Abstract
The last two decades have witnessed the exponential development of DNA as a generic material instead of just a genetic material. The biological function, nanoscale geometry, biocompatibility, biodegradability, and molecular recognition capacity of DNA make it a promising candidate for the construction of novel functional nanomaterials. As a result, DNA has been recognized as one of the most appealing and versatile nanomaterial building blocks. Scientists have used DNA in this way to construct various amazing nanostructures, such as ordered lattices, origami, supramolecular assemblies, and even three‐dimensional objects. In addition, DNA has been utilized as a guide and template to direct the assembly of other nanomaterials including nanowires, free‐standing membranes, and crystals. Furthermore, DNA can also be used as structural components to construct bulk materials such as DNA hydrogels, demonstrating its ability to behave as a unique polymer. Overall, these novel DNA materials have found applications in various areas in the biomedical field in general, and nanomedicine in particular. In this review, we summarize the development of DNA assemblies, describe the innovative progress of multifunctional and bulk DNA materials, and highlight some real‐world nanomedical applications of these DNA materials. We also show our insights throughout this article for the future direction of DNA materials. WIREs Nanomed Nanobiotechnol 2010 2 648–669
This article is categorized under:
-
1
Diagnostic Tools > Biosensing
-
2
Nanotechnology Approaches to Biology > Cells at the Nanoscale
-
3
Implantable Materials and Surgical Technologies > Nanomaterials and Implants
-
4
Nanotechnology Approaches to Biology > Nanoscale Systems in Biology
About 30 years ago, DNA was regarded only as a genetic molecule, and the concept of using DNA beyond biology was unheard of until Seeman's demonstration of the first artificial DNA junctions and lattices.1 Today, this alternative view of DNA has become widely accepted as scientists have used DNA to fabricate a myriad of materials and put these so‐called ‘DNA materials’ to work for different applications. DNA, the biomolecule of life, possesses many unique properties in comparison with synthetic polymers due to billions of years of evolution. Specifically, the molecular recognition capability that gives rise to its biological functions has been extensively exploited for the structural directing of nanocomponents to generate materials and devices with precise control at nanometer scale.2, 3, 4, 5, 6, 7 Thus DNA has bridged the gap between molecular biology and materials science and engineering, paving the way for novel nanomaterial synthesis.2
In general, DNA molecules can be single stranded (ssDNA) or double stranded (dsDNA). Each DNA strand is made of a phosphate‐deoxyribose backbone plus one of four bases: adenine (A), guanine (G), cytosine (C), or thymine (T). Two complementary ssDNA strands are hybridized into a double helix structure by obeying the Watson–Crick base‐pairing rule: A with T and C with G (Figure 1(a)). The pairing is also reversible: dsDNA can be denatured into two ssDNA strands by heating or exposure to high pH. G–C pairing is more stable than A–T pairing because three hydrogen bonds form between G and C, whereas only two between A and T. Consequently, DNA double helices with more G–C pairings are more stable against heating and pH, which is an important design factor in the construction of DNA sequences for self‐assembly.
Figure 1.
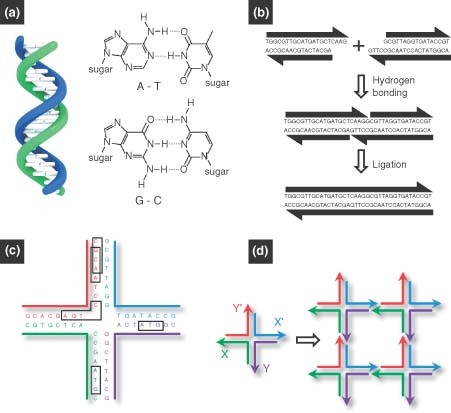
Basic principles of DNA self‐assembly.5 (a) Watson–Crick base‐pairing rules: the double helix is assembled from two single‐stranded DNA by following the Watson–Crick base‐paring rules: adenine (A) with thymine (T) and guanine (G) with cytosine (C). (b) Sticky‐end cohesion. Linear double‐stranded DNA (dsDNA) can be designed with sticky‐end overhangs that are complementary to each other. The two dsDNA can be connected by the hybridization of sticky ends. The nick between the two duplexes can be chemically sealed by an enzyme, resulting in a single duplex. (c) A branched DNA motif must have base‐pair asymmetry to be stable enough to function as a component for DNA assembly. (d) Sticky‐end assembly of branched DNA motifs. A branched DNA motif is shown on the left with four sticky ends: X complementary to X′ and Y complementary to Y′. The four motifs assemble to form a quadrilateral, shown on the right, with further sticky ends on the outside, further allowing the structure to form a 2D lattice..
The distinctive properties of DNA make it an excellent building block, template, and guide for the assembly of nanomaterials. The first and most important property is the molecular recognition capability afforded by Watson–Crick base pairing, which makes the DNA hybridization highly programmable,8 and hence the desired DNA materials rationally designable. The second key property is that the DNA double helix is an inherently nanoscale object, having a diameter of about 2 nm, a base separation of 0.34 nm, and a helical periodicity of 10–10.5 nucleotide pairs, or 3.5 nm per turn.
Equally attractive is the third property of DNA. As a polymeric chain, the rigidity and flexibility of DNA can be easily tailored by simply adjusting the numbers of base pairs. Although the DNA double helix is actually a flexible polymeric chain in biological systems such as plasmids and chromosomes, this flexibility does not apply to short double strands. The persistence length of DNA is about 50 nm (which corresponds to 150 bp), and below this length the DNA strands can be considered rigid rods. On the other hand, ssDNA is much more flexible (the persistence length of ssDNA is about 1 nm), and therefore can easily form loop and curved structures. Hence, the rigidity and flexibility can be designed and tailored by a combination of ssDNA and dsDNA: the rigid dsDNA double helix can be linked by relatively flexible ssDNA strands to form stable motifs with a desired topology.
The fourth property of DNA is the most unique property of all: molecular biology provides rich toolkits for the synthesis, manipulation, and modification of DNA. For example, bulk DNA can be purified from cultured organisms such as bacteria or yeast. Individual genes or DNA fragments with desired sequences can be preserved in a plasmid, and shorter DNA sequences (oligonucleotides) can be synthesized chemically de novo. DNA can also be amplified via the polymerase chain reaction, an amplification technology that exponentially increases the amount of a specific DNA sequence. Nature also provides us with more than 4000 different enzymes for DNA manipulation: for example, endonucleases function as ‘scissors’ to cut DNA strands at internal sites; exonucleases function as ‘sanders’ to cut DNA strands from terminal ends; restriction enzymes can recognize and cleave specific DNA sequences; ligases function as permanent ‘glue’ to covalently connect DNA together; polymerases can elongate DNA chains; and nonspecific nucleases can degrade DNA.2
Finally, as expected, DNA molecules are nontoxic and biocompatible, making them suitable materials for both in vitro and in vivo applications. In fact, no other materials can provide all the aforementioned advantages, making DNA an attractive, versatile, and highly manipulatable building block for the construction of novel nanoscale materials.
Fabrication of DNA materials is mainly based on two assembly processes: (1) self‐assembly and (2) enzyme‐assisted assembly. The simplest DNA self‐assembly method involves the use of two complementary ssDNA strands spontaneously hybridizing together to form a dsDNA double helix (Figure 1(a)). The process is driven by several non‐covalent interactions, including hydrogen bonds, electrostatics, base stacking, hydrophobic interactions, and van der Waals forces, which strictly follow the thermodynamically favorable Watson–Crick base‐paring rules. The use of single‐stranded overhangs at the end of dsDNA (called ‘sticky ends’) allows two dsDNA chains to connect. In this case, two dsDNA strands with complementary sticky ends will hybridize, and nicks in the backbones can be sealed by a DNA ligase, resulting in a single integrated duplex (Figure 1(b)). However, these linear DNA chains alone cannot yield more complicated nanostructures. As a result, branched DNA motifs have been created to extend the diversity of DNA assembly. Such branched motifs can be found in nature: for example, the four‐arm Holliday junction is an intermediate in genetic recombination.9,10 The main concern from a structural viewpoint has been that the symmetry of the Holliday junction results in branch point migration, making this unstable motif unusable as a building block to construct stable nanoarchitectures. As a solution, various asymmetrical branched motifs were synthetically designed and characterized.7,11 Figure 1(c) shows an example of a stable asymmetrical Holliday junction design. The ability to form stable branched DNA motifs allows for the bottom‐up construction of nanoscale objects. Therefore, the basic prerequisites behind the self‐assembly of DNA materials are complementary sticky ends and stable branched DNA molecules with asymmetric base pairing. Figure 1(d) shows the conceptual proof of sticky‐end assembly of stable branched DNA: four motifs with stickynds assembled to form a quadrilateral with additional sticky ends on the outside, further allowing the structure to form a two‐dimensional (2D) lattice. Enzyme‐assisted assembly is another type of assembly process which is a unique to DNA and is not possible with other synthetic polymers. The detailed examples of enzyme‐assisted assembly of DNA materials are given in the later sections of this article.
The history of DNA materials' fabrication began in the early 1980s.1 Seeman first envisioned the possibility of combining branched DNA with sticky ends to construct 2D arrays, which was later experimentally realized by his group.12 Branched DNA junctions are fairly flexible, so the assembly of these building blocks often did not yield the desired stable nanostructures. To solve this problem, Seeman and coworkers constructed a group of branched complexes called crossover tiles with greater rigidity, such as double‐crossover (DX) tiles,12, 13, 14, 15, 16 triple‐crossover (TX) tiles,17, 18, 19 and paranemic‐crossover (PX) tiles.20, 21, 22 Following this work, well‐defined 2D DNA assemblies with different morphologies have evolved a great deal in the DNA‐nanotechnology community. Another milestone in DNA structure design is when Rothemund realized the concept of DNA origami to increase the complexity and versatility in DNA assembly.23 DNA origami can be used to engineer almost any arbitrary patterns, such as a smiley face and DNA box with a controlled lid.23,24 As the field begins to mature with the knowledge that has enabled 2D assemblies, three‐dimensional (3D) DNA assemblies can now be fabricated in a controllable and elegant manner.25,26
Because DNA can be conjugated with other biomolecules and metal nanoparticles, DNA assemblies were used as templates to construct biomolecules and nanoparticle arrays.27, 28, 29 In addition to these arrays, the successful fabrication of bulk, non‐ordered, non‐repeated DNA materials such as DNA gel opened new opportunities to integrate DNA materials into real‐world applications.30, 31, 32 In addition, anisotropic and multivalent DNA building blocks were created from which multifunctional materials have been synthesized, with potential applications in biomedical diagnostics.33,34
Although there have been a myriad of excellent reviews about DNA nanotechnology,3,5, 6, 7,11,35, 36, 37, 38, 39, 40, 41, 42, 43 this review will focus on recent progress of the fabrication of novel DNA materials for nanomedical applications. For other applications that will not be included, such as DNA devices, nanomachines, and electric/optical applications, readers may refer to other reviews.38,43, 44, 45, 46, 47, 48 Due to the limited space, we apologize in advance for not including other significant papers and works.
DNA MATERIALS
One‐Dimensional DNA Materials
One‐dimensional (1D) DNA materials, including nanotubes and nanowires, can be fabricated through the programed self‐assembly of DNA tiles, which have the potential to template the growth of nanowires19,49,50 and induce alignment of membrane proteins.51 Endo et al. 52 reported a method to prepare structurally controlled DNA nanotubes that used a DNA tile system, with the assistance of a four‐way‐branched DNA connector. The connector was a DNA–porphyrin conjugate, Porph‐(Tc)4, in which 10‐mer DNA strands were connected to four spacers of a tetraphenylporphyrin derivative. These connectors allowed planar DNA tiles to assemble into 2D array structures by taking advantage of the geometry of the 2.5 helical turns between two DNA tiles (Figure 2(a), left). The 2D arrays were assembled into tube structures in the presence of Porph‐(Tc)4, as shown by atomic force microscopy (AFM) characterization (Figure 2(a), right). These fiber‐like structures could reach lengths of over 2 µm. A simplified approach for 1D fabrication was demonstrated in which the self‐assembly of DNA nanotubes resulted from one single strand of DNA,53 which resembled the self‐assembly of natural protein nanofilaments. In this procedure, a DX‐like structure was first formed from two identical, 52‐nt long strands with palindromic segments; regions with single‐stranded overhangs allowed the complexes to further assemble into 2D lattices, which were then folded into 2D sheets and eventually DNA nanotubes (Figure 2(b)). This one‐stranded assembly can provide a foundation for nanofabrication protocols, templated assembly of metallic nanowires, and even building tracks for nanomotors. Similarly, Kuzuya et al. 54 prepared DNA nanotubes assembled from half‐tube components. Two different species were produced: a six‐helix bundle and an eight‐helix bundle. AFM analysis indicated that long half‐tubes readily self‐assembled to full tubes (Figure 2(c)). This study proved that larger tubes could be built from subcomponents that span only a portion of the circumference. Yin et al. 55 prepared DNA tubes with monodisperse and programmable circumferences. They tailored DNA tube circumferences by specifying the complementarity relationship between modular regions in a 42‐base ssDNA motif. Following single‐step annealing, long tubes were assembled with specific monodisperse circumferences. To achieve a high level of dimensional control, DNA lattices were built using flexible ssDNA motifs that were much simpler than previous methods based on multistranded rigid tiles. By pairing specific modular domains in the motif, DNA lattices could be formed from parallel DNA helices connected by half‐crossovers, and from these lattices full DNA tubes could be produced with a variety of predetermined circumferences (Figure 2(d), left). Overall, the modularity and standardization of the ssDNA motif enables easy codification of lattice design, which allowed one to ignore molecular structure and interpret the lattice design in simple abstract forms. Tubes with 4, 5, 6, 7, 8, 10, and 20 helices could be controllably prepared, resulting in width of open tubes that was proportional to the helix number (Figure 2(d), right).
Figure 2.
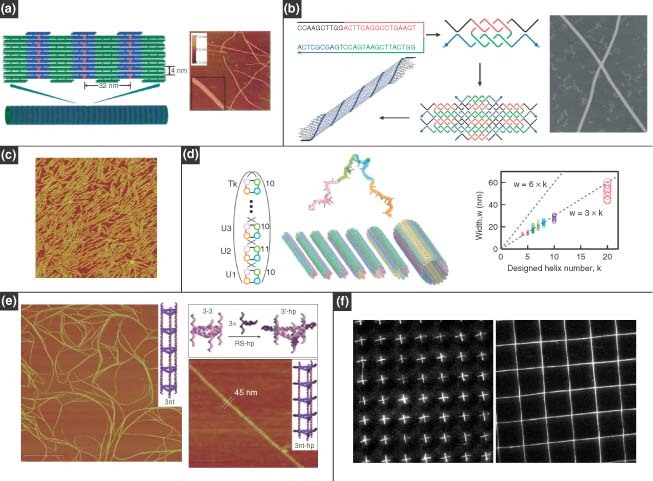
One‐dimensional DNA nanostructures. (a) Nanotubes assembled from double‐crossover DNA tiles with the assistance of a four‐way‐branched DNA connector. (Reprinted with permission from Ref 52. Copyright 2005 Wiley‐VCH Verlag GmbH & Co. KGaA). (b) Nanotubes assembled from a single strand containing four palindromic segments. (Reprinted with permission from Ref 53. Copyright 2006 Wiley‐VCH Verlag GmbH & Co. KGaA). (c) Nanotubes assembled from four half‐tubes. The atomic force microscopy image shows four‐helix bundles. (Reprinted with permission from Ref 54. Copyright 2007 American Chemical Society). (d) Monodisperse DNA tubes with programed circumferences: the molecular program (left) and width plot of opened tubes showing the monodispersity of their circumferences (right). (Reprinted with permission from Ref 55. Copyright 2008 AAAS). (e) DNA nanotubes with well‐defined triangular and square geometries based on a modular approach. (Reprinted with permission from Ref 56. Copyright 2009 Nature Publishing Group). (f) Arrays of DNA nanostrands patterned by molecular combing and soft‐lithography micropatterning. (Reprinted with permission from Ref 58. Copyright 2005 PNAS)..
The aforementioned methods for the construction of DNA nanotubes were specifically for producing cylindrical geometries. Recently, Aldaye et al. 56 prepared DNA nanotubes with well‐defined triangular and square geometries based on a modular approach. In addition, this group was the first to construct nanotube assemblies that can exist in single‐ and double‐stranded forms with significantly different stiffness, in contrast to previously reported DNA nanotubes that were entirely double‐stranded. The assembly protocol began with the formation of single‐stranded and cyclic DNA building blocks with rigid vertices in triangular or square shapes. The blocks were longitudinally assembled by linking strands to form ‘rungs’ in a nanotube format. The rung structure dictated the final geometry of the nanotube, and the linking strands determined the stiffness and the persistence length. The nanotubes can extend over several micrometers and have a uniform periodicity along the longitudinal direction (Figure 2(e)). This method allowed for the creation of ‘designer’ multifunctional nanotubes, whose properties (geometry, stiffness, and single‐ or double‐stranded character) could be directly programed into the assembly process.
DNA nanotubes often incorporate a self‐assembly approach for fabrication of 1D nanostructures, but there are also nanofabrication approaches for directing the assembly of ordered arrays of 1D DNA nanostrands.57, 58, 59 For example, Guan et al. 58 used modified molecular combing and soft‐lithography micropatterning tools to create arrays of DNA nanostrands. The modified molecular combing method used polydimethylsiloxane (PDMS) stamps with microwells, which, after stamping onto a droplet of DNA strands, were peeled away to produce arrays of stretched DNA molecules that were subsequently transferred via microcontact printing to other solid surfaces. Crosses and crossbars could be prepared by double printing of PDMS stamps (Figure 2(f)).
Two‐Dimensional DNA‐Ordered Assembly
As discussed in the introduction, to achieve complicated DNA assemblies such as 2D or 3D nanostructures, stable motifs must first be designed. A general rule in the sequence design for DNA motifs is to minimize the sequence symmetry in the branched structure to avoid possible undesired pairing between strands and mobility of the junction points. Several reviews have summarized the general rules and programs for rational design of oligonucleotide sequences, stable motifs, and DNA assembly.5,7,60,61
In the past decade, a large variety of 2D ordered assemblies with different geometries and topologies have been achieved with excellent yield. The most important branched DNA molecule in 2D DNA self‐assembly is DX DNA (Figure 3(a)). DX molecules are analogues of intermediates in meiosis that consist of two side‐by‐side double‐stranded helices linked at two crossover junctions.62 The junction points were constrained to a single orientation as opposed to being flexible as in the case of the four‐arm junction, which made the DX motif suitable as a structural building block for larger DNA complexes.63 Winfree et al. 12 first realized the assembly of 2D lattices from DX motifs (Figure 3(a)).
Figure 3.
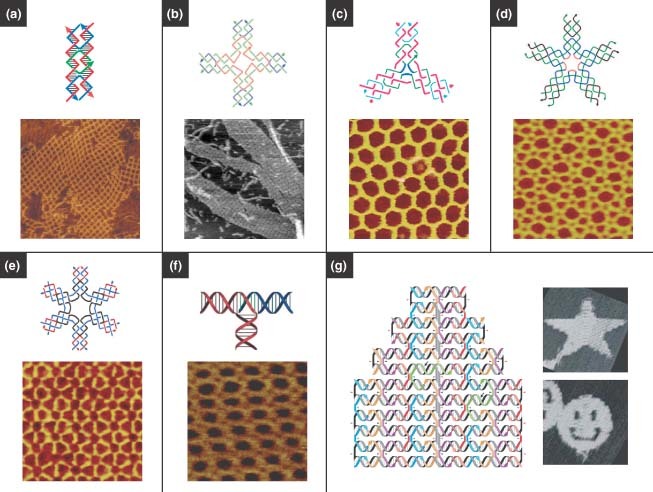
Two‐dimensional (2D) DNA nanostructures. (a) 2D crystals formed from double‐crossover motifs. (Reprinted with permission from Refs 4, 12. Copyright 1998, 2003 Nature Publishing Group). (b) Square lattices formed from four‐way junctions. (Reprinted with permission from Ref 49. Copyright 2003 AAAS). (c) Hexagonal lattices formed from three‐point‐star motifs. (Reprinted with permission from Ref 65. Copyright 2005 American Chemical Society). (d) Complex lattices assembled from five‐point‐star motifs. (Reprinted with permission from Ref 66. Copyright 2008 PNAS). (e) 2D periodic arrays assembled from symmetric six‐point‐star motifs. (Reprinted with permission from Ref 67. Copyright 2006 American Chemical Society). (f) 2D periodic arrays assembled from T‐junctions. (Reprinted with permission from Ref 71. Copyright 2009 Wiley‐VCH Verlag GmbH & Co. KGaA). (g) 2D patterns made by DNA origami, in which a long single strand is folded with the help of smaller ‘staple strands’. (Reprinted with permission from Ref 23. Copyright 2006 Nature Publishing Group)..
Inspired by the DX motif, several other motifs were engineered to provide more options for generating 2D structures, for example, TX motifs,17 the DNA parallelogram,64 four‐by‐four‐structures,49 and 3‐, 5‐, and 6‐point‐star motifs (Figure 3(b)–(e)).65, 66, 67 Those motifs with greater flexibility, rigidity, and tensegrity (tension and integrity) led to successful formation of 2D arrays. For example, Yan et al. 49 designed a four‐arm junction to assemble uniform‐width nanoribbons and 2D nanogrids (Figure 3(b)). He et al. 65 prepared porous and hexagonal 2D arrays assembled from three‐point‐star motifs (Figure 3(c)). Previously, it was believed that there were exactly three regular tilings of the plane: one composed of triangles, one of squares, and one of hexagons.68 However, 2D DNA self‐assembled arrays can incorporate the formation of multiple tilings and are not restricted to only one type of tiling. For example, Zhang et al. 66 prepared 2D ordered lattices composed of both triangles and squares by using the five‐point‐star motif (Figure 3(d)).
Mao and coworkers developed a concept in which DNA sequence symmetry could be used to design DNA building blocks for larger self‐assembled structures.67,69,70 For example, they used symmetric six‐point‐star motifs to prepare stable and robust 2D periodic arrays (Figure 3(e)). The DNA sequence symmetry had the advantages of using fewer DNA strands and sizes of unique sequences, thus avoiding some unpredictable distortions in DNA nanostructures. This further allowed 2D DNA arrays to grow into much larger sizes, as large as 1 mm, compared to the micrometer‐scale arrays reported previously.65,69 It should be noted that the ‘sequence symmetry’ here is between different strands in the same DNA motif at the symmetric positions, but within each individual strand the base‐pairing symmetry is still avoided.
Very recently, Hamada et al. 71 developed a new motif, an interconnected single‐duplex‐based T‐junction, to assemble into various structures in solution, such as orthogonal coordinated ladders, lattices, and polar coordinated wheels (Figure 3(f)). Unlike the crossover junction with the same geometry, the T‐junction provides a right‐angled geometry to expand the varieties of DNA nanostructures. Furthermore, T‐junctions enable higher structural resolution because they have a shorter interconnection length (6 or 7 bp) between the motifs compared to crossover‐based motifs (10.5 bp at least). This could enable the formation of smaller ‘nanoscopic’ details in DNA nanostructures. Also, the T‐junction limits the electrostatic repulsion among DNA backbones avoiding distortion that arises from helices aligned in parallel.72 Moreover, these structures could grow consistently over the whole substrate surface in a substrate‐assisted assembly method. Sun et al. 73 also reported similar substrate‐mediated DNA self‐assembly.
DNA origami, a landmark work developed by Rothemund and Winfree, has brought much attention to the field of DNA nanotechnology.23,74 In contrast to the conventional crossover strategy, which creates single building blocks that self‐assemble into larger structures in a ‘two‐step’ process, DNA origami provides a versatile and simple ‘one‐pot’ method using numerous short single strands of DNA (staple strands) to direct the folding of a long, single strand of DNA into the desired shape (Figure 3(g)).23 DNA nanostructures assembled by the DNA origami method can be very complex, and it is believed that any arbitrary shape could be achieved. However, one critical challenge in DNA origami is scaling up the size of DNA origami structures. Fortunately, Zhao et al. 75 developed a new strategy that used rectangular‐shaped DNA tiles instead of traditional staple strands to construct larger scale 2D DNA origami: about four times the size of Rothemund's origami structure using the same length scaffold. In addition, the authors also suggested that their strategy could be combined with other scale‐up techniques, such as hierarchical DNA assembly or surface mediated self‐assembly.
Three‐Dimensional DNA‐Ordered Assembly
3D DNA materials have tremendous potential applications in drug delivery systems, tissue engineering, and protein engineering. The construction of these 3D DNA nanostructures has become a hot topic in recent years based on this potential. Scientists have demonstrated the capability of fabricating a broad array of 2D DNA‐ordered nanostructures, but 3D DNA nanostructures are significantly more challenging to design than 2D structures. Conceptually, there is no difference between 2D and 3D assembly, which means that the same principles of 2D assembly should apply to 3D systems. However, early attempts at preparing 3D structures according to these principles were hampered by the limited rigidity of DNA building blocks. With the development of rigid motifs and assembling strategies, a large number of rationally designed 3D nanomaterials were produced. The early examples of DNA cubes and truncated octahedrons were achieved by Seeman and coworkers.76,77 After that, different unique 3D DNA motifs were produced, which made the design and fabrication of 3D structures more feasible, especially in recent years.24,78, 79, 80, 81, 82, 83, 84
Shih et al. 78 built a rigid and chiral DNA octahedron by folding a 1.7‐kb ssDNA, which consisted of five double‐crossover struts and seven PX struts joined at six four‐way junctions (Figure 4(a), left). They employed transmission electron microscopy (cryo‐TEM) to observe the morphology of the assembly products, which clearly revealed the successful folding of the DNA strands into an octahedron structure (Figure 4(a), right). Turberfield and coworkers constructed a series of DNA tetrahedra79 (Figure 4(b)). These structures were assembled exclusively through hybridization of participating ssDNA molecules (i.e., no ligation was necessary) and featured high yield (>95% at a DNA concentration of 50 nM).
Figure 4.
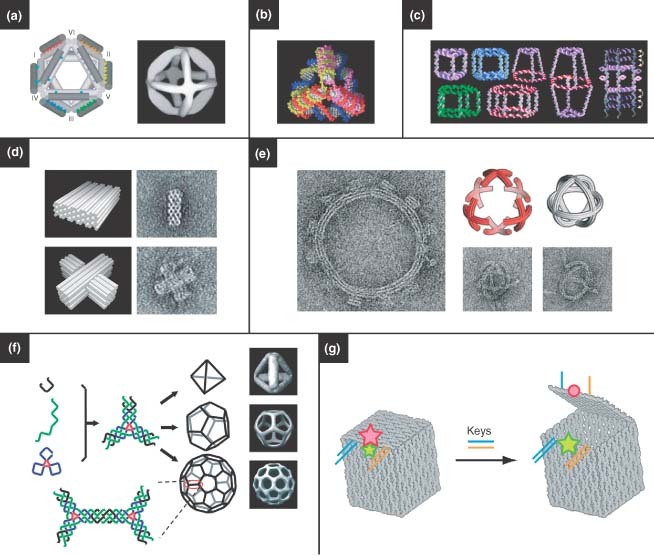
Three‐dimensional (3D) DNA nanostructures. (a) Octahedrons, (Reprinted with permission from Ref 78. Copyright 2004 Nature Publishing Group). (b) tetrahedrons, (Reprinted with permission from Ref 79. Copyright 2005 AAAS). (c) a variety of polyhedrons, (Reprinted with permission from Ref 85. Copyright 2007 American Chemical Society). and (d, e) 3D DNA origami. (Reprinted with permission from Refs 82, 83. Copyright 2009 Nature Publishing Group and AAAS). (f) Hollow polyhedrons have been assembled by engineering flexibility into three‐point‐star motifs, where the curvature dictates the resulting shape. (Reprinted with permission from Ref 81. Copyright 2008 Nature Publishing Group). (g) A DNA box based on DNA origami. The box can be opened by the introduction of two ‘key’ oligonucleotides, which is indicated by a fluorescence signal. (Reprinted with permission from Ref 24. Copyright 2009 Nature Publishing Group)..
Aldaye and Sleiman introduced a novel stepwise assembly method to construct 3D DNA prisms.85 They first prepared cyclic ssDNA molecules by solid‐phase DNA synthesis and subsequent DNA‐templated chemical ligation. The geometry of these cyclic ssDNA molecules could be designed and controlled in the synthesis process. As a result, a library of DNA polygon structures with ssDNA sides and rigid organic molecules as vertices was generated (Figure 4(c)). In the next step, the DNA polygons served as the top and bottom faces of 3D DNA prisms, both of which were connected by linking strands, and the vertical edges were additionally strengthened by rigidifying strands. This modular design and combinatorial assembly strategy enabled the fabrication of a large number of 3D DNA cages, including DNA cube and homo‐, hetero‐, and bi‐prisms. Dynamic DNA cages were realized by the addition and displacement of rigidifying strands with different lengths.
Shih and coworkers recently extended DNA origami to 3D nanoconstruction.82,83 They reported a honeycomb‐pleated origami approach that proceeded in a one‐pot reaction between a scaffold strand and hundreds of oligonucleotide staple strands which are used to direct its folding into the desired shape. 3D objects with different morphologies were rationally designed, including monoliths, square nuts, railed bridges, slotted crosses, and stacked crosses; two representative images are shown in Figure 4(d).82 Using the same strategy, they expanded the DNA‐origami shapes to incorporate a rich diversity of nanostructures with designed twist and curvature. They not only prepared twisted DNA bundles and curved shapes with well‐defined bend angles, but also assembled twisted and curved objects into more complicated structures such as nanogears and spherical wireframes83 (Figure 4(e)).
Hollow 3D nanostructures, like 2D lattices, are assembled from simpler DNA subunits, and are particularly interesting due to their cargo‐carrying potential. He et al. 81 developed a robust, one‐pot assembly strategy for assembling unique 3D structures. Three‐point‐star motifs, which have typically been used to assemble into 2D arrays, could be designed to have a more flexible structure through careful engineering of the junction. This flexibility could promote curvature in the subunits that led to the formation of closed polyhedral structures, including tetrahedrons, dodecahedrons, and buckyballs (Figure 4(f)). Similarly, Zhang et al. 66 used a symmetric five‐point‐star motif to construct icosahedra and nanocages. Andersen et al. 24 recently employed the DNA origami approach in designing 3D boxes with controllable lids. Here, six DNA origami faces were designed to assemble into a hollow 3D box (Figure 4(g)). The lid of the box is dynamically ‘opened’ and ‘closed’ by introducing ‘key’ oligonucleotide sequences that displaced DNA duplexes holding the lid closed. Opening of the box lid could be observed by changes in fluorescence resonance energy transfer between two fluorescent dyes placed on the DNA structure at strategic locations. The opening of the DNA box could be programed to require two oligonucleotide keys to unlock it, and thus the authors propose that there is a possibility for using these structures for logic‐mediated cargo release.
Bulk DNA Materials: Dendrimers, Networks, and Multifunctional DNA Materials
The base‐pairing complementarity and programmability of DNA‐based structures can be exploited to create DNA shapes that act as building blocks for the design of multifunctional materials.86,87 Because of the designability of DNA structures and the employment of DNA enzymes, the construction of multifunctional materials assembled from DNA should be more straightforward, more efficient, and more controllable than other polymers. Inspired by conventional dendritic polymers, branched or dendritic structures can be designed from DNA, and these building blocks can then be assembled, via an efficient DNA enzyme (T4 ligase), into complex structures in a highly controlled fashion. Dendritic DNA nanostructures have been envisioned for a long time.88,89 Recently, our group realized the controlled assembly of dendrimer‐like DNA (DL‐DNA) from the enzymatic ligation of Y‐shaped DNA (Y‐DNA) building blocks.90 More specifically, the Y‐DNA was synthesized from three single oligonucleotides, each of which has partial complementary sequences to the other two oligonucleotides (Figure 5(a), left). For additional complexity, ‘generations’ of branches can be extended from an initial ‘seed’ (or ‘core’) DL‐DNA. In short, the ends of a first‐generation DL‐DNA are ligated to six Y‐DNA, resulting in a second‐generation DL‐DNA (Figure 5(a), center). Higher generations of DL‐DNA can be formed in a similar manner. To confirm the generation of DL‐DNA species, the fourth‐generation DL‐DNA was visualized by AFM (Figure 5(a), right). In addition to high yield and purity, this approach has a number of other advantages including unidirectional and stepwise growth and nonreversible, efficient synthesis. Moreover, DL‐DNA can be manipulated further at the molecular length scale by DNA enzymes and the conjugation of different functional groups. Overall, these DL‐DNA structures could serve as templates for fabrication and synthesis of multifunctional structures.
Figure 5.
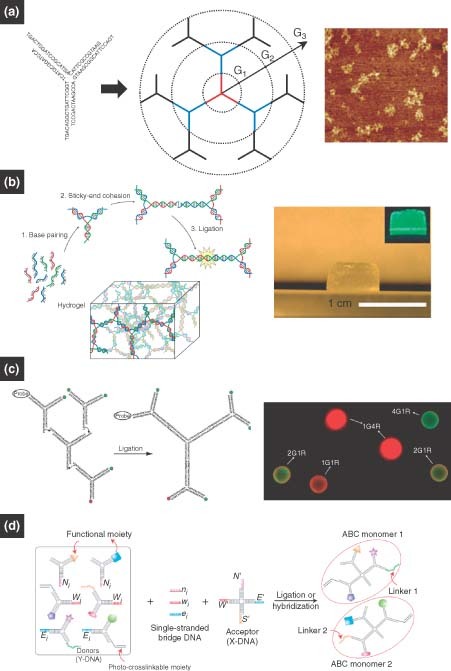
(a) Dendrimer‐like DNA (DL‐DNA): Y‐shaped DNA with carefully designed overhangs can be enzymatically assembled into DL‐DNA structures one generation at a time.90 (b) DNA hydrogel: palindromic overhangs, on the other hand, will result in a bulk hydrogel on the addition of DNA ligase. (Reprinted with permission from Ref 30. Copyright 2006 Nature Publishing Group). (c, d) DNA nanobarcodes and ABC monomers: monomeric subunits with various modifications, such as molecular probes, fluorescent dyes, and photocrosslinkable moieties, can be rationally assembled into anisotropic dendrimers.33, 34.
By connecting these dendritic or branched DNA motifs together, one can realize the fabrication of 3D network structures such as a 3D DNA gel, resembling the synthesis of polymeric hydrogel. Using branched building blocks, which acted as both monomers and crosslinkers, our group designed a DNA hydrogel (Figure 5(b), right).30 This DNA hydrogel differs from other hydrogels in that crosslinking is performed through efficient, ligase‐mediated reactions under physiological conditions. The basic synthesis began with the formation of branched DNA through base‐pairing interactions among oligonucleotides; it then underwent the sticky‐end ligation of branched DNA in the presence of T4 ligase (Figure 5(b), left). Proteins, drugs, and live cells can be encapsulated by the hydrogel in situ. The micromorphology of DNA hydrogels can be easily tailored by changing the concentrations and types of DNA motifs, which allows for the manipulation of hydrogel properties for applications such as controlled drug delivery, tissue engineering, and 3D cell culture.
As multibranched DNA structures can now be fabricated, it is easy to decorate different arms with different functional groups, resulting in multifunctional DNA nanostructures. Our group prepared DL‐DNA‐based, fluorescence‐intensity‐encoded nanobarcodes that contained a built‐in, color‐ratio code for multiplexed molecular sensing.33,91 The DNA nanobarcodes were prepared using Y‐shaped DNA to construct DL‐DNA with different arms decorated with different dyes or molecular probes. In more detail, Y‐shaped DNA structures were designed as follows: two arms carried a combination of fluorescent dyes or a fluorescent dye and a molecular probe; the other free arm contained a sticky end for ligation. Mixing these Y‐DNA structures together with the addition of DNA ligase produced the desired nanobarcode (Figure 5(c)). A polystyrene microbead decorated with nanobarcodes can preserve the programed fluorescence ratios (Figure 5(c), right). The use of these DNA nanobarcodes in multiplexed diagnostics will be discussed later.
In addition, DNA nanomaterials modified by functional moieties can be used to construct multifunctional architectures for applications in nanoelectronics, intelligent sensing, and targeted drug delivery. One of the requirements for these systems is that building block needs to be multivalent, anisotropic, and universally applicable. To address this goal, our group recently created novel, DNA‐based, anisotropic, branched, and crosslinkable monomers (termed ‘ABC monomers’) for the assembly of multifunctional nanoarchitectures for highly sensitive pathogen detection.34 The ABC monomers represent a modular ‘plug‐and‐play’ approach to multifunctional nanomaterials design. The creation of ABC monomers has enabled an entirely new polymerization scheme—‘target‐driven polymerization’—where DNA‐based ABC monomers are polymerized only in the presence of a specific pathogen DNA target. As the polymer is much larger and thus easier to detect than the monomers by themselves, and as signals within each monomers are aggregated and thus amplified within one polymer, the target‐driven polymerization gives rise to an unprecedented sensitivity and specificity in molecular sensing. In more detail, the ABC monomers are formed from ‘donor’ Y‐DNA and ‘acceptor’ X‐DNA. Both donors and acceptors are linked together through ssDNA bridges (bridge sequences are complementary to regions of both Y‐ and X‐DNA). The Y‐DNA can be functionalized with different modules such as DNA capture probes, fluorescent dyes, quantum dots (QDs), gold nanoparticles (AuNPs), and photocrosslinkable moieties (e.g., polyethylene glycol monoacrylate). These conjugations allow the ABC monomers to have almost unlimited diversity of anisotropy and multifunctionality (Figure 5(d)). To further demonstrate the specificity of multivalent conjugations, the ABC monomers were designed to have different predetermined ratios of fluorescent dyes within a single monomer. Through gel electrophoresis it was found that the fluorescent colors of the ABC monomers correspond specifically to the different combinations of donor Y‐DNAs as designed for each monomer.
APPLICATIONS
DNA as Template for Self‐Assembly
One of the key challenges in nanotechnology is assembling interconnected, spatially addressable nanoscale components in a controlled fashion. As top‐down lithographic patterning is approaching its feature‐size limits, DNA scaffolds are being investigated as a promising bottom‐up alternative and enhancement to nanofabrication because they allow precise positioning of nanoscale materials with a resolution lower than 10 nm. In addition, the assembly process is simplified because these scaffolds can self‐assemble from rationally designed DNA subunits. Hence, by circumventing the limitations of current lithographic techniques, DNA scaffolds are likely to play a promising role in the assembly of spatially addressable nanoscale components in future optoelectronic devices.
For the purpose of assembling nanowires, linear arrays of nanoparticles have been demonstrated by several groups. Nanoparticle wires have been achieved using electrostatic binding of ligand‐capped AuNPs to the DNA backbone,92,93 intercalation of the nanoparticle capping ligand into the DNA template,94 and by capturing streptavidin‐functionalized nanoparticles on biotinylated linear DNA tile arrays.95 In a recent report by Stearns et al.,96 DNA nanotubes with surface‐immobilized peptide–DNA conjugates were used to generate linear nanoparticle arrays. Here, gold salt was reduced in the presence of the peptides, resulting in the nucleation and growth of AuNPs at the regions in which the peptide was displayed.
By incorporating functional moieties into their architectures, 2D DNA lattices can serve as scaffolds for organizing nanoscale materials, such as nanoparticles and macromolecules. Xiao et al. 97 reported DNA tile grids made from DX DNA tiles. Nanoparticles were covalently attached to DNA within one of the tiles. In a later approach, AuNPs were capped with ssDNA and were subsequently tethered to 2D DX DNA tile arrays by means of complementary overhangs that extended from the tiles.98,99 In addition to tile arrays, periodic square lattices assembled from four‐armed branched junctions have been used to arrange DNA‐functionalized AuNPs. Careful design of the lattice tiles such that a single nanoparticle could be immobilized at each junction resulted in arrays with precise positioning of the nanoparticles100,101 (Figure 6(a)). Zheng et al. 102 demonstrated the periodic arrangement of nanoparticles of different sizes (Figure 6(b)). In addition to AuNPs, DNA scaffolds are capable of arranging other nanoscale components, such as QDs103 and even carbon nanotubes.104 Recently, DNA origami‐based strategies have been used to organize AuNPs. In a hybrid approach combining lithography and self‐assembly, Hung et al. 105 were able to selectively deposit DNA origami structures on a lithographically patterned surface. The DNA structures were designed to hybridize with DNA‐capped AuNPs, demonstrating a versatile approach for large‐scale nanoparticle patterning.
Figure 6.
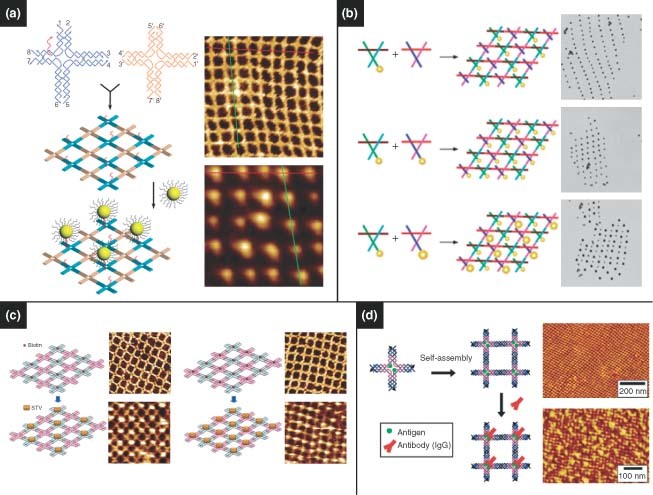
DNA nanostructures serve as template for assembling nanoscale components. (a) Two‐dimensional tile arrays of gold nanoparticles. (Reprinted with permission from Ref 100. Copyright 2006 American Chemical Society). (b) Programmable subunits enable the arrangement of different‐sized nanoparticles. (Reprinted with permission from Ref 102. Copyright 2006 American Chemical Society). (c) Streptavidin arrays. (Reprinted with permission from Ref 106. Copyright 2005 American Chemical Society). (d) Antibody arrays. (Reprinted with permission from Ref 108. Copyright 2006 American Chemical Society)..
A variety of templating techniques have been applied toward spatially addressable macromolecule patterning. Precise patterning of proteins, in particular, allows for fundamental protein–protein interactions to be investigated, which will aid in future drug design and biomolecular diagnostics. Yan et al. 106 first reported the programmable assembly of periodic protein arrays by means of self‐assembled DNA lattices (Figure 6(c)). By incorporating protein‐binding aptamer sequences into both DX DNA tiles and DNA origami, Chhabra et al. 107 reported a versatile and facile technique for generating well‐defined linear protein arrays. Similar approaches have been developed for developing highly ordered antibody arrays for investigating protein–protein interaction108,109 (Figure 6(d)). Numajiri et al. 110 demonstrated the potential of DNA origami in selectively forming protein–DNA complexes. Here, DNA origami structures were designed to have well‐defined cavities that enabled size‐selective capture of streptavidin, allowing selective asymmetric modification. Voigt et al. 111 used a DNA origami approach to perform spatially addressable single‐molecule chemical reactions. Formation and cleavage of chemical bonds were observed by AFM of the resulting biotin–streptavidin complexes that formed as products. Such strategies allow DNA scaffolds to act as breadboards for controlling molecular interactions.
DNA as Guide for Self‐Assembly
DNA‐guided assembly, unlike predefined scaffolds, is based on the real‐time assembly of nanoscale components by virtue of DNA's molecular recognition capabilities. The success of such self‐assembly strategies has led to great advances in biomolecular sensing, and it is expected to facilitate the design of novel materials with unique optical, magnetic, and electronic properties. Mirkin and Alivisatos were the first to report programmable aggregation of DNA‐capped AuNPs112, 113, 114 (Figure 7(a)). These programmable strategies have since been employed to develop ‘nanoparticle molecules’115,116 (Figure 7(b) and (c)) as well as 3D nanoparticle crystals.117, 118, 119 Park et al. 119 were able to program nanoparticle–DNA conjugates that could assemble into different crystal lattice arrangements, including single‐component and multicomponent systems, by varying the DNA linkers (Figure 7(d)). Maye et al. 120 demonstrated switchable states of nanoparticle crystals in which the crystals respond to the presence of free ssDNA by changes in lattice spacing.
Figure 7.
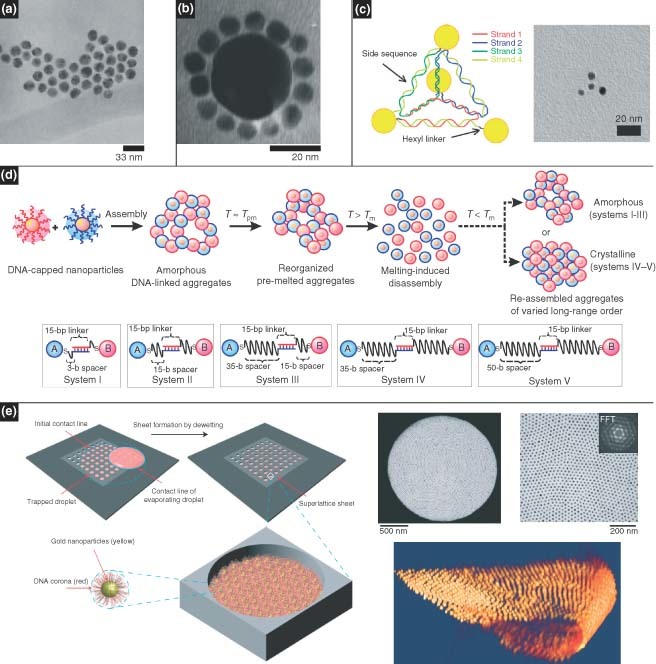
DNA nanostructures serve as guide for assembling nanoscale components. (a, b) DNA programmable nanoparticles have been shown to form nanoparticle networks. (Reprinted with permission from Refs 112 and 116. Copyright 1996 and 1998 Nature Publishing Group and American Chemical Society). (c) Molecule‐like nanoparticle arrangements. (Reprinted with permission from Ref 115. Copyright 2009 American Chemical Society). (d) The formation of amorphous or crystalline systems can be controlled by adjusting the DNA spacers.118 (e) Two‐dimensional nanoparticle superlattices have been achieved based on solvent evaporation. (Reprinted with permission from Ref 118. Copyright 2008 Nature Publishing Group)..
In addition to programmability, high‐precision nanoparticle order has also been achieved using DNA as a generic spacer without base‐pairing requirements. We demonstrated patterning of nanoparticle superlattices based on controlled drying of a colloidal solution on a solid surface with a microfabricated stamp.121 Developing this drying‐mediated strategy further, free‐standing nanoparticle membranes were formed within micropatterned holes of a silicon nitride membrane122 (Figure 7(e)). Here, it was shown that simply varying the DNA ligand length could tune the interparticle spacing, which in turn varied the mechanical and optical properties. 3D crystallization of these soft nanoparticles was also observed in real time and in situ by means of small‐angle X‐ray scattering.123 These studies revealed the formation of crystallites during the evaporation process, which exhibited a continuum of lattice spacings. It was also shown that the crystals could expand and contract by varying the humidity.
Medical Diagnostics
Medical diagnostic techniques use signals to detect and identify diseases or medical conditions. In the past several decades, various new and novel materials, such as nanoparticles, nanowires, and QDs, have been incorporated into conventional diagnostic systems to improve the sensitivity, selectivity, and practicality of assays.124 Although the process and principle of diagnostics is similar, namely to identify the target based on specific molecular interactions and transduce the interactions into detectable signals, there is no universal method for every application‐specific system. DNA materials possess a unique molecular recognition capability and can be rationally designed, and thus DNA‐based nanostructures have shown outstanding performance for sensing in medical diagnostics systems.125
DNA is a desirable material for sensing not only because of its excellent molecular recognition ability, but also because of its high physicochemical stability. This means that it can be synthesized, processed, and stored under a wide range of temperatures and conditions, which is necessary for the widespread dissemination of medical technologies. Furthermore, the ability to control self‐assembly, as represented by the assembly of branched anisotropic DNA nanostructures, provides a path toward achieving high levels of signal amplification and multiplexed detection.33,34 Additionally, chemical modifications and enzyme operations endow DNA the capacity for linking with different functional materials such as proteins and nanoparticles.
DNA–nanoparticle conjugates were the first demonstrated DNA‐based materials in diagnostics. The system was principally based on a landmark work in the assembly of oligonucleotide‐modified nanoparticle driven by DNA hybridization.126,127 After that, Mirkin and coworkers used DNA‐modified AuNPs to develop a highly selective, colorimetric DNA detection technique for DNA sensing. In this system, target DNA acted as a hybridization bridge between surface‐immobilized oligonucleotides, producing a red to purple color shift due to the plasmon coupling of the resulting AuNP aggregates.128,129 Since then, colorimetric methods based on DNA–nanoparticle conjugates have been used in various detection systems, such as proteins,130 mercuric ions,131 and DNA sequences.129
Colorimetric technology presented a simple and inexpensive diagnostic method. Moreover, it was able to detect targets with sensitivity down to the zeptomole scale without target or signal amplification.132 In addition to color changes in DNA–nanoparticle‐based diagnostics, Mirkin's group developed a series of signal transduction strategies, including light scattering,133 electrochemistry,134 and Raman spectroscopy.135 In addition to nanoparticles, nanowires and nanotubes have also been investigated as signal transduction motifs for their unique electronic properties.136,137 The assays are based on changes in the conduction of DNA‐modified nanowires or nanotubes after hybridization with target DNA.
Multiplexed detection is highly desirable in medical diagnostics for providing high‐throughput assays and more economical detection. Mirkin's group developed a well‐known nanoparticle‐based method, called the ‘bio‐barcode assay’, for multiplexed detection of proteins and nucleic acids.130,138,139 This method is based on the ability of target DNA to bridge AuNPs and magnetic microparticles, each of which are modified by ssDNA that are complementary to opposite ends of the target. Magnetic microparticles complexed with target DNA and functionalized AuNPs are collected by applying a magnetic field, leaving behind all unbound nanoparticle probes that did not sense any targets. Barcode DNA is isolated from the collected nanoparticles and is subsequently used in a scanometric amplification assay for target identification.140
Branched DNA nanostructures also have potential in multiplexed diagnostics, as they offer the ability to tailor specific functionalities onto unique arms, resulting in rationally designed anisotropic structures. As mentioned previously, our group developed DNA nanobarcodes for multiplexed detection.33 Multicolor nanobarcodes could be fabricated by precisely controlling both the types and the numbers of the fluorescent labels. DNA nanobarcodes enabled the simultaneous detection of several different pathogenic DNA. The readouts were flexible using: fluorescence microscopy, dot blotting, flow cytometry, or even a digital camera. The sensitivity reached down to the attomole scale with a very rapid detection time. For example, Figure 8(a) shows the results of nanobarcodes that were used for dot blotting diagnostics. Six samples, including the controls of 27‐mer ssDNA with irrelevant sequences and a 6.1‐kb plasmid DNA, were first blotted onto a membrane; five types of nanobarcodes were then used to hybridize onto target DNA for detection (Figure 8(a), top scheme). Simultaneous detection of four colors on the resultant membrane indicated four DNA targets (Bacillus anthracis, Francisella tularensis, Ebola virus, and SARS coronavirus), which were subsequently identified by referring to a preassigned color‐ratio decoding library. It was found that no fluorescent signals were detected in the two control spots, suggesting nanobarcode‐based detection is highly specific (Figure 8(a), lower image).
Figure 8.
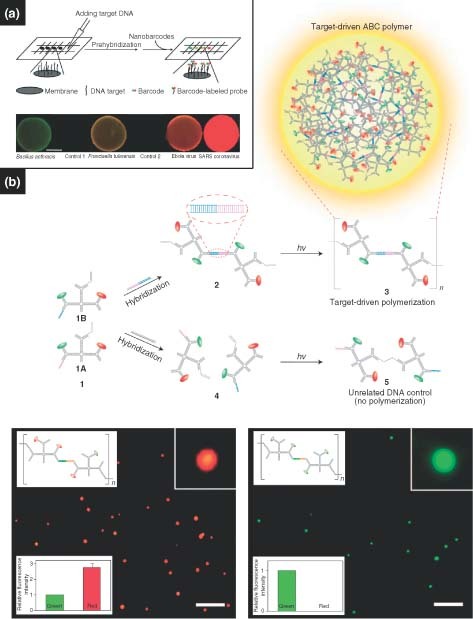
(a) Nanobarcodes and (b) a target‐driven polymerization of ABC monomers used in a multiplexed detection strategy.33, 34.
Our group further developed a target‐driven polymerization strategy based on branched DNA‐based ABC monomers.34 As mentioned previously, subunits of ABC monomers were labeled with various moieties including two QDs, one photocrosslinkable group, and one single‐stranded oligonucleotide probe that was complementary to a specific pathogen DNA (HIV DNA here, also see 1A and 1B monomer in Figure 8(b), top scheme). These ABC monomers formed dimers only in the presence of a specific pathogen DNA because the pathogen DNA served as a complementary linker DNA. On exposure to a short‐wavelength UV illumination, these dimers polymerized into polymer aggregates (polymeric spheres), with diameters of about 400 nm (from 1 to 2, and to 3 in Figure 8(b), top scheme). However, if the target pathogen DNA was absent, polymerization did not occur (from 1 to 4, and to 5 in Figure 8(b), top scheme). This target‐DNA‐driven polymerization enabled rapid signal amplification to detect pathogens with high specificity and sensitivity. Figure 8(b) shows the fluorescence microscopic images of the detection: B. anthracis (left) and Ebola pathogen (right). Interestingly, it was found that the concentration of ABC polymer spheres was linearly proportional to the log of concentration of target DNA, so that the concentration of pathogen DNA could be read out easily when plotted against the number of spherical aggregates. The limit of detection was about 100 fM.
Armitage and coworkers developed fluorescent DNA nanotags using branched DNA nanostructures as a template to obtain high‐resolution images and sensitive detection.141,142 Here, DNA nanostructures served as carriers for the assembly of multiple intercalating dyes within a small and well‐defined region. The compact structure of the nanotag allowed efficient energy transfer to remote acceptor groups, resulting in bright multichromophore assemblies. In the systems, a branched double‐helical DNA nanostructure (e.g., Y‐shaped DNA) was used as the template, and the dyes were bound noncovalently to the DNA by intercalation between base pairs. The resulting self‐assembled multichromophore arrays exhibited high effective extinction coefficients and efficient energy transfer between co‐intercalated dyes or from intercalated dyes to covalently attached acceptor fluorophores. Furthermore, they developed 3D DNA tetrahedron nanostructures as the template of fluorescent DNA nanotags, which showed improved photostability and enhanced resistance to nuclease digestion. The compact nature of the supramolecular DNA tetrahedron provided protection for the fluorophores.
DNA Gel as a Scaffold for Cell‐Free Protein Production
High‐yield protein production is an essential task in protein expression and protein engineering. Traditional animal‐ or cell‐based systems for protein production are generally labor‐intensive, costly, and time‐consuming. Moreover, many proteins are difficult to be produced. As an alternative, cell‐free protein production systems can surmount the difficulties associated with traditional animal‐ or cell‐based systems. Cell‐free systems use purified cell extracts (lysates) to transcribe and translate DNA, which allows faster and simplified protein production. Current cell‐free protein expression systems developed over the past 50 years have led to an increased volumetric yield in the micrograms per milliliter range, but seldom reaching the milligrams per milliliter level.143, 144, 145, 146, 147, 148, 149, 150, 151, 152 Almost all cell‐free systems are solution phase systems (SPS), in which the gene templates are dispersed in solution. However, the gene format (the incorporation of the gene into larger structures or through surface fixation) is an alternate way that can be modified to improve the yield and efficiency.
The merits of DNA gel, such as programmability and biocompatibility, make it a promising candidate as a scaffold material for biomedical applications in general, and protein expression in particular. Though applications regarding macro‐scale DNA materials have been somewhat limited, our group was the first to successfully utilize DNA gels for cell‐free protein expression. Here, DNA hydrogel was used as a scaffold for cell‐free protein production (called ‘P‐gel’), resulting in a 300‐fold protein yield over commercially available cell‐free systems (Figure 9).31 The gel is composed of X‐DNA that have sticky ends complementary to the overhangs of the linearized plasmid. Such a design allows the formation of covalent bonds between the linearized plasmid and the DNA network scaffold through ligation. Three mechanisms were proposed for the higher yield: first, the plasmid is linked with X‐DNA, which protects genes from degradation and denaturing, and subsequently leads to increased stability and prolonged expression; second, local gene concentration on P‐gel is higher than that in SPS; third, because the gene templates are confined, the enzymatic turnover rate increases and facilitates a higher rate of protein production. So far, this P‐gel system has been employed to successfully express more than several dozens of different proteins, including reporter proteins, membrane proteins, kinases, and toxic proteins with molecular weights ranging from 16 to 110 kDa. All proteins were functional with total protein yields of up to 5 mg/mL. It is expected that the P‐gel system would be compatible with high‐throughput formats such as microfluidics for large‐scale protein screening or production.
Figure 9.
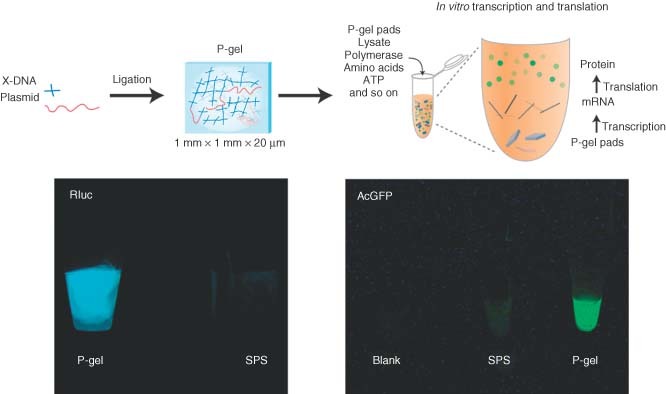
Protein‐producing DNA hydrogel (‘P‐gel’). The plasmid is ligated into a DNA hydrogel matrix, resulting in gene‐encapsulated P‐gel pads. The pads are then added to a cell lysate to express proteins. A comparison of P‐gel to a solution phase system shows greater expression of two fluorescent proteins by P‐gel.31.
SUMMARY AND OUTLOOK
In this review we have summarized the development of DNA‐based nanomaterials and their biomedical applications. To date, scientists from multiple disciplines, including biology, chemistry, and materials science, have fabricated a large category of DNA‐based novel materials through either self‐assembly or enzyme‐assisted assembly in a rationally designed and controlled manner. It is certain that almost any arbitrary‐shaped DNA material can be achieved in the future.
The developmental history of materials suggests that only materials that have real‐world applications and impact and benefit human kind will continue to be investigated. Currently, DNA materials are only in the initial stage of this pathway. But what is the fate of DNA materials in the future? Although DNA has shown great potential as the ‘bricks and concrete’ in the construction of nanomaterials, much progress must be made before its real‐world applications can be fully realized. From a production perspective, limiting factors include difficulty in scaling‐up, the time to produce these materials, and the cost. From a functional perspective, other important hurdles include robustness and durability, optimization of error prevention, ease of design and fabrication, and the ability to interface DNA nanomaterials with other materials. Now is the time to investigate ways to overcome these challenges. Thanks to the endeavors of many researchers in this field, various remarkable applications have already been demonstrated. We envision that through collective efforts, DNA materials will be ultimately employed for a myriad of real‐world applications, but before this we expect that their impact will be felt most strongly in the field of nanomedicine.
REFERENCES
- 1. Seeman NC. Nucleic‐Acid junctions and lattices. J Theor Biol 1982; 99: 237–247. [DOI] [PubMed] [Google Scholar]
- 2. Luo D. The road from biology to materials. Mater Today 2003; 6: 38–43. [Google Scholar]
- 3. LaBean TH, Li HY. Constructing novel materials with DNA. Nano Today 2007; 2: 26–35. [Google Scholar]
- 4. Seeman NC. DNA in a material world. Nature 2003; 421: 427–431. [DOI] [PubMed] [Google Scholar]
- 5. Seeman NC, Lukeman PS. Nucleic acid nanostructures: bottom‐up control of geometry on the nanoscale. Rep Prog Phys 2005; 68: 237–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin C, Liu Y, Yan H. Designer DNA nanoarchitectures. Biochemistry 2009; 48: 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feldkamp U, Niemeyer CM. Rational design of DNA nanoarchitectures. Angew Chem Int Edit 2006; 45: 1856–1876. [DOI] [PubMed] [Google Scholar]
- 8. Watson JD, Crick FHC. Molecular structure of nucleic acids—a Structure for deoxyribose nucleic acid. Nature 1953; 171: 737–738. [DOI] [PubMed] [Google Scholar]
- 9. Holliday R. A mechanism for gene conversion in fungi. Genet Res 1964; 5: 282–304. [DOI] [PubMed] [Google Scholar]
- 10. Meselson MS, Radding CM. General model for genetic recombination. Proc Natl Acad Sci U S A 1975; 72: 358–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin CX, Liu Y, Rinker S, Yan H. DNA tile based self‐assembly: building complex nanoarchitectures. ChemPhysChem 2006; 7: 1641–1647. [DOI] [PubMed] [Google Scholar]
- 12. Winfree E, Liu FR, Wenzler LA, Seeman NC. Design and self‐assembly of two‐dimensional DNA crystals. Nature 1998; 394: 539–544. [DOI] [PubMed] [Google Scholar]
- 13. Fu TJ, Seeman NC. DNA double‐crossover molecules. Biochemistry 1993; 32: 3211–3220. [DOI] [PubMed] [Google Scholar]
- 14. Li XJ, Yang XP, Qi J, Seeman NC. Antiparallel DNA double crossover molecules as components for nanoconstruction. J Am Chem Soc 1996; 118: 6131–6140. [Google Scholar]
- 15. Liu FR, Sha RJ, Seeman NC. Modifying the surface features of two‐dimensional DNA crystals. J Am Chem Soc 1999; 121: 917–922. [Google Scholar]
- 16. Reishus D, Shaw B, Brun Y, Chelyapov N, Adleman L. Self‐assembly of DNA double‐double crossover complexes into high‐density, doubly connected, planar structures. J Am Chem Soc 2005; 127: 17590–17591. [DOI] [PubMed] [Google Scholar]
- 17. LaBean TH, Yan H, Kopatsch J, Liu FR, Winfree E, et al. Construction, analysis, ligation, and self‐assembly of DNA triple crossover complexes. J Am Chem Soc 2000; 122: 1848–1860. [Google Scholar]
- 18. Liu Y, Lin CX, Li HY, Yan H. Protein nanoarrays—aptamer‐directed self‐assembly of protein arrays on a DNA nanostructure. Angew Chem Int Edit 2005; 44: 4333–4338. [DOI] [PubMed] [Google Scholar]
- 19. Liu D, Park SH, Reif JH, LaBean TH. DNA nanotubes self‐assembled from triple‐crossover tiles as templates for conductive nanowires. Proc Natl Acad Sci U S A 2004; 101: 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan H, Zhang XP, Shen ZY, Seeman NC. A robust DNA mechanical device controlled by hybridization topology. Nature 2002; 415: 62–65. [DOI] [PubMed] [Google Scholar]
- 21. Shen ZY, Yan H, Wang T, Seeman NC. Paranemic crossover DNA: a generalized Holliday structure with applications in nanotechnology. J Am Chem Soc 2004; 126: 1666–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu WY, Wang X, Wang T, Sha RJ, Seeman NC. PX DNA triangle oligomerized using a novel three‐domain motif. Nano Lett 2008; 8: 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature 2006; 440: 297–302. [DOI] [PubMed] [Google Scholar]
- 24. Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, et al. Self‐assembly of a nanoscale DNA box with a controllable lid. Nature 2009; 459: 73–76. [DOI] [PubMed] [Google Scholar]
- 25. Simmel FC. Three‐dimensional nanoconstruction with DNA. Angew Chem Int Edit 2008; 47: 5884–5887. [DOI] [PubMed] [Google Scholar]
- 26. Zheng JP, Birktoft JJ, Chen Y, Wang T, Sha RJ, et al. From molecular to macroscopic via the rational design of a self‐assembled 3D DNA crystal. Nature 2009; 461: 74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Niemeyer CM, Sano T, Smith CL, Cantor CR. Oligonucleotide‐directed self‐assembly of proteins—semisynthetic DNA streptavidin hybrid molecules as connectors for the generation of macroscopic arrays and the construction of supramolecular bioconjugates. Nucleic Acids Res 1994; 22: 5530–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Niemeyer CM, Ceyhan B, Hazarika P. Oligofunctional DNA‐gold nanoparticle conjugates. Angew Chem Int Edit 2003; 42: 5766–5770. [DOI] [PubMed] [Google Scholar]
- 29. Niemeyer CM, Simon U. DNA‐based assembly of metal nanoparticles. Eur J Inorg Chem 2005; 3641–3655. [Google Scholar]
- 30. Um SH, Lee JB, Park N, Kwon SY, Umbach CC, Luo D. Enzyme‐catalysed assembly of DNA hydrogel. Nat Mater 2006; 5: 797–801. [DOI] [PubMed] [Google Scholar]
- 31. Park N, Um SH, Funabashi H, Xu JF, Luo D. A cell‐free protein‐producing gel. Nat Mater 2009; 8: 432–437. [DOI] [PubMed] [Google Scholar]
- 32. Cheng EJ, Xing YZ, Chen P, Yang Y, Sun YW, et al. A pH‐triggered, fast‐responding DNA hydrogel. Angew Chem Int Edit 2009; 48: 7660–7663. [DOI] [PubMed] [Google Scholar]
- 33. Li YG, Cu YTH, Luo D. Multiplexed detection of pathogen DNA with DNA‐based fluorescence nanobarcodes. Nat Biotechnol 2005; 23: 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee JB, Roh YH, Um SH, Funabashi H, Cheng WL, et al. Multifunctional nanoarchitectures from DNA‐based ABC monomers. Nat Nanotechnol 2009; 4: 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gothelf KV, LaBean TH. DNA‐programmed assembly of nanostructures. Org Biomol Chem 2005; 3: 4023–4037. [DOI] [PubMed] [Google Scholar]
- 36. Li HY, Carter JD, LaBean TH. Nanofabrication by DNA self‐assembly. Mater Today 2009; 12: 24–32. [Google Scholar]
- 37. He Y, Liu HP, Chen Y, Tian Y, Deng ZX, et al. DNA‐based nanofabrications. Microsc Res Tech 2007; 70: 522–529. [DOI] [PubMed] [Google Scholar]
- 38. Niemeyer CM. Functional devices from DNA and proteins. Nano Today 2007; 2: 42–52. [Google Scholar]
- 39. Seeman NC. An overview of structural DNA nanotechnology. Mol Biotechnol 2007; 37: 246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seeman NC. DNA engineering and its application to nanotechnology. Trends Biotechnol 1999; 17: 437–443. [DOI] [PubMed] [Google Scholar]
- 41. Seeman NC. Nucleic acid nanostructures and topology. Angew Chem Int Edit 1998; 37: 3220–3238. [DOI] [PubMed] [Google Scholar]
- 42. Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science 2008; 321: 1795–1799. [DOI] [PubMed] [Google Scholar]
- 43. Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol 2007; 2: 275–284. [DOI] [PubMed] [Google Scholar]
- 44. Niemeyer CM, Adler M. Nanomechanical devices based on DNA. Angew Chem Int Edit 2002; 41: 3779–3783. [DOI] [PubMed] [Google Scholar]
- 45. Simmel FC, Dittmer WU. DNA nanodevices. Small 2005; 1: 284–299. [DOI] [PubMed] [Google Scholar]
- 46. Liedl T, Sobey TL, Simmel FC. DNA‐based nanodevices. Nano Today 2007; 2: 36–41. [Google Scholar]
- 47. Liu H, Liu DS. DNA nanomachines and their functional evolution. Chem Commun 2009; 2625–2636. [DOI] [PubMed] [Google Scholar]
- 48. Kwon YW, Lee CH, Choi DH, Jin JI. Materials science of DNA. J Mater Chem 2009; 19: 1353–1380. [Google Scholar]
- 49. Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA‐templated self‐assembly of protein arrays and highly conductive nanowires. Science 2003; 301: 1882–1884. [DOI] [PubMed] [Google Scholar]
- 50. Fischler M, Simon U, Nir H, Eichen Y, Burley GA, et al. Formation of bimetallic Ag‐Au nanowires by metallization of artificial DNA duplexes. Small 2007; 3: 1049–1055. [DOI] [PubMed] [Google Scholar]
- 51. Douglas SM, Chou JJ, Shih WM. DNA‐nanotube‐induced alignment of membrane proteins for NMR structure determination. Proc Natl Acad Sci U S A 2007; 104: 6644–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Endo M, Seeman NC, Majima T. DNA tube structures controlled by a four‐way‐branched DNA connector. Angew Chem Int Edit 2005; 44: 6074–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu HP, Chen Y, He Y, Ribbe AE, Mao CD. Approaching the limit: can one DNA oligonucleotide assemble into large nanostructures?. Angew Chem Int Edit 2006; 45: 1942–1945. [DOI] [PubMed] [Google Scholar]
- 54. Kuzuya A, Wang RS, Sha RJ, Seeman NC. Six‐helix and eight‐helix DNA nanotubes assembled from half‐tubes. Nano Lett 2007; 7: 1757–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yin P, Hariadi RF, Sahu S, Choi HMT, Park SH, et al. Programming DNA tube circumferences. Science 2008; 321: 824–826. [DOI] [PubMed] [Google Scholar]
- 56. Aldaye FA, Lo PK, Karam P, McLaughlin CK, Cosa G, Sleiman HF. Modular construction of DNA nanotubes of tunable geometry and single‐ or double‐stranded character. Nat Nanotechnol 2009; 4: 349–352. [DOI] [PubMed] [Google Scholar]
- 57. Nakao H, Gad M, Sugiyama S, Otobe K, Ohtani T. Transfer‐printing of highly aligned DNA nanowires. J Am Chem Soc 2003; 125: 7162–7163. [DOI] [PubMed] [Google Scholar]
- 58. Guan JJ, Lee J. Generating highly ordered DNA nanostrand arrays. Proc Natl Acad Sci U S A 2005; 102: 18321–18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Deng ZX, Mao CD. DNA‐templated fabrication of 1D parallel and 2D crossed metallic nanowire arrays. Nano Lett 2003; 3: 1545–1548. [Google Scholar]
- 60. Brenneman A, Condon A. Strand design for biomolecular computation. Theor Comput Sci 2002; 287: 39–58. [Google Scholar]
- 61. Condon A. Designed DNA molecules: principles and applications of molecular nanotechnology. Nat Rev Genet 2006; 7: 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 1995; 83: 783–791. [DOI] [PubMed] [Google Scholar]
- 63. Seeman NC. Nanotechnology and the double helix. Sci Am 2004; 290: 64–75. [DOI] [PubMed] [Google Scholar]
- 64. Mao CD, Sun WQ, Seeman NC. Designed two‐dimensional DNA Holliday junction arrays visualized by atomic force microscopy. J Am Chem Soc 1999; 121: 5437–5443. [Google Scholar]
- 65. He Y, Chen Y, Liu HP, Ribbe AE, Mao CD. Self‐assembly of hexagonal DNA two‐dimensional (2D) arrays. J Am Chem Soc 2005; 127: 12202–12203. [DOI] [PubMed] [Google Scholar]
- 66. Zhang C, Su M, He Y, Zhao X, Fang PA, et al. Conformational flexibility facilitates self‐assembly of complex DNA nanostructures. Proc Natl Acad Sci U S A 2008; 105: 10665–10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. He Y, Tian Y, Ribbe AE, Mao CD. Highly connected two‐dimensional crystals of DNA six‐point‐stars. J Am Chem Soc 2006; 128: 15978–15979. [DOI] [PubMed] [Google Scholar]
- 68. Chelyapov N, Brun Y, Gopalkrishnan M, Reishus D, Shaw B, Adleman L. DNA triangles and self‐assembled hexagonal tilings. J Am Chem Soc 2004; 126: 13924–13925. [DOI] [PubMed] [Google Scholar]
- 69. He Y, Tian Y, Chen Y, Deng ZX, Ribbe AE, Mao CD. Sequence symmetry as a tool for designing DNA nanostructures. Angew Chem Int Edit 2005; 44: 6694–6696. [DOI] [PubMed] [Google Scholar]
- 70. Zhang C, Ko SH, Su M, Leng YJ, Ribbe AE, et al. Symmetry Controls the Face Geometry of DNA Polyhedra. J Am Chem Soc 2009; 131: 1413–1415. [DOI] [PubMed] [Google Scholar]
- 71. Hamada S, Murata S. Substrate‐Assisted Assembly of Interconnected Single‐Duplex DNA Nanostructures. Angew Chem Int Edit 2009; 48: 6820–6823. [DOI] [PubMed] [Google Scholar]
- 72. Liu HP, He Y, Ribbe AE, Mao CD. Two‐dimensional (2D) DNA crystals assembled from two DNA strands. Biomacromolecules 2005; 6: 2943–2945. [DOI] [PubMed] [Google Scholar]
- 73. Sun XP, Ko SH, Zhang CA, Ribbe AE, Mao CD. Surface‐Mediated DNA Self‐Assembly. J Am Chem Soc 2009; 131: 13248–13249. [DOI] [PubMed] [Google Scholar]
- 74. Rothemund PWK, Papadakis N, Winfree E. Algorithmic self‐assembly of DNA Sierpinski triangles. PLoS Biol 2004; 2: 2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhao Z, Yan H, Liu Y. A route to scale up DNA origami using DNA tiles as folding staples. Angew Chem Int Edit 2010; 49: 1414–1417. [DOI] [PubMed] [Google Scholar]
- 76. Chen JH, Seeman NC. Synthesis from DNA of molecule with the connectivity of a cube. Nature 1991; 350: 631–633. [DOI] [PubMed] [Google Scholar]
- 77. Zhang YW, Seeman NC. Construction of a DNA‐truncated octahedron. J Am Chem Soc 1994; 116: 1661–1669. [Google Scholar]
- 78. Shih WM, Quispe JD, Joyce GF. A 1.7‐kilobase single‐stranded DNA that folds into a nanoscale octahedron. Nature 2004; 427: 618–621. [DOI] [PubMed] [Google Scholar]
- 79. Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 2005; 310: 1661–1665. [DOI] [PubMed] [Google Scholar]
- 80. Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN, Turberfield AJ. Reconfigurable, braced, three‐dimensional DNA nanostructures. Nat Nanotechnol 2008; 3: 93–96. [DOI] [PubMed] [Google Scholar]
- 81. He Y, Ye T, Su M, Zhang C, Ribbe AE, et al. Hierarchical self‐assembly of DNA into symmetric supramolecular polyhedra. Nature 2008; 452: 198–201. [DOI] [PubMed] [Google Scholar]
- 82. Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. Self‐assembly of DNA into nanoscale three‐dimensional shapes. Nature 2009; 459: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science 2009; 325: 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ke YG, Sharma J, Liu MH, Jahn K, Liu Y, Yan H. Scaffolded DNA origami of a DNA tetrahedron molecular container. Nano Lett 2009; 9: 2445–2447. [DOI] [PubMed] [Google Scholar]
- 85. Aldaye FA, Sleiman HF. Modular access to structurally switchable 3D discrete DNA assemblies. J Am Chem Soc 2007; 129: 13376–13377. [DOI] [PubMed] [Google Scholar]
- 86. Feldkamp U, Sacca B, Niemeyer CM. Dendritic DNA building blocks for amplified detection assays and biomaterials. Angew Chem Int Edit 2009; 48: 5996–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Caminade AM, Turrin CO, Majoral JP. Dendrimers and DNA: combinations of two special topologies for nanomaterials and biology. Chem Eur J 2008; 14: 7422–7432. [DOI] [PubMed] [Google Scholar]
- 88. Nilsen TW, Grayzel J, Prensky W. Dendritic nucleic acid structures. J Theor Biol 1997; 187: 273–284. [DOI] [PubMed] [Google Scholar]
- 89. Wang J, Jiang M, Nilsen TW, Getts RC. Dendritic nucleic acid probes for DNA biosensors. J Am Chem Soc 1998; 120: 8281–8282. [Google Scholar]
- 90. Li YG, Tseng YD, Kwon SY, D'Espaux L, Bunch JS, et al. Controlled assembly of dendrimer‐like DNA. Nat Mater 2004; 3: 38–42. [DOI] [PubMed] [Google Scholar]
- 91. Um SH, Lee JB, Kwon SY, Li Y, Luo D. Dendrimer‐like DNA‐based fluorescence nanobarcodes. Nat Protoc 2006; 1: 995–1000. [DOI] [PubMed] [Google Scholar]
- 92. Warner MG, Hutchison JE. Linear assemblies of nanoparticles electrostatically organized on DNA scaffolds. Nat Mater 2003; 2: 272–277. [DOI] [PubMed] [Google Scholar]
- 93. Nakao H, Shiigi H, Yamamoto Y, Tokonami S, Nagaoka T, et al. Highly ordered assemblies of Au nanoparticles organized on DNA. Nano Lett 2003; 3: 1391–1394. [Google Scholar]
- 94. Patolsky F, Weizmann Y, Lioubashevski O, Willner I. Au‐nanoparticle nanowires based on DNA and polylysine templates. Angew Chem Int Ed 2002; 41: 2323–2327. [DOI] [PubMed] [Google Scholar]
- 95. Li H, Park SH, Reif JH, LaBean TH, Yan H. DNA‐templated self‐assembly of protein and nanoparticle linear arrays. J Am Chem Soc 2004; 126: 418–419. [DOI] [PubMed] [Google Scholar]
- 96. Stearns LA, Chhabra R, Sharma J, Liu Y, Petuskey WT, et al. Template‐directed nucleation and growth of inorganic nanoparticles on DNA scaffolds. Angew Chem Int Ed 2009; 48: 8494–8496. [DOI] [PubMed] [Google Scholar]
- 97. Xiao S, Liu F, Rosen AE, Hainfeld JF, Seeman NC, et al. Selfassembly of metallic nanoparticle arrays by DNA scaffolding. J Nanoparticle Res 2002; 4: 313–317. [Google Scholar]
- 98. Le JD, Pinto Y, Seeman NC, Musier‐Forsyth K, Taton TA, Kiehl RA. DNA‐templated self‐assembly of metallic nanocomponent arrays on a surface. Nano Lett 2004; 4: 2343–2347. [DOI] [PubMed] [Google Scholar]
- 99. Pinto YY, Le JD, Seeman NC, Musier‐Forsyth K, Taton TA, Kiehl RA. Sequence‐encoded self‐assembly of multiple‐nanocomponent arrays by 2D DNA scaffolding. Nano Lett 2005; 5: 2399–2402. [DOI] [PubMed] [Google Scholar]
- 100. Zhang J, Liu Y, Ke Y, Yan H. Periodic square‐like gold nanoparticle arrays templated by self‐assembled 2D DNA nanogrids on a surface. Nano Lett 2006; 6: 248–251. [DOI] [PubMed] [Google Scholar]
- 101. Sharma J, Chhabra R, Liu Y, Ke Y, Yan H. DNA‐templated self‐assembly of two‐dimensional and periodical gold nanoparticle arrays. Angew Chem Int Ed 2006; 45: 730–735. [DOI] [PubMed] [Google Scholar]
- 102. Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. Two‐dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett 2006; 6: 1502–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sharma J, Ke Y, Lin C, Chhabra R, Wang Q, et al. DNA‐tile‐directed self‐assembly of quantum dots into two‐dimensional nanopatterns. Angew Chem Int Ed 2008; 47: 5157–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Maune HT, Han S‐P, Barish RD, Bockrath M, Goddard WA III, et al. Self‐assembly of carbon nanotubes into two‐dimensional geometries using DNA origami templates. Nat Nanotechnol 2010; 5: 61–66. [DOI] [PubMed] [Google Scholar]
- 105. Hung AM, Micheel CM, Bozano LD, Osterbur LW, Wallraff GM, Cha JN. Large‐area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat Nanotechnol 2010; 5: 121–126. [DOI] [PubMed] [Google Scholar]
- 106. Park SH, Yin P, Liu Y, Reif JH, LaBean TH, Yan H. Programmable DNA self‐assemblies for nanoscale organization of ligands and proteins. Nano Lett 2005; 5: 729–733. [DOI] [PubMed] [Google Scholar]
- 107. Chhabra R, Sharma J, Ke Y, Liu Y, Rinker S, et al. Spatially addressable multiprotein nanoarrays templated by aptamer‐tagged DNA nanoarchitectures. J Am Chem Soc 2007; 129: 10304–10305. [DOI] [PubMed] [Google Scholar]
- 108. He Y, Ribbe AE, Mao C. Antibody nanoarrays with a pitch of ∼20 nanometers. J Am Chem Soc 2006; 128: 12664–12665. [DOI] [PubMed] [Google Scholar]
- 109. Williams BAR, Lund K, Liu Y, Yan H, Chaput JC. Self‐assembled peptide nanoarrays: an approach to studying protein‐protein interactions. Angew Chem Int Ed 2007; 46: 3051–3054. [DOI] [PubMed] [Google Scholar]
- 110. Numajiri K, Kuzuya A, Komiyama M. Asymmetric secondary and tertiary streptavidin/DNA complexes selectively formed in a nanometer‐scale DNA well. Bioconjugate Chem 2010; 21: 338–344. [DOI] [PubMed] [Google Scholar]
- 111. Voigt NV, Tørring T, Rotaru A, Jacobsen MF, Ravnsbæk JB, et al. Single‐molecule chemical reactions on DNA origami. Nat Nanotechnol 2010; 5: 200–203. [DOI] [PubMed] [Google Scholar]
- 112. Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA‐based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996; 382: 607–609. [DOI] [PubMed] [Google Scholar]
- 113. Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, et al. Organization of ‘nanocrystal molecules’ using DNA. Nature 1996; 382: 609–611. [DOI] [PubMed] [Google Scholar]
- 114. Loweth CJ, Caldwell WB, Peng X, Alivisatos AP, Schultz PG. DNA‐based assembly of gold nanocrystals. Angew Chem Int Ed 1999; 38: 1808–1812. [DOI] [PubMed] [Google Scholar]
- 115. Mastroianni AJ, Claridge SA, Alivisatos AP. Pyramidal and chiral groupings of gold nanocrystals assembled using DNA scaffolds. J Am Chem Soc 2009; 131: 8455–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Mucic RC, Storhoff JJ, Mirkin CA, Letsinger RL. DNA‐directed synthesis of binary nanoparticle network materials. J Am Chem Soc 1998; 120: 12674–12675. [Google Scholar]
- 117. Maye MM, Nykypanchuk D, van der Lelie D, Gang O. DNA‐regulated micro‐ and nanoparticle assembly. Small 2007; 3: 1678–1682. [DOI] [PubMed] [Google Scholar]
- 118. Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA‐guided crystallization of colloidal nanoparticles. Nature 2008; 451: 549–552. [DOI] [PubMed] [Google Scholar]
- 119. Park SY, Lytton‐Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. DNA‐programmable nanoparticle crystallization. Nature 2008; 451: 553–556. [DOI] [PubMed] [Google Scholar]
- 120. Maye MM, Kumara MT, Nykypanchuk D, Sherman WB, Gang O. Switching binary states of nanoparticle superlattices and dimer clusters by DNA strands. Nat Nanotechnol 2010; 5: 116–120. [DOI] [PubMed] [Google Scholar]
- 121. Cheng WL, Park N, Walter MT, Hartman MR, Luo D. Nanopatterning self‐assembled nanoparticle superlattices by moulding microdroplets. Nat Nanotechnol 2008; 3: 682–690. [DOI] [PubMed] [Google Scholar]
- 122. Cheng WL, Campolongo MJ, Cha JJ, Tan SJ, Umbach CC, et al. Free‐standing nanoparticle superlattice sheets controlled by DNA. Nat Mater 2009; 8: 519–525. [DOI] [PubMed] [Google Scholar]
- 123. Cheng WL, Hartman MR, Smilgies D‐M, Long R, Campolongo MJ, et al. Probing in real time the soft crystallization of DNA‐capped nanoparticles. Angew Chem Int Ed 2010; 49: 380–384. [DOI] [PubMed] [Google Scholar]
- 124. Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev 2005; 105: 1547–1562. [DOI] [PubMed] [Google Scholar]
- 125. Lee JB, Campolongo MJ, Kahn JS, Roh YH, Hartman MR, Luo D. DNA‐based nanostructures for molecular sensing. Nanoscale 2010; 2: 188–197. [DOI] [PubMed] [Google Scholar]
- 126. Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA‐based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996; 382: 607–609. [DOI] [PubMed] [Google Scholar]
- 127. Alivisatos AP, Johnsson KP, Peng XG, Wilson TE, Loweth CJ, et al. Organization of ‘nanocrystal molecules’ using DNA. Nature 1996; 382: 609–611. [DOI] [PubMed] [Google Scholar]
- 128. Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance‐dependent optical properties of gold nanoparticles. Science 1997; 277: 1078–1081. [DOI] [PubMed] [Google Scholar]
- 129. Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One‐pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc 1998; 120: 1959–1964. [Google Scholar]
- 130. Nam JM, Thaxton CS, Mirkin CA. Nanoparticle‐based bio‐bar codes for the ultrasensitive detection of proteins. Science 2003; 301: 1884–1886. [DOI] [PubMed] [Google Scholar]
- 131. Lee JS, Han MS, Mirkin CA. Colorimetric detection of mercuric ion (Hg2+) in aqueous media using DNA‐functionalized gold nanoparticles. Angew Chem Int Edit 2007; 46: 4093–4096. [DOI] [PubMed] [Google Scholar]
- 132. Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat Biotechnol 2004; 22: 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Taton TA, Lu G, Mirkin CA. Two‐color labeling of oligonucleotide arrays via size‐selective scattering of nanoparticle probes. J Am Chem Soc 2001; 123: 5164–5165. [DOI] [PubMed] [Google Scholar]
- 134. Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol 2003; 21: 1192–1199. [DOI] [PubMed] [Google Scholar]
- 135. Cao YWC, Jin RC, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 2002; 297: 1536–1540. [DOI] [PubMed] [Google Scholar]
- 136. Hahm J, Lieber CM. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett 2004; 4: 51–54. [Google Scholar]
- 137. Woolley AT, Guillemette C, Cheung CL, Housman DE, Lieber CM. Direct haplotyping of kilobase‐size DNA using carbon nanotube probes. Nat Biotechnol 2000; 18: 760–763. [DOI] [PubMed] [Google Scholar]
- 138. Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew Chem Int Edit 2006; 45: 3303–3306. [DOI] [PubMed] [Google Scholar]
- 139. Nam JM, Stoeva SI, Mirkin CA. Bio‐bar‐code‐based DNA detection with PCR‐like sensitivity. J Am Chem Soc 2004; 126: 5932–5933. [DOI] [PubMed] [Google Scholar]
- 140. Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science 2000; 289: 1757–1760. [DOI] [PubMed] [Google Scholar]
- 141. Benvin AL, Creeger Y, Fisher GW, Ballou B, Waggoner AS, Armitage BA. Fluorescent DNA nanotags: Supramolecular fluorescent labels based on intercalating dye arrays assembled on nanostructured DNA templates. J Am Chem Soc 2007; 129: 2025–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ozhalici‐Unal H, Armitage BA. Fluorescent DNA nanotags based on a self‐assembled DNA tetrahedron. ACS Nano 2009; 3: 425–433. [DOI] [PubMed] [Google Scholar]
- 143. DeVries JK, Zubay G. DNA‐directed peptide synthesis. II. The synthesis of the alpha‐fragment of the enzyme beta‐galactosidase. Proc Natl Acad Sci U S A 1967; 57: 1010–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Spirin AS, Baranov VI, Ryabova LA, Ovodov SY, Alakhov YB. A continuous cell‐free translation system capable of producing polypeptides in high yield. Science 1988; 242: 1162–1164. [DOI] [PubMed] [Google Scholar]
- 145. Kim DM, Choi CY. A semicontinuous prokaryotic coupled transcription/translation system using a dialysis membrane. Biotechnol Prog 1996; 12: 645–649. [DOI] [PubMed] [Google Scholar]
- 146. Kim DM, Swartz JR. Prolonging cell‐free protein synthesis with a novel ATP regeneration system. Biotechnol Bioeng 1999; 66: 180–188. [PubMed] [Google Scholar]
- 147. Kim DM, Swartz JR. Regeneration of adenosine triphosphate from glycolytic intermediates for cell‐free protein synthesis. Biotechnol Bioeng 2001; 74: 309–316. [PubMed] [Google Scholar]
- 148. Kim DM, Swartz JR. Efficient production of a bioactive, multiple disulfide‐bonded protein using modified extracts of Escherichia coli . Biotechnol Bioeng 2004; 85: 122–129. [DOI] [PubMed] [Google Scholar]
- 149. DiTursi MK, Cha J, Newman MR, Dordick JS. Simultaneous in vitro protein synthesis using solid‐phase DNA template. Biotechnol Prog 2004; 20: 1705–1709. [DOI] [PubMed] [Google Scholar]
- 150. Ghosh D, Ramakanth M, Bhaumik A, Faure N, Rondelez F, Chatterji D. Transcription of T7 DNA immobilised on latex beads and Langmuir–Blodgett film. J Biochem Biophys Methods 2005; 62: 51–62. [DOI] [PubMed] [Google Scholar]
- 151. Yang J, Kanter G, Voloshin A, Michel‐Reydellet N, Velkeen H, et al. Rapid expression of vaccine proteins for B‐cell lymphoma in a cell‐free system. Biotechnol Bioeng 2005; 89: 503–511. [DOI] [PubMed] [Google Scholar]
- 152. Mei Q, Fredrickson CK, Simon A, Khnouf R, Fan ZH. Cell‐free protein synthesis in microfluidic array devices. Biotechnol Prog 2007; 23: 1305–1311. [DOI] [PubMed] [Google Scholar]


