Abstract
Viral nucleocapsid proteins (NCs) enwrap the RNA genomes of viruses to form NC–RNA complexes, which act as a template and are essential for viral replication and transcription. Beyond packaging viral RNA, NCs also play important roles in virus replication, transcription, assembly, and budding by interacting with viral and host cellular proteins. Additionally, NCs can inhibit interferon signaling response and function in cell stress response, such as inducing apoptosis. Finally, NCs can be the target of vaccines, benefiting from their conserved gene sequences. Here, we summarize important findings regarding the additional functions of NCs as much more than structural RNA‐binding proteins, with specific emphasis on (1) their association with the viral life cycle, (2) their association with host cells, and (3) as ideal candidates for vaccine development. WIREs RNA 2016, 7:213–226. doi: 10.1002/wrna.1326
This article is categorized under:
-
1
RNA Interactions with Proteins and Other Molecules > RNA–Protein Complexes
-
2
RNA Interactions with Proteins and Other Molecules > Protein–RNA Interactions: Functional Implications
-
3
Translation > Translation Regulation
INTRODUCTION
The nucleocapsid proteins (NCs) of RNA viruses encapsulate the viral RNA genome to form a helical nucleocapsid containing an NC–RNA complex, which usually serves as a template for viral transcription and replication1, 2, 3, 4 (Figure 1). In addition, NCs can interact with various other macromolecules, including those of viral and cellular origins. For some negative‐stranded RNA viruses, NCs can interact with themselves and phosphoproteins (Ps),5, 6 and NC–P complexes can form cytoplasmic inclusion bodies (IBs) that contain viral RNA and may function as virus replication ‘factories.’5 NC of vesicular stomatitis virus (VSV) alone binds to RNA with high affinity but with little or no sequence specificity, and the P protein associates with a nascent NC to form an NC0–P (free of RNAs) complex, thereby preventing the NC from binding to cellular RNAs.6 While Hantavirus NC can specifically recognize the viral RNA panhandle and high‐affinity interactions between trimeric NC and RNA panhandles are conserved within the genus Hantavirus.7 Furthermore, NC–RNA complexes can protect viral RNA from nuclease digestion and NCs that lose their RNA‐binding activity are no longer able to protect viral RNA from nuclease digestion.
Figure 1.
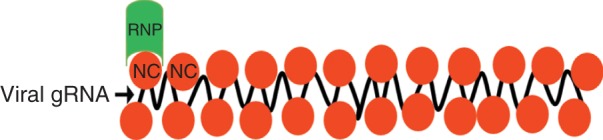
Nucleocapsid proteins (NCs) package the viral genome RNAs (gRNAs). For RNA viruses, NCs enwrap the viral gRNAs to form NC–RNA complexes, which act as a template for viral replication and transcription. NCs also interact with viral polymerase complexes to form ribonucleoprotein (RNP) to initiate the viral replication and transcription.
In the late stage of viral life cycle, to initiate the assembly and release of virions, NCs can prevent generating redundant viral genomes or viral proteins by regulating transcription and replication8; therefore, in some viruses, NCs regulate the genome template switching from production nonstructural proteins to structural proteins,9 or change model for the switch from transcription to replication.10, 11 In addition, viruses also can use and bind to cellular replication complexes to facilitate their own replication.8 In the process of virus budding, the interaction of NCs with matrix (M) protein is responsible for the incorporation of NC–RNA into virions, and, in some instances, NCs can enhance or are required for the release of virus‐like particles (VLPs) in which M protein is a key factor.12
The functions of NCs as a key adaptor molecule between virus and host cell processes are diversified. NCs inhibit cellular antiviral immune response by disturbing the function of key proteins in antiviral immune signaling pathways, such as TANK‐binding kinase 1 (TBK1) and IFN‐regulated factor 3 (IRF3).13 Furthermore, NCs directly interfere with the host cell cycle, and trigger apoptosis14 and autophagy.15
Furthermore, NCs conserved genome sequence and critical role in the viral life cycle make NCs an ideal target for vaccine development, and vaccines based on NCs are safe and effective inducing immunodominant response and in clinical therapy.
THE FUNCTION OF NC IN VIRUS LIFE CYCLE
NC Regulates Viral Transcription and Replication
The primary role of NCs in virus life cycle is to encapsulate the RNA genome to form a helical NC–RNA complex. For most viruses belonging to paramyxoviridae family, genome length is a multiple of six (the ‘rule of six’) because NC monomer exactly associates with six nucleotides.16, 17 Recently, a novel study showed that the structure of parainfluenza virus 5 (PIV5) NC–RNA complex at 3.11‐Å resolution, suggesting that six nucleotides bind to per NC, consistent with the ‘rule of six’ and high amino acid conservation exist in the RNA‐binding pocket and for the scheme of RNA packaging18 within NC. In addition to this function, NCs also play critical roles in virus transcription and replication (Figure 1).
To find the critical amino acids in NCs which is responsible for viral transcription and replication, mutations within NCs have been made to explore the effect of NCs on transcription and replication, 15 amino acid substitutions in NCs of influenza A virus (IAV) decreased the transcription and replication of the viral genome and also dramatically weakened the growth of viruses, as evidenced by comprehensive mutational analysis and in vitro replication assay.19 Another study showed that several mutations in NCs affected the packaging the multiple viral genome RNA (gRNA) segments into progeny virions.20 Alternations of nine amino acids in bunyamwera orthobunyavirus NC severely impaired RNA synthesis, furthermore, 57 viable recombinant bunyamwera orthobunyavirus viruses with mutations in NC were recovered via reverse genetics and displayed a range of plaque sizes and titers in cell culture, and a number of viruses were shown to be temperature sensitive.21 For coronavirus mouse hepatitis virus, NC interaction with nsp3, a component of the viral replicase complex,22 is indispensable for the enhancement of the infectivity of genomic RNA, suggesting that NC–nsp3 interaction serves as a dock to recruit the viral genome to the replicase complex for the initiation of viral infection.23
In addition, NCs also can associate with translation initiation complex for viral replication. NC of hantavirus was shown to have a critical role in directly mediating viral translation initiation via two ways: (1) The NC interacts with the 43S preinitiation complex and replaces eIF4G (which links the mRNA cap with the 43S preinitiation complex) and eIF4A (a helicase necessary for translation initiation) to facilitate the loading of ribosomes onto capped mRNA and ensure the efficient translation of viral mRNA,24 and (2) the NC specifically interacts with ribosomal protein 19 (a structural component of the 40S subunit), which causes the host translation machinery to preferentially bind to the viral transcripts by facilitating ribosome loading on capped viral mRNAs owing to the higher affinity of the NC to bind to ribosomal protein S19.25, 26 The cellular protein eEF1A interacts with the respiratory syncytial virus (RSV) replication complex by binding to the NC and consequently facilitates gRNA synthesis and viral production.27 An eEF1A plays a similar role in tobacco mosaic virus28, 29 and tomato bushy stunt virus30, 31 via interaction with the NCs. Another example of NC association with cellular protein is that human immunodeficiency virus (HIV) NC binds to Tat and significantly reduces the level of Tat but not mRNA expression via the proteasomal pathway, which results in the decrease of transcriptional activation at the late stage of the viral infection cycle.32 Furthermore, both poly [ADP–ribose] polymerase 1 and poly (A)‐binding protein (a host cellular protein that enhances translational efficiency) play an important role in the Porcine reproductive and respiratory syndrome virus (PRRSV) life cycle via their interaction with the NC; the former increases the propagation of PRRSV, and the latter enhances viral replication.33, 34
In addition to being required for gRNAs packaging into the virion,35 NCs also highly regulate the replication of coronavirus gRNA. In a previous study, the transcription of coronavirus gRNA was regulated by the template switching to generate shorter, longer subgenomic (sg) mRNAs and gRNAs through a unique discontinuous transcription mechanism36 (Figure 2(a)). The phosphorylation of NC‐regulated glycogen synthase kinase‐3 (GSK3) is required for the template to switch from discontinuous to continuous transcription, and NC–GSK3 interaction is the base of the direct phosphorylation effect of GSK3 on NCs. Furthermore, phosphorylated NCs can recruit the RNA helicase DEAD (Asp‐Glu‐Ala‐Asp) box helicase 1 (DDX1) to the phosphorylated NC‐containing complex through directly interaction with DDX1,9 which is critical for the production of longer several subgenomic mRNAs (sgmRNAs) and gRNAs.36 Therefore, GSK3 inhibition, DDX1 knockdown, or helicase activity substitution reduces the production of longer sgmRNAs and gRNAs but not short sgmRNAs.36 As a result, viral RNA synthesis and replication were also decreased, suggesting that NC phosphorylation mediated by GSK3 plays an important role in regulating the viral life cycle.8
Figure 2.
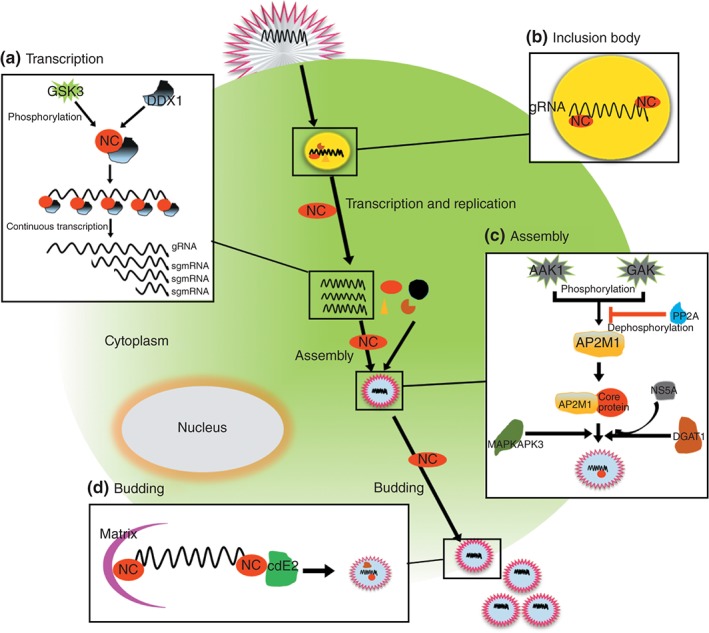
Viral nucleocapsid proteins (NCs) function in several stages of the virus life cycle. After virus entry into the cell, inclusion bodies (IBs) were formed to be the center of transcription and replication, and the NC was one of indispensable partners of IBs. NCs package the viral RNA, to protect the viral RNA from digestion by RNase, and together with other viral components into viral particles, which then mature and yield. (a) NC of coronavirus can be phosphorylated by Glycogen synthase kinase‐3 (GSK3), which enhance the interaction of NC and DDX1 to facilitate template switching from discontinuous to continuous transcription for producing longer sgmRNAs and genome RNAs (gRNAs); (b) NCs package the viral RNA and associate with other viral proteins to form viral replication ‘factory’; (c) Hepatitis C virus (HCV) core protein interacts with the μ subunit of clathrin adaptor protein complex 2 (AP2M1) which is phosphorylated by AP2 associated kinase 1 (AAK1) and cyclin G associated kinase (GAK), and dephosphorylated by Phosphatase 2A (PP2A), and phosphorylated AP2M1 helps NC for assembly; diacylglycerol acyltransferase 1 (DGAT1) can recruit core protein to lipid droplets to facilitate the interaction of core with HCV non‐structural protein 5A (NS5A) protein; mitogen‐activated protein kinase‐activated protein kinase 3 (MAPKAPK3) increased HCV internal ribosome entry site (IRES)‐mediated translation and this activity was further enhanced by core protein; (d) NCs interact with M protein and cdE2 to facilitate the viral budding.
NC Regulates Viral Assembly and Budding
The core protein of hepatitis C virus (HCV) has a general feature of the NC, which mediates RNA packaging to form the viral nucleocapsid. The core protein of HCV also affects the viral life cycle by interacting with various cellular proteins. Previous studies have shown that the association of the core protein of HCV with AP2M1 is essential for viral assembly but not for HCV RNA replication. The phosphorylation of AP2M1 by the serine/threonine kinases AAK137 and GAK38, 39increases the binding of core protein to AP2M1; conversely, dephosphorylation of AP2M1 by PP2A40 impaired its interaction with core protein and HCV assembly. Furthermore, pharmacologic inhibition of core protein‐AP2M1 interaction was shown to negatively regulate HCV assembly41 (Figure 2(c)). In addition, triglyceride‐synthesizing enzyme acyl CoA: diacylglycerol acyltransferase 1 can recruit core protein to lipid droplets to facilitate the interaction between core and NS5A proteins42 (Figure 2(c)). Core protein was also found to interact with mitogen‐activated protein kinase‐activated protein kinase 3 (MAPKAPK3), a serine/threonine protein kinase that is activated by stress. HCV infection can increase MAPKAPK3 expression at both the RNA and protein levels. Knockdown of MAPKAPK3 resulted in a reduction of viral proteins and HCV propagation but not HCV RNA levels. MAPKAPK3 increased HCV IRES‐mediated translation, and this activity was further increased by core protein, suggesting that HCV core protein facilitates its own propagation by modulating MAPKAPK343 (Figure 2(c)).
The release process of VLPs can mimic virion budding. The formation of VLPs is critically dependent on the presence of the viral matrix (M) proteins, and, in some instances, NCs can also be incorporated into VLPs. Furthermore, for some other viruses, the NC is absolutely required for VLP formation and release. Our previous studies showed that expression of the human parainfluenza virus type 3 (HPIV3) M protein alone is sufficient to initiate the release of VLPs,44 and our unpublished data further showed that the NC can be incorporated into VLPs via interaction with M protein, suggesting that the NC mediates transcription or replication complexes into virions by interacting with M protein. Another study showed that M protein is necessary and sufficient for Newcastle disease virus VLP budding (Figure 2(d)). Furthermore, that group suggested that M–NC interactions are responsible for the incorporation of the NC into VLPs and that F protein is incorporated indirectly owing to interactions with NC and HN protein.45 Similarly, in Mopeia virus, the incorporation of the NC into viral VLPs is highly selective39, 46 and induced by matrix Z protein; the same is true for Nipah virus47, 48 and HPIV1.49
However, for PIV5, not only M protein, but also the NC and F or HN protein are critically required for VLPs formation and release, and the results derived from CsCl density gradient centrifugation indicated that almost the entire NC in the cells had assembled into nucleocapsid‐like structures.50 For Ebola virus, the C‐terminal 50 amino acids of the NC may interact with VP40 and enhance the release of VP40 VLPs.12 Furthermore, for Mumps virus, M induces the release of only a small quantity of VLPs when expressed alone, but when coexpressed with the NC and F protein, maximum VLPs were obtained.51 Similarly, for coronaviruses such as mouse hepatitis coronavirus,52 severe acute respiratory syndrome coronavirus (SARS)53 and infectious bronchitis virus (IBV),54 the NC also can greatly increase the VLP yield.
In addition, for most enveloped viruses, budding is initiated from the plasma membrane. The critical process for budding is the interaction of viral matrix with transmembrane glycoprotein and NCs participate and play an important function in this process. Alphavirus NC binds to the cytoplasmic domain of the E2 glycoprotein (cdE2) to facilitate virus budding55 (Figure 2(d)).
NC Regulates Viral RNA Stability
Our recent study revealed a new role for the NC of VSV beyond its role in RNA encapsidation. Using a well‐established VSV minireplicon assay, we found that three triple‐amino acid substitutions (TVK4‐6A3, RII7‐9A3, and VIV13‐15A3) and one single‐amino acid substitution (R7A) within the N‐terminal 21 amino acids of the NC resulted in RNA synthesis loss, whereas all the mutants maintained the ability to oligomerize and encapsidate viral RNA. Furthermore, Northern blotting analysis of nuclease sensitivity of gRNAs encapsidated by the NC or mutants showed that all the mutants failed to protect viral RNAs from nuclease digestion, suggesting that NCs are also involved in the protection of viral gRNA from nuclease digestion for the formation of a functional viral RNA template.56 Similarly, a recent study showed that the positively charged and polar amino acids in the surface cleft of the NC of tomato spotted wilt virus are critical for viral RNA template function, and a subsequent study found that the NC of tomato spotted wilt virus was indeed able to protect the RNA from the degradation of RNase; the mutants of NC, R94A/R95A and K183A/Y184A, which lost the RNA‐binding activity, were no longer able to protect the RNA from RNase degradation.57
NC Regulates Inclusion Body Formation
IBs are characteristic features generated by some negative‐stranded RNA viruses in which viral proteins, genomes, and host factors are concentrated together for efficient replication of virus.58 NCs are indispensible for the formation of IBs, or are incorporated into IBs. Human metapneumovirus NC and P proteins provide the minimal viral requirements for human metapneumovirus IB formation, and the N‐terminal 28 amino acids of NC do not need to bind to P but are necessary for the recruitment NCs to IBs and formation of cytoplasmic IBs.59 Marburg virus (MGBV)‐induced IBs contain four proteins (NC, VP35, VP30, and L), which may represent the key components of the MGBV transcription and replication machinery.60 The results from immunoelectron microscopy showed that MGBV NCs assemble into large aggregates that localize on the membranes of the rough endoplasmic reticulum (ER), and NCs formed tubule‐like structures similar to MGBV‐induced IBs.61
Our studies also showed that the association of the NC with P protein of HPIV3 is sufficient for the formation of IBs, which contain viral RNA, P, and polymerase in HPIV3 infected cells (Figure 2(b)); the NC mutant protein (NCL478A) was unable to form IBs when coexpressed with P protein because NCL478A lost the ability to interact with P; whereas coexpressed NC with NCL478A rescued the IB forming ability and replication activity, thereby suggesting that complexes formed by the NC and NCL478A are functional and competent to form IBs.5 Similarly, RSV NC and P were also sufficient to form cytoplasmic IBs and further research showed that at early RSV infection, melanoma differentiation‐associated gene 5 (MDA5) colocalized with viral genomic RNA and the NC, and at the later of infection, MDA5 and mitochondrial antiviral signaling (MAVS) were packaged into large viral IBs. When the NC and P were coexpressed or infection with RSV had begun, the NC was always infinitely close to MDA5 and MAVS in IBs throughout, which significantly resulted in a decrease in the expression of interferon (IFN) β mRNA, suggesting that NC regulates the formation of IBs and prevents antiviral signaling by interacting with MDA5 and localizing close proximity to MAVS.62 In addition, it was also found that M protein of RSV can associate with cytoplasmic IBs for assembly. If a target for novel antiviral therapy can block the association of M with NC, inhibiting viral assembly may be possible.63 Other findings suggested that F protein interaction with IBs is an important step in the virion assembly process.62
THE EFFECT OF NC ON HOST CELL
NC Regulates Cell Cycle
NC of SARS virus can be modified by sumoylation, and this modification plays a critical role in the NC‐mediated interference of host cell division.64 In addition, NCs can be phosphorylated by cyclin‐dependent kinase (CDK) and phosphorylated NCs directly inhibit the activity of the cyclin–CDK complex, which plays a critical role in cell cycle regulation.65 Both studies suggest that NCs can deregulate the cell cycle by multiple mechanisms to facilitate viral replication. Herpes simplex virus,66, 67 cytomegalovirus,68, 69 and Epstein‐Barr virus70 can arrest the cell cycle at G1 phase. An IBV infection was shown to induce cell cycle arrest at both S and G(2)/M phases for the enhancement of viral replication and progeny production through the interaction of nsp3 and DNA polymerase δ and induce cellular DNA damage response (Table 1).72
Table 1.
Host Cellular Proteins Interact with Viral NCs
| Virus | Host Cellular Proteins | Function in Viral Life Cycle | References |
|---|---|---|---|
| Coronaviruses | GSK3 | Template switching from discontinuous to continuous transcription | 7, 8, 16 |
| DDX1 | Production of longer sgmRNAs and gRNAs | ||
| Hantavirus | eIF4G | Ensure the efficient translation of viral mRNAs | 19 |
| eIF4A | |||
| Ribosomal protein 19 | Help host translation machinery preferentially bind to the viral transcripts | 20, 21 | |
| RSV | eEF1A | Facilitate gRNA synthesis and virus production | 22 |
| TMV | 23, 24 | ||
| TBSV | 25, 26 | ||
| PRRSV | Poly [ADP–ribose] polymerase 1 | Increases the propagation of PRRSV | 28, 29 |
| Poly (A)‐binding protein | Enhance viral replication | ||
| HCV | AP2M1 | Essential for vial assembly | 30, 31, 32, 33, 34 |
| DGAT1 | Recruit core protein to lipid | 35 | |
| MAPKAPK3 | Essential for HCV propagation | 36 | |
| Human myeloid cell factor 1 | Proapoptotic property | 71 | |
| PEDV | TBK1 | Inhibits IFN‐β production | 10 |
| LCMV | IKKε | 69 |
DGAT1, diacylglycerol acyltransferase 1; GSK3, glycogen synthase kinase‐3; HCV, hepatitis C virus; IKKε, IκB kinase‐related kinase; MAPKAPK3, mitogen‐activated protein kinase‐activated protein kinase 3; NCs, nucleocapsid proteins; PRRSV, Porcine reproductive and respiratory syndrome virus; RSV, respiratory syncytial virus; TBK1, TANK‐binding kinase 1; TBSV, tomato bushy stunt virus; TMV, Tobacco mosaic virus.
NC Regulates Innate Immunity
RNA viral infection of cells can be recognized by two cytoplasmic RNA helicases: RIG‐I and MDA5 (termed RIG‐like receptors).73 The binding of an RNA ligand to these pathogen recognition receptors leads to phosphorylation and dimerization of IRF3, which subsequently induces type I IFN production.74 A recent study showed that RIG‐I is capable of recognizing incoming NC‐packaged viral genomes with a 5′ppp double‐stranded RNA ‘panhandle’ structure which triggers IFN activation and is independent of viral RNA synthesis, suggesting that perhaps RIG‐I thereby directly interacts with the panhandle of viral genome or the ribonucleoprotein complex and triggers the activation of IRF375 (Figure 3). Cytoplasmic entry of NCs is the proximal RIG‐I‐sensitive step during infection, and viral NCs with a 5′ppp double‐stranded RNA panhandle act as a RIG‐I activator.73 However, IFN induction and production are also inhibited by the NCs of many RNA viruses via various strategies.
Figure 3.
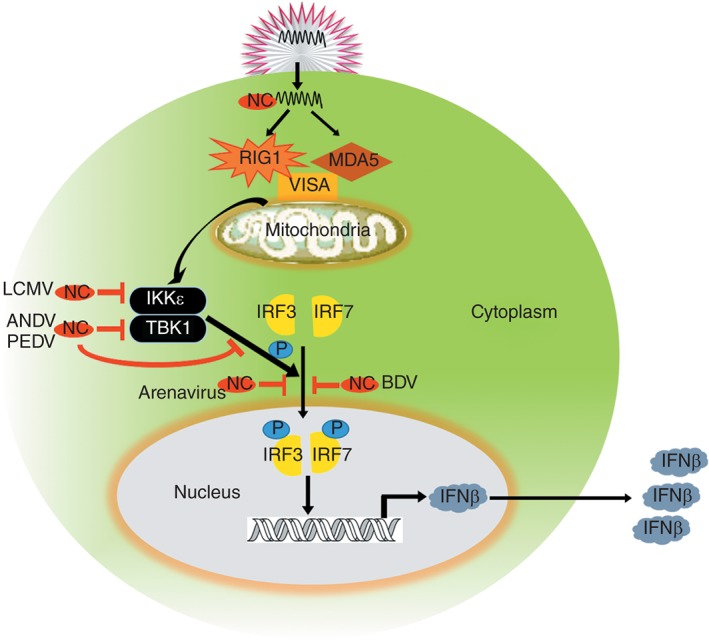
Viral nucleocapsid proteins (NCs) interfere with several stages of innate immune signaling. When RNA viruses infect cells, retinoic acid‐inducible gene I (RIG1) and melanoma differentiation‐associated gene 5 (MDA5) respond to the viral RNA and then activate the adaptor protein located on the mitochondria virus‐induced signaling adapter (VISA), and VISA passes the signal to TANK‐binding kinase 1 (TBK1) and IκB kinase‐related kinase (IKKε), both of which are phosphokinases that can phosphosphorylate IFN‐regulated factor 3 (IRF3) and interferon regulatory factor 7 (IRF7). Finally, phosphorylated IRF3 and IRF7 translocate into the nucleus to activate interferon (IFN) promoter and lead to the production of IFN. Several viral NCs can block the IFN signal transfer; lymphocytic choriomeningitis virus (LCMV) NCs can block the IFN signal transfer through interfere the function of IKKε; NCs of Andes virus (ANDV) and porcine epidemic diarrhea virus (PEDV) inhibit IFN signal by interfering with function of TBK1 and IRF3; Arenavirus NCs prevent IRF3 from localizing to nucleus and Borna diseae virus (BDV) NCs prevent IRF7 from localizing to nucleus.
For negative‐strand RNA viruses, NC of Andes virus (ANDV) can uniquely inhibit IFN signaling responses by interfering with TBK1 autophosphorylation and activation and by inhibiting TBK1‐directed IRF3 phosphorylation and NF‐κB activation.76 In addition, the NC of ANDV acts as a new protein kinase R (PKR) inhibitor by inhibiting PKR dimerization, which leads to PKR autophosphorylation inactivation. By destroying the PKR antiviral response, the NC of ANDV ensures the continuous synthesis of viral proteins required for efficient viral replication and survival in infected hosts77 (Figure 3). Other studies showed that the lymphocytic choriomeningitis virus NC binds to the kinase domain of IκB kinase‐related kinase (IKKε) to block IKKε autocatalytic activity and the ability to phosphorylate IRF3, and the NC‐IKKε interaction plays a crucial role in arenavirus‐host interaction.77 With the exception of Tacaribe virus, all the NCs of arenaviruses from both Old World and New World antigenic groups disrupt innate antiviral defense by inhibiting the activation of IRF‐3 as well as the nuclear translocation of IRF‐3,78 and several residues spanning residues 382–386 of the NC of prototypic arenavirus lymphocytic choriomeningitis virus have been found to contribute to the inhibition of type I IFN production.79 For Lassa virus, an Old World arenavirus, a conserved DEDDH RNase domain within the NC is involved in type IFN suppression, thus allowing the establishment of a productive early viral infection and possibly contributing to the process of viral RNA replication.80 All the catalytic mutants D389A, E391A, D466A, D533A, and H528A showed a complete loss function of suppressing IFN.81, 82, 83 Furthermore, natural killer (NK) cells are strongly activated, and antigen‐presenting cell‐mediated NK cell responses were also significantly increased as evidenced by the responses of NK cells to dendritic cells and macrophages when infected by recombinant Lassa virus with mutations at residues D389A in the NC.84 The NC of borna disease virus, a nonsegmented, negative‐stranded RNA virus, inhibits IFN induction by preventing the nuclear localization of IRF785 (Figure 3). A recent study also showed that the NC of borna disease virus inhibits the processing of NF‐κB1p105 into p50 through its ankyrin‐like domain, leading to the suppression of IKK /NF‐κB1 pathway activation.86
For positive‐strand RNA viruses, NC of porcine epidemic diarrhea virus also inhibits sendai virus‐induced IFN‐β production by directly interacting with TBK1, and this interaction sequesters the connection between TBK1 and IRF313 (Figure 3).
NC Regulates Production of Cytokine/Chemokine
Previous studies showed that interleukin‐10 (IL‐10), an immunoregulatory cytokine, plays an important role in PRRSV‐induced immunosuppression.87 A recent study showed that the NC of type 2 PRRSV significantly increased IL‐10 expression in 3D4/2 macrophages, and alanine substitution mutation analysis indicated that amino acid 33–37 of the NC play important roles in IL‐10 induction. Furthermore, recombinant PRRSV carrying mutations at residues 33–37 in the NC induced significantly lower levels of IL‐10 production in infected monocyte‐derived dendritic cells.88
For RSV, the NC could be at least partially responsible for inhibiting T‐cell activation by expressing at the surface of infected dendritic cells. Furthermore, NCs were also shown to interfere with pMHC–T‐cell receptor interactions and impair the assembly of T‐cell immunological synapse.89
NC Regulates Apoptosis and Autophagy
NC of Hantaan virus is capable of decreasing p53 levels through a post‐translational mechanism that might be associated with the inhibition of apoptosis.90 NC of IAV induces cell death and heterologous expression of NC of IAV alone can induce apoptosis in human airway epithelial cells, NC interacts with Clusterin (CLU), a protein which inhibits the intrinsic apoptosis pathway by binding to Bax and by inhibiting Bax movement into the mitochondria, and attenuates the association of CLU with Bax and hence induce apoptosis.71 Recently a novel function of IAV NC in inducing host cell death was also identified via a yeast two‐hybrid screen, the researchers identified RING finger protein 43, a RING‐type E3 ubiquitin ligase, as a novel interactor of NC and as an important partner of NC to modulate p53 ubiquitination levels, which causes p53 stabilization and enhances apoptosis level in IAV‐infected cells.91
HCV core protein also can induce apoptosis in mature dendritic cells, as evidenced by DNA fragmentation and annexin V‐propidium iodide staining.92 Furthermore, expression of HCV core protein induces ER stress and ER calcium depletion in vitro and in vivo, suggesting that HCV core protein may trigger apoptosis through ER stress and the modification of calcium signaling.93 Moreover, a previous study showed that a Bcl‐2 homology 3 domain in the core protein is essential for its proapoptotic property and ability to interact with human myeloid cell factor 1, a prosurvival member of the Bcl‐2 family, and this interaction contributes to the induction of apoptosis during HCV infection.94 Another study found that using a broad spectrum caspase inhibitor zVAD‐Fmk only partially inhibited apoptosis induced by HCV core protein, suggesting HCV core protein also induces a partially caspase‐independent apoptosis.95 However, a study contrarily showed that the core protein of HCV could inhibit H2O2‐induced apoptosis mediated by p53. H2O2 can induce ROS and, consequently, apoptotic cell death, which can eliminate tumorigenic species through the p14–MDM2–p53 pathway. To overcome H2O2‐induced apoptotic cell death, HCV core protein inhibits p14 expression via promoter hypermethylation and promotes virus survival and hepatocellular carcinoma formation.96 Although one research group found that that HCV core protein can regulate some cellular microRNAs in Huh7 cells, they also found that microRNA‐345 was upregulated when HCV core protein was expressed, leading to the downregulation of p21 (Waf1/Cip1) and inhibition of curcumin‐induced apoptosis.97 In addition to acting as a cell stress for inducing or inhibiting apoptosis, HCV core protein has also been reported to induce complete autophagy, resulting in the fusion of autophagosomes with lysosomes to form autolysosomes. Elsewhere, HCV core protein was shown to induce ER stress, leading to the activation of EIF2AK3 and ATF6, but not the ERN1–XPB1 pathways, which results in upregulation of ATG12 via ATF4 and DDIT3 increase of LC3B expression by direct binding of the LC3B promoter region, the key proteins in the development of autophagy.15
NC of SARS coronavirus, when expressed in COS‐1 monkey kidney cells, induced apoptosis in the absence of growth factors by downregulating extracellular signal‐regulated kinase, phospho‐Akt, and Bcl‐2 levels, upregulating c‐Jun N‐terminal kinase and p38 MAPK pathways, and activating caspases 3 and 7.14 Another study showed that expression of the SARS coronavirus NC in COS‐1 cells under starvation resulted in an increase in reactive oxygen species (ROS), and a decrease in both mitochondrial membrane potential and cytochrome C release into cytosol, suggesting that NC can induce apoptosis of COS‐1 cells by activating the mitochondrial pathway.98 Furthermore work is needed to confirm that this phenomenon is actually recapitulated in vivo, and the detailed mechanism remains to be elucidated. The NC of transmissible gastroenteritis virus, an enveloped virus containing a positive‐sense and single‐stranded RNA genome, is known to cause cell cycle arrest and apoptosis via activation of p53 signaling. Likewise, NC expression in PK‐15 cells reduces cell viability and induces S and G2/M‐phase arrest and apoptosis.99, 100, 101, 102
NC Regulates Stress Granule Formation
Stress granules are environmental stress‐inducible cytoplasmic granules containing various mRNAs and proteins. RSV replication can induce stress response, and the formation of stress granules enhances RSV replication. Studies have shown that the stress granules can transiently interact with IBs formed by NC and P during RSV infection, suggesting a functional relationship between stress granules and IBs during RSV infection.103 Additional research is required to identify the exact relations and roles of stress granules and IBs during RSV infection.
THE FUNCTION OF NC IN VACCINE DEVELOPMENT
Neutralizing antibodies bind to the viral surface glycoproteins such as hemagglutinin and neuraminidase to prevent the virus from entering host cells, but frequent mutation of viral glycoproteins abolishes antibody‐mediated immunity. The NC can be an important antigen for early diagnosis and for the development of vaccines for many viruses because its gene sequence is conserved104, 105 (Table 2).
Table 2.
Vaccine Development Dependent on NCs
| Virus | Design | Responses | References |
|---|---|---|---|
| Influenza virus | Immunodominant epitopes | Immunodominant CD4(+)/CD8(+) T‐cell responses | 97, 98 |
| Recombinant PIV5 encoding NC | Humoral and T‐cell responses | 96 | |
| HIV | Zinc inhibitors | Strong antiviral ability but quite toxic | 99 |
| Noncovalent NC inhibitors | Great specificity and less toxic | 99, 100, 101, 102, 103, 104, 105 | |
| SARS‐CoV | DNA vaccine expressing NC fusing with calreticulin | B‐cell and T‐cell and specific humoral response | 106 |
| DNA vaccine of NC fused with lysosome‐associated membrane protein | Long‐lasting T‐cell memory response | 107 | |
| DNA vaccine plasmids expressing NC and IL‐12 | IL‐12 plays an immunoadjuvant and better specific humoral and cellular immunity | 108 | |
| Immunodominant epitopes | B‐cell and T‐cell responses | 109, 110 | |
| Expressing from Lactobacillus lactis | Significant NC‐specific IgG in the sera | 111 | |
| Expressing in tobacco | Strong humoral and cellular responses | 112 |
HIV, human immunodeficiency virus; IL‐12, interleukin‐12; NCs, nucleocapsid proteins; PIV5, parainfluenza virus 5; SARS, severe acute respiratory syndrome coronavirus.
Antigenic drift and shift of influenza viruses are among the most significant challenges to the development of a vaccine that can provide broad protection against various types of influenza viruses, and more importantly, the NCs among all influenza viruses are well conserved with over 90% homology of amino acid residues.104 A recent report showed that the NC of IAV is a major target of immunodominant CD8(+) T‐cell responses, and six novel immunodominant epitopes were clustered in the carboxyl terminal in the NC and were highly conserved, suggesting that the epitope‐rich NC of IAV is a promising target for the vaccine mediated by T‐cell responses.106 In addition, CD4(+) T‐cell responses are also critical for efficient CD8(+) T‐cell response and have a protective effect against influenza virus. The two most dominant regions (amino acids 457–480 and amino acids 397–420) within the NC have been identified and are the important dominant targets of the immunodominant CD4(+) T‐cell responses.107 Therefore, it is critical for the design of T cell‐based influenza vaccines targeted to NCs. Three recombinant PIV5 encoding NCs of H5N1 (A/Vietnam/1203/2004), between HN and L (PIV5‐NP‐HN/L), between F and SH (PIV5‐NP‐F/SH), or between SH and HN (PIV5‐NP‐SH/HN), induced humoral and T‐cell responses in mice and provided protection against lethal H5N1 virus challenge.105
The NC of SARS‐CoV is also a major antigen, and vaccination in rhesus macaques with an NC antigen induced T‐cell responses against the NC.108 A DNA vaccine expressing NC can induce B‐cell and T‐cell, and humoral responses, and when the NC is fused with calreticulin, NC‐specific humoral and cellular immunity are stronger.108 Also DNA vaccine of NC fused with lysosome‐associated membrane protein can enhance immunization and a long‐lasting T‐cell memory response.109 Furthermore, when inoculating DNA vaccine plasmids expressing NC and interleukin 2 (IL‐2) were injected into mice, IL2 played an immunoadjuvant role to induce better specific humoral and cellular immunity.110 In addition, peptides derived from NCs can also be candidates for vaccine development. Several conserved B‐cell and T‐cell immunodominant epitopes of the NC of the coronavirus in mice, monkeys, and humans have been identified and are significant for developing SARS diagnostic kits and vaccines.111 Moreover, a new epitope N1 (QFKDNVILL) of the SARS NC was identified, and its crystal structure showed that two intrachain hydrogen bonds increase the interaction of the epitope and the T‐cell receptor.112
In another interesting report, the NC of SARS‐CoV was expressed from Lactobacillus lactis to develop a mucosal vaccine and induced significant NC‐specific immunoglobulin in the sera.113 With the progress of the protein expression system, plants are now considered a promising ‘factory’ for producing pharmaceutical proteins with the advantages of safety, low cost, and post‐translational modifications. The NC of SARS‐CoV expressed in tobacco induces strong humoral and cellular responses in mice.114
CONCLUSION
In the past few years, many advancements have been made toward identifying the functions of NCs beyond packaging viral RNA. NCs are becoming more and more important not only in the viral life cycle but also in the process of associating with host cells. In this review, we aimed to summarize the multifunctional nature viral NCs.
Further research should be done to explore the specific functions of NCs and mechanisms by which they interact with cellular proteins, induce autophagy, apoptosis, and stress granules, and inhibit IFN signaling responses. Different viruses use different strategies to associate with the viral life cycle and host cells. The more we learn about the functions of NCs, the more we can use NCs to develop antiviral therapies.
ACKNOWLEDGMENTS
This work was supported by a grant from the China Natural Science Foundation (grant 81471939, 81271816) and Major State Basic Research Development Program (973 Program) (2012CB518906).
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
FURTHER READING
- Lifland AW, Jung J, Alonas E, Zurla C, Crowe JE Jr, Santangelo PJ. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J Virol 2012, 86:8245–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 1. Banerjee AK, Barik S, De BP. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol Ther 1991, 51:47–70. [DOI] [PubMed] [Google Scholar]
- 2. Banerjee AK. The transcription complex of vesicular stomatitis virus. Cell 1987, 48:363–364. [DOI] [PubMed] [Google Scholar]
- 3. Baric RS, Nelson GW, Fleming JO, Deans RJ, Keck JG, Casteel N, Stohlman SA. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol 1988, 62:4280–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Green TJ, Cox R, Tsao J, Rowse M, Qiu S, Luo M. Common mechanism for RNA encapsidation by negative‐strand RNA viruses. J Virol 2014, 88:3766–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang S, Chen L, Zhang G, Yan Q, Yang X, Ding B, Tang Q, Sun S, Hu Z, Chen M. An amino acid of human parainfluenza virus type 3 nucleoprotein is critical for template function and cytoplasmic inclusion body formation. J Virol 2013, 87:12457–12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen L, Zhang S, Banerjee AK, Chen M. N‐terminal phosphorylation of phosphoprotein of vesicular stomatitis virus is required for preventing nucleoprotein from binding to cellular RNAs and for functional template formation. J Virol 2013, 87:3177–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mir MA, Brown B, Hjelle B, Duran WA, Panganiban AT. Hantavirus N protein exhibits genus‐specific recognition of the viral RNA panhandle. J Virol 2006, 80:11283–11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu CH, Yeh SH, Tsay YG, Shieh YH, Kao CL, Chen YS, Wang SH, Kuo TJ, Chen DS, Chen PJ. Glycogen synthase kinase‐3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem 2009, 284:5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu CH, Chen PJ, Yeh SH. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe 2014, 16:462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vidal S, Kolakofsky D. Modified model for the switch from Sendai virus transcription to replication. J Virol 1989, 63:1951–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang WH, Hwang CK, Hu WS, Gorelick RJ, Pathak VK. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo . J Virol 2002, 76:7473–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Licata JM, Johnson RF, Han Z, Harty RN. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus‐like particles. J Virol 2004, 78:7344–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes β interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 2014, 88:8936–8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Surjit M, Liu B, Jameel S, Chow VT, Lal SK. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS‐1 cells in the absence of growth factors. Biochem J 2004, 383:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J, Kang R, Huang H, Xi X, Wang B, Wang J, Zhao Z. Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway‐mediated MAP1LC3B and ATG12 expression. Autophagy 2014, 10:766–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol 1993, 67:4822–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Durbin AP, Siew JW, Murphy BR, Collins PL. Minimum protein requirements for transcription and RNA replication of a minigenome of human parainfluenza virus type 3 and evaluation of the rule of six. Virology 1997, 234:74–83. [DOI] [PubMed] [Google Scholar]
- 18. Alayyoubi M, Leser GP, Kors CA, Lamb RA. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein‐RNA complex. Proc Natl Acad Sci USA 2015, 112:E1792–E1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mena I, Jambrina E, Albo C, Perales B, Ortin J, Arrese M, Vallejo D, Portela A. Mutational analysis of influenza A virus nucleoprotein: identification of mutations that affect RNA replication. J Virol 1999, 73:1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Z, Watanabe T, Hatta M, Watanabe S, Nanbo A, Ozawa M, Kakugawa S, Shimojima M, Yamada S, Neumann G, et al. Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein. J Virol 2009, 83:4153–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eifan SA, Elliott RM. Mutational analysis of the Bunyamwera orthobunyavirus nucleocapsid protein gene. J Virol 2009, 83:11307–11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. An interaction between the nucleocapsid protein and a component of the replicase‐transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol 2010, 84:10276–10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hurst KR, Koetzner CA, Masters PS. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase‐transcriptase complex. J Virol 2013, 87:9159–9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mir MA, Panganiban AT. A protein that replaces the entire cellular eIF4F complex. EMBO J 2008, 27:3129–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng E, Haque A, Rimmer MA, Hussein IT, Sheema S, Little A, Mir MA. Characterization of the Interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19). J Biol Chem 2011, 286:11814–11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haque A, Mir MA. Interaction of hantavirus nucleocapsid protein with ribosomal protein S19. J Virol 2010, 84:12450–12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wei T, Li D, Marcial D, Khan M, Lin MH, Snape N, Ghildyal R, Harrich D, Spann K. The eukaryotic elongation factor 1A is critical for genome replication of the paramyxovirus respiratory syncytial virus. PLoS One 2014, 9:e114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamaji Y, Kobayashi T, Hamada K, Sakurai K, Yoshii A, Suzuki M, Namba S, Hibi T. In vivo interaction between Tobacco mosaic virus RNA‐dependent RNA polymerase and host translation elongation factor 1A. Virology 2006, 347:100–108. [DOI] [PubMed] [Google Scholar]
- 29. Yamaji Y, Sakurai K, Hamada K, Komatsu K, Ozeki J, Yoshida A, Yoshii A, Shimizu T, Namba S, Hibi T. Significance of eukaryotic translation elongation factor 1A in tobacco mosaic virus infection. Arch Virol 2010, 155:263–268. [DOI] [PubMed] [Google Scholar]
- 30. Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, Nagy PD. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus‐strand synthesis. PLoS Pathog 2010, 6:e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co‐factor. Virology 2009, 385:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hong HW, Lee SW, Myung H. Induced degradation of Tat by nucleocapsid (NC) via the proteasome pathway and its effect on HIV transcription. Viruses 2013, 5:1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu L, Lear Z, Hughes DJ, Wu W, Zhou EM, Whitehouse A, Chen H, Hiscox JA. Resolution of the cellular proteome of the nucleocapsid protein from a highly pathogenic isolate of porcine reproductive and respiratory syndrome virus identifies PARP‐1 as a cellular target whose interaction is critical for virus biology. Vet Microbiol 2015, 176:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X, Bai J, Zhang L, Wang X, Li Y, Jiang P. Poly(A)‐binding protein interacts with the nucleocapsid protein of porcine reproductive and respiratory syndrome virus and participates in viral replication. Antiviral Res 2012, 96:315–323. [DOI] [PubMed] [Google Scholar]
- 35. Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol 2007, 81:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pasternak AO, Spaan WJ, Snijder EJ. Regulation of relative abundance of arterivirus subgenomic mRNAs. J Virol 2004, 78:8102–8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ricotta D, Conner SD, Schmid SL, von Figura K, Honing S. Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J Cell Biol 2002, 156:791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Korolchuk VI, Banting G. CK2 and GAK/auxilin2 are major protein kinases in clathrin‐coated vesicles. Traffic 2002, 3:428–439. [DOI] [PubMed] [Google Scholar]
- 39. Zhang CX, Engqvist‐Goldstein AE, Carreno S, Owen DJ, Smythe E, Drubin DG. Multiple roles for cyclin G‐associated kinase in clathrin‐mediated sorting events. Traffic 2005, 6:1103–1113. [DOI] [PubMed] [Google Scholar]
- 40. Ricotta D, Hansen J, Preiss C, Teichert D, Honing S. Characterization of a protein phosphatase 2A holoenzyme that dephosphorylates the clathrin adaptors AP‐1 and AP‐2. J Biol Chem 2008, 283:5510–5517. [DOI] [PubMed] [Google Scholar]
- 41. Neveu G, Barouch‐Bentov R, Ziv‐Av A, Gerber D, Jacob Y, Einav S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog 2012, 8:e1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Camus G, Herker E, Modi AA, Haas JT, Ramage HR, Farese RV Jr, Ott M. Diacylglycerol acyltransferase‐1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J Biol Chem 2013, 288:9915–9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ngo HT, Pham LV, Kim JW, Lim YS, Hwang SB. Modulation of mitogen‐activated protein kinase‐activated protein kinase 3 by hepatitis C virus core protein. J Virol 2013, 87:5718–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang G, Zhang S, Ding B, Yang X, Chen L, Yan Q, Jiang Y, Zhong Y, Chen M. A leucine residue in the C terminus of human parainfluenza virus type 3 matrix protein is essential for efficient virus‐like particle and virion release. J Virol 2014, 88:13173–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pantua HD, McGinnes LW, Peeples ME, Morrison TG. Requirements for the assembly and release of Newcastle disease virus‐like particles. J Virol 2006, 80:11062–11073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shtanko O, Imai M, Goto H, Lukashevich IS, Neumann G, Watanabe T, Kawaoka Y. A role for the C terminus of Mopeia virus nucleoprotein in its incorporation into Z protein‐induced virus‐like particles. J Virol 2010, 84:5415–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ciancanelli MJ, Basler CF. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol 2006, 80:12070–12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Patch JR, Crameri G, Wang LF, Eaton BT, Broder CC. Quantitative analysis of Nipah virus proteins released as virus‐like particles reveals central role for the matrix protein. Virol J 2007, 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Coronel EC, Murti KG, Takimoto T, Portner A. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus‐like particles containing nucleocapsid‐like structures. J Virol 1999, 73:7035–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmitt AP, Leser GP, Waning DL, Lamb RA. Requirements for budding of paramyxovirus simian virus 5 virus‐like particles. J Virol 2002, 76:3952–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus‐like particles. J Virol 2009, 83:7261–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boscarino JA, Logan HL, Lacny JJ, Gallagher TM. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J Virol 2008, 82:2989–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, Tsao SW, Nicholls JM, Altmeyer R, Peiris JS, et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus‐like particles. J Virol 2008, 82:11318–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ruch TR, Machamer CE. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J Virol 2011, 85:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jose J, Przybyla L, Edwards TJ, Perera R, Burgner JW 2nd, Kuhn RJ. Interactions of the cytoplasmic domain of Sindbis virus E2 with nucleocapsid cores promote alphavirus budding. J Virol 2012, 86:2585–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen L, Yan Q, Lu G, Hu Z, Zhang G, Zhang S, Ding B, Jiang Y, Zhong Y, Gong P, et al. Several residues within the N‐terminal arm of vesicular stomatitis virus nucleoprotein play a critical role in protecting viral RNA from nuclease digestion. Virology 2015, 478:9–17. [DOI] [PubMed] [Google Scholar]
- 57. Li J, Feng Z, Wu J, Huang Y, Lu G, Zhu M, Wang B, Mao X, Tao X. Structure and function analysis of nucleocapsid protein of tomato spotted wilt virus interacting with RNA using homology modeling. J Biol Chem 2015, 290:3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res 2007, 70:101–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Derdowski A, Peters TR, Glover N, Qian R, Utley TJ, Burnett A, Williams JV, Spearman P, Crowe JE Jr. Human metapneumovirus nucleoprotein and phosphoprotein interact and provide the minimal requirements for inclusion body formation. J Gen Virol 2008, 89:2698–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Becker S, Rinne C, Hofsass U, Klenk HD, Muhlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology 1998, 249:406–417. [DOI] [PubMed] [Google Scholar]
- 61. Kolesnikova L, Muhlberger E, Ryabchikova E, Becker S. Ultrastructural organization of recombinant Marburg virus nucleoprotein: comparison with Marburg virus inclusions. J Virol 2000, 74:3899–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baviskar PS, Hotard AL, Moore ML, Oomens AG. The respiratory syncytial virus fusion protein targets to the perimeter of inclusion bodies and facilitates filament formation by a cytoplasmic tail‐dependent mechanism. J Virol 2013, 87:10730–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li D, Jans DA, Bardin PG, Meanger J, Mills J, Ghildyal R. Association of respiratory syncytial virus M protein with viral nucleocapsids is mediated by the M2‐1 protein. J Virol 2008, 82:8863–8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li FQ, Xiao H, Tam JP, Liu DX. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett 2005, 579:2387–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Surjit M, Kumar R, Mishra RN, Reddy MK, Chow VT, Lal SK. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14‐3‐3‐mediated translocation. J Virol 2005, 79:11476–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hobbs WE 2nd, DeLuca NA. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J Virol 1999, 73:8245–8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lomonte P, Everett RD. Herpes simplex virus type 1 immediate‐early protein Vmw110 inhibits progression of cells through mitosis and from G(1) into S phase of the cell cycle. J Virol 1999, 73:9456–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wiebusch L, Hagemeier C. Human cytomegalovirus 86‐kilodalton IE2 protein blocks cell cycle progression in G(1). J Virol 1999, 73:9274–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lu M, Shenk T. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J Virol 1999, 73:676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cayrol C, Flemington EK. The Epstein‐Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin‐dependent kinase inhibitors. EMBO J 1996, 15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 71. Tripathi S, Batra J, Cao W, Sharma K, Patel JR, Ranjan P, Kumar A, Katz JM, Cox NJ, Lal RB, et al. Influenza A virus nucleoprotein induces apoptosis in human airway epithelial cells: implications of a novel interaction between nucleoprotein and host protein Clusterin. Cell Death Dis 2013, 4:e562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu LH, Huang M, Fang SG, Liu DX. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase delta. J Biol Chem 2011, 286:39546–39559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kato H, Takahasi K, Fujita T. RIG‐I‐like receptors: cytoplasmic sensors for non‐self RNA. Immunol Rev 2011, 243:91–98. [DOI] [PubMed] [Google Scholar]
- 74. Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. The NEMO adaptor bridges the nuclear factor‐κB and interferon regulatory factor signaling pathways. Nat Immunol 2007, 8:592–600. [DOI] [PubMed] [Google Scholar]
- 75. Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, et al. Incoming RNA virus nucleocapsids containing a 5′‐triphosphorylated genome activate RIG‐I and antiviral signaling. Cell Host Microbe 2013, 13:336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cimica V, Dalrymple NA, Roth E, Nasonov A, Mackow ER. An innate immunity‐regulating virulence determinant is uniquely encoded by the Andes virus nucleocapsid protein. MBio 2014, 5:e01088–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pythoud C, Rodrigo WW, Pasqual G, Rothenberger S, Martinez‐Sobrido L, de la Torre JC, Kunz S. Arenavirus nucleoprotein targets interferon regulatory factor‐activating kinase IKK epsilon. J Virol 2012, 86:7728–7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Martinez‐Sobrido L, Giannakas P, Cubitt B, Garcia‐Sastre A, de la Torre JC. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol 2007, 81:12696–12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Martinez‐Sobrido L, Emonet S, Giannakas P, Cubitt B, Garcia‐Sastre A, de la Torre JC. Identification of amino acid residues critical for the anti‐interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 2009, 83:11330–11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Huang Q, Shao J, Lan S, Zhou Y, Xing J, Dong C, Liang Y, Ly H. In vitro and in vivo characterizations of pichinde viral nucleoprotein exoribonuclease functions. J Virol 2015, 89:6595–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 2010, 468:779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA‐specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci USA 2011, 108:2396–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Reynard S, Russier M, Fizet A, Carnec X, Baize S. Exonuclease domain of the Lassa virus nucleoprotein is critical to avoid RIG‐I signaling and to inhibit the innate immune response. J Virol 2014, 88:13923–13927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Russier M, Reynard S, Carnec X, Baize S. The exonuclease domain of Lassa virus nucleoprotein is involved in antigen‐presenting‐cell‐mediated NK cell responses. J Virol 2014, 88:13811–13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Song W, Kao W, Zhai A, Qian J, Li Y, Zhang Q, Zhao H, Hu Y, Li H, Zhang F. Borna disease virus nucleoprotein inhibits type I interferon induction through the interferon regulatory factor 7 pathway. Biochem Biophys Res Commun 2013, 438:619–623. [DOI] [PubMed] [Google Scholar]
- 86. Makino A, Fujino K, Parrish NF, Honda T, Tomonaga K. Borna disease virus possesses an NF‐kB inhibitory sequence in the nucleoprotein gene. Sci Rep 2015, 5:8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mateu E, Diaz I. The challenge of PRRS immunology. Vet J 2008, 177:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu X, Fan B, Bai J, Wang H, Li Y, Jiang P. The N‐N non‐covalent domain of the nucleocapsid protein of type 2 porcine reproductive and respiratory syndrome virus enhances induction of IL‐10 expression. J Gen Virol 2015, 96:1276–1286. [DOI] [PubMed] [Google Scholar]
- 89. Cespedes PF, Bueno SM, Ramirez BA, Gomez RS, Riquelme SA, Palavecino CE, Mackern‐Oberti JP, Mora JE, Depoil D, Sacristan C, et al. Surface expression of the hRSV nucleoprotein impairs immunological synapse formation with T cells. Proc Natl Acad Sci USA 2014, 111:E3214–E3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Park SW, Han MG, Park C, Ju YR, Ahn BY, Ryou J. Hantaan virus nucleocapsid protein stimulates MDM2‐dependent p53 degradation. J Gen Virol 2013, 94:2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nailwal H, Sharma S, Mayank AK, Lal SK. The nucleoprotein of influenza A virus induces p53 signaling and apoptosis via attenuation of host ubiquitin ligase RNF43. Cell Death Dis 2015, 6:e1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Siavoshian S, Abraham JD, Thumann C, Kieny MP, Schuster C. Hepatitis C virus core, NS3, NS5A, NS5B proteins induce apoptosis in mature dendritic cells. J Med Virol 2005, 75:402–411. [DOI] [PubMed] [Google Scholar]
- 93. Benali‐Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, Buscail L, Bartenschlager R, Ichas F, Rizzuto R, et al. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 2005, 24:4921–4933. [DOI] [PubMed] [Google Scholar]
- 94. Mohd‐Ismail NK, Deng L, Sukumaran SK, Yu VC, Hotta H, Tan YJ. The hepatitis C virus core protein contains a BH3 domain that regulates apoptosis through specific interaction with human Mcl‐1. J Virol 2009, 83:9993–10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Berg CP, Schlosser SF, Neukirchen DK, Papadakis C, Gregor M, Wesselborg S, Stein GM. Hepatitis C virus core protein induces apoptosis‐like caspase independent cell death. Virol J 2009, 6:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Seo YL, Heo S, Jang KL. Hepatitis C virus core protein overcomes H2O2‐induced apoptosis by downregulating p14 expression via DNA methylation. J Gen Virol 2015, 96:822–832. [DOI] [PubMed] [Google Scholar]
- 97. Shiu TY, Huang SM, Shih YL, Chu HC, Chang WK, Hsieh TY. Hepatitis C virus core protein down‐regulates p21(Waf1/Cip1) and inhibits curcumin‐induced apoptosis through microRNA‐345 targeting in human hepatoma cells. PLoS One 2013, 8:e61089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98. Zhang L, Wei L, Jiang D, Wang J, Cong X, Fei R. SARS‐CoV nucleocapsid protein induced apoptosis of COS‐1 mediated by the mitochondrial pathway. Artif Cells Blood Substit Immobil Biotechnol 2007, 35:237–253. [DOI] [PubMed] [Google Scholar]
- 99. Ding L, Huang Y, Du Q, Dong F, Zhao X, Zhang W, Xu X, Tong D. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem Biophys Res Commun 2014, 445:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ding L, Xu X, Huang Y, Li Z, Zhang K, Chen G, Yu G, Wang Z, Li W, Tong D. Transmissible gastroenteritis virus infection induces apoptosis through FasL‐ and mitochondria‐mediated pathways. Vet Microbiol 2012, 158:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ding L, Huang Y, Dai M, Zhao X, Du Q, Dong F, Wang L, Huo R, Zhang W, Xu X, et al. Transmissible gastroenteritis virus infection induces cell cycle arrest at S and G2/M phases via p53‐dependent pathway. Virus Res 2013, 178:241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Huang Y, Ding L, Li Z, Dai M, Zhao X, Li W, Du Q, Xu X, Tong D. Transmissible gastroenteritis virus infection induces cell apoptosis via activation of p53 signalling. J Gen Virol 2013, 94:1807–1817. [DOI] [PubMed] [Google Scholar]
- 103. Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE Jr. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J Virol 2010, 84:12274–12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA‐binding protein pivotal to virus replication. J Gen Virol 2002, 83:723–734. [DOI] [PubMed] [Google Scholar]
- 105. Li Z, Gabbard JD, Mooney A, Gao X, Chen Z, Place RJ, Tompkins SM, He B. Single‐dose vaccination of a recombinant parainfluenza virus 5 expressing NP from H5N1 virus provides broad immunity against influenza A viruses. J Virol 2013, 87:5985–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Grant E, Wu C, Chan KF, Eckle S, Bharadwaj M, Zou QM, Kedzierska K, Chen W. Nucleoprotein of influenza A virus is a major target of immunodominant CD8+ T‐cell responses. Immunol Cell Biol 2013, 91:184–194. [DOI] [PubMed] [Google Scholar]
- 107. Chen L, Zanker D, Xiao K, Wu C, Zou Q, Chen W. Immunodominant CD4+ T‐cell responses to influenza A virus in healthy individuals focus on matrix 1 and nucleoprotein. J Virol 2014, 88:11760–11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kim TW, Lee JH, Hung CF, Peng S, Roden R, Wang MC, Viscidi R, Tsai YC, He L, Chen PJ, et al. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol 2004, 78:4638–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yang K, Sun K, Srinivasan KN, Salmon J, Marques ET, Xu J, August JT. Immune responses to T‐cell epitopes of SARS CoV‐N protein are enhanced by N immunization with a chimera of lysosome‐associated membrane protein. Gene Ther 2009, 16:1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hu H, Tao L, Wang Y, Chen L, Yang J, Wang H. Enhancing immune responses against SARS‐CoV nucleocapsid DNA vaccine by co‐inoculating interleukin‐2 expressing vector in mice. Biotechnol Lett 2009, 31:1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Liu SJ, Leng CH, Lien SP, Chi HY, Huang CY, Lin CL, Lian WC, Chen CJ, Hsieh SL, Chong P. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 2006, 24:3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Liu J, Wu P, Gao F, Qi J, Kawana‐Tachikawa A, Xie J, Vavricka CJ, Iwamoto A, Li T, Gao GF. Novel immunodominant peptide presentation strategy: a featured HLA‐A*2402‐restricted cytotoxic T‐lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J Virol 2010, 84:11849–11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pei H, Liu J, Cheng Y, Sun C, Wang C, Lu Y, Ding J, Zhou J, Xiang H. Expression of SARS‐coronavirus nucleocapsid protein in Escherichia coli and Lactococcus lactis for serodiagnosis and mucosal vaccination. Appl Microbiol Biotechnol 2005, 68:220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zheng N, Xia R, Yang C, Yin B, Li Y, Duan C, Liang L, Guo H, Xie Q. Boosted expression of the SARS‐CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine 2009, 27:5001–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]


