les plus fréquents (14,0% et 12,6% des épisodes, respectivement). La sensibilité pour la détection de tout virus dans les échantillons postés était de 67,2% (IC 95%: 55,4–78,9), comparéà 65,6% (IC95%: 53,7–77,5) pour les spécimens congelés.
Conclusion: L’envoi de prélèvements nasaux non congelés à partir des collectivités éloignées ne compromet pas la viabilité de l’échantillon pour les études virales.
Keywords: virus respiratoires, transport d’échantillon, distance, infections respiratoires
Aplicación exitosa de un método simple de transporte de muestras para realizar una vigilancia de virus respiratorios en comunidades indígenas remotas en Australia
Objetivo: Los programas de vigilancia e investigación de infecciones respiratorias agudas en comunidades remotas de Aborígenes tienen dificultades con el almacenaje y transporte de muestras congeladas hasta laboratorios urbanos para ser evaluadas. En este estudio se ha evaluado la sensibilidad de un método simple para transportar muestras respiratorias desde un emplazamiento remoto, comparado con muestras congeladas, para realizar PCR virales.
Métodos: Hemos tomado muestras de cada individuo que se presentó en alguna clínica comunitaria Aborigen remota, en una estación no epidémica para infecciones respiratorias. Se recolectaron dos frotis nasales anteriores de cada participante. La muestra de la fosa nasal izquierda se envió por correo al laboratorio mediante el servicio postal rutinario. La muestra de la fosa nasal derecha se transportó congelada. Se realizaron pruebas para 15 virus mediante PCR multiplex a tiempo‐real.
Resultados: Se incluyeron 140 participantes que contribuyeron con 150 visitas dentro del estudio. Las enfermedades respiratorias explicaban un 10% de las razones por las cuales buscaban ayuda. Se identificaron 61 virus en 50 (33.3%) presentaciones para 40 (28.6%) individuos; los más comunes eran bocavirus y rhinovirus (14.0% y 12.6% de los episodios respectivamente). La sensibilidad para cualquier virus detectado en las muestras enviadas por correo era del 67.2% (95%IC 55.4, 78.9) comparado con 65.6% (95%IC 53.7, 77.5) para las muestras congeladas.
Conclusión: El envío por correo de muestras nasales sin congelar desde comunidades remotas no compromete la viabilidad de las muestras para estudios de virus.
Introduction
The rates of acute and chronic infections of the upper and lower respiratory tract in Indigenous children in remote communities in Australia are among the highest reported worldwide (O’Grady et al. 2010). Furthermore, repeat infections in childhood are thought to contribute to the high rates of chronic lung disease in both adolescents and adults in these communities (Valery et al. 2004).
Despite the excess burden of disease, with the exception of ear disease (Leach et al. 1994; Morris et al. 2005), there are no studies that have addressed the aetiology and epidemiology of these infections at the community level. One of the major impediments to this work has been the limited capacity for the collection, handling and transport of respiratory specimens. There is a critical need for studies to both better understand respiratory disease epidemiology and to evaluate the efficacy of interventions such as vaccines. Given the uncertainty about the interplay between viruses and bacteria in the disease process, viral studies should be a mandatory component of this research.
To inform the design of surveillance and intervention studies addressing respiratory infections in remote communities, we compared the sensitivity of a simple, cost‐efficient method for transporting respiratory samples from a remote setting for viral real‐time PCR with transport using frozen specimens. Our primary hypothesis was that there was no difference in the proportion of viruses detected between mailed and frozen discordant pairs.
Methods
Setting
The study was conducted in a remote desert community in Central Australia over a period of 3 weeks from late August to mid‐September 2009. The community is approximately 5 h drive from the nearest urban centre, with almost half of that drive being on unsealed roads that can become inaccessible during rainy periods. It has an estimated resident population of 580, although this fluctuates throughout the year. Daily presentation at the community clinic ranges from 20 to 50 people.
Study participants
We convenience‐sampled any person presenting to the local health clinic who had spent the majority of the previous 14 days in the community. Individuals could either be presenting with a medical complaint, visiting, or accompanying a friend or family member. Individuals could participate in the study multiple times during the course of the 3 weeks.
Data collection
We collected basic demographic and clinical data from enrolled subjects which included Indigenous status, reason for presentation at the clinic, clinic diagnosis for those seen by clinic staff, current respiratory and general symptoms, temperature, heart rate, oxygen saturation, respiratory rate, medications taken within the previous 7 days and vaccination history.
Clinical specimens
Two anterior nose swabs (from the nasal vestibule just inside the nares), one from each nostril, were collected by one of two study investigators using the Virocult® collection system (MW950, Medical Wire & Equipment, England) consisting of a rayon swab on a plastic shaft, with viral transport medium (VTM)‐soaked foam pad in the base of the transport tube. The efficiency of this system for detection of influenza virus has been compared favourably to nasopharyngeal nylon‐flocked swabs in combination with a universal transport medium (Esposito et al. 2010). Swabs taken from the left nostril were placed in the clinic refrigerator and transported by normal post (air transport) in standard padded postbags from the community once a week as exempt human specimens in accordance with Australia Post’s Dangerous and Prohibited Goods and Packaging requirements (Australia Post, 2009). Posted specimens were transported at ambient temperature for the entire journey to the Queensland Paediatric Infectious Diseases (Qpid) Laboratory, Royal Children’s Hospital, Brisbane. Specimens left the clinic on Thursday morning and arrived at Qpid by midday the following Monday where they were immediately frozen at −70 °C. Swabs taken from the right nostril were placed in a portable freezer with a minimum temperature capacity of within −50 °C of the current ambient temperature (Engel‐MT60F‐G4‐S). At the end of 3 weeks, the freezer was transported by car to Darwin using a portable battery power supply (Calibre‐PLU127904). Specimens were not thawed in Darwin. They were stored at −20 °C until transported to Qpid on dry ice by overnight air transport (total transport time = 16 h) where specimens were frozen on arrival in a −70 °C freezer and stored until they were batch tested for viruses at the end of the study. The fridge and freezer were temperature monitored throughout the study using Escort Junior loggers (Escort Data Logging Systems, Auckland) with twice daily visual inspections of temperature, and 5‐minutely electronic logging of temperature. A logger was sent with the final post of specimens to monitor the temperature range experienced by specimens in transit.
Laboratory methods
Prior to extraction, 2 ml of VTM was added to the swab transport tube and thoroughly mixed by a vortex. Samples (200 μl) were extracted using the Qiagen X‐tractor Gene (Qiagen, Australia) and the Qiagen DX Xtraction kit (Qiagen) according to manufacturer’s instructions. Sample extracts were stored at −70 °C prior to testing. Extraction efficiency was assessed by spiking specimens with a known concentration of Equine Herpes virus (EHV) before extraction (Bialasiewicz et al. 2009). Specimens were tested using previously validated and reported real‐time PCR assays with reverse transcription for RNA viruses. We tested for 16 viruses: human rhinoviruses (Lu et al. 2008; Arden & Mackay 2010) influenza A, influenza B, RSV, adenoviruses, HMPV, parainfluenza viruses I, II, and III (Lambert et al. 2008b), bocavirus (Tozer et al. 2009), hPyV‐WU, hPyV‐KI (Bialasiewicz et al. 2007), and human coronaviruses: OC43, 229E, NL63 (Gunson et al. 2005) and HKU1 (Dare et al. 2007). Appropriate positive and negative controls were included on each PCR run. For hPyV‐WU, specimens were initially tested using a duplex assay with two targets (WU‐B, WU‐C) (Bialasiewicz et al. 2007). If either of these provided a positive signal, a third assay targeting a different region was used (WU‐E). WU‐E assay details are: TTTCTCCAGGAGATTTAGGCATTTC (forward primer), ATACCCACATATGGTACCCCAGACT (reverse primer), and TCTTCCCAGTAGGAATTAAACTGAGACCACCAGT (probe labelled with FAM fluorophore). The reaction mix consisted of 12.5 μl Quantitect Probe PCR Mix (Qiagen), 10 pmol of each primer, 4 pmol of probe and 5 μl of template in a 25‐μl final reaction was performed in a Rotorgene 3000 or 6000 (Qiagen) under the following conditions: 15 min incubation at 95 °C, followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min. Specimens needed to be positive in either WU‐B or WU‐C, and then WU‐E to be considered positive for hPyV‐WU.
We assessed the quality of specimen collection by evaluating for the presence of marker of human genomic DNA, ERV3, using a modified version of a previously published assay (Yuan et al. 2001). The assay modification involved adjusting PCR master mix and cycling conditions to match those described earlier for the WU‐E assay.
Data analysis
We performed descriptive analyses of demographic and clinical data and are presented as proportions of all participants or all paired specimens collected. Given the sensitivity and specificity of real‐time PCR diagnosis, we considered a specimen from either nostril positive for any virus to represent a true‐positive, similar to previous studies (Lambert et al. 2008b; Meerhoff et al. 2010). This approach means that the specificity of either specimen type for any virus will be, by definition, 100%. Using this standard, we calculated sensitivity overall and by specific viruses with 95% confidence intervals (CIs). Binomial probability was used to test our primary hypothesis of no difference in the likelihood of virus detection in either the mailed or frozen sample for discordant specimens, with α = 0.05 (two‐sided test). Cycle threshold (Ct) values (cycle number at the threshold level of log‐based fluorescence) are a semi‐quantitative marker of, and are indirectly proportional to, nucleic acid load. A 3.3 cycle difference represents approximately a one log difference in nucleic acid load (Whiley et al. 2010). We used t‐tests to compare differences in: the mean ERV3 and detected virus Ct values between paired specimens and the mean ERV3 Ct value between discordant specimens. Specimens where ERV3 was not detected were arbitrarily assigned a Ct value of 50 for the purpose of mean Ct value calculation. All data were analysed using Stata 10 for Windows (Stata Corp, College Station, TX).
Ethical conduct
We obtained signed informed consent from adult participants, assent from 10‐ to 18‐year‐olds, and parent or guardian consent for children up to 18 years of age. The project was approved by the Central Australia Health Research Ethics Committee, which includes an Indigenous research ethics subcommittee, and by the Board of the region’s Aboriginal Community Controlled Health Service.
Results
Over the 3 weeks of the study, we recruited 140 participants (82% of those screened) who contributed 153 study visits. Three study visits were excluded from further analysis because of inadequate specimen identification. Of the participants, 82% were Aboriginal, with the remainder being Caucasian, 60% were women and 21% were younger than 5 years (Figure 1). Eight individuals were seen twice and three were seen three times.
Figure 1.
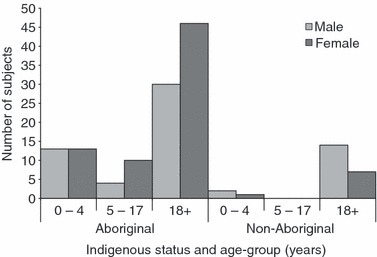
Study participants (n = 140) by Indigenous status, sex and age group.
The reason for presentation at the clinic was for an acute upper respiratory illness in 10 (6.7%) visits, acute lower respiratory illness in 3 (2.0%) visits, chronic respiratory illness in two visits (1.4%), other medical conditions in 43 (28.7%) and visiting or accompanying another person in 92 (61.3%) visits. There were no hospitalizations although one child with an acute lower respiratory illness was monitored overnight at the clinic. While respiratory illnesses accounted for 10% of the overall reported reasons for presentation, a respiratory symptom was present in 78 (52%) of all visits; a runny nose in 58 (74.4%) of these and moist cough in 31 (40.3%). A self‐report of fever within the previous 24 h was available from three visits, although no participant was febrile at the time of the study visit. Clinical signs (heart rate, respiratory rate and oxygen saturation) were all within the normal ranges for age.
During the conduct of the study at the clinic, the freezer temperature range for right nare specimens was −22.7 to −15.2 °C; the range for the refrigerator with the left nare specimens was 4.5 to 6.7 °C. Left nare specimens were transported weekly from the clinic at 10 a.m. on a Thursday morning for postage and reached the laboratory by midday on the following Monday; the temperature range for these specimens during transport was 12.8–30.5 °C. Right nare specimens were transported on dry ice at the end of the study from Darwin to Brisbane by overnight courier; the maximum temperature reached for these specimens was −5 °C.
A total of 61 viruses were identified in 50 (33.3%) presentations for 40 (28.6%) individuals. Of these 50 episodes, the reason for presentation at the clinic was ALRI in one presentation (2.0%), acute URI in 7 (14.0%), chronic URI in 1 (2.0%), other medical condition in 11 (22.0%) and as an accompanying person or visitor in 30 (60.0%). Table 1 presents virus‐positive episodes by age group, clinical presentation and presence or absence of respiratory symptoms. There was no difference in the proportion of visitors to the clinic who were virus positive when compared to those presenting for any medical reason (32.6%vs. 34.4%, P = 0.81). After excluding clinic presentations for non‐respiratory illnesses, a higher proportion of those presenting with a respiratory illness were virus positive than visitors (60%vs. 32.6%, P = 0.06). Forty‐three per cent of episodes in which any respiratory symptom was present (regardless of reason for presentation) were virus positive compared to 22% of those in which no respiratory symptoms were present (P = 0.006).
Table 1.
Demographic characteristics and clinical presentations for virus‐positive and virus‐negative episodes
| Any virus positive (N = 50) n (%) | Virus negative (N = 100) n (%) | |
|---|---|---|
| Age group | ||
| 0–4 | 20 (40) | 17 (17) |
| 5–17 | 6 (12) | 75 (75) |
| 18+ | 24 (48) | 8 (8) |
| Aboriginal | 38 (76) | 87 (87) |
| Non‐Aboriginal | 12 (24) | 13 (13) |
| Presenting reason | ||
| Acute LRI | 1 (2) | 2 (2) |
| Acute URI | 7 (14) | 3 (3) |
| Chronic LRI | 0 (0) | 1 (1) |
| Chronic URI | 1 (2) | 0 (0) |
| Other condition | 11 (22) | 32 (32) |
| Visitor | 30 (60) | 62 (62) |
| Respiratory symptoms | ||
| Any symptom* | 34 (68) | 44 (44) |
| No symptoms | 16 (32) | 56 (56) |
*Any one or more of the following: fever, cough, runny nose, chills, headache, fatigue, muscle aches, wheeze, tachypnoea, shortness of breath, chest indrawing and sore throat.
In the 50 episodes where any virus was identified, 41 had one virus (82%), 7 (14%) had two viruses and 2 (4%) had three viruses (Table 2). There were 41 (27.3%) discordant specimen pairs; 21 were positive only in the mailed specimen and 20 were positive only in the frozen specimen (Table 2). There was no statistical evidence to support a difference between the two transport methods (P = 0.87). The overall sensitivity for any virus detected in mailed specimens was 67.2% (95%CI 55.4, 78.9) against 65.6% (95%CI 53.7, 77.5) for frozen specimens (absolute difference, 1.6%, 95%CI 1.5, 1.8) (Table 3). The sensitivity of the two methods varied by virus identified (Table 3).
Table 2.
Viruses detected in 150 paired nasal swabs
| Parameter | N | % |
|---|---|---|
| Concordant mailed/frozen paired specimens | 109 | 72.7 |
| Discordant mailed/frozen paired specimens | 41 | 27.3 |
| No virus identified | 100 | 66.7 |
| Single virus only identified | 41 | 27.3 |
| Concordant | ||
| Rhinovirus | 6 | |
| Parainfluenza 2 | 2 | |
| HCoV 229E | 3 | |
| Bocavirus | 4 | |
| Discordant | ||
| Rhinovirus (mailed: 4; frozen: 3) | 7 | |
| Parainfluenza 2 (mailed: 2) | 2 | |
| Adenovirus (mailed: 3) | 3 | |
| WU (mailed: 2) | 2 | |
| HCoV‐229E (mailed: 1) | 1 | |
| Bocavirus (mailed: 5; frozen: 6) | 11 | |
| Two viruses identified | 6 | 4.0 |
| Concordant | ||
| Rhinovirus and HCoV‐229E | 1 | |
| Rhinovirus and adenovirus | 1 | |
| Discordant | ||
| Bocavirus (mailed) and rhinovirus (frozen) | 1 | |
| WU (mailed) and bocavirus (frozen) | 1 | |
| Parainfluenza 2 (mailed) and bocavirus (frozen) | 1 | |
| Influenza A (mailed) and bocavirus (frozen) | 1 | |
| Three viruses identified | 2 | 1.3 |
| Discordant | ||
| Rhinovirus (mailed/frozen), adenovirus (mailed) and bocavirus (mailed) | 1 | |
| Rhinovirus (frozen), parainfluenza 3 (frozen) and HCoV‐229E (frozen) | 1 | |
Table 3.
Sensitivity for the detection of different viruses according to specimen transport method
| Virus | Episodes with virus identified | Mailed (left nare) | Frozen (right nare) | ||
|---|---|---|---|---|---|
| Sensitivity % | 95%CI* | Sensitivity % | 95%CI* | ||
| Rhinovirus | 19 | 68.4 | 43.4–87.4 | 78.9 | 54.4–93.9 |
| Influenza A† | 1 | 100.0 | 25.0–100.0 | 0 | – |
| Adenovirus | 5 | 100.0 | 54.1–100.0 | 20.0 | 0.5–71.6 |
| WU | 3 | 66.7 | 9.4–99.2 | 33.3 | 0.8–90.6 |
| Parainfluenza 2 | 5 | 100.0 | 59.0–100.0 | 40.0 | 5.2–85.3 |
| Parainfluenza 3 | 1 | 0 | – | 100.0 | 25.0–100.0 |
| HCoV‐229E | 6 | 66.7 | 22.2–95.7 | 100.0 | 47.8–100.0 |
| Bocavirus | 21 | 52.4 | 31.0–76.7 | 66.7 | 46.5–86.8 |
| Total | 50 | 67.2 | 55.4–78.9 | 65.6 | 53.7–77.5 |
*Where sensitivity is 100%, a 1‐sided, 97.5% lower CI is provided.
†The single influenza A specimen was confirmed as 2009 pandemic influenza (H1N1) using a previously described swine flu–specific assay (Whiley et al. 2009).
ERV3 Ct values were higher (lower nucleic acid load) in mailed specimens (mean: 33.1, 95%CI 33.0, 33.2) than in frozen specimens (mean: 31.5, 95%CI 31.4, 31.6), although for a study of this nature, we do not consider the absolute difference between the two to be clinically meaningful (1.6, 95%CI 1.5, 1.7). Among discordant pairs containing one specimen negative for all viruses, there was no difference in mean ERV3 Ct values between any virus‐positive and all virus‐negative specimens (positive: 32.4, negative: 31.2, absolute difference 1.21, 95%CI ‐1.12, 3.55). There were, however, two bocavirus discordant pairs in which ERV3 was not detected in the negative specimen. There was one concordant virus‐negative pair in which ERV3 was not detected in either specimen.
In specimen pairs concordant‐positive for any DNA virus (n = 5, 4 bocavirus, 1 adenovirus, Table 1), the absolute difference in the mean Ct values between mailed (mean 31.5, 95%CI 19.8, 43.1) and frozen (mean 30.2 95%CI 19.8, 40.6) specimens was 1.3 (95%CI ‐11.8, 14.2, P = 0.83). In specimen pairs concordant‐positive for any RNA virus (n = 15, 9 rhinovirus, 2 PIV2, 4 HCoV229E, Table 1), the absolute difference in the mean Ct values between mailed (mean 34.3, 95%CI 31.6, 37.0) and frozen (mean 34.6 95%CI 31.5, 37.7) specimens was 0.3 (95%CI ‐4.1, 3.7, P = 0.9).
Discussion
Determining the aetiology and burden of viral respiratory infections in remote communities has to date been limited by the inability to store and transport clinical specimens requiring freezing/refrigeration to urban laboratories. So difficult is the task of collecting specimens for viral identification in remote locations, that a paper from Guinea‐Bissau suggested a one‐third reduction in sensitivity for RSV identification might be reasonable in epidemiological and vaccine studies if the method was acceptable to parents, easy to implement and had affordable costs (Stensballe et al. 2002). Our study, using sensitive real‐time PCR assays, has shown that diagnostic studies in remote settings can be conducted by simply mailing specimens through routine postal systems.
Our use of ERV3 as a marker of nucleic acid load in specimens provided evidence to support the use of the surface mail method. While the mean ERV3 Ct value was significantly lower (higher nucleic acid load) in frozen than in mailed specimens, the difference was not clinically meaningful representing a less than one log reduction in load. ERV3 Ct values also demonstrated that the quality of specimen collection appeared to play a limited role in failure to identify a virus. For discordant paired specimens with one specimen negative for any virus, the mean ERV3 Ct value was no lower in positive samples than in the negative samples. The mean Ct value for RNA viruses, typically thought to be less robust than DNA viruses, (Jerome et al. 2002) in concordant pairs was lower (higher load) in the mailed than in the frozen specimens. Further, the difference in the mean Ct values for mailed and frozen specimens in concordant pairs for RNA viruses was less than the same difference in DNA viruses, although the number of DNA viruses available for this calculation was small (n = 5).
A study of the viral aetiology of severe pneumonia among 759 infants and children at a rural hospital in Kenya using real‐time PCR methods on nasal wash samples found one or more respiratory viruses in 56% of children; RSV in 34% and other respiratory viruses in 29%, the most common being HCoV‐229E (6.7%) and Influenza A (5.8%) (Berkley et al. 2010). Twenty‐eight per cent of well controls were positive for any virus. The method of storage and transport to the laboratory in South Africa was not described. We did not detect RSV in our study population; however, the time the study was conducted was outside of the known RSV season in central Australia.
A potential limitation of our findings is that the study was conducted at the beginning of spring, when temperatures were considerably lower than the extreme heat that can be reached in desert communities in central Australia during the summer months. It is not uncommon for daytime temperatures to exceed 40 °C for extended periods. Mail bags may sit on airstrip tarmacs for extended periods. The effect of these temperatures on the viability and sensitivity of viral specimens needs further investigation. A further limitation is the convenience sampling strategy which may result in an inaccurate representation of the prevalence of viruses in the community. Similarly, the lack of adolescents (aged 5–17) means that an important section of the population was not adequately represented in our study population.
We propose that this method, combining standard clinic refrigeration and weekly surface mailing of specimens combined with real‐time PCR, can be used for viral respiratory research in remote locations. In the absence of any other diagnostic service during a community‐wide respiratory outbreak, this may also be appropriate as a diagnostic tool in some settings, particularly to guide public health responses to outbreaks of respiratory illness. In a Melbourne cohort study of pre‐school‐aged children, a nasal swab from both nares appeared just as likely to identify any virus as a swab from both nares combined with an oropharyngeal swab (Lambert et al. 2008a). In future studies, to maximize virus detection, we recommend sampling from each nare with the one swab. Combined with sensitive molecular techniques, this approach will help us understand the epidemiology of respiratory viruses and bacteria in remote locations, where recurrent infections and persistent carriage may result in chronic lung disease.
Acknowledgements
We thank the staff, residents and Health Council of the community in which this study was conducted. The study was funded by the Centre for Clinical Research Excellence in Child and Adolescent Immunisation, GlaxoSmithKline Biologicals and through ‘Ochre’– a fundraising initiative of the Menzies School of Health Research. KOG is supported by a NHMRC Post‐Doctoral Training Fellowship in Indigenous Health. The funding agencies had no role in the design, analysis, interpretation or reporting of the study.
References
- Arden KE & Mackay IM (2010) Newly identified human rhinoviruses: molecular methods heat up the cold viruses. Reviews in Medical Virology 20, 156–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Australia Post (2009) Post Guide: Dangerous and Prohibited Goods and Packaging. Australia Post, Melbourne. [Google Scholar]
- Berkley JA, Munywoki P, Ngama M et al. (2010) Viral etiology of severe pneumonia among Kenyan infants and children. JAMA 303, 2051–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialasiewicz S, Whiley DM, Lambert SB, Gould A, Nissen MD & Sloots TP (2007) Development and evaluation of real‐time PCR assays for the detection of the newly identified KI and WU polyomaviruses. Journal of Clinical Virology 40, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialasiewicz S, Whiley DM, Buhrer‐Skinner M et al. (2009) A novel gel‐based method for self‐collection and ambient temperature postal transport of urine for PCR detection of Chlamydia trachomatis. Sexually Transmitted Infections 85, 102–105. [DOI] [PubMed] [Google Scholar]
- Dare RK, Fry AM, Chittaganpitch M, Sawanpanyalert P, Olsen SJ & Erdman DD (2007) Human coronavirus infections in rural Thailand: a comprehensive study using real‐time reverse‐transcription polymerase chain reaction assays. Journal of Infectious Diseases 196, 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S, Molteni CG, Daleno C et al. (2010) Comparison of nasopharyngeal nylon flocked swabs with universal transport medium and rayon‐bud swabs with a sponge reservoir of viral transport medium in the diagnosis of paediatric influenza. Journal of Medical Microbiology 59, 96–99. [DOI] [PubMed] [Google Scholar]
- Gunson RN, Collins TC & Carman WF (2005) Real‐time RT‐PCR detection of 12 respiratory viral infections in four triplex reactions. Journal of Clinical Virology 33, 341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome KF, Huang ML, Wald A, Selke S & Corey L (2002) Quantitative stability of DNA after extended storage of clinical specimens as determined by real‐time PCR. Journal of Clinical Microbiology 40, 2609–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SB, Allen KM & Nolan TM (2008a) Parent‐collected respiratory specimens‐‐a novel method for respiratory virus and vaccine efficacy research. Vaccine 26, 1826–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SB, Whiley DM, O’Neill NT et al. (2008b) Comparing nose‐throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real‐time polymerase chain reaction. Pediatrics 122, e615–e620. [DOI] [PubMed] [Google Scholar]
- Leach AJ, Boswell JB, Asche V, Nienhuys TG & Mathews JD (1994) Bacterial colonization of the nasopharynx predicts very early onset and persistence of otitis media in Australian aboriginal infants. Pediatric Infectious Disease Journal 13, 983–989. [DOI] [PubMed] [Google Scholar]
- Lu X, Holloway B, Dare RK et al. (2008) Real‐time reverse transcription‐PCR assay for comprehensive detection of human rhinoviruses. Journal of Clinical Microbiology 46, 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerhoff TJ, Houben ML, Coenjaerts FE et al. (2010) Detection of multiple respiratory pathogens during primary respiratory infection: nasal swab versus nasopharyngeal aspirate using real‐time polymerase chain reaction. European Journal of Clinical Microbiology and Infectious Diseases 29, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PS, Leach AJ, Silberberg P et al. (2005) Otitis media in young Aboriginal children from remote communities in Northern and Central Australia: a cross‐sectional survey. BMC Pediatrics 5, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady KF, Torzillo PJ & Chang AB (2010) Hospitalisation of Indigenous children in the Northern Territory for lower respiratory illness in the first year of life. Medical Journal of Australia 192, 586–590. [DOI] [PubMed] [Google Scholar]
- Stensballe LG, Trautner S, Kofoed PE et al. (2002) Comparison of nasopharyngeal aspirate and nasal swab specimens for detection of respiratory syncytial virus in different settings in a developing country. Tropical Medicine and International Health 7, 317–321. [DOI] [PubMed] [Google Scholar]
- Tozer SJ, Lambert SB, Whiley DM et al. (2009) Detection of human bocavirus in respiratory, fecal, and blood samples by real‐time PCR. Journal of Medical Virology 81, 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valery PC, Torzillo PJ, Mulholland K, Boyce NC, Purdie DM & Chang AB (2004) Hospital‐based case‐control study of bronchiectasis in indigenous children in Central Australia. Pediatric Infectious Disease Journal 23, 902–908. [DOI] [PubMed] [Google Scholar]
- Whiley DM, Bialasiewicz S, Bletchly C et al. (2009) Detection of novel influenza A(H1N1) virus by real‐time RT‐PCR. Journal of Clinical Virology 45, 203–204. [DOI] [PubMed] [Google Scholar]
- Whiley DM, Goire N, Ray ES et al. (2010) Neisseria gonorrhoea multi‐antigen sequence typing using non‐cultured clinical specimens. Sexually Transmitted Infections 86, 51–55. [DOI] [PubMed] [Google Scholar]
- Yuan CC, Miley W & Waters D (2001) A quantification of human cells using an ERV‐3 real time PCR assay. Journal of Virological Methods 91, 109–117. [DOI] [PubMed] [Google Scholar]


