Summary
Outbreaks of porcine epidemic diarrhoea (PED) were reported across Europe during the 1980s and 1990s, but only sporadic outbreaks occurred in recent years. PED virus (PEDV) spread for the first time into the USA in 2013 and has caused severe economic losses. Retrospectively, it was found that two different strains of PEDV have been introduced into the United States, both are closely related to strains circulating in China where a new wave of the disease occurred from 2010 onwards. Since autumn 2014, new outbreaks of PED have occurred in Europe. In this study, weaned piglets were inoculated with an early European isolate (Br1/87) or faecal/intestinal suspensions derived from pigs infected with a recent European strain of PEDV (from Germany) or a US strain of PEDV. No evidence for infection resulted from inoculation of pigs with the German sample that contained high levels of PEDV RNA; there were no clinical signs, excretion of viral RNA or anti‐PEDV antibody production. In contrast, all the pigs in the other two groups showed evidence of infection. Mild clinical signs of disease, mainly diarrhoea, occurred in piglets inoculated with the Br1/87 and US PEDV strains. PEDV RNA was detected throughout the intestine in euthanized animals at 4 days post‐inoculation. In addition, within these animals, low levels of viral RNA were detected in lungs and livers with higher levels in spleens. Seroconversion against PEDV occurred in all surviving infected animals within 10 days. PEDV RNA excretion occurred for at least 2 weeks. The US PEDV RNA was detected at low levels in serum samples on multiple days. It is apparent that current diagnostic systems can detect infection by the different virus strains.
Keywords: coronavirus, virus excretion, seroconversion, clinical signs, virus detection
Introduction
Porcine epidemic diarrhoea (PED) was initially recognized in the UK in 1971 and is caused by infection with a coronavirus (PED virus), a member of the Alphacoronavirus genus. The disease is characterized by diarrhoea and vomiting leading to severe dehydration with high mortality among young piglets and hence severe economic losses. During the 1980s and 1990s, the disease spread within Europe and also to Asia and caused significant problems (reviewed in Jung and Saif, 2015). A new wave of PED virus (PEDV) infections occurred from 2010 onwards in China and adjacent countries (Sun et al., 2012) and the virus appeared for the first time in the USA in 2013 (Chen et al., 2014). Retrospectively, it has been shown that two closely related variants of PEDV, termed ‘INDEL’ and ‘non‐INDEL’ that reflect the presence or absence of specific insertions and deletions within the S‐gene sequence, were introduced into the United States at that time (Huang et al., 2013; Vlasova et al., 2014; Wang et al., 2014). Recently, new cases of PED have occurred within several countries in Europe and the circulating viruses are genetically similar to the ‘INDEL’ strains found in the USA and China (Hanke et al., 2015); these viruses have been suggested to cause less severe disease than the ‘non‐INDEL’ strains (Wang et al., 2014) and this has been supported by recent experimental studies (Yamamoto et al., 2015). However, it is apparent that the nature and severity of the disease can vary significantly even with apparently very similar virus strains and thus, other factors must also determine the outcome of the infection. These can include the age, immune status and general health status (EFSA (European Food Standards Agency), AHAW Panel (EFSA Panel on Animal Health and Welfare), 2014). Sequence analysis of the recent PED viruses circulating in the United States, China and Europe has been reported (Chen et al., 2014; Vlasova et al., 2014; Wang et al., 2014; Hanke et al., 2015). Some initial animal studies using the recent PEDV strains to determine the characteristics of the infection have been performed (Jung et al., 2014; Madson et al., 2014; Crawford et al., 2015; Jung et al., 2015; Yamamoto et al., 2015) and also previously animal experiments with an early European strain have been described (Debouck et al., 1981). The experiments described here extend these studies and also demonstrate the efficacy of current diagnostic systems to detect and monitor infections by distinct PEDVs viruses.
Materials and Methods
All animal work was approved and conducted according to the requirements of the Danish Animal Experiment Inspectorate which meet the provisions of the ‘European Union Directive 2010/63/EU’.
Weaned piglets (ca. 5 weeks old) were divided, at random, into groups 1, 2 and 3, each with 5 animals. The pigs were inoculated (orally), on post‐infection day (PID) 0, with a virus suspension (2 mls) within secure biocontainment facilities. Group 1 received the Vero cell culture‐grown European isolate of PEDV Br1/87 (very closely related to the prototypic CV777 strain (Bridgen et al., 1993; Wang et al., 2014)) that was obtained from the Central Veterinary Laboratory, Weybridge, U.K., in 1987. It was subsequently passaged three times in Vero cells in the presence of trypsin (10 μg/ml); the virus stock used in these studies had a titre of 106.8 TCID/ml, as measured by immune staining of infected Vero cells. Group 2 was inoculated with a suspension derived from a sample of faeces that was homogenized (10% w/v suspension) and clarified by centrifugation. The sample originated from a PEDV‐infected pig that was submitted to a diagnostic laboratory for routine examination and was kindly supplied by Sandra Blome (Friedrich Loeffler Institute, Germany). Pigs in Group 3 were inoculated with a 10% (w/v) clarified homogenate of intestine and faeces from a pig in Iowa infected with PEDV (a ‘non‐INDEL’ strain); this material was kindly provided by APHIS, Ames, IA, USA. The ‘non‐INDEL’ status was verified by sequencing the S gene (data not shown). Each inoculum contained high levels of PEDV RNA as determined using a modified real‐time RT‐qPCR assay. Briefly, RNA was extracted from samples using a Magna Pure LC robot with a Magna Pure LC Total Nucleic Acid kit (Roche) and eluted in nuclease‐free water (50 μl). RT‐qPCR assays used a one‐step RT‐PCR kit (Qiagen) with the primers and probe described by Kim et al. (2007) together with the primers and probe described by Chen et al. (2014) (note: the reverse primers are identical in each primer set and both probes were labelled with the same fluorophores). The assays were run for 40 cycles.
Throughout the period of the experiment, the rectal temperatures of the pigs were measured and recorded on a daily basis. In addition, following inoculation, the animals were monitored daily for clinical signs and a score evaluated using seven parameters relevant to PED infection: (i) well‐being: 0 = normal, 1 = slightly depressed/hesitates to rise, 2 = depressed/lies down, 3 = lethargic, (ii) appetite: 0 = greedy/hungry, 1 = eats slowly/only tastes food, 2 = does not eat, (iii) body tension: 0 = relaxed/straight back, 1 = stiffness/bent back, (iv) body shape: 0 = full stomach/round body, 1 = empty stomach/thin body, (v) defaecation: 0 = soft faeces/normal amount, 1 = thin/non‐formed faeces, 2 = watery diarrhoea, (vi) vomiting: 0 = no, 1 = yes, (vii) leftovers in feeding trough: 0 = trough empty/clean, 1 = food only partially eaten, 2 = trough still full/nothing eaten. In total, a maximum score of 12 could be assigned to an individual pig on a single day.
Selected pigs (2 animals from each Group) were euthanized on PID 4. A third pig from Group 3 was also euthanized at the same time for welfare reasons due to accidental injury and lameness unrelated to the PED infection. Tissue samples were collected separately, at necropsy, from the jejunum, ileum and colon of each pig. In addition, samples from the liver, lung and spleen of each pigs were collected and stored frozen (−80°C) until analysed further (see below). The surviving pigs were maintained until PID 28, and blood samples were collected on pre‐selected days (PID 0, 7, 10, 14, 17, 21, 24 and 28). The presence of anti‐PEDV antibodies in the sera was determined using an in‐house blocking ELISA (analogous to that used for PRRSV (Sørensen et al., 1997) using cell culture‐grown PEDV (Br1/87) as antigen and a biotin‐conjugated pig anti‐PEDV polyclonal antibody (B. Strandbygaard and A. Bøtner, unpublished data). The bound biotinylated antibody was detected using avidin‐conjugated horseradish peroxidase (eBioscience) plus 3,3′,5,5′‐tetramethylbenzidine substrate and the OD measured at 450/630 nm. The cut‐off value for a positive reaction is set at 40% blocking.
Sera collected from the PEDV‐infected pigs on PID 21 were used in an immunoperoxidase staining test (IPT) to verify their ability to cross‐recognize PEDV antigens within Vero cells infected with the Br1/87 strain. These assays were performed essentially as described for the detection of anti‐PRRSV antibodies (Bøtner et al., 1994).
The presence of PEDV RNA in faecal swab (fs), serum and tissue (10% (w/v) homogenate) samples was determined by RT‐qPCR using the assay described above. Selected samples were also assayed for the presence of rotavirus RNA using an assay described previously (Pang et al., 2004) with an AgPath ID kit (Applied Biosystems).
Results
Prior to inoculation of the pigs, the virus suspensions were assayed for the presence of PEDV RNA using a single real‐time RT‐qPCR assay that efficiently recognizes both the early European PEDV RNA and recent US PEDV RNA (see Table S1). The assay using the primers and probe described by Kim et al. (2007) shows a markedly greater sensitivity for the early European PEDV than for the US PEDV, while, in contrast, the assay using the reagents described by Chen et al. (2014) recognizes the US PEDV more efficiently than the early European PEDV. The combined assay, as used here, recognizes the two different types of PEDV with a similar efficiency as the optimal single assays. This combined assay produced Ct values of about 20, 13 and 15 for the Br1/87, German and US samples, respectively (i.e. there is a high level of viral RNA in each sample and especially the German and US samples). The virus suspensions derived from intestinal and faecal material were also assayed for the presence of rotavirus RNA; a low level of rotavirus RNA was detected in the US sample (see below); however, no rotavirus RNA was detected within the German inoculum.
Clinical signs of infection
Following inoculation of the pigs, the animals were closely monitored. As described above, two or three pigs from each Group were euthanized on PID 4 and tissue samples were collected at necropsy for further analysis. The remaining animals were maintained until PID 28, with collection of blood samples (on pre‐selected days) and faecal swab samples. At PID 28, these pigs were also euthanized. Throughout the course of the study, rectal temperatures remained fairly constant and none of the pigs developed fever (data not shown).
Among the pigs in Group 1 (numbered 1‐5), inoculated with the Br1/87 strain, 3 showed slight depression on PID 4 and, of these, 2 had diarrhoea of varying consistency, these (pigs 1 and 3) were selected for euthanasia. The highest clinical score for any one pig in this Group was 4 (pig 3 only) and this was reached on PID 4. In contrast, pig 5 showed no signs of disease at any time until euthanized on PID 28. At PID 4, necroscopic examination of the two euthanized pigs (1 and 3) from Group 1 showed that the small intestine (jejunum) was dilated by gas or fluid and there was an accumulation of watery contents. Furthermore, within the colon region, watery fluid was observed in one of the pigs (pig 3).
No signs of clinical disease were apparent at any stage in pigs from Group 2 (inoculated with German sample). No unusual signs were observed during necroscopic examination of the pigs (6 and 10) from Group 2 that were euthanized at PID 4, consistent with the lack of clinical signs of disease.
In pigs inoculated with the US PEDV sample (Group 3, numbered 11‐15), all 5 animals showed slight depression on PID 3 and PID 4. The pigs 11, 13 and 14 had watery diarrhoea on PID 4 and reached a clinical score of 4; each of these pigs were euthanized on that day. As with the pigs from Group 1, at PID 4, necroscopic examination of these three euthanized pigs from Group 3 showed that the jejunum was dilated by gas or fluid and there was an accumulation of watery contents. In addition, the colon of all three pigs contained watery fluid as well. No further disease was apparent in the surviving pigs kept until PID 28.
Serology
Serum samples were obtained from each animal on pre‐selected days. As expected, no antibodies against PEDV were detected in any of the pigs at PID 0. One of the animals in groups 1 and 3 had seroconverted against PEDV by PID 7 and all surviving pigs in these Groups had done so by PID 10 and remained seropositive at PID 28 (Fig. 1a and c). In contrast, pigs inoculated with the German sample did not seroconvert against PEDV (Fig. 1b), consistent with the lack of clinical disease and suggesting a lack of infection.
Figure 1.
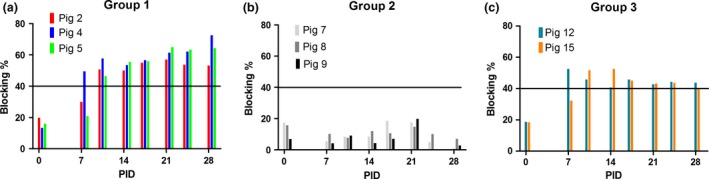
Detection of anti‐PEDV antibodies in pigs following oral inoculation. Serum samples were collected on the indicated days from pigs in Group 1 (Br1/87, panel a), Group 2 (German sample, panel b) and Group 3 (US PEDV, panel c) that were kept for the whole duration of the experiment and assayed for the presence of anti‐PEDV antibodies using an in‐house blocking ELISA. The threshold for a positive reaction is set at 40% blocking (indicated by horizontal line).
Excretion of viral RNA
The presence of PEDV RNA in faecal swab (fs) samples was determined by RT‐qPCR (Table 1).
Table 1.
Detection of PEDV RNA in samples from inoculated pigs
| Br1/87 PEDV (Group 1) | US (non‐INDEL) PEDV (Group 3) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pig 1 | Pig 2 | Pig 3 | Pig 4 | Pig 5 | Pig 11 | Pig 12 | Pig 13 | Pig 14 | Pig 15 | |||||||||||
| PID | fs | ser | fs | ser | fs | ser | fs | ser | fs | ser | fs | ser | fs | ser | fs | ser | fs | ser | fs | ser |
| 0 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 1 | – | – | – | – | – | – | – | – | 29 | 38 | ||||||||||
| 2 | – | – | – | – | 32 | 38 | – | – | – | – | 16 | 39 | 24 | 31 | 27 | – | 17 | 36 | 23 | – |
| 3 | 33 | 33 | 17 | 28 | 30 | 17 | 22 | 20 | 18 | 30 | ||||||||||
| 4 | 27 | – | 32 | – | 17 | – | 30 | – | 33 | – | 15 | – | 16 | 32 | 16 | – | 18 | 39 | 24 | 29 |
| 6 | – | 26 | 36 | 17 | 16 | |||||||||||||||
| 7 | 35 | 34 | 36 | 35 | 35 | |||||||||||||||
| 8 | 35 | 36 | 26 | 25 | 18 | |||||||||||||||
| 10 | 36 | – | 32 | 33 | 32 | |||||||||||||||
| 14 | 36 | – | – | – | – | – | 34 | 37 | 27 | 39 | ||||||||||
| 16 | – | 40 | – | 34 | 39 | |||||||||||||||
| 18 | 36 | – | – | 37 | 33 | |||||||||||||||
| 21 | – | – | – | – | – | |||||||||||||||
| 24 | – | – | – | – | – | |||||||||||||||
| 28 | – | – | – | – | – | |||||||||||||||
RNA was extracted from faecal swab (fs) and serum (ser) samples collected from the individual pigs, in groups 1 and 3, on the indicated days post‐infection (PID) and assayed by RT‐qPCR for the presence of PEDV RNA. Samples giving no Ct value (no PEDV RNA detected) are indicated by a (–), and Ct values are given to the nearest integer. The grey blocks indicate that the animals were euthanized on PID 4. Empty cells indicate no samples were collected on that day. Similar samples were collected and analysed from Group 2 (pigs 6–10), but all were negative throughout the study, and so the results are not shown.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
No PEDV RNA was present in fs samples from the pigs (6–10) in Group 2 at any stage (results not shown), again indicative of a lack of infection by PEDV.
As expected, no PEDV RNA was present in fs samples at PID 0 from the pigs in groups 1 (inoculated with Br1/87) and 3 (inoculated with the US PEDV) either (Table 1). However, the detection of Br1/87 RNA was achieved in fs samples from pig 3 on PID 2 and from all 5 animals within Group 1 on PID 3. The highest level of PEDV RNA in fs samples from this Group was found in pigs 1 and 3 on PID 4; these pigs also showed the clearest clinical disease. Lower levels of PEDV RNA were excreted in faeces from one pig in this Group until at least PID 18. The US PEDV RNA was detectable in fs samples from two animals (pig 14 and pig 15, in Group 3) on PID 1 and was found at higher levels (Ct < 28) in samples from all 5 animals from PID 2 onwards (Table 1). Viral RNA continued to be excreted from the surviving pigs until at least PID 18, albeit at a reduced level.
The virus suspensions, derived from infected pigs, used for the inoculation and also some of the fs samples generated in this experiment were also tested for the presence of rotavirus RNA. Although rotavirus RNA was found by RT‐qPCR, at just detectable levels, in the US PEDV inoculum as given to pigs in Group 3, no excretion of rotavirus RNA could be detected in faecal samples from pigs 11–15 (collected between PID 0 and PID 8) but was detected in the ileum and jejunum of pig 14 (but not from pigs 11 and 13) when they were euthanized at PID 4. As indicated above, no rotavirus RNA was detected within the inoculum (German sample) given to pigs in Group 2.
PEDV RNA in tissues
Pigs 1 and 3 (from Group 1) and pigs 11, 13 and 14 (from Group 3) that each showed clear clinical signs of disease were euthanized on PID 4. Two animals (pigs 6 and 10) from Group 2 (without any clinical disease) were also euthanized at this stage. Tissues from the euthanized animals were analysed for the presence of PEDV RNA (Table 2). High levels of viral RNA were detected throughout the intestine of most of the inoculated pigs in groups 1 and 3 (although not in the jejunum of pig 1). Low levels of viral RNA were detected in the lungs and livers of these animals and rather higher levels (indicated by lower Ct values) were found in some spleen samples (from pigs 11, 13 and 14, in Group 3). No PEDV RNA was detected in the tissues from pigs 6 and 10 in Group 2 (data not shown), again consistent with the apparent lack of infection in this Group.
Table 2.
Detection of PEDV in tissues of inoculated pigs at PID 4
| Tissue | Br1/87 PEDV (Group 1) | US (non‐INDEL) PEDV (Group 3) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pig 1 | Pig 2 | Pig 3 | Pig 4 | Pig 5 | Pig 11 | Pig 12 | Pig 13 | Pig 14 | Pig 15 | |
| Ileum | 32 | 20 | 19 | 17 | 16 | |||||
| Jejunum | – | 16 | 17 | 16 | 16 | |||||
| Colon | 39 | 19 | 17 | 18 | 18 | |||||
| Lungs | – | – | 39 | 37 | 37 | |||||
| Liver | – | 38 | 36 | 35 | – | |||||
| Spleen | – | 37 | 31 | 34 | 33 | |||||
Pigs with clinical signs indicative of PED were euthanized on PID 4; RNA was extracted from the indicated tissue sample homogenates (10% (w/v) suspension) and assayed, by RT‐qPCR, for the presence of PEDV RNA. Samples with no detectable PEDV RNA (no Ct) are indicated by a (–). Ct values were rounded to the nearest integer. Grey blocks indicate animals that were maintained until PID 28, and thus, no tissue samples were available at PID 4. Similar samples were collected on PID 4 from two pigs (6 and 10) within Group 2, although these did not show any clinical signs. None of these samples contained detectable PEDV RNA (no Ct value), and thus, the results are not shown.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
PEDV RNA detection in sera
As expected, no PEDV RNA was detected in the sera at PID 0 in any of the pigs from groups 1 and 3 (Table 1). Furthermore, no PEDV RNA was found in the sera from animals in Group 2 at any stage, pre‐ or post‐inoculation (data not shown). However, significant levels of the US PEDV RNA were detectable in the sera of pigs 12 and 15 (Ct < 32) at PID 4 and indeed 4 of the 5 pigs in this Group had PEDV RNA detectable in sera at some stage (note: serum samples were collected on up to 8 separate occasions during the 28‐day duration of the experiment, as indicated in Table 1). Indeed, PEDV RNA was also present in the serum at PID 7 and PID 14 in pigs 12 and 15, but no viral RNA was detected in sera from any of the pigs at PID 21 or later. In contrast, Br1/87 RNA was detected in sera collected on a single day (PID 7) from pigs 2, 4 and 5 and a low level of viral RNA was also detected in serum from pig 3 on PID 2.
Cross‐recognition of Br1/87 PEDV antigens by anti‐PEDV antibodies using a cell staining assay
Sera collected on PID 21 from pig 5 (inoculated with Br1/87) and from pig 15 (inoculated with the US PEDV) were used to detect PEDV antigens in Vero cells infected with the Br1/87 strain using an immunoperoxidase staining test. Clear staining of the PEDV‐infected cells was observed using both sera (Fig. 2a and b), whereas no specific staining was observed with a control, uninfected, pig serum (Fig. 2c). Thus, consistent with the ELISA data (see above), the infection of pigs with either the Br1/87 strain or the US PEDV induced antibodies which react with Br1/87 antigens in the immunostaining test.
Figure 2.
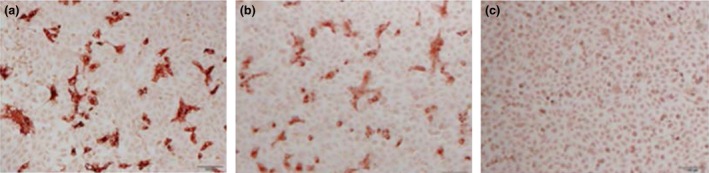
Immunostaining of PEDV antigens using sera from PEDV‐infected pigs. Sera collected on PID 21 from pig 5 (inoculated with Br1/87) (panel a) and from pig 15 (inoculated with the US PEDV) (panel b) and a negative control serum (panel c) were used to detect PEDV antigens produced by infection of Vero cells with the Br1/87 strain of PEDV using immunoperoxidase staining as described in the Methods section.
Discussion
This study has demonstrated the efficient experimental infection of young pigs with an early European strain of PEDV (Br1/87) and a recent, non‐INDEL, strain of PEDV from the USA. Clear evidence of infection occurred in all 5 pigs inoculated with each of these virus samples. In contrast, unfortunately, no apparent infection was achieved within Group 2 using an inoculum derived from a faecal sample from a recent case of PED in Germany despite the presence of high levels of PEDV RNA in this sample. No information on the collection and storage conditions of the faecal sample prior to submission to the diagnostic laboratory is available, and various factors may have been deleterious for the infectivity of the PEDV in the sample. It should be noted that the US PEDV sample was a homogenate of intestine, as well as faecal material, and this difference compared with the German sample may, or may not, be significant for its ability to cause infection.
Neither excretion of PEDV RNA nor induction of anti‐PEDV antibodies was detected in the pigs within Group 2. Furthermore, it is clear that the diagnostic assays produced negative results with the samples taken from all the experimental animals prior to inoculation consistent with the lack of any history of PEDV infection in Denmark. The relative level of infectivity within the different samples used as inoculum is not known as attempts to isolate PEDV in cell culture from the supplied faecal/intestinal samples were not successful (data not shown). However, it is clear that sufficient virus was present in the cell culture‐grown Br1/87 isolate and in the US PEDV sample to cause infection in all of the inoculated animals. These infections were demonstrated by a variety of means including the appearance of fairly mild clinical disease, the presence of high levels of PEDV RNA in the intestine (at PID 4) or the prolonged excretion of PEDV RNA and the induction of anti‐PEDV antibodies within 10 days post‐inoculation. The time course of seroconversion is consistent with previous results using the US PEDV (Madson et al., 2014; Crawford et al., 2015).
In the current study, faecal shedding of PEDV RNA was observed through to PID 18 in both infected Groups and faecal shedding of PEDV RNA has been reported to occur until PID 24 and even PID 42 but it is not known whether infectious virus is shed beyond PID 21 (Madson et al., 2014; Crawford et al., 2015).
PEDV RNA was detected in serum samples collected from pigs in Group 3 (US PEDV) on multiple days from PID 2 through to PID 14. A more restricted presence of PEDV RNA was apparent in the sera from the Br1/87 inoculated pigs (Group 1) with viral RNA mainly detected at PID 7. The presence of PEDV RNA in serum from acutely infected pigs has been reported previously (Jung et al., 2014, 2015). A consistent observation is that the viral RNA was found at a very much lower level in serum than in faecal material and the RNA that is present in serum has not yet been shown to be within infectious virus.
In pigs euthanized at PID 4, both the Br1/87 and the US PEDV were found throughout the intestine. PEDV RNA was also detected in the spleens of some of these pigs (even in the absence of virus RNA in the serum) and the level of viral RNA in the spleen was higher than in liver or lungs. Thus, it does not seem possible to account for the level of viral RNA within the spleen by the presence of virus in the blood. In earlier studies, PEDV antigens were not detected by immunostaining within the spleen, liver or lungs of CV777‐infected pigs (Debouck et al., 1981) or in lungs of PEDV PC21A‐infected pigs (Jung et al., 2014), but the sensitivity of the RT‐qPCR assays used here is likely higher than the sensitivity of the immunostaining procedures. It has been reported (Park and Shin, 2014) that PEDV can replicate in pulmonary macrophages in vivo and thus extra‐intestinal replication of PEDV seems possible (Jung and Saif, 2015). The results presented here indicate that the US PEDV produced slightly more severe clinical signs than the cell culture‐grown Br1/87 strain. Furthermore, the duration and extent of virus excretion together with the presence of viremia were all more marked in the US PEDV inoculated Group. However, these differences may be due to use of the cell culture‐grown virus (potentially adapted) compared with the field virus or possible differences in the level of infectious virus used for the inoculation rather than a real difference in properties of the strains.
Conclusions
Infection of piglets by the early European isolate of PEDV (Br1/87, a close relative of the CV777 strain) and by a US non‐INDEL strain of the virus has been performed and it has been possible to monitor infection by each strain using a range of diagnostic assays for the presence of viral RNA and the induction of anti‐PEDV antibodies. The mild clinical outcome observed in this study may be related to the age of piglets used. Newborn piglets appear most severely affected by the infection in the field, and hence, experimental infection of such piglets can be expected to produce a more severe disease and even mortality. Indeed, the outcome of PEDV infections in pigs appears to be dependent upon a range of factors (see EFSA, AHAW Panel (EFSA Panel on Animal Health and Welfare), 2014). Potential differences in the pathogenicity of different strains of the virus require side‐by‐side comparisons with defined virus isolates under carefully defined conditions.
Supporting information
Table S1. Detection of PEDV RNA by RT‐qPCR assays.
Acknowledgements
We are grateful for the excellent technical assistance from Jane Borch, Carina Becke, Helle Rasmussen, Eva Rasmussen and Pernille Manley Hansen. Likewise, Marion Petersen and Janni Oxfeldt are thanked for taking excellent care of the animals and for their assistance during the experiment. We also thank Sandra Blome (FLI, Germany) and APHIS, Ames, IA, USA, for providing samples from PEDV‐infected pigs. The work was funded from internal resources of the DTU National Veterinary Institute.
References
- Bøtner, A. , Nielsen J., and Bille‐Hansen V., 1994: Isolation of porcine reproductive and respiratory syndrome (PRRS) virus in a Danish swine herd and experimental infection of pregnant gilts with the virus. Vet. Microbiol. 40, 351–360. [DOI] [PubMed] [Google Scholar]
- Bridgen, A. , Duarte M., Tobler K., Laude H., and Ackermann M., 1993: Sequence determination of the nucleocapsid protein gene of the porcine epidemic diarrhoea virus confirms that this virus is a coronavirus related to human coronavirus 229E and porcine transmissible gastroenteritis virus. J. Gen. Virol. 74, 1795–1804. [DOI] [PubMed] [Google Scholar]
- Chen, Q. , Li G., Stasko J., Thomas J. T., Stensland W. R., Pillatzki A. E., Gauger P. C., Schwartz K. J., Madson D., Yoon K. J., Stevenson G. W., Burrough E. R., Harmon K. M., Main R. G., and Zhang J., 2014: Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 52, 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, K. , Lager K., Miller L., Opriessnig T., Gerber P., and Hesse R., 2015: Evaluation of porcine epidemic diarrhea virus transmission and the immune response in growing pigs. Vet. Res. 46, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck, P. , Pensaert M., and Coussement W., 1981: The pathogenesis of an enteric infection in pigs, experimentally induced by the coronavirus–like agent, CV‐777. Vet. Micro. 6, 157–165. [Google Scholar]
- EFSA, AHAW Panel (EFSA Panel on Animal Health and Welfare) , 2014: Scientific Opinion on porcine epidemic diarrhoea and emerging pig deltacoronavirus. EFSA J. 12, 3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke, D. , Jenckel M., Petrov A., Ritzmann M., Stadler J., Akimkin V., Blome S., Pohlmann A., Schirrmeier H., Beer M., and Höper D., 2015: Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerg. Infect. Dis. 21, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. W. , Dickerman A., Piñeyro P., Li L., Fang L., Kiehne R., Opriessnig T., and Meng X. J., 2013: Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio 4, e00737–00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K. , and Saif L. J., 2015: Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 204, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K. , Wang Q., Scheuer K. A., Lu Z., Zhang Y., and Saif L. J., 2014: Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 20, 662–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K. , Annamalai T., Lu Z., and Saif L. J., 2015: Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9‐day‐old nursing piglets vs. 26‐day‐old weaned pigs. Vet. Microbiol. 178, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. H. , Kim I. J., Pyo H. M., Tark D. S., Song J. Y., and Hyun B. H., 2007: Multiplex real‐time RT‐PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods 146, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson, D. M. , Magstadt D. R., Arruda P. H., Hoang H., Sun D., Bower L. P., Bhandari M., Burrough E. R., Gauger P. C., Pillatzki A. E., Stevenson G. W., Wilberts B. L., Brodie J., Harmon K. M., Wang C., Main R. G., Zhang J., and Yoon K. J., 2014: Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3‐week‐old weaned pigs. Vet. Microbiol. 174, 60–68. [DOI] [PubMed] [Google Scholar]
- Pang, X. L. , Lee B., Boroumand N., Leblanc B., Preiksaitis J. K., and Yu Ip C. C., 2004: Increased detection of rotavirus using a real time reverse transcription‐polymerase chain reaction (RT‐PCR) assay in stool specimens from children with diarrhea. J. Med. Virol. 72, 496–501. [DOI] [PubMed] [Google Scholar]
- Park, J. E. , and Shin H. J., 2014: Porcine epidemic diarrhea virus infects and replicates in porcine alveolar macrophages. Virus Res. 191, 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen, K. J. , Bøtner A., Madsen E. S., Strandbygaard B., and Nielsen J., 1997: Evaluation of a blocking ELISA for screening of antibodies against porcine reproductive and respiratory syndrome (PRRS) virus. Vet. Microbiol. 56, 1–8. [DOI] [PubMed] [Google Scholar]
- Sun, R. Q. , Cai R. J., Chen Y. Q., Liang P. S., Chen D. K., and Song C. X., 2012: Outbreak of porcine epidemic diarrhea in suckling piglets. China. Emerg. Infect. Dis. 18, 161–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova, A. N. , Marthaler D., Wang Q., Culhane M. R., Rossow K. D., Rovira A., Collins J., and Saif L. J., 2014: Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 20, 1620–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Byrum B., and Zhang Y., 2014: New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 20, 917–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, R. , Soma J., Nakanishi M., Yamaguchi R., and Niinuma S., 2015: Isolation and experimental inoculation of an S INDEL strain of porcine epidemic diarrhea virus in Japan. Res. Vet. Sci. 103, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Detection of PEDV RNA by RT‐qPCR assays.


