Abstract
BACKGROUND
Middle East respiratory syndrome‐coronavirus (MERS‐CoV) is a novel zoonotic pathogen. Although the potential for MERS‐CoV transmission through blood transfusion is not clear, MERS‐CoV was recognized as a pathogen of concern for the safety of the blood supply especially after its detection in whole blood, serum, and plasma of infected individuals. Here we investigated the efficacy of amotosalen and ultraviolet A light (UVA) to inactivate MERS‐CoV in fresh‐frozen plasma (FFP).
STUDY DESIGN AND METHODS
Pooled FFP units were spiked with a recent clinical MERS‐CoV isolate. Infectious and genomic viral titers were determined in plasma before and after inactivation with amotosalen/UVA treatment by plaque assay and reverse transcription–quantitative polymerase chain reaction, respectively. In addition, residual replicating or live virus after inactivation was examined by passaging in the permissive Vero E6 cells.
RESULTS
The mean MERS‐CoV infectious titer in pretreatment samples was 4.67 ± 0.25 log plaque‐forming units (pfu)/mL, which was reduced to undetectable levels after inactivation with amotosalen/UVA demonstrating a mean log reduction of more than 4.67 ± 0.25 pfu/mL. Furthermore, inoculation of inactivated plasma on Vero E6 cells did not result in any cytopathic effect (CPE) even after 7 days of incubation and three consecutive passages, nor the detection of MERS RNA compared to pretreatment samples which showed complete CPE within 2 to 3 days postinoculation and log viral RNA titer ranging from 9.48 to 10.22 copies/mL in all three passages.
CONCLUSION
Our data show that amotosalen/UVA treatment is a potent and effective way to inactivate MERS‐CoV infectious particles in FFP to undetectable levels and to minimize the risk of any possible transfusion‐related MERS‐CoV transmission.
ABBREVIATIONS
- CPE
cytopathic effect
- IBS
INTERCEPT Blood System
- MERS‐CoV
Middle East respiratory syndrome‐coronavirus
- pfu
plaque‐forming units
- RT‐qPCR
reverse transcription–quantitative polymerase chain reaction
- SARS‐CoV
severe acute respiratory syndrome‐coronavirus
Transfusion of blood components saves millions of lives by controlling bleeding due to accidents, surgeries, or other disease complications. However, transmission of pathogens is one of the biggest risks of transfusion of labile blood components. Therefore, a key mission of blood transfusion services is to provide safe blood and blood products. Screening of blood products has reduced the spread of known blood‐borne pathogens such as hepatitis B and C viruses (HBV and HCV), human immunodeficiency virus (HIV), and human T‐lymphotropic virus (HTLV).1, 2 However, other known or unknown pathogens pose a threat to the blood supply with regional differences that is difficult to address. The number of pathogens screened in blood banks is limited by the number of blood screening assays that are commercially available. Therefore, pathogen inactivation offers an appealing alternative to blood screening (serology or nucleic acid testing [NAT]) because of its proactive nature and the broad spectrum of protection it offers without a priori characterization of unknown pathogens. Indeed, pathogen inactivation technologies have been developed to provide safe blood products while reducing the need for the implementation of additional screening assays.3
The Middle East respiratory syndrome‐coronavirus (MERS‐CoV) is a zoonotic pathogen that is endemic in Saudi Arabia and other countries in the Arabian Peninsula since 2012.4 As of February 24, 2017, MERS‐CoV has been responsible for 1905 confirmed infections including 677 deaths (approx. 35.5% mortality rate) in 27 countries.5 Most cases (1552 confirmed infections) were reported in Saudi Arabia with a mortality rate of approximately 41.9%.6 MERS‐CoV causes respiratory illness in humans varying from asymptomatic or mild to severe pneumonia.4, 7 The elderly and patients suffering from comorbidities are the most at risk for death outcome after development of severe clinical symptoms such as severe pneumonia and extrapulmonary manifestations.8 The major route of transmission for MESR‐CoV is via droplets, fomites, and person‐to‐person contact through the respiratory system as well as via direct or indirect contact with zoonotic sources. So far, most MERS cases are linked to residence in or travel to Saudi Arabia.9 Several hospital and household outbreaks have been reported in Saudi Arabia as well as South Korea due to nosocomial transmission and close contact with patients.9, 10 However, the number of asymptomatic cases is estimated to be much higher than the reported cases,11 which together with mild cases may increase the spread of the virus and pose a significant risk for blood safety. Indeed, presymptomatic and asymptomatic donors may be allowed to donate blood while their donation has the potential to carry the virus, although there is no report on MERS‐CoV detection in blood from presymptomatic and asymptomatic patients. While most of the reported MERS‐CoV cases were due to human‐to‐human transmissions especially under inadequate infection control measures, dromedary camels are believed to be the reservoir host and an important source of human infection.9, 12, 13, 14, 15, 16, 17 Specifically, primary or index cases that had no contact history with infected individuals are more likely to have had direct or indirect contact with dromedaries. Interestingly, current regulations of blood donation do not consider recent history of contact with dromedaries as a reason for deferral of blood donation.
MERS‐CoV has been detected in a variety of samples from infected patients including respiratory samples, serum, plasma, urine, and stool.18, 19, 20, 21, 22 While detection of MERS‐CoV RNA is most frequent and persistent in respiratory secretions with high viral loads, detection of viral RNA in plasma or serum was documented in up to one‐third of the patients and showed association with disease severity.20, 21, 22 This is reminiscent of the severe acute respiratory syndrome‐coronavirus (SARS‐CoV) outbreak which started in China in 2003 and led to global human‐to‐human spread with more than 8000 confirmed cases and approximately 10% death rate. During the SARS outbreak, viral RNA was detected in serum from several patients but no transfusion‐associated transmission was reported.23, 24 While SARS‐CoV disappeared quickly, MERS‐CoV continues to be endemic in the Arabian Peninsula for more than 4 years now. Similar to SARS‐CoV, there is no proven evidence so far of transfusion‐transmitted MERS‐CoV infections,25 but the presence of viral RNA in plasma and serum of acute patients raises this concern especially in endemic areas like Saudi Arabia. In fact, it is not known whether presence of viral RNA in blood products could lead to transmission of MERS‐CoV or not. Therefore, it is important to find a method to mitigate the risk associated with MERS‐CoV RNA presence in blood components to proactively prevent any possible transmission of MERS‐CoV through contaminated blood products in endemic regions and elsewhere.
The INTERCEPT Blood System (IBS) inactivates pathogens by forming crosslinks or monoadducts on nucleic acids using amotosalen, a photoactive psoralen, after illumination with low‐energy ultraviolet A (UVA) light. During this highly specific targeted process, an adduct is formed at high frequency in the nucleic acids, thus inhibiting transcription and replication.26 The IBS is a Food and Drug Administration–approved pathogen reduction system that has been shown to inactivate a broad spectrum of viruses, bacteria, and parasites in plasma as well as other blood products27, 28, 29, 30 without affecting the plasma efficacy and patient safety as demonstrated in clinical evaluation31 and by hemovigilance data from multiple countries.32, 33 Until now there has been no data available on the inactivation of MERS‐CoV with IBS. However, the inactivation of SARS‐CoV with IBS, which is related to MERS‐CoV, was successfully shown,27 raising the expectation of a sufficient inactivation efficacy. Therefore, the aim of this study was to evaluate the efficacy of amotosalen/UVA treatment to inactivate MERS‐CoV in human apheresis plasma concentrates.
MATERIALS AND METHODS
Cell line and MERS‐CoV virus
African Green monkey kidney‐derived Vero E6 cells (ATCC #1568) were grown and maintained as previously described in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS).34 The human MERS‐CoV isolate used in this study was MERS‐CoV/Hu/Taif/SA/2015, which was characterized previously.34 All experiments involving live virus were conducted in a Biosafety Level 3 facility at the Special Infectious Agents Unit at King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Kingdom of Saudi Arabia, following the recommended safety precautions and measures.
MERS‐CoV culture
MERS‐CoV amplification and culture was done as previously described.35 Virus was inoculated on 90% to 95% confluent Vero E6 cells in a T175 tissue culture flask at multiplicity of infection of 1 and incubated in a humidified 5% CO2 incubator at 37°C for 1 hour with gentle shaking every 15 to 20 minutes. Subsequently, the inoculum was replaced by 25 mL of viral inoculation medium (DMEM with 2% FBS, 1% penicillin/streptomycin, and 10 mmol/L HEPES [pH 7.2]) and the cells were incubated in a humidified 5% CO2 incubator at 37°C for 72 hours or until 80% to 90% of cells showed a cytopathic effect (CPE). Supernatant was collected and centrifuged to remove cellular debris for 5 minutes at 500 × g at room temperature. Virus was then aliquoted and stored at –80°C, and the titer was determined by plaque assay.
Plasma preparation
Whole blood units (450 mL ± 10%) were collected and prepared at King Abdulaziz University Hospital, Transfusion Services, Jeddah, Kingdom of Saudi Arabia, from voluntary donors. Briefly, blood units were centrifuged at 544 × g for 10 minutes to separate the platelet (PLT)‐rich plasma. The PLT rich plasma was then centrifuged at 2305 × g at 20°C for 10 minutes. The plasma was then separated into the plasma bag and kept at not more than –18°C. All blood units were screened routinely for HCV antibody or antigen, HBsAg, HBc antibody, HIV (1/2) antibody, HTLV (1/2) antibody, and syphilis as well as HCV, HBV, and HIV by NAT.
MERS‐CoV inactivation studies
Two units of fresh‐frozen plasma (≥210 mL each) were pooled for each experiment. The count of red blood cells was less than 4 × 106/mL in the pooled plasma. In total, four pools (n = 4) were used in this study. Pooled plasma units were then inoculated with approximately 4 mL MERS‐CoV stock (1:100 dilution). Plasma units were inactivated with the INTERCEPT processing set for plasma and the INTERCEPT illuminator according to the manufacturer's instructions. Residual amotosalen and the free photoproducts were removed with an INTERCEPT compound adsorption device according to the manufacturer's instructions. Samples collected for testing were as follows (Fig. 1): a positive control sample from the virus stock, a negative control sample from the plasma pool before inoculation, a 2‐ to 3‐mL pretreatment sample from the inoculated plasma pool, and an inactivated sample from the treated pooled plasma. All samples were stored at –80°C until testing by plaque and reverse transcription–quantitative polymerase chain reaction (RT‐qPCR) assays.
Figure 1.
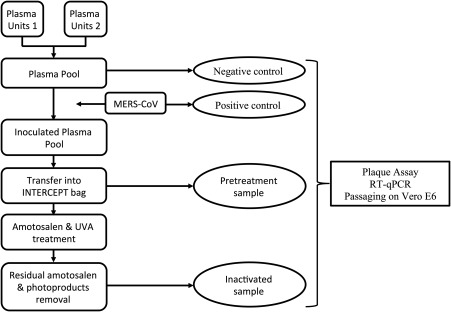
Schematic experimental overview.
Detection of MERS‐CoV replication
For the detection of MERS‐CoV replication, all samples were diluted at 1:10 dilution in DMEM with 10% FBS, inoculated on 90% confluent Vero E6 cells in six‐well plates, and incubated for 3 to 7 days at 37°C. Supernatants were collected, diluted 1:10 in DMEM with 10% FBS, and blindly passaged for two more times. Each passage was observed for CPE and all collected supernatants were used for viral RNA quantification by RT‐qPCR.
Plaque assay
Vero E6 cells were prepared at 1 × 105/mL in DMEM growth medium and 2 mL were seeded in each well in six‐well plates and incubated overnight at 37°C. Samples was then serially diluted in inoculation DMEM starting from 10−1, and 200 μL of each dilution were applied to the confluent Vero E6 cell monolayers in each well and incubated for 1 hour at 37°C with gentle shaking every 15 to 20 minutes. After 1 hour, inoculum was removed and replaced with overlay medium (DMEM with 0.8% agarose) and incubated for 3 to 4 days at 37°C. After incubation, the overlay was removed and cells were fixed with 4% paraformaldehyde for 15 minutes at room temperature and stained with crystal violet stain. Plaques were examined and counted to calculate titer as plaque‐forming units (pfu)/mL as previously described.35 In some experiments, 15 to 30 mL of IBS‐treated plasma was tested in plaque assay to detect any residual infectious virus and to increase the dynamic range of the assay in which 200 μL of neat plasma were inoculated in each well.
RT‐qPCR quantitation
For all samples (positive, negative, pretreatment, and inactivated samples) and cell supernatants, RNA was extracted as previously described12 using viral RNA mini kit (Qiagen) according to the manufacturer's instructions. Real‐time RT‐qPCR was performed using primers and probe targeting MERS‐CoV N gene as previously described.36 Specifically, the following forward primer 5′‐CAAAACCTTCCCTAAGAAGGAAAAG‐3′, reverse primer 5′‐GCTCCTTTGGAGGTTCAGACAT‐3′, and probe 5′‐FAM‐ACAAAAGGCACCAAAAGAAGAATCAACAGACC‐BHQ1‐3′ were used. Real‐time RT‐qPCR was performed in 96‐well plates on a fast real‐time PCR system (Model 7500, Applied Biosystems) using an RT‐PCR kit (QuantiTect Probe, Qiagen) according to the manufacturer's instructions in a total reaction volume of 25 μL. RNA transcript corresponding to the same target region was generated using a cDNA synthesis kit (Superscript RT III, Invitrogen) from a plasmid containing the MERS‐CoV N gene according to the manufacturer's instructions and used to generate a standard curve to estimate the viral RNA copy number in each sample as previously described.37, 38 Each run included a positive viral template control and no‐template negative control. Each sample was tested in duplicate and the mean is reported as log copies/mL. Samples were reported as negative if the cycle threshold values were higher than 36 cycles.
Institutional review board approval
The study was approved by the biomedical ethics committee unit of King Abdulaziz University Hospital (Approval 257‐16).
RESULTS
Inactivation of MERS‐CoV by amotosalen and UVA
To evaluate the potential of the IBS in inactivating MERS‐CoV in pooled plasma, MERS‐CoV virus was spiked into pooled plasma units and treated with amotosalen and UVA light. The viral titer in pretreatment samples ranged from 4.51 to 5.04 log pfu/mL with mean titer of 4.67 ± 0.25 log pfu/mL (Table 1). The treatment resulted in no detection of viable viruses in inactivated samples by plaque assay with a mean reduction of 4.67 ± 0.25 log pfu/mL in viral infectivity titer from four independent experiments (Experiments A‐D) and Fig. 2 shows representative results from the plaque assay. Therefore, larger volumes from inactivated samples were tested to increase the sensitivity and the dynamic range of the assay as shown in Table 1. Nonetheless, no viable virus was detected post pathogen inactivation treatment even when 15 to 30 mL of plasma was tested. The viral genomic titer was not reduced post treatment as expected (Table 2). Together, these data show that amotosalen/UVA light pathogen inactivation technology inactivated all MERS‐CoV infectious particles in the spiked plasma units.
Table 1.
Reduction in MERS‐CoV titers after inactivation by amotosalen and UVA in pooled plasmaa
| Viral load (log pfu/mL) | Log reduction | ||||
|---|---|---|---|---|---|
| Experiment | Positive control | Negative control | Pretreatment sample | Inactivated sample | |
| Ac | 7.85 | ND | 4.52 | NDb | >4.52 |
| B | 8.18 | ND | 4.51 | NDc | >4.51 |
| C | 7.60 | ND | 5.04 | NDd | >5.04 |
| D | 7.60 | ND | 4.60 | NDd | >4.60 |
| Mean ± SD | 7.80 ± 0.27 | ND | 4.67 ± 0.25 | ND | >4.67 ± 0.25 |
Data are shown as log pfu/mL.
No infectious virus was detected in 1.5‐mL assayed volume.
No infectious virus was detected in 15‐mL assayed volume.
No infectious virus was detected in 30‐mL assayed volume.
ND = not detected.
Figure 2.
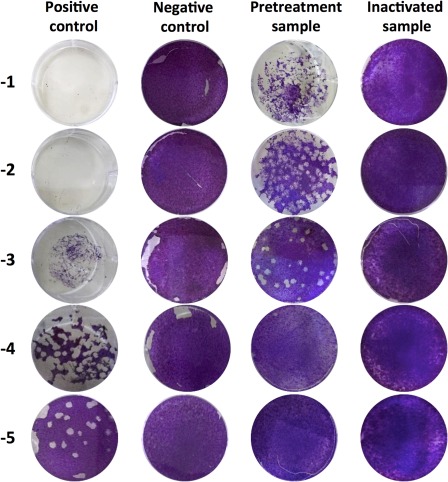
Inhibition of MERS‐CoV in plasma by amotosalen and UVA treatment. Representative plaque assay is shown for samples collected, which includes positive, negative, pretreatment, and inactivated samples.
Table 2.
MERS‐CoV genomic titers before and after inactivation by amotosalen and UVA in pooled plasma*†
| Experiment | Positive control | Negative control | Pretreatment sample | Inactivated sample |
|---|---|---|---|---|
| A | 10.34 | ND | 7.80 | 6.82 |
| B | 11.32 | ND | 7.53 | 7.07 |
| C | 10.23 | ND | 7.61 | 7.23 |
| D | 10.15 | ND | 7.60 | 7.39 |
| Mean ± SD | 10.51 ± 0.55 | ND | 7.64 ± 0.12 | 7.13 ± 0.24 |
Data are shown as log RNA copies/mL.
Genomic titer was determined from the same samples used in Table 1.
ND = not detected.
Detection of replicative MERS‐CoV in treated pooled plasma
Next, we analyzed the presence of any potential low level of replicating MERS‐CoV in inactivated plasma units taking into consideration that genomic viral titer did not change before and after treatment (Table 2). To this end, Vero E6 cells were inoculated with plasma samples of pathogen‐inactivated units and incubated for 3 days, followed by two additional passages with 3 days of incubation. No CPE was observed from the negative control or inactivated samples even on Day 7 postinoculation or after passaging for three times compared to the positive or pretreatment samples, which completely destroyed all cells by Day 3 (data not shown). To further confirm these results, the viral genomic RNA titer was measured by RT‐qPCR from supernatant collected on Day 3 from all samples in the three consecutive passages. As shown in Table 3, viral genomic titer in positive control and pretreatment samples ranged from 9.03 to 10.51 and 9.48 to 10.22 log RNA copies/mL, respectively, in the three passages indicating presence of replicating viruses in these samples. On the other hand, no viral RNA was detected in the supernatant collected from any of the three passages from the negative control or inactivated samples, confirming the complete inactivation of the infectious MERS‐CoV in plasma.
Table 3.
Replication of MERS‐CoV in Vero E6 cells before and after inactivation of spiked pooled plasma*†
| Experiment | Passage 1 | Passage 2 | Passage 3 | |
|---|---|---|---|---|
| A | ||||
| Positive control | 10.51 | 9.98 | 9.81 | |
| Negative control‡ | ND | ND | ND | |
| Pretreatment sample | 9.96 | 9.94 | 10.22 | |
| Inactivated sample | ND | ND | ND | |
| B | ||||
| Positive control | 9.74 | 9.90 | 9.75 | |
| Negative control | ND | ND | ND | |
| Pretreatment sample | 10.16 | 10.07 | 10.18 | |
| Inactivated sample | ND | ND | ND | |
| C | ||||
| Positive control | 9.88 | 10.16 | 9.44 | |
| Negative control | ND | ND | ND | |
| Pretreatment sample | 10.09 | 10.08 | 9.72 | |
| Inactivated sample | ND | ND | ND | |
| D | ||||
| Positive control | 9.73 | 10.09 | 9.03 | |
| Negative control | ND | ND | ND | |
| Pretreatment sample | 10.12 | 9.96 | 9.48 | |
| Inactivated sample | ND | ND | ND | |
Data are shown as log RNA copies/mL.
Samples used in Table 1 were used in this experiment. Samples were used at 1:10 dilution and titer was determined on Day 3 postinoculation.
ND = not detected.
DISCUSSION
Currently, there are more than 70 known pathogens that have been recognized by the AABB as pathogens of concern for blood transfusion;25 however, blood donations are only screened for a small number of pathogens on a regular basis. Although such specific pathogen screening and testing of blood products have significantly reduced transfusion‐related infections, transmission of well‐recognized pathogens such as HIV, HBV, and HCV is still being reported in many parts of the world.39, 40 Furthermore, emerging and/or reemerging pathogens which are usually of zoonotic nature pose a high risk for blood safety.41 The unpredictable nature of such outbreaks and the erratic dynamics of their spread represent a challenging hurdle to preparedness plans and implementation of safety measures. A recent example is the emergence of Zika virus, a mosquito‐borne flavivirus that is transmissible through blood transfusion.42
Outbreaks of coronaviruses with high mortality in humans have been described for SARS‐CoV in China and Southeast Asia23, 24 and MERS‐CoV in the Middle East and South Korea.9 The detection of viral RNA in the blood of patients infected with SARS‐CoV and MERS‐CoV suggests a potential risk for their transmission through transfusion.21, 22, 23, 24 During a MERS‐CoV outbreak in South Korea, levels of viral load ranged from 3.3 to 4.2 log genomic copies/mL in whole blood and from 2.7 to 4.0 log genomic copies/mL in serum,21 while another study reported up to 6 logs of RNA copies/mL in serum from infected individuals.20 That corresponds to an approximate infectious titer of less than 1.2 log in whole blood and less than 3 logs in serum according to our data which shows 3‐log difference between genomic and infectious titers (Tables 1 and 2). Using IBS, we were able to show a mean reduction of 4.67 ± 0.25 log in MERS‐CoV infectious titers (ranging from 4.51 to 5.04) in plasma (Table 1) which is approximately 1.5 logs higher than the minimum reduction titer recommended by the European Committee of Blood Transfusion,43 suggesting an inactivation capacity of MERS‐CoV in plasma with a high safety margin. In contrast to the infectious titer, the genomic titer was only affected slightly by the pathogen inactivation treatment (Table 2). The amotosalen/UVA process effectively crosslinks nucleic acids, but does not break the strands. Therefore, to ensure that there are no remaining amounts of infectious virus particles, the collected supernatants were passaged three times after inactivation for 3 days. The absence of CPE even after 7 days of incubation in cultures inoculated with the posttreatment samples suggests that there is no infectious MERS‐CoV remaining in these samples. No viral genomes were detected in the supernatant of cell cultures inoculated with posttreatment samples even after multiple passages mostly due to nonreplicating viral genome and degradation of viral RNA during the incubation period. This is in contrast to the pretreatment and the positive control samples that showed a high level of viral RNA after incubation in all three passages. This demonstrates the loss of infectious virus through passaging and the successful inactivation of infectious particles by the IBS treatment.
So far, there have been no reports of any transmission of MERS‐CoV or SARS‐CoV via blood transfusion despite the detection of their viral RNA in serum from infected individuals.20 Nonetheless, the AABB still lists both of these viruses as pathogens of concern for blood safety.25 MERS‐CoV RNA detection persisted in patient serum up to 14 days after diagnosis,20 and there is evidence that MERS‐CoV could infect and replicate in macrophages, dendritic cells, and T cells from the peripheral blood.36, 44, 45 Therefore, it is not really clear whether presence of MERS‐CoV RNA in blood, serum, or plasma could have any consequences on the transmission risks of MERS‐CoV during transfusion.
Apart from that, a recent large blood virome study found that blood from 42% of the study population (8240 healthy blood donors) contained genetic material from 19 human viruses including human herpesviruses (HSV‐1, EBV, CMV, HHV‐6A/B, HHV‐7, HHV‐8), human papillomaviruses, HIV‐1/2, HTLV‐1/2, HBV, HCV, human parvovirus B19, human adenovirus, human polyomaviruses (Merkel cell polyomavirus and Trichodysplasia spinulosa polyomavirus), Anellovirus (Torque teno virus [TTV] and TTV‐like mini virus), and influenza virus.46 Many of these viruses are of great importance in transfusion medicine although their detection in blood does not necessarily imply infectivity since only genetic material was detected. Furthermore, 75 other nonhuman viral species have also been detected in blood from many individuals, which were attributed to contamination from commercial reagents or the environment.46 Nonetheless, identification of other viruses such as the sewage‐associated gemycircularvirus was suggested to be due to contamination during phlebotomy or plasma pooling processing. Therefore, targeted testing of blood products might not be the best economic or logistic option to ensure blood safety.
Here, we were able to show efficient inactivation of MERS‐CoV in therapeutic human plasma units of 4.67 logs, far above the expected viral load of MERS‐CoV in human blood and serum described until now. A recent study investigated the inactivation of MERS‐CoV in plasma using riboflavin and UV light.47 The study showed a reduction of at least 4.07 and at least 4.42 log in viral titers for pooled and individual donor plasma, respectively. However, passaging experiments after pathogen reduction were not conducted, so complete inactivation of all infectious particles cannot be taken for granted. Several previous studies have also proved the efficiency of IBS in inactivating a large number of pathogens (viruses, bacteria, and parasites). The study described here adds MERS‐CoV to the list of pathogens that can be inactivated by IBS. Taken together with previously published work, our data show that IBS could represent an economically and logistically efficient protective solution to reduce the risk associated with the circulation of MERS‐CoV in the Arabian peninsula and the potential transmission of not only MERS‐CoV but also other known or unknown pathogens for which there is no commercially available screening assays.
CONFLICT OF INTEREST
The authors have disclosed no conflicts of interest.
ACKNOWLEDGMENT
We thank Edwin Raj from the blood transfusion services, King Abdulaziz University Hospital, King Abdulaziz University, Jeddah, Saudi Arabia, for the technical help with plasma collection and screening.
This study was partially funded by Cerus Corporation. The support was in the form of supplies and reagents needed for the study.
REFERENCES
- 1. Torane VP, Shastri JS. Comparison of ELISA and rapid screening tests for the diagnosis of HIV, hepatitis B and hepatitis C among healthy blood donors in a tertiary care hospital in Mumbai. Indian J Med Microbiol 2008;26:284‐5. [DOI] [PubMed] [Google Scholar]
- 2. Tessema B, Yismaw G, Kassu A, et al. Seroprevalence of HIV, HBV, HCV and syphilis infections among blood donors at Gondar University Teaching Hospital, Northwest Ethiopia: declining trends over a period of five years. BMC Infect Dis. England 2010;10:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paty MC. [The expansion of vector‐borne diseases and the implications for blood transfusion safety: the case of West Nile virus, dengue and chikungunya]. Transfus Clin Biol 2013;20:165‐73. [DOI] [PubMed] [Google Scholar]
- 4. Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012;367:1814‐20. [DOI] [PubMed] [Google Scholar]
- 5.Middle East respiratory syndrome coronavirus (MERS‐CoV) [Internet]. Geneva: World Health Organization; 2017 [cited 2017 Feb 24]. Available from: http://www.who.int/emergencies/mers-cov/en/.
- 6.Statistics [Internet]. Riyadh: Saudi Ministry of Health, Command & Control Center (CCC) . 2014 [cited 2017 Feb 24]. Available from: http://www.moh.gov.sa/en/CCC/PressReleases/Pages/default.aspx.
- 7. Mackay IM, Arden KE. Middle East respiratory syndrome: an emerging coronavirus infection tracked by the crowd. Virus Res 2015;202:60‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Almekhlafi GA, Albarrak MM, Mandourah Y, et al. Presentation and outcome of Middle East respiratory syndrome in Saudi intensive care unit patients. Crit Care 2016;20:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arabi YM, Balkhy HH, Hayden FG, et al. Middle East respiratory syndrome. N Engl J Med 2017;376:584‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shehata MM, Gomaa MR, Ali MA, et al. Middle East respiratory syndrome coronavirus: a comprehensive review. Front Med 2016;10:120‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lessler J, Salje H, Van Kerkhove MD, et al. Estimating the severity and subclinical burden of Middle East respiratory syndrome coronavirus infection in the Kingdom of Saudi Arabia. Am J Epidemiol 2016;183:657‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Azhar EI, El‐Kafrawy SA, Farraj SA, et al. Evidence for camel‐to‐human transmission of MERS coronavirus. N Engl J Med 2014;370:2499‐505. [DOI] [PubMed] [Google Scholar]
- 13. Müller MA, Corman VM, Jores J, et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983‐1997. Emerg Infect Dis 2014;20:2093‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hemida MG, Perera RA, Al Jassim RA, et al. Seroepidemiology of Middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Euro Surveill 2014;19. pii: 20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alagaili AN, Briese T, Mishra N, et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio 2014;5:e00884‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sabir JS, Lam TT, Ahmed MM, et al. Co‐circulation of three camel coronavirus species and recombination of MERS‐CoVs in Saudi Arabia. Science 2016;351:81‐4. [DOI] [PubMed] [Google Scholar]
- 17. Alraddadi BM, Watson JT, Almarashi A, et al. Risk factors for primary Middle East respiratory syndrome coronavirus illness in humans, Saudi Arabia, 2014. Emerg Infect Dis 2016;22:49‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Memish ZA, Assiri AM, Al‐Tawfiq JA. Middle East respiratory syndrome coronavirus (MERS‐CoV) viral shedding in the respiratory tract: an observational analysis with infection control implications. Int J Infect Dis 2014;29:307‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shalhoub S, Farahat F, Al‐Jiffri A, et al. IFN‐α2a or IFN‐β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother 2015;70:2129‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corman VM, Albarrak AM, Omrani AS, et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis 2016;62:477‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim SY, Park SJ, Cho SY, et al. Viral RNA in blood as indicator of severe outcome in Middle East respiratory syndrome coronavirus infection. Emerg Infect Dis 2016;22:1813‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Min CK, Cheon S, Ha NY, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep 2016;6:25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967‐76. [DOI] [PubMed] [Google Scholar]
- 24. Berger A, Drosten C, Doerr HW, et al. Severe acute respiratory syndrome (SARS)—paradigm of an emerging viral infection. J Clin Virol 2004;29:13‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stramer SL. Current perspectives in transfusion‐transmitted infectious diseases: emerging and re‐emerging infections. ISBT Sci Ser 2014;9:30‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Irsch J, Lin L. Pathogen inactivation of platelet and plasma blood components for transfusion using the INTERCEPT Blood System™. Transfus Med Hemother 2011;38:19‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pinna D, Sampson‐Johannes A, Clementi M, et al. Amotosalen photochemical inactivation of severe acute respiratory syndrome coronavirus in human platelet concentrates. Transfus Med 2005;15:269‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Musso D, Richard V, Broult J, et al. Inactivation of dengue virus in plasma with amotosalen and ultraviolet A illumination. Transfusion 2014;54:2924‐30. [DOI] [PubMed] [Google Scholar]
- 29. Aubry M, Richard V, Green J, et al. Inactivation of Zika virus in plasma with amotosalen and ultraviolet A illumination. Transfusion 2016;56:33‐40. [DOI] [PubMed] [Google Scholar]
- 30. Schlenke P. Pathogen inactivation technologies for cellular blood components: an update. Transfus Med Hemother 2014;41:309‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mintz PD, Neff A, MacKenzie M, et al. A randomized, controlled Phase III trial of therapeutic plasma exchange with fresh‐frozen plasma (FFP) prepared with amotosalen and ultraviolet A light compared to untreated FFP in thrombotic thrombocytopenic purpura. Transfusion 2006;46:1693‐704. [DOI] [PubMed] [Google Scholar]
- 32. Cazenave JP, Waller C, Kientz D, et al. An active hemovigilance program characterizing the safety profile of 7483 transfusions with plasma components prepared with amotosalen and UVA photochemical treatment. Transfusion 2010;50:1210‐9. [DOI] [PubMed] [Google Scholar]
- 33. Bost V, Chavarin P, Boussoulade F, et al. Independent evaluation of tolerance of therapeutic plasma inactivated by amotosalen‐HCl‐UVA (Intercept ™) over a 5‐year period of extensive delivery. Vox Sang 2015;109:414‐6. [DOI] [PubMed] [Google Scholar]
- 34. Al‐amri SS, Abbas AT, Siddiq LA, et al. Immunogenicity of candidate MERS‐CoV DNA vaccines based on the spike protein. Sci Rep 2017;7:44875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Coleman CM, Frieman MB. Growth and quantification of MERS‐CoV infection. Curr Protoc Microbiol 2015;37:15E.2.1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou J, Chu H, Li C, et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis 2014;209:1331‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Corman VM, Eckerle I, Bleicker T, et al. Detection of a novel human coronavirus by real‐time reverse‐transcription polymerase chain reaction. Euro Surveill 2012;17. pii: 20285. [DOI] [PubMed] [Google Scholar]
- 38. Corman VM, Müller MA, Costabel U, et al. Assays for laboratory confirmation of novel human coronavirus (hCoV‐EMC) infections. Euro Surveill 2012;17. pii: 20334. [DOI] [PubMed] [Google Scholar]
- 39. Centers for Disease Control and Prevention (CDC) . HIV transmission through transfusion—Missouri and Colorado, 2008. MMWR Morb Mortal Wkly 2010;59:1335‐9. [PubMed] [Google Scholar]
- 40. Bloch EM, Vermeulen M, Murphy E. Blood transfusion safety in Africa: a literature review of infectious disease and organizational challenges. Transfus Med Rev 2012;26:164‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woolhouse ME, Haydon DT, Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol 2005;20:238‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Motta IJ, Spencer BR, Cordeiro da Silva SG, et al. Evidence for transmission of Zika virus by platelet transfusion. N Engl J Med 2016;375:1101‐3. [DOI] [PubMed] [Google Scholar]
- 43. European Directorate for the Quality of Medicines and Healthcare . Guide to the preparation, use, and quality assurance of blood components. 17th ed. Strasbourg: Council of Europe; 2013.
- 44. Chu H, Zhou J, Wong BH, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte‐derived dendritic cells modulates innate immune response. Virology 2014;454‐455:197‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chu H, Zhou J, Wong BH, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis 2016;213:904‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moustafa A, Xie C, Kirkness E, et al. The blood DNA virome in 8,000 humans. PLoS Pathog 2017;13:e1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keil SD, Bowen R, Marschner S. Inactivation of Middle East respiratory syndrome coronavirus (MERS‐CoV) in plasma products using a riboflavin‐based and ultraviolet light‐based photochemical treatment. Transfusion 2016;56:2948‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]


