Summary
Hepatitis E is an acute human liver disease in healthy individuals which may eventually become chronic. It is caused by the hepatitis E virus (HEV) and can have a zoonotic origin. Nearly 57,000 people die yearly from hepatitis E‐related conditions. The disease is endemic in both developing and developed countries with distinct epidemiologic profiles. In developing countries, the disease is associated with inadequate water treatment, while in developed countries, transmission is associated with animal contact and the ingestion of raw or uncooked meat, especially liver. All human HEV are grouped into at least four genotypes, while HEV or HEV‐related viruses have been identified in an increasing number of domestic and wild animal species. Despite a high genetic diversity, only one single HEV serotype has been described to date for HEV genotypes 1–4. The discovery of new HEV or HEV‐related viruses leads to a continuing increase in the number of genotypes. In addition, the genome organization of all these viruses is variable with overlapping open reading frames (ORF) and differences in the location of ORF3. In spite of the role of some domestic and wild animals as reservoir, the origin of HEV and HEV‐related viruses in humans and animals is still unclear. This review discusses aspects of the detection, molecular virology, zoonotic transmission and origin of HEV and HEV‐related viruses in the context of ‘One Health’ and establishes a link between the previous and the new taxonomy of this growing virus family.
Keywords: hepatitis E virus, new taxonomy, infected species, zoonosis, origin
Introduction
Hepatitis E is an acute human liver disease caused by the hepatitis E virus (HEV) and can have a zoonotic origin. Hepatitis E is usually a mild self‐limiting disease; however, it may be fatal, especially among pregnant women in developing countries, or become chronic in immunocompromised individuals. Besides the infection of humans, HEV or HEV‐related viruses have been identified in an increasing number of domestic and wild animal species (Smith et al., 2013). Nearly 57,000 people die yearly from hepatitis E‐related causes (WHO, 2013). According to the World Health Organization (WHO), 20 million people get infected with HEV and three million develop acute hepatitis every year (WHO, 2013). The disease is endemic in both developing and developed countries although the epidemiological profile varies between countries according to the level of development. In developed countries, hepatitis E is more associated with animal contact and the ingestion of raw or uncooked meat, especially liver (swine, wild boar and deer) (Wichmann et al., 2008; Lewis et al., 2010). In developing countries, hepatitis E is still linked to poor sanitary conditions ((CDC), C. f. D. C. a. P., 2013).
The first epidemiological study of hepatitis E came from India in the early fifties (Viswanathan, 1957) and described a waterborne infection due to sewage contamination of the Yamuna River (Viswanathan, 1957; Khuroo, 2011). The unknown viral agent was only identified in the eighties and named ‘enterically transmitted non‐A and non‐B hepatitis’ (ET‐NANBH; Francis and Maynard, 1979; Sreenivasan et al., 1984). Later in the nineties, the ET‐NANBH virus was inoculated into cynomolgus monkeys, HEV cDNA was isolated for the first time, and the name ‘HEV’ was proposed (Reyes et al., 1990). Since then the number of reports on HEV infections in the human population has increased considerably, showing that HEV was present in many different countries (Aye et al., 1992; Huang et al., 1992; Tsarev et al., 1992; Yin et al., 1994).
Natural infection of HEV was detected in pigs for the first time in 1995 in Nepal (Clayson et al., 1995). Further studies showed that the porcine HEV strain is closely related to but distinct from the human HEV strains observed so far and represents the HEV genotype 3 (HEV‐3) (Meng et al., 1997). Later, genotype 4 (HEV‐4) was also found in humans and swine (Wang et al., 1999, 2002).
Current Nomenclature of HEV
Hepatitis E virus was initially classified in the Picornaviridae family with hepatitis A virus, based on clinical and epidemiological characteristics (Sreenivasan et al., 1984). However, due to morphological features and similarities to noroviruses in genome organization, HEV was thereafter repositioned as a member of the Caliciviridae, in the genus Hepevirus (Bradley et al., 1988). Later on, based on molecular analyses, HEV was placed as the sole species of the family Hepeviridae, genus Hepevirus and viruses were grouped into four major genotypes (1–4) (Emerson et al., 2005; Meng et al., 2011). In the meantime, other HEV‐related viruses were detected (e.g. in rats), but not assigned until the most recent proposed classification (Table 1; Meng et al., 2011; Smith et al., 2014).
Table 1.
Comparison between previous and current Hepeviridae taxonomy
| Previous taxonomy (ninth ICTV report) | New taxonomy (Smith et al., 2014) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Species | Genus | Family | Predominant host species | Reference strain | GenBank accession | Family | Genus | Species | Genotype |
| 1 | HEV‐1 | Hepevirus | Hepeviridae | Human | Burma | M73218 | Hepeviridae | Orthohepevirus | Orthohepevirus A | HEV‐1 |
| 2 | HEV‐2 | Human | Mexico | M74506 | HEV‐2 | |||||
| 3 | HEV‐3 | Human, suids, cervids, mongoose, rabbit | Meng | AF082843 | HEV‐3 | |||||
| 4 | HEV‐4 | Human, suids | T1 | AJ272108 | HEV‐4 | |||||
| Unclassified | Wild boar | JBOAR135‐Shiz09 | AB573435 | HEV‐5 | ||||||
| Wild boar | wbJOY_06 | AB602441 | HEV‐6 | |||||||
| Camel | DcHEV‐178C | KJ496143 | HEV‐7 | |||||||
| Avian HEV‐1 | / | Hepeviridae | Chicken | F93‐5077 | AM943647 | Orthohepevirus B | ||||
| Avian HEV‐2 | Chicken | AY535004 | ||||||||
| Avian HEV‐3 | Chicken | AM943646 | ||||||||
| / | Hepevirus | Rat | R63 | GU345042 | Orthohepevirus C | HEV‐C1 | ||||
| Unclassified | Ferret | FRHEV4 | JN998606 | HEV‐C2 | ||||||
| Bat | BatHEV/BS 7/GE/2009 | JQ001749 | Orthohepevirus D | |||||||
| Trout | Heenan lake | HQ731075 | Piscihepevirus | Piscihepevirus A | ||||||
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
HEV genotypes
Recently, the ICTV Hepeviridae Study Group proposed a new consensual classification, placing all the HEV and HEV‐related viruses into two genera: Orthohepevirus with four species (A–D) and Piscihepevirus with a single species (A) (Smith et al., 2014). The criteria proposed for this new classification were based on phylogeny and host range. Accordingly, Orthohepevirus A comprises sequences found in humans, pigs, wild boar, rabbit, deer, mongoose and camel; Orthohepevirus B contains viruses found in chickens; Orthohepevirus C encompasses sequences found in rats (HEV‐C1) and ferrets (HEV‐C2); and Orthohepevirus D, the bat virus (Table 1). Genotypes were also proposed for Orthohepevirus A: genotypes HEV‐1 and HEV‐2 have only been identified in humans, genotypes HEV‐3 and HEV‐4 have been reported in both humans and different animal species and are associated with the zoonotic cases. Genotypes HEV‐5 and HEV‐6 have been found in wild boar in Japan and genotype HEV‐7 in dromedary camels in Dubai (Table 1; Smith et al., 2014; Pavio et al., 2010).
Partial sequences from other potential members of the family were recently identified in moose, fox and mink. They show similarities to HEV in genome organization, size and phylogenetic analyses, suggesting they could represent new members of the Hepeviridae family. The moose virus appears to cluster closely to genotypes 1–7, while HEV isolated from mink is closely related to the ferret virus (HEV‐C2), and the virus found in foxes is related to that in rats (HEV‐C1) virus (Fig. 1; Lin et al., 2013; Woo et al., 2014; Batts et al., 2011; Takahashi et al., 2011; Raj et al., 2012; Drexler et al., 2012). These viruses have not been placed in any specific genotype because complete genomic sequences are required to place these viruses definitively.
Figure 1.
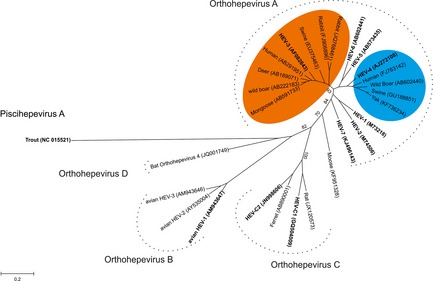
Phylogenetic tree based on nucleotide sequences of complete capsid protein from different hepatitis E virus (HEV) and HEV‐related viruses. Phylogenetic tree was inferred using the maximum likelihood method. A bootstrap analysis of 1000 replicates was included, and the results displayed on the interior branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA6. Sequences in bold represent reference sequence listed by Smith et al., 2014 (Table 1). Accession numbers are shown in brackets. Hepatitis E virus found in bovids and molluscs is not presented here. Genotypes HEV‐3 and HEV‐4 are, respectively, coloured in orange and blue. [Colour figure can be viewed at http://wileyonlinelibrary.com].
HEV subgenotypes
Classification under the genotype level is very useful and important for both clinical and epidemiological studies. The most frequently used classification was proposed by Lu et al. (2006) and encompasses 24 subtypes. Genotypes 1–4 are split into five (a–e), two (a–b), 10 (a–j) and seven (a–g) subtypes, respectively (Lu et al., 2006). This classification is controversial and is not accepted by all researchers in the field. For instance, there are a number of publications including partial and complete genomic sequences of HEV with no differentiation under the genotype level (Takahashi et al., 2003; Tei et al., 2003; Sonoda et al., 2004; Wibawa et al., 2004). The main concern is related to the reliability of the subtype separation and, consequently, the usefulness of subtyping (Oliveira‐Filho et al., 2013; Smith et al., 2013). The ICTV study group has not defined a permanent system for classification under genotype level (subgenotype or subtypes); however, it has recommended the use of approaches based on labelling clades according to tree topology (Smith et al., 2014). Thus, genotype HEV‐3 has been divided into three subgenotypes 3.1 (or G3 group 1), 3.2 (or G3 group 2) and 3.3 (Oliveira‐Filho et al., 2013; Ijaz et al., 2014), and genotype HEV‐4 has been divided into seven subgenotypes 4 a–g (Dai et al., 2013).
So far all the different viruses infecting humans from genotypes HEV‐1 to HEV‐4 are grouped in a single serotype. The serological diversity of the other hepeviruses remains uncertain, although it is known that specific assays can distinguish between viruses from Orthohepevirus B and HEV‐C1 and the HEV‐1–HEV‐4 (Dremsek et al., 2012; Liu et al., 2014).
Structure
Hepatitis E virus is a small non‐enveloped virus with a diameter of approximately 27–34 nm and an icosahedral capsid. The genome consists of a single‐stranded positive RNA of 6.6–7.3 kb in length, polyadenylated at its 3′‐end and 5′‐capped with a 7‐methylguanine. It contains three partly overlapping open reading frames (ORFs; Fig. 2) (Mushahwar, 2008). In addition, subgenomic viral RNA is also synthesized (Graff et al., 2006).
Figure 2.
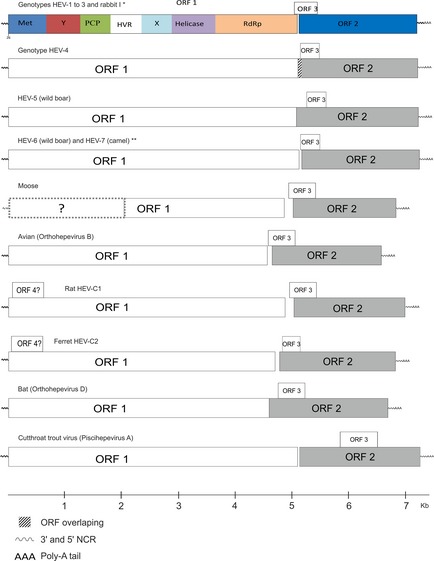
Genome organization of genotypes 1, 3 and 4 and hepatitis E virus (HEV)‐related viruses from wild boar, rat, bat, ferret, moose, avian and cutthroat trout. The complete genomic sequence from the moose virus was not available. The true frame usage of the open reading frames (ORFs) is not shown in this figure. Sequences used in this figure were obtained from GenBank with following accession numbers: HEV‐1 (M73218), HEV‐2 (M74506), HEV‐3 (AF082843) and rabbit (FJ906895); HEV‐4 (AJ272108); avian (AM943647); rat (GQ504009); bat (NC_018382); CTV (NC_015521); ferret (AB890001); moose (KF951328); wild boar HEV‐5 (AB602441), camel (KJ496143) and wild boar HEV‐6 (AB573435). ORF1 encodes methyltransferase (Met), Y, papain‐like cysteine protease (PCP), hypervariable region (HVR), X (macro domain), helicase and RNA‐dependent RNA polymerase (RdRp) domains.* based on AF082843 ** based on AB602441. [Colour figure can be viewed at http://wileyonlinelibrary.com].
The 5′ end of the genome contains a short non‐coding region (NCR), 26–28 nucleotides in length. ORF1 has a size of approximately 1693 amino acids (aa). This region encodes a polyprotein which is probably cleaved into the viral non‐structural proteins including methyltransferase, papain‐like cysteine protease, macro domain, helicase and RNA‐dependent RNA polymerase (RdRp); these enzymes are involved in viral replication, transcription and polyprotein cleavage (Reyes et al., 1990; Kaur et al., 1992; Koonin et al., 1992; Holla et al., 2013).
Open reading frames 2 encodes the structural capsid protein and has a size of approximately 660 aa for genotypes 1–3 and 675 aa for members of genotype 4. This protein is highly immunogenic and is responsible for various functions such as assembly and host interaction. Due to the high aa homogeneity of this region and the presence of only one reported HEV serotype, it is used both for diagnostic tests and vaccine development (Tsarev et al., 1997; Zhang et al., 2001; Engle et al., 2002; Koff, 2007; Panda et al., 2007).
Open reading frames 3 encodes a small phosphorylated protein with a size of approximately 120 aa which binds to the hepatocellular cytoskeleton to form a complex together with the capsid protein. Other possible ORF3 functions may be related to the regulation of cell signalling pathway and infectivity in vivo (Graff et al., 2005; Jiménez de Oya et al., 2007; Panda et al., 2007; Khuroo, 2008).
A fourth ORF (ORF4) was reported from HEV‐C2 (ferret) and two HEV‐C1 (rat) sequences (Johne et al., 2012; Raj et al., 2012). In these Orthohepviruses C, ORF4 has 183 aa and overlaps with the 5′ end of ORF1. Its function is not yet known, but it has been located into the methyltransferase domain. Two additional ORFs were reported for one sequence found in rats, however, without the presence of a conventional start codon (Johne et al., 2012). A HEV‐related sequence found in moose has been partially sequenced. It contains a 3′ end with poly A tail, ORF2, ORF3 and partial ORF1 (Lin et al., 2013). More details regarding the genome organization of the different HEV and HEV‐related viruses are available in Fig. 2.
Direct and Indirect Detection of HEV‐Related Viruses in Animals
Hepatitis E virus and HEV‐related strains have been genetically detected in naturally infected domestic and wild animals (Fig. 3). In addition, antibodies against HEV have also been detected in many animal species such as dog, cat, sheep, goat, horse, rodents, cattle, duck, pigeon and non‐human primates (Arankalle et al., 2001; Vitral et al., 2005; Mochizuki et al., 2006; Zhang et al., 2008a; Pavio et al., 2010). The antibody presence in these species raises a puzzling question and suggests that these animals may have been exposed to the HEV or to an antigenically related agent.
Figure 3.
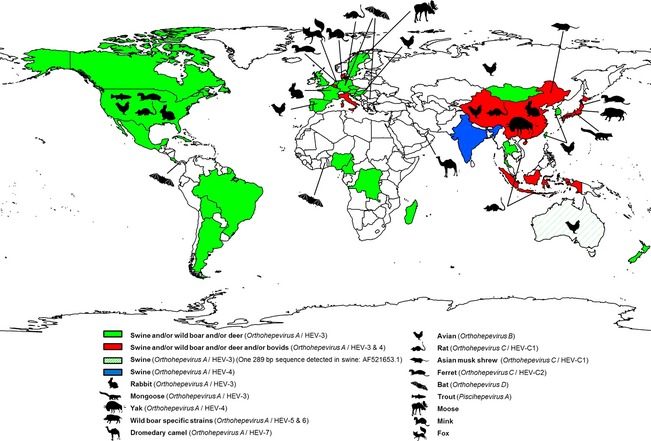
Geographical distribution of animal hepatitis E virus (HEV) and HEV‐related viruses (without non‐human primates). References used in this figure are shown in Table S1. [Colour figure can be viewed at http://wileyonlinelibrary.com].
HEV in Mammals
HEV infection of non‐human primates
The first study reporting the infection of non‐human primates with genotype 1 HEV (previously called ET‐NANBH) showed that the cynomolgus monkey was susceptible to infection and suitable for use in experimental infection studies (Bradley et al., 1987). Several other species such as tamarins, rhesus monkey, three‐striped night monkey and the common chimpanzee were also successfully infected with HEV (Bradley et al., 1987; Gupta et al., 1990; Ticehurst et al., 1992; McCaustland et al., 2000). Recently, it was demonstrated that the cynomolgus monkey is susceptible to infection with human and swine genotype 3 HEV from Argentina, Brazil and the Netherlands (de Carvalho et al., 2013). These studies were very important in understanding different aspects of HEV infection in humans. Naturally occurring anti‐HEV antibodies were reported in both free‐living and captive non‐human primates, and their role as reservoir has been discussed (Arankalle et al., 1994; Hirano et al., 2003b). However, neither HEV nor HEV‐related viruses have been found naturally in non‐human primates (Meng, 2011).
HEV infection of Suidae
Since the first sequence was reported in swine in 1997 in the USA (Meng et al., 1997), HEV genotypes 3 and 4 have been detected and seem to be enzootic in domestic pigs across the five continents (Fig. 3).
In Japan, HEV was detected in wild boar for the first time in 2004 (Sonoda et al., 2004). Subsequently, it has been detected in several countries (Pavio et al., 2010). The majority of HEV strains detected from wild boar belong to genotype 3 (Meng, 2010b). In contrast to domestic swine, high detection rates of HEV RNA have been reported not only in young animals but also in adult wild boar (de Deus et al., 2008; Martelli et al., 2008; Oliveira‐Filho et al., 2014). In addition, it seems that viral genetic heterogeneity is higher in wild boar populations. Different subtypes have been reported within the same populations from Germany and Sweden (Adlhoch et al., 2009; Widén et al., 2011; Oliveira‐Filho et al., 2014). In Japan, sequences from both genotypes 3 and 4 have been detected in wild boar as well as sequences belonging to the genotypes HEV‐5 and HEV‐6 (Sato et al., 2011; Takahashi et al., 2011; Smith et al., 2014).
HEV infection of Cervidae
Molecular and serological evidence of HEV and HEV‐related viruses in cervids has been reported in Sika deer, red deer, tufted deer, Reeve's muntjac, roe deer and moose (Tei et al., 2003; Matsuura et al., 2007; Yu et al., 2007; Zhang et al., 2008b; Reuter et al., 2009; Boadella et al., 2010; Forgách et al., 2010; Rutjes et al., 2010; Lin et al., 2013). The first report of HEV‐related sequences in deer demonstrated a close phylogenetic relationship with genotype 3. In contrast to the other viruses found in cervids, the HEV‐related virus detected in Swedish moose does not cluster within genotypes HEV‐1 to HEV‐7. Phylogenetic analysis based on the complete capsid protein indicates that the virus found in moose clusters in a separate branch (Fig. 1).
HEV infection of Bovidae
Antibodies against HEV have been detected in cattle in several studies (Arankalle et al., 2001; Wang et al., 2002; Vitral et al., 2005; Zhang et al., 2008a). However, only one study from China has genetic confirmation, obtained through the amplification of a 189 bp sequence of ORF2 from eight cow faeces. These sequences showed homology of 96–100% to each other and 76–86%, 82–84%, 79–85% and 84–96% homology with genotypes 1–4, respectively. Accordingly, the HEV sequences found in bovines were assigned to genotype 4 (Hu and Ma, 2010). The same short HEV ORF2 sequence of 189 bp was amplified from six faecal samples of sheep by the same research group in China. These sequences showed homology of 99–100% between themselves and 79–85%, 81–83%, 79–84% and 85–95% homology with genotypes 1, 2, 3 and 4, respectively, and clustered into genotype 4 (Wang and Ma, 2010). However, additional studies must be conducted to confirm the presence of viruses in these animal species, and longer sequences need to be isolated to confirm the genotype assignment. Recently, full genome sequences closely related to HEV‐4 were detected in yak in China (Fig. 1; Xu et al., 2014).
HEV infection of Camelidae
Recently, HEV‐related virus was recovered in the faeces of three dromedary camels in Dubai. The isolates were called DcHEV (178 and 180C) and show more than 20% nucleotide divergence from the other HEVs (Woo et al., 2014). These viruses are most similar to viruses in the genus, Orthohepevirus A, and have been placed within this genus as genotype HEV‐7 (Smith et al., 2014).
HEV infection of Leporidae
In 2009, HEV‐related sequences were found in Chinese farmed rabbits. Phylogenetic analysis based on the complete genome sequence suggests that they were genetically closely related to human and porcine genotype HEV‐3. The sequences of rabbit HEV have about 85% nucleotide homology between themselves and 74%, 73%, 78–79%, 74–75% and 46–47% homology with genotypes 1–4 of mammals and avian HEV, respectively. Later on, more sequences were reported in China, the USA and France, including closely related sequences found in humans (Cossaboom et al., 2011, 2012; Izopet et al., 2012; Han et al., 2014).
The sequences found in rabbits clustered in a new distinct branch related to genotype 3 and were initially suggested to form another genotype (Zhao et al., 2009; Geng et al., 2011; Izopet et al., 2012). Further studies have shown that the HEV strains found in rabbits cluster at the edge of genotype 3 sequence space and could indeed represent a new genotype or a new genotype HEV‐3 clade (Oliveira‐Filho et al., 2013; Smith et al., 2013). According to the ICTV Hepevirus study group, the HEV strains found in rabbits are considered a distant variant in genotype HEV‐3 (Smith et al., 2014).
HEV infection of Muridae
The presence of antibodies against HEV has been reported in different rodent species (Kabrane‐Lazizi et al., 1999; Favorov et al., 2000; Arankalle et al., 2001; Hirano et al., 2003a; Easterbrook et al., 2007). Hepatitis E virus antigens were detected in different organs after experimental infection of Wistar rats with human virus (genotype 1) (Maneerat et al., 1996). Nevertheless, none of the above‐mentioned studies succeeded in recovering viral genome.
In 2008, fragments of HEV‐related viruses were sequenced from faeces of Rattus norvegicus from Germany. The HEV‐related sequences detected in rats have a sequence homology of about 60% and 50% with human and avian HEV strains, respectively. Subsequently, the complete genomes of the two HEV strains from rats were determined and it was suggested that they might belong to a new species within Hepeviridae (Johne et al., 2010). Since then, new HEV‐C1 sequences have been reported in Germany, USA, China, Vietnam and Indonesia (Johne et al., 2012; Li et al., 2013b,2013d) and specific HEV‐C1 sequences were included in the species Orthohepevirus C genotype HEV‐C1 (Smith et al., 2014). Recently, several liver tissue samples from Rattus rattus and R. norvegicus in museum collections were tested and found to contain HEV genotype 3 (Lack et al., 2012).
HEV infection of Soricidae
Sequences closely related to HEV‐C1 were detected from Asian musk shrews which shared the environment with wild rats in China (Guan et al., 2013).
HEV infection of Mustelidae
The first HEV‐related virus found in Mustelidae was reported in the Netherlands in 2012. Two years later, HEV was detected in a ferret breeding colony in the USA and in Japan (Li et al., 2014). Phylogenetic analysis based on complete genomic sequences showed that HEV‐related sequences found in ferrets were separate from genotypes 1 to 4 and clustered in a separate branch near to sequences found in rats (HEV‐C1) (Raj et al., 2012; Li et al., 2014). Recently, partial genomic sequences of HEV‐related viruses, genetically close to viruses found in ferrets (HEV‐C2), were detected in 4 farmed mink from Denmark in 2013 (Krog et al., 2013).
HEV infection of Herpestidae
The only HEV‐related virus in Herpestidae was detected in 2006, in a mongoose from Japan (Nakamura et al., 2006). The whole genome sequence of this strain was determined: it was HEV genotype 3 closely related to a Japanese porcine HEV sequence.
HEV infection of Canidae
In 2013, fragments of HEV‐related virus were sequenced from faeces of two foxes in the Netherlands. These fragments were related to HEV‐C1, but it is unknown, whether this virus circulates naturally in foxes or if detection is due to the consumption of prey such as rats (Bodewes et al., 2013).
HEV infection of Chiroptera
In 2012, a study analysed 3869 bat faecal and blood samples across five continents to detect HEV‐related sequences. This study included 85 species of bats. Viruses were detected in 3 bat families (Hipposideridae, Vespertilionidae and Phyllostomidae) from Africa, Central America and Europe. There was a high genetic diversity between HEV‐related viruses found in bats, comparable to that observed between sequences found in Orthohepevirus A (Drexler et al., 2012). Currently, all the HEV sequences found in bats have been assigned to the new species Orthohepevirus D (Table 1; Smith et al., 2014).
HEV Infection of Birds
In 1999, a virus related to HEV associated to big liver and spleen disease (BLS) was identified in chickens from Australia. This virus has about 62% homology with HEV nucleotide sequences from genotypes 1–4 (Payne et al., 1999). In 2001, another virus was detected from bile samples of White Leghorn chickens from the USA with hepato‐splenomegaly (HS) syndrome (Haqshenas et al., 2001). Later, the virus was designated avian HEV, and the viruses causing both BLS and HS were found to be distinct variant strains of the same virus (Meng, 2010a). Since then, they have been found to be circulating among North American, European and Asian chicken flocks (Huang et al., 2002; Sun et al., 2004a; Peralta et al., 2009; Kwon et al., 2012; Zhao et al., 2013; Hsu and Tsai, 2014). Currently, all strains found in chicken are considered as belonging to the same specie Orthohepevirus B.
Contrary to other animal species where HEV infections are usually asymptomatic, Orthohepeviruse B infection is associated with symptomatic disease. Besides the enlargement of spleen and liver, both ovarian regression and presence of red fluid in the abdomen are commonly associated with the HS syndrome (Payne et al., 1999; Haqshenas et al., 2001). Experimental infection studies have also described several other clinical signs such as anorexia and diarrhoea (Billam et al., 2005). In addition, it was suggested that heterogeneity into the distinct strains may be related to different pathogeneses (Billam et al., 2009).
Orthohepevirus B shares several antigenic epitopes with HEV found in humans and swine, although at least one specific epitope has been reported among the viruses found in birds (Haqshenas et al., 2002; Guo et al., 2006). Further experimental infections have demonstrated that the Orthohepevirus B can infect and produce low viremia in turkeys, but is not infectious for rhesus monkeys (Huang et al., 2004; Sun et al., 2004b).
HEV infection of Fish
In 1988, a new virus, infecting trout without causing disease, was isolated and named cutthroat trout virus (CTV; Hedrick et al., 1991). After two decades, the complete genome of this virus was obtained and the organization was shown to be similar to HEV. Phylogenetic analysis based on partial RdRp aa sequences indicates proximity to the Hepeviridae family (Batts et al., 2011), and currently, it has been assigned as the sole species into the genus Piscihepevirus (Smith et al., 2014).
HEV infection of Bivalve Molluscs
The presence of HEV RNA was reported on several occasions in bivalve molluscs (Li et al., 2007; Crossan et al., 2012; Donia et al., 2012). However, this is most likely due to passive filtration of HEV from contaminated water rather than to HEV replication occurring in molluscs. Indeed, experimental bio accumulation of HEV has been demonstrated in oysters (Grodzki et al., 2014).
Zoonotic Concerns of HEV and HEV‐Related Viruses
Before the first detection of HEV in swine, the occurrence of hepatitis E was associated with lack of access to clean water and contamination of drinking water due to unreliable sewage systems and hepatitis E was considered a disease of developing countries (Viswanathan, 1957; Aggarwal and Naik, 2009). Due to this, from the late eighties to the early nineties, it was improbable that clinicians would diagnose autochthonous hepatitis E in industrialized countries (Scharschmidt, 1995). However, the presence of HEV antibodies in healthy individuals and blood donors in Europe and North America could not always be associated with travel to an endemic region (Skaug et al., 1994). Thus, several autochthonous cases were reported in the USA, Europe, Australia and New Zealand (Mast et al., 1996).
Later on, the viruses were revealed to be present in domestic swine and wild boar populations in both developing and developed countries (Meng et al., 1997; Meng, 2010a; Thiry et al., 2014). The genetic proximity raised the possibility of an animal reservoir and phylogenetic analysis clearly revealed that the autochthonous cases occurred more frequently than had been previously recognized in developed countries (Clemente‐Casares et al., 2003).
Nowadays, viruses from the Hepeviridae family and HEV‐related viruses are recognized to be present in mammals, avian and fish. Evidence suggesting zoonotic infections has been reported from strains circulating in domestic swine, wild boars, deer and rabbits (Meng et al., 1997; Hsieh et al., 1999; Tei et al., 2003; Izopet et al., 2012). To date, limited information is available concerning the ability of these viruses to infect different species.
Strong Evidence of Foodborne Transmission of HEV
In 2003, zoonotic transmission was reported for the first time. Accordingly, four human cases were linked to the consumption of uncooked wild boar liver and Sika deer meat in Japan. Confirmation was only possible from the deer meat, as no wild boar liver remained to be tested (Matsuda et al., 2003; Tei et al., 2003). A nucleotide homology of 99.7–100% was observed between fragments of 326 nucleotides obtained from the patients who consumed raw deer sushi or sashimi and from the frozen deer meat used to prepare these meals (Tei et al., 2003).
In 2004, another study showed 99.7% homology between the full viral genome sequences found in wild boar and deer hunted in the same forest as the Sika deer consumed by the four patients above (Takahashi et al., 2004). Another report from Japan in 2005 demonstrated transmission via ingestion of wild boar meat. Phylogenetic analysis based on the complete capsid sequences showed 99.95% homology between HEV genotype 3 sequences obtained from the patient serum and the frozen wild boar meat consumed by them (Li et al., 2005).
Recently, high homology between sequences detected in consumed pork meat and an acute hepatitis E patient was observed in Spain (Riveiro‐Barciela et al., 2015). Another strong evidence of HEV foodborne transmission was reported in a patient who consumed figatelli (raw pig liver sausage) in France (Renou et al., 2014).
HEV Transmission through Direct Contact or Indirect Evidence
Epidemiological investigations or clinical evidence has pointed out the consumption of wild boar or domestic pig meat as sources of several human HEV infections with genotypes HEV‐3 and HEV‐4 (Matsuda et al., 2003; Tamada et al., 2004; Wichmann et al., 2008). In addition, a number of studies have reported the presence of HEV in pig and wild boar meat products with sequences either similar or closely related to the ones found in humans (Feagins et al., 2007; Colson et al., 2010, 2012; Wenzel et al., 2011; Moal et al., 2012).
Geographically, human–swine co‐exposure could be related to a higher incidence in humans (Bouquet et al., 2011). Alternatively, if the area with a high incidence of human cases is far from regions with high pig density, eating habits could be a likely explanation (Matsuda et al., 2003; Wibawa et al., 2004; Colson et al., 2010; Bouquet et al., 2011).
Compared to unexposed groups, the HEV antibody prevalence is higher in diverse population groups with higher occupational exposure to HEV. These groups include swine workers (butchers, farmers) and veterinarians. Regardless of exposure, no increase of associated hepatic disease was observed in these populations (Galiana et al., 2008; Chang et al., 2009; Pourpongporn et al., 2009; Krumbholz et al., 2012, 2014). HEV was detected in both the bile of slaughtered pigs and effluent from the slaughterhouse, indicating that pigs can shed HEV at the time of slaughter (Casas et al., 2011; dos Santos et al., 2011). Indeed, a hepatitis E case has already been reported in a slaughterhouse worker (Pérez‐Gracia et al., 2007).
Role of Animals as HEV Reservoirs
The phylogenetic relationship of human zoonotic genotype HEV sequences with those of swine and wild boar (HEV‐3 and HEV‐4) suggests their involvement in the transmission cycle of HEV infection in humans. Rabbits may also represent a reservoir of HEV for humans (Izopet et al., 2012). In addition, cynomolgus macaques and pigs have been experimentally infected with rabbit HEV (Cossaboom et al., 2012; Liu et al., 2013). It is important to note that human HEV does not formally need a reservoir as faecal–oral transmission is effective in maintaining the infection within a human population, as occurs in non‐industrialized regions. However, secondary reservoirs in pigs and wild boar contribute to the maintenance of the infection (HEV genotypes 3 and 4) in developed countries with a high sanitary level. In addition, the presence of the virus in other wild species such as deer may contribute to the maintenance of the virus in the natural environment.
Rodents have long been believed to be a potential reservoir of human HEV. However, the susceptibility of rats to human HEV genotypes is very controversial. HEV‐C1 does not infect rhesus monkeys (Purcell et al., 2011), and laboratory rats do not appear to be susceptible to genotypes 1, 3 and 4 (Li et al., 2013a,c), although one study reported partial genotype 3 sequences in rat liver specimens (Lack et al., 2012). Viruses found in birds could not be transmitted experimentally to primates (Huang et al., 2004). The transmission of rat or avian HEV to humans cannot be completely ruled out, but these species are not currently considered to be reservoirs of human HEV. The possibility that bats may play a role as a reservoir of HEV in mammals, as reported for many viruses such as coronaviruses or paramyxoviruses, was examined. Phylogenetic analysis suggested that viruses found in bats belong to the Hepeviridae family and seem to be the most divergent mammalian hepeviruses described so far. Therefore, it is unlikely that bats transmit the virus to humans (Drexler et al., 2012). It is also unlikely that the transmission from trout to humans occurs due to the phylogenetic gaps found between HEV and CTV sequences. The human susceptibility for moose and dromedary camel HEV‐related viruses is still unknown and transmission to humans cannot be excluded.
Hypothesis on HEV Origin
The origin of HEV in animals and in humans is still unknown. The presence of animal reservoirs can be considered at two non‐mutually exclusive levels: animals as reservoirs for hepevirus ancestors of human HEV or current reservoirs interacting with human populations for zoonotic transmission.
The role played by animals in the emergence of HEV in humans is still uncertain. If the ancestor of genotypes 1–4 is hypothesized to be a human virus transmitted from humans to suids, animals would be the recent onset reservoirs with a role of maintenance of infection and reinfection of other mammals including humans. For instance, the presence of HEV more specific to wild boar (Sato et al., 2011; Takahashi et al., 2011) may be indicative of the existence of a specific viral evolution, reflecting the maintenance of viral infection in this animal population. The difference between HEV sequences from wild boars and domestic pigs might be related to a dissimilarity in the structure of the two animal populations that could be explained by a higher turnover in domestic pig populations than in wild boar, despite a possible conservation of viruses in reproductive pig herds in sows and boars. This might lead to a lower capacity of the virus to evolve in the domestic swine population. Instead, environmental and behavioural differences of the wild boar may allow a greater stabilization of the virus in this population and promote selection and maintenance of new viruses. A similar evolution may have occurred with the rabbit strain, starting from a related genotype 3 ancestor, but with a separate evolution in the rabbit population. The higher HEV prevalence in domestic swine than in wild animals could be explained by the continuous exchange of viruses between the human and the swine populations. In addition, the intensive contact among confined populations or farmed swine may increase the potential for infection.
The roots of sequences from genotypes HEV‐1 to HEV‐4 and HEV‐C1 rat were calculated (Fig. 4), and results suggested the ancestor to be of animal origin (Purdy and Khudyakov, 2010). This Bayesian analysis considered neither recombination events nor host species evolution. However, hosts of Orthohepevirus C variants have been detected from Carnivora, Rodentia and Soricomorpha, and Orthohepevirus A variants have been detected from Artiodactyla, Carnivora, Lagomorpha and Primates. Such diversification of host orders suggests that HEV did not co‐evolve with its hosts, but is more likely an opportunistic pathogen (Smith et al., 2014). The division that might have led to the ancestors of human and swine hepevirus occurred at about the same time. The appearance of the HEV‐C1 suggests that mammalian HEV could have been adapted to different mammalian species over time (Purdy and Khudyakov, 2010). There is evidence that genotype 1 or an anthropotropic ancestor of genotype 1 has been responsible for acute hepatitis infections among adults since the last decade of the 18th century (Teo, 2012). This indicates that either estimates for the time for the most recent common ancestor for genotype 1 are underestimated or genotype 1 has gone through an evolutionary bottleneck and the ancestor is a survivor of that bottleneck. The further discovery of other mammalian HEV‐related viruses may help to improve the understanding of Hepeviridae evolution. However, the lack of fossil viruses, preventing a confirmation of statistical analyses conducted so far (Purdy and Khudyakov, 2010), poses a challenge regarding the elucidation of the HEV (and HEV‐related viruses) origin. Therefore, Hepeviridae origin analysis will remain speculative but allows a clarification of the current situation.
Figure 4.
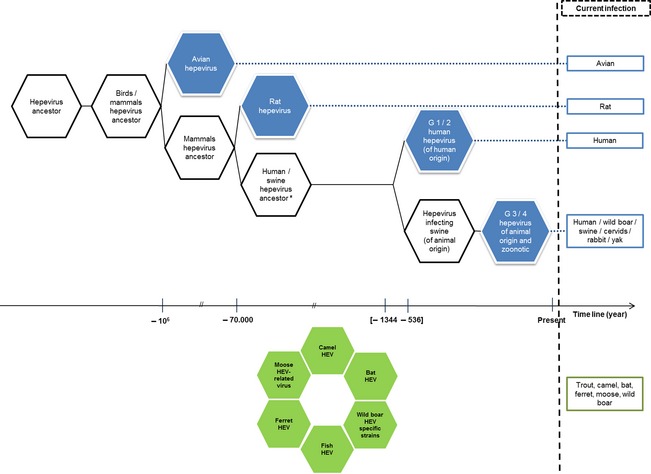
From hepevirus ancestors to current hepeviruses. In blue, viruses currently responsible for infections. In green, viruses currently responsible for infections, but not time classified. Based on data adapted from Purdy and Khudyakov (2010), Drexler et al. (2012) and Smith et al. (2013, 2014). This figure does not consider any recombinational event. *Infecting mammals of unknown species. [Colour figure can be viewed at http://wileyonlinelibrary.com].
Conclusion
Since the first characterized hepatitis E epidemic in 1955, several outbreaks have been described and the discovery of HEV‐related viruses has increased significantly during the last 15 years. Rather than representing a true emergence, this trend is probably due to the increasing interest in public health and food safety as well as the use of improved diagnostic tools. As their discovery, the taxonomy of the Hepeviridae family has been in perpetual evolution due to the introduction of newly identified viruses.
The high HEV prevalence in animals worldwide raises the question of transmission to humans. Indeed, studies have shown transmission of HEV via deer, swine and wild boar meat in Europe and Japan. High seroprevalence rates and similarities between the nucleotide sequences studied also raise the question of possible transmission between pigs and wild boar; particularly, when contact between these animals is possible, for example in the context of outdoor breeding pigs. Even if zoonotic transmission from different animal species is accepted in non‐endemic regions, strong molecular evidence is scarce and the human susceptibility for bat, rat, ferret, dromedary camel and moose HEV‐related viruses is still unknown.
Origins of the virus and pathways of spread through time are still largely unknown due, among other things, to the lack of fossil records. Research to date cannot show the exact roles of humans and animals in Hepeviridae evolutionary history.
Disclaimer
This information is distributed solely for the purpose of pre‐dissemination peer review under applicable information quality guidelines. It has not been formally disseminated by the Centers for Disease Control and Prevention/Agency for Toxic Substances and Disease Registry. It does not represent and should not be construed to represent any agency determination or policy. Use of trade names is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The authors declare that they have no conflict of interest.
Supporting information
[Correction added on 20 June 2015, after first online publication: Legend of Table S1 has been corrected.]
Table S1. Detection of HEV or HEV‐related viruses in animals.
Acknowledgements
We thank Annette Dougall and Louisa Ludwig for the careful English correction of the manuscript. D. Thiry is supported by the Belgium Federal Public Service, Health, Food Chain Safety and Environment and E.F. Oliveira‐Filho by CNPq (Brazilian National Council for Scientific and Technological Development).
References
- Adlhoch, C. , Wolf A., Meisel H., Kaiser M., Ellerbrok H., and Pauli G., 2009: High HEV presence in four different wild boar populations in East and West Germany. Vet. Microbiol. 139, 270–278. [DOI] [PubMed] [Google Scholar]
- Aggarwal, R. , and Naik S., 2009: Epidemiology of hepatitis E: current status. J. Gastroenterol. Hepatol. 24, 1484–1493. [DOI] [PubMed] [Google Scholar]
- Arankalle, V. A. , Goverdhan M. K., and Banerjee K., 1994: Antibodies against hepatitis E virus in old world monkeys. J. Viral Hepat. 1, 125–129. [DOI] [PubMed] [Google Scholar]
- Arankalle, V. A. , Joshi M. V., Kulkarni A. M., Gandhe S. S., Chobe L. P., Rautmare S. S., Mishra A. C., and Padbidri V. S., 2001: Prevalence of anti‐hepatitis E virus antibodies in different Indian animal species. J. Viral Hepat. 8, 223–227. [DOI] [PubMed] [Google Scholar]
- Aye, T. T. , Uchida T., Ma X. Z., Iida F., Shikata T., Zhuang H., and Win K. M., 1992: Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986–1988) of China. Nucleic Acids Res. 20, 3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batts, W. , Yun S., Hedrick R., and Winton J., 2011: A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res. 158, 116–123. [DOI] [PubMed] [Google Scholar]
- Billam, P. , Huang F. F., Sun Z. F., Pierson F. W., Duncan R. B., Elvinger F., Guenette D. K., Toth T. E., and Meng X. J., 2005: Systematic pathogenesis and replication of avian hepatitis E virus in specific‐pathogen‐free adult chickens. J. Virol. 79, 3429–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billam, P. , LeRoith T., Pudupakam R. S., Pierson F. W., Duncan R. B., and Meng X. J., 2009: Comparative pathogenesis in specific‐pathogen‐free chickens of two strains of avian hepatitis E virus recovered from a chicken with Hepatitis‐Splenomegaly syndrome and from a clinically healthy chicken. Vet. Microbiol. 139, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boadella, M. , Casas M., Martín M., Vicente J., Segalés J., de la Fuente J., and Gortázar C., 2010: Increasing contact with hepatitis E virus in red deer, Spain. Emerg. Infect. Dis. 16, 1994–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes, R. , van der Giessen J., Haagmans B. L., Osterhaus A. D., and Smits S. L., 2013: Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J. Virol. 87, 7758–7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquet, J. , Tessé S., Lunazzi A., Eloit M., Rose N., Nicand E., and Pavio N., 2011: Close similarity between sequences of hepatitis E virus recovered from humans and swine, France, 2008–2009. Emerg. Infect. Dis. 17, 2018–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, D. W. , Krawczynski K., Cook E. H., McCaustland K. A., Humphrey C. D., Spelbring J. E., Myint H., and Maynard J. E., 1987: Enterically transmitted non‐A, non‐B hepatitis: serial passage of disease in cynomolgus macaques and tamarins and recovery of disease‐associated 27‐ to 34‐nm viruslike particles. Proc. Natl Acad. Sci. U S A 84, 6277–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, D. , Andjaparidze A., Cook E. H., McCaustland K., Balayan M., Stetler H., Velazquez O., Robertson B., Humphrey C., and Kane M., 1988: Aetiological agent of enterically transmitted non‐A, non‐B hepatitis. J. Gen. Virol. 69, 731–738. [DOI] [PubMed] [Google Scholar]
- de Carvalho, L. G. , Marchevsky R. S., Dos Santos D. R., de Oliveira J. M., de Paula V. S., Lopes L. M., Van der Poel W. H., González J. E., Munné M. S., Moran J., Cajaraville A. C., Pelajo‐Machado M., Cruz O. G., and Pinto M. A., 2013: Infection by Brazilian and Dutch swine hepatitis E virus strains induces haematological changes in Macaca fascicularis. BMC Infect. Dis. 13, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas, M. , Cortés R., Pina S., Peralta B., Allepuz A., Cortey M., Casal J., and Martín M., 2011: Longitudinal study of hepatitis E virus infection in Spanish farrow‐to‐finish swine herds. Vet. Microbiol. 148, 27–34. [DOI] [PubMed] [Google Scholar]
- (CDC), C. f. D. C. a. P. , 2013: Investigation of hepatitis E outbreak among refugees – Upper Nile, South Sudan, 2012–2013. MMWR Morb. Mortal. Wkly Rep. 62, 581–586. [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. , Wang L., Geng J., Zhu Y., Fu H., Ren F., Li L., Wang X., and Zhuang H., 2009: Zoonotic risk of hepatitis E virus (HEV): a study of HEV infection in animals and humans in suburbs of Beijing. Hepatol. Res. 39, 1153–1158. [DOI] [PubMed] [Google Scholar]
- Clayson, E. T. , Innis B. L., Myint K. S., Narupiti S., Vaughn D. W., Giri S., Ranabhat P., and Shrestha M. P., 1995: Detection of hepatitis E virus infections among domestic swine in the Kathmandu Valley of Nepal. Am. J. Trop. Med. Hyg. 53, 228–232. [DOI] [PubMed] [Google Scholar]
- Clemente‐Casares, P. , Pina S., Buti M., Jardi R., MartIn M., Bofill‐Mas S., and Girones R., 2003: Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 9, 448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, P. , Borentain P., Queyriaux B., Kaba M., Moal V., Gallian P., Heyries L., Raoult D., and Gerolami R., 2010: Pig liver sausage as a source of hepatitis E virus transmission to humans. J. Infect. Dis. 202, 825–834. [DOI] [PubMed] [Google Scholar]
- Colson, P. , Romanet P., Moal V., Borentain P., Purgus R., Benezech A., Motte A., and Gérolami R., 2012: Autochthonous infections with hepatitis E virus genotype 4, France. Emerg. Infect. Dis. 18, 1361–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossaboom, C. M. , Córdoba L., Dryman B. A., and Meng X. J., 2011: Hepatitis E virus in rabbits, Virginia, USA. Emerg. Infect. Dis. 17, 2047–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossaboom, C. M. , Córdoba L., Sanford B. J., Piñeyro P., Kenney S. P., Dryman B. A., Wang Y., and Meng X. J., 2012: Cross‐species infection of pigs with a novel rabbit, but not rat, strain of hepatitis E virus isolated in the United States. J. Gen. Virol. 93, 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossan, C. , Baker P. J., Craft J., Takeuchi Y., Dalton H. R., and Scobie L., 2012: Hepatitis E virus genotype 3 in shellfish, United Kingdom. Emerg. Infect. Dis. 18, 2085–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. , Dong C., Zhou Z., Liang J., Dong M., Yang Y., Fu J., Tian H., Wang S., Fan J., Meng J., and Purdy M. A., 2013: Hepatitis E virus genotype 4, Nanjing, China, 2001–2011. Emerg. Infect. Dis. 19, 1528–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Deus, N. , Peralta B., Pina S., Allepuz A., Mateu E., Vidal D., Ruiz‐Fons F., Martín M., Gortázar C., and Segalés J., 2008: Epidemiological study of hepatitis E virus infection in European wild boars (Sus scrofa) in Spain. Vet. Microbiol. 129, 163–170. [DOI] [PubMed] [Google Scholar]
- Donia, D. , Dell'Amico M. C., Petrinca A. R., Martinucci I., Mazzei M., Tolari F., and Divizia M., 2012: Presence of hepatitis E RNA in mussels used as bio‐monitors of viral marine pollution. J. Virol. Methods 186, 198–202. [DOI] [PubMed] [Google Scholar]
- Dremsek, P. , Wenzel J. J., Johne R., Ziller M., Hofmann J., Groschup M. H., Werdermann S., Mohn U., Dorn S., Motz M., Mertens M., Jilg W., and Ulrich R. G., 2012: Seroprevalence study in forestry workers from eastern Germany using novel genotype 3‐ and rat hepatitis E virus‐specific immunoglobulin G ELISAs. Med. Microbiol. Immunol. 201, 189–200. [DOI] [PubMed] [Google Scholar]
- Drexler, J. F. , Seelen A., Corman V. M., Fumie Tateno A., Cottontail V., Melim Zerbinati R., Gloza‐Rausch F., Klose S. M., Adu‐Sarkodie Y., Oppong S. K., Kalko E. K., Osterman A., Rasche A., Adam A., Müller M. A., Ulrich R. G., Leroy E. M., Lukashev A. N., and Drosten C., 2012: Bats worldwide carry hepatitis E‐related viruses that form a putative novel genus within the family Hepeviridae. J. Virol. 86, 9134–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterbrook, J. D. , Kaplan J. B., Vanasco N. B., Reeves W. K., Purcell R. H., Kosoy M. Y., Glass G. E., Watson J., and Klein S. L., 2007: A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol. Infect. 135, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, S. , Anderson D., Arankalle A., Meng X.‐J., Purdy M., Schlauder G., and Purcell R., 2005: Hepevirus In: Fauquet C., Mayo M., Maniloff J., Desselberger U. and Ball L. A. (eds), Virus Taxonomy, VIIIth Report of the ICTV. pp. 853–857. Elsevier/Academic Press, London. [Google Scholar]
- Engle, R. E. , Yu C., Emerson S. U., Meng X. J., and Purcell R. H., 2002: Hepatitis E virus (HEV) capsid antigens derived from viruses of human and swine origin are equally efficient for detecting anti‐HEV by enzyme immunoassay. J. Clin. Microbiol. 40, 4576–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favorov, M. O. , Kosoy M. Y., Tsarev S. A., Childs J. E., and Margolis H. S., 2000: Prevalence of antibody to hepatitis E virus among rodents in the United States. J. Infect. Dis. 181, 449–455. [DOI] [PubMed] [Google Scholar]
- Feagins, A. R. , Opriessnig T., Guenette D. K., Halbur P. G., and Meng X. J., 2007: Detection and characterization of infectious Hepatitis E virus from commercial pig livers sold in local grocery stores in the USA. J. Gen. Virol. 88, 912–917. [DOI] [PubMed] [Google Scholar]
- Forgách, P. , Nowotny N., Erdélyi K., Boncz A., Zentai J., Szucs G., Reuter G., and Bakonyi T., 2010: Detection of hepatitis E virus in samples of animal origin collected in Hungary. Vet. Microbiol. 143, 106–116. [DOI] [PubMed] [Google Scholar]
- Francis, D. P. , and Maynard J. E., 1979: The transmission and outcome of hepatitis A, B, and non‐A, non‐B: a review. Epidemiol. Rev. 1, 17–31. [DOI] [PubMed] [Google Scholar]
- Galiana, C. , Fernández‐Barredo S., García A., Gómez M. T., and Pérez‐Gracia M. T., 2008: Occupational exposure to hepatitis E virus (HEV) in swine workers. Am. J. Trop. Med. Hyg. 78, 1012–1015. [PubMed] [Google Scholar]
- Geng, Y. , Zhao C., Song A., Wang J., Zhang X., Harrison T. J., Zhou Y., Wang W., and Wang Y., 2011: The serological prevalence and genetic diversity of hepatitis E virus in farmed rabbits in China. Infect. Genet. Evol. 11, 476–482. [DOI] [PubMed] [Google Scholar]
- Graff, J. , Nguyen H., Yu C., Elkins W. R., St Claire M., Purcell R. H., and Emerson S. U., 2005: The open reading frame 3 gene of hepatitis E virus contains a cis‐reactive element and encodes a protein required for infection of macaques. J. Virol. 79, 6680–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff, J. , Torian U., Nguyen H., and Emerson S. U., 2006: A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 80, 5919–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzki, M. , Schaeffer J., Piquet J. C., Le Saux J. C., Chevé J., Ollivier J., Le Pendu J., and Le Guyader F. S., 2014: Bioaccumulation efficiency, tissue distribution, and environmental occurrence of hepatitis E virus in bivalve shellfish from France. Appl. Environ. Microbiol. 80, 4269–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, D. , Li W., Su J., Fang L., Takeda N., Wakita T., Li T. C., and Ke C., 2013: Asian musk shrew as a reservoir of rat hepatitis E virus, China. Emerg. Infect. Dis. 19, 1341–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. , Zhou E. M., Sun Z. F., Meng X. J., and Halbur P. G., 2006: Identification of B‐cell epitopes in the capsid protein of avian hepatitis E virus (avian HEV) that are common to human and swine HEVs or unique to avian HEV. J. Gen. Virol. 87, 217–223. [DOI] [PubMed] [Google Scholar]
- Gupta, H. , Joshi Y. K., Varma A., Shenoy S., Sriramchari S., Iyenger B., and Tandon B. N., 1990: Transmission of enteric non‐A, non‐B hepatitis virus in Macaca mulatta monkeys by intraportal route: subsequent passages of HEV virus. J. Gastroenterol. Hepatol. 5, 608–615. [DOI] [PubMed] [Google Scholar]
- Han, J. , Zeng H., Wang L., Liu P., Liu L., Xia J., Zhang Y., and Zhuang H., 2014: Hepatitis E virus infection in farmed rabbits and swine in the Eastern Chinese city Lianyungang: showing no potential interspecies transmission. J. Med. Virol. 86, 1898–1904. [DOI] [PubMed] [Google Scholar]
- Haqshenas, G. , Shivaprasad H. L., Woolcock P. R., Read D. H., and Meng X. J., 2001: Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis‐splenomegaly syndrome in the United States. J. Gen. Virol. 82, 2449–2462. [DOI] [PubMed] [Google Scholar]
- Haqshenas, G. , Huang F. F., Fenaux M., Guenette D. K., Pierson F. W., Larsen C. T., Shivaprasad H. L., Toth T. E., and Meng X. J., 2002: The putative capsid protein of the newly identified avian hepatitis E virus shares antigenic epitopes with that of swine and human hepatitis E viruses and chicken big liver and spleen disease virus. J. Gen. Virol. 83, 2201–2209. [DOI] [PubMed] [Google Scholar]
- Hedrick, R. P. , Yun S., and Wingfield W. H., 1991: A small RNA virus isolated from salmonid fishes in California, USA. Can. J. Fish Aquat. Sci. 48, 99–104. [Google Scholar]
- Hirano, M. , Ding X., Li T. C., Takeda N., Kawabata H., Koizumi N., Kadosaka T., Goto I., Masuzawa T., Nakamura M., Taira K., Kuroki T., Tanikawa T., Watanabe H., and Abe K., 2003a: Evidence for widespread infection of hepatitis E virus among wild rats in Japan. Hepatol. Res. 27, 1–5. [DOI] [PubMed] [Google Scholar]
- Hirano, M. , Ding X., Tran H. T., Li T. C., Takeda N., Sata T., Nakamura S., and Abe K., 2003b: Prevalence of antibody against hepatitis E virus in various species of non‐human primates: evidence of widespread infection in Japanese monkeys (Macaca fuscata). Jpn. J. Infect. Dis. 56, 8–11. [PubMed] [Google Scholar]
- Holla, R. P. , Ahmad I., Ahmad Z., and Jameel S., 2013: Molecular virology of hepatitis E virus. Semin. Liver Dis. 33, 3–14. [DOI] [PubMed] [Google Scholar]
- Hsieh, S. Y. , Meng X. J., Wu Y. H., Liu S. T., Tam A. W., Lin D. Y., and Liaw Y. F., 1999: Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J. Clin. Microbiol. 37, 3828–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, I. W. , and Tsai H. J., 2014: Avian hepatitis E virus in chickens, Taiwan, 2013. Emerg. Infect. Dis. 20, 149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, G. D. , and Ma X., 2010: Detection and sequences analysis of bovine hepatitis E virus RNA in Xinjiang autonomous region. Bing Du Xue Bao 26, 27–32. [PubMed] [Google Scholar]
- Huang, C. C. , Nguyen D., Fernandez J., Yun K. Y., Fry K. E., Bradley D. W., Tam A. W., and Reyes G. R., 1992: Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV). Virology 191, 550–558. [DOI] [PubMed] [Google Scholar]
- Huang, F. F. , Haqshenas G., Shivaprasad H. L., Guenette D. K., Woolcock P. R., Larsen C. T., Pierson F. W., Elvinger F., Toth T. E., and Meng X. J., 2002: Heterogeneity and seroprevalence of a newly identified avian hepatitis e virus from chickens in the United States. J. Clin. Microbiol. 40, 4197–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F. F. , Sun Z. F., Emerson S. U., Purcell R. H., Shivaprasad H. L., Pierson F. W., Toth T. E., and Meng X. J., 2004: Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J. Gen. Virol. 85, 1609–1618. [DOI] [PubMed] [Google Scholar]
- Ijaz, S. , Said B., Boxall E., Smit E., Morgan D., and Tedder R. S., 2014: Indigenous hepatitis E in England and wales from 2003 to 2012: evidence of an emerging novel phylotype of viruses. J. Infect. Dis. 209, 1212–1218. [DOI] [PubMed] [Google Scholar]
- Izopet, J. , Dubois M., Bertagnoli S., Lhomme S., Marchandeau S., Boucher S., Kamar N., Abravanel F., and Guérin J. L., 2012: Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg. Infect. Dis. 18, 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez de Oya, N. , Escribano‐Romero E., Blázquez A. B., and Saiz J. C., 2007: Hepatitis E virus: zoonotic implications. Gastroenterol. Hepatol. 30, 408–418. [DOI] [PubMed] [Google Scholar]
- Johne, R. , Plenge‐Bönig A., Hess M., Ulrich R. G., Reetz J., and Schielke A., 2010: Detection of a novel hepatitis E‐like virus in faeces of wild rats using a nested broad‐spectrum RT‐PCR. J. Gen. Virol. 91, 750–758. [DOI] [PubMed] [Google Scholar]
- Johne, R. , Dremsek P., Kindler E., Schielke A., Plenge‐Bönig A., Gregersen H., Wessels U., Schmidt K., Rietschel W., Groschup M. H., Guenther S., Heckel G., and Ulrich R. G., 2012: Rat hepatitis E virus: geographical clustering within Germany and serological detection in wild Norway rats (Rattus norvegicus). Infect. Genet. Evol. 12, 947–956. [DOI] [PubMed] [Google Scholar]
- Kabrane‐Lazizi, Y. , Fine J. B., Elm J., Glass G. E., Higa H., Diwan A., Gibbs C. J., Meng X. J., Emerson S. U., and Purcell R. H., 1999: Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am. J. Trop. Med. Hyg. 61, 331–335. [DOI] [PubMed] [Google Scholar]
- Kaur, M. , Hyams K. C., Purdy M. A., Krawczynski K., Ching W. M., Fry K. E., Reyes G. R., Bradley D. W., and Carl M., 1992: Human linear B‐cell epitopes encoded by the hepatitis E virus include determinants in the RNA‐dependent RNA polymerase. Proc. Natl Acad. Sci. U S A 89, 3855–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuroo, M. S. , 2008: Hepatitis E virus. Curr. Opin. Infect Dis. 21, 539–543. [DOI] [PubMed] [Google Scholar]
- Khuroo, M. S. , 2011: Discovery of hepatitis E: the epidemic non‐A, non‐B hepatitis 30 years down the memory lane. Virus Res. 161, 3–14. [DOI] [PubMed] [Google Scholar]
- Koff, R. S. , 2007: Review article: vaccination and viral hepatitis ‐ current status and future prospects. Aliment. Pharmacol. Ther. 26, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Koonin, E. V. , Gorbalenya A. E., Purdy M. A., Rozanov M. N., Reyes G. R., and Bradley D. W., 1992: Computer‐assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive‐strand RNA plant and animal viruses. Proc. Natl Acad. Sci. U S A 89, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krog, J. S. , Breum S., Jensen T. H., and Larsen L. E., 2013: Hepatitis E virus variant in farmed mink, Denmark. Emerg. Infect. Dis. 19, 2028–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumbholz, A. , Mohn U., Lange J., Motz M., Wenzel J. J., Jilg W., Walther M., Straube E., Wutzler P., and Zell R., 2012: Prevalence of hepatitis E virus‐specific antibodies in humans with occupational exposure to pigs. Med. Microbiol. Immunol. 201, 239–244. [DOI] [PubMed] [Google Scholar]
- Krumbholz, A. , Joel S., Dremsek P., Neubert A., Johne R., Dürrwald R., Walther M., Müller T. H., Kühnel D., Lange J., Wutzler P., Sauerbrei A., Ulrich R. G., and Zell R., 2014: Seroprevalence of hepatitis E virus (HEV) in humans living in high pig density areas of Germany. Med. Microbiol. Immunol. 203, 273–282. [DOI] [PubMed] [Google Scholar]
- Kwon, H. M. , Sung H. W., and Meng X. J., 2012: Serological prevalence, genetic identification, and characterization of the first strains of avian hepatitis E virus from chickens in Korea. Virus Genes 45, 237–245. [DOI] [PubMed] [Google Scholar]
- Lack, J. B. , Volk K., and Van Den Bussche R. A., 2012: Hepatitis E virus genotype 3 in wild rats, United States. Emerg. Infect. Dis. 18, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, H. C. , Wichmann O., and Duizer E., 2010: Transmission routes and risk factors for autochthonous hepatitis E virus infection in Europe: a systematic review. Epidemiol. Infect. 138, 145–166. [DOI] [PubMed] [Google Scholar]
- Li, T. C. , Chijiwa K., Sera N., Ishibashi T., Etoh Y., Shinohara Y., Kurata Y., Ishida M., Sakamoto S., Takeda N., and Miyamura T., 2005: Hepatitis E virus transmission from wild boar meat. Emerg. Infect. Dis. 11, 1958–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. C. , Miyamura T., and Takeda N., 2007: Detection of hepatitis E virus RNA from the bivalve Yamato‐Shijimi (Corbicula japonica) in Japan. Am. J. Trop. Med. Hyg. 76, 170–172. [PubMed] [Google Scholar]
- Li, T. C. , Ami Y., Suzaki Y., Takeda N., and Takaji W., 2013a: No evidence for hepatitis E virus genotype 3 susceptibility in rats. Emerg. Infect. Dis. 19, 1343–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. C. , Ami Y., Suzaki Y., Yasuda S. P., Yoshimatsu K., Arikawa J., Takeda N., and Takaji W., 2013b: Characterization of full genome of rat hepatitis E virus strain from Vietnam. Emerg. Infect. Dis. 19, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. C. , Yoshizaki S., Ami Y., Suzaki Y., Yasuda S. P., Yoshimatsu K., Arikawa J., Takeda N., and Wakita T., 2013c: Susceptibility of laboratory rats against genotypes 1, 3, 4, and rat hepatitis E viruses. Vet. Microbiol. 163, 54–61. [DOI] [PubMed] [Google Scholar]
- Li, W. , Guan D., Su J., Takeda N., Wakita T., Li T. C., and Ke C. W., 2013d: High prevalence of rat hepatitis E virus in wild rats in China. Vet. Microbiol. 165, 275–280. [DOI] [PubMed] [Google Scholar]
- Li, T. C. , Yang T., Ami Y., Suzaki Y., Shirakura M., Kishida N., Asanuma H., Takeda N., and Takaji W., 2014: Complete genome of hepatitis E virus from laboratory ferrets. Emerg. Infect. Dis. 20, 709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. C. , Yonemitsu K., Terada Y., Takeda N., Wakita T., and Maeda K., 2015: Ferret hepatitis E virus infection in Japan. Jpn. J. Infect. Dis. 68, 60–62. [DOI] [PubMed] [Google Scholar]
- Lin, J. , Norder H., Uhlhorn H., Belák S., and Widén F., 2013: Novel hepatitis E like virus found in Swedish moose. J. Gen. Virol. 95, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P. , Bu Q. N., Wang L., Han J., Du R. J., Lei Y. X., Ouyang Y. Q., Li J., Zhu Y. H., Lu F. M., and Zhuang H., 2013: Transmission of hepatitis E virus from rabbits to cynomolgus macaques. Emerg. Infect. Dis. 19, 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Zhao Q., Sun Y., Wang X., Zhao J., Du T., Wang C., Xiao S., Mu Y., Zhang G., Luo J., Hsu W. H., and Zhou E. M., 2014: Development of a blocking ELISA for detection of antibodies against avian hepatitis E virus. J. Virol. Methods 204, 1–5. [DOI] [PubMed] [Google Scholar]
- Lu, L. , Li C., and Hagedorn C. H., 2006: Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 16, 5–36. [DOI] [PubMed] [Google Scholar]
- Maneerat, Y. , Clayson E. T., Myint K. S., Young G. D., and Innis B. L., 1996: Experimental infection of the laboratory rat with the hepatitis E virus. J. Med. Virol. 48, 121–128. [DOI] [PubMed] [Google Scholar]
- Martelli, F. , Caprioli A., Zengarini M., Marata A., Fiegna C., Di Bartolo I., Ruggeri F. M., Delogu M., and Ostanello F., 2008: Detection of hepatitis E virus (HEV) in a demographic managed wild boar (Sus scrofa scrofa) population in Italy. Vet. Microbiol. 126, 74–81. [DOI] [PubMed] [Google Scholar]
- Mast, E. E. , Purdy M. A., and Krawczynski K., 1996: Hepatitis E. Baillieres Clin. Gastroenterol. 10, 227–242. [DOI] [PubMed] [Google Scholar]
- Matsuda, H. , Okada K., Takahashi K., and Mishiro S., 2003: Severe hepatitis E virus infection after ingestion of uncooked liver from a wild boar. J. Infect. Dis. 188, 944. [DOI] [PubMed] [Google Scholar]
- Matsuura, Y. , Suzuki M., Yoshimatsu K., Arikawa J., Takashima I., Yokoyama M., Igota H., Yamauchi K., Ishida S., Fukui D., Bando G., Kosuge M., Tsunemitsu H., Koshimoto C., Sakae K., Chikahira M., Ogawa S., Miyamura T., Takeda N., and Li T. C., 2007: Prevalence of antibody to hepatitis E virus among wild Sika deer, Cervus nippon, in Japan. Arch. Virol. 152, 1375–1381. [DOI] [PubMed] [Google Scholar]
- McCaustland, K. A. , Krawczynski K., Ebert J. W., Balayan M. S., Andjaparidze A. G., Spelbring J. E., Cook E. H., Humphrey C., Yarbough P. O., Favorov M. O., Carson D., Bradley D. W., and Robertson B. H., 2000: Hepatitis E virus infection in chimpanzees: a retrospective analysis. Arch. Virol. 145, 1909–1918. [DOI] [PubMed] [Google Scholar]
- Meng, X. J. , 2010a: Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140, 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. J. , 2010b: Recent advances in Hepatitis E virus. J. Viral Hepat. 17, 153–161. [DOI] [PubMed] [Google Scholar]
- Meng, X. J. , 2011: From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 161, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. J. , Purcell R. H., Halbur P. G., Lehman J. R., Webb D. M., Tsareva T. S., Haynes J. S., Thacker B. J., and Emerson S. U., 1997: A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl Acad. Sci. U S A 94, 9860–9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. J. , Anderson D. A., Arankalle V. A., Emerson S. U., Harrison T. J., Jameel S., and Okamoto H., 2011: Family Hepeviridae In: King A. M., Adams M. J., Carstens E. B., and Lefkowitz E. J. (eds), Virus Taxonomy ‐ Ninth Report of the International Committee on Taxonomy of Viruses, 1st edn Elservier, San‐Diego, USA. [Google Scholar]
- Moal, V. , Gerolami R., and Colson P., 2012: First human case of co‐infection with two different subtypes of hepatitis E virus. Intervirology 55, 484–487. [DOI] [PubMed] [Google Scholar]
- Mochizuki, M. , Ouchi A., Kawakami K., Ishida T., Li T. C., Takeda N., Ikeda H., and Tsunemitsu H., 2006: Epidemiological study of hepatitis E virus infection of dogs and cats in Japan. Vet. Rec. 159, 853–854. [PubMed] [Google Scholar]
- Mushahwar, I. K. , 2008: Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J. Med. Virol. 80, 646–658. [DOI] [PubMed] [Google Scholar]
- Nakamura, M. , Takahashi K., Taira K., Taira M., Ohno A., Sakugawa H., Arai M., and Mishiro S., 2006: Hepatitis E virus infection in wild mongooses of Okinawa, Japan: Demonstration of anti‐HEV antibodies and a full‐genome nucleotide sequence. Hepatol. Res. 34, 137–140. [DOI] [PubMed] [Google Scholar]
- Oliveira‐Filho, E. F. , König M., and Thiel H. J., 2013: Genetic variability of HEV isolates: Inconsistencies of current classification. Vet. Microbiol. 165, 148–154. [DOI] [PubMed] [Google Scholar]
- Oliveira‐Filho, E. F. , Bank‐Wolf B. R., Thiel H. J., and König M., 2014: Phylogenetic analysis of hepatitis E virus in domestic swine and wild boar in Germany. Vet. Microbiol. 174, 233–238. [DOI] [PubMed] [Google Scholar]
- Panda, S. K. , Thakral D., and Rehman S., 2007: Hepatitis E virus. Rev. Med. Virol. 17, 151–180. [DOI] [PubMed] [Google Scholar]
- Pavio, N. , Meng X. J., and Renou C., 2010: Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet. Res. 41, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, C. J. , Ellis T. M., Plant S. L., Gregory A. R., and Wilcox G. E., 1999: Sequence data suggests big liver and spleen disease virus (BLSV) is genetically related to hepatitis E virus. Vet. Microbiol. 68, 119–125. [DOI] [PubMed] [Google Scholar]
- Peralta, B. , Biarnés M., Ordóñez G., Porta R., Martín M., Mateu E., Pina S., and Meng X. J., 2009: Evidence of widespread infection of avian hepatitis E virus (avian HEV) in chickens from Spain. Vet. Microbiol. 137, 31–36. [DOI] [PubMed] [Google Scholar]
- Pérez‐Gracia, M. T. , Mateos M. L., Galiana C., Fernández‐Barredo S., García A., Gómez M. T., and Moreira V., 2007: Autochthonous hepatitis E infection in a slaughterhouse worker. Am. J. Trop. Med. Hyg. 77, 893–896. [PubMed] [Google Scholar]
- Pourpongporn, P. , Samransurp K., Rojanasang P., Wiwattanakul S., and Srisurapanon S., 2009: The prevalence of anti‐hepatitis E in occupational risk groups. J. Med. Assoc. Thai. 92, 38–42. [PubMed] [Google Scholar]
- Purcell, R. H. , Engle R. E., Rood M. P., Kabrane‐Lazizi Y., Nguyen H. T., Govindarajan S., St Claire M., and Emerson S. U., 2011: Hepatitis E virus in rats, Los Angeles, California, USA. Emerg. Infect. Dis. 17, 2216–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy, M. A. , and Khudyakov Y. E., 2010: Evolutionary history and population dynamics of hepatitis E virus. PLoS ONE 5, e14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, V. S. , Smits S. L., Pas S. D., Provacia L. B., Moorman‐Roest H., Osterhaus A. D., and Haagmans B. L., 2012: Novel hepatitis E virus in ferrets, the Netherlands. Emerg. Infect. Dis. 18, 1369–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renou, C. , Afonso A. M., and Pavio N., 2014: Foodborne transmission of hepatitis e virus from raw pork liver sausage, france. Emerg. Infect. Dis. 20, 1945–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, G. , Fodor D., Forgách P., Kátai A., and Szucs G., 2009: Characterization and zoonotic potential of endemic hepatitis E virus (HEV) strains in humans and animals in Hungary. J. Clin. Virol. 44, 277–281. [DOI] [PubMed] [Google Scholar]
- Reyes, G. R. , Purdy M. A., Kim J. P., Luk K. C., Young L. M., Fry K. E., and Bradley D. W., 1990: Isolation of a cDNA from the virus responsible for enterically transmitted non‐A, non‐B hepatitis. Science 247, 1335–1339. [DOI] [PubMed] [Google Scholar]
- Riveiro‐Barciela, M. , Mínguez B., Gironés R., Rodriguez‐Frías F., Quer J., and Buti M., 2015: Phylogenetic demonstration of hepatitis E infection transmitted by pork meat ingestion. J. Clin. Gastroenterol. 49, 165–168. [DOI] [PubMed] [Google Scholar]
- Rutjes, S. A. , Lodder‐Verschoor F., Lodder W. J., van der Giessen J., Reesink H., Bouwknegt M., and de Roda Husman A. M., 2010: Seroprevalence and molecular detection of hepatitis E virus in wild boar and red deer in The Netherlands. J. Virol. Methods 168, 197–206. [DOI] [PubMed] [Google Scholar]
- dos Santos, D. R. , de Paula V. S., de Oliveira J. M., Marchevsky R. S., and Pinto M. A., 2011: Hepatitis E virus in swine and effluent samples from slaughterhouses in Brazil. Vet. Microbiol. 149, 236–241. [DOI] [PubMed] [Google Scholar]
- Sato, Y. , Sato H., Naka K., Furuya S., Tsukiji H., Kitagawa K., Sonoda Y., Usui T., Sakamoto H., Yoshino S., Shimizu Y., Takahashi M., Nagashima S., Jirintai T. Nishizawa., and Okamoto H., 2011: A nationwide survey of hepatitis E virus (HEV) infection in wild boars in Japan: identification of boar HEV strains of genotypes 3 and 4 and unrecognized genotypes. Arch. Virol. 156, 1345–1358. [DOI] [PubMed] [Google Scholar]
- Scharschmidt, B. F. , 1995: Hepatitis E: a virus in waiting. Lancet 346, 519–520. [DOI] [PubMed] [Google Scholar]
- Skaug, K. , Hagen I. J., and von der Lippe B., 1994: Three cases of acute hepatitis E virus infection imported into Norway. Scand. J. Infect. Dis. 26, 137–139. [DOI] [PubMed] [Google Scholar]
- Smith, D. B. , Purdy M. A., and Simmonds P., 2013: Genetic variability and the classification of hepatitis E virus. J. Virol. 87, 4161–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. B. , Simmonds P., Jameel S., Emerson S. U., Harrison T. J., Meng X. J., Okamoto H., Van der Poel W. H., and Purdy M. A., 2014: Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 95, 2223–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda, H. , Abe M., Sugimoto T., Sato Y., Bando M., Fukui E., Mizuo H., Takahashi M., Nishizawa T., and Okamoto H., 2004: Prevalence of hepatitis E virus (HEV) Infection in wild boars and deer and genetic identification of a genotype 3 HEV from a boar in Japan. J. Clin. Microbiol. 42, 5371–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan, M. A. , Arankalle V. A., Sehgal A., and Pavri K. M., 1984: Non‐A, non‐B epidemic hepatitis: visualization of virus‐like particles in the stool by immune electron microscopy. J. Gen. Virol. 65, 1005–1007. [DOI] [PubMed] [Google Scholar]
- Sun, Z. F. , Larsen C. T., Dunlop A., Huang F. F., Pierson F. W., Toth T. E., and Meng X. J., 2004a: Genetic identification of avian hepatitis E virus (HEV) from healthy chicken flocks and characterization of the capsid gene of 14 avian HEV isolates from chickens with hepatitis‐splenomegaly syndrome in different geographical regions of the United States. J. Gen. Virol. 85, 693–700. [DOI] [PubMed] [Google Scholar]
- Sun, Z. F. , Larsen C. T., Huang F. F., Billam P., Pierson F. W., Toth T. E., and Meng X. J., 2004b: Generation and infectivity titration of an infectious stock of avian hepatitis E virus (HEV) in chickens and cross‐species infection of turkeys with avian HEV. J. Clin. Microbiol. 42, 2658–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M. , Nishizawa T., Miyajima H., Gotanda Y., Iita T., Tsuda F., and Okamoto H., 2003: Swine hepatitis E virus strains in Japan form four phylogenetic clusters comparable with those of Japanese isolates of human hepatitis E virus. J. Gen. Virol. 84, 851–862. [DOI] [PubMed] [Google Scholar]
- Takahashi, K. , Kitajima N., Abe N., and Mishiro S., 2004: Complete or near‐complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology 330, 501–505. [DOI] [PubMed] [Google Scholar]
- Takahashi, M. , Nishizawa T., Sato H., Sato Y., Jirintai S. Nagashima., and Okamoto H., 2011: Analysis of the full‐length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J. Gen. Virol. 92, 902–908. [DOI] [PubMed] [Google Scholar]
- Tamada, Y. , Yano K., Yatsuhashi H., Inoue O., Mawatari F., and Ishibashi H., 2004: Consumption of wild boar linked to cases of hepatitis E. J. Hepatol. 40, 869–870. [DOI] [PubMed] [Google Scholar]
- Tei, S. , Kitajima N., Takahashi K., and Mishiro S., 2003: Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362, 371–373. [DOI] [PubMed] [Google Scholar]
- Teo, C. G. , 2012: Fatal outbreaks of jaundice in pregnancy and the epidemic history of hepatitis E. Epidemiol. Infect. 140, 767–787. [DOI] [PubMed] [Google Scholar]
- Thiry, D. , Mauroy A., Saegerman C., Thomas I., Wautier M., Miry C., Czaplicki G., Berkvens D., Praet N., van der Poel W., Cariolet R., Brochier B., and Thiry E., 2014: Estimation of hepatitis E virus (HEV) pig seroprevalence using ELISA and Western blot and comparison between human and pig HEV sequences in Belgium. Vet. Microbiol. 172, 407–414. [DOI] [PubMed] [Google Scholar]
- Ticehurst, J. , Rhodes L. L., Krawczynski K., Asher L. V., Engler W. F., Mensing T. L., Caudill J. D., Sjogren M. H., Hoke C. H., and LeDuc J. W., 1992: Infection of owl monkeys (Aotus trivirgatus) and cynomolgus monkeys (Macaca fascicularis) with hepatitis E virus from Mexico. J. Infect. Dis. 165, 835–845. [DOI] [PubMed] [Google Scholar]
- Tsarev, S. A. , Emerson S. U., Reyes G. R., Tsareva T. S., Legters L. J., Malik I. A., Iqbal M., and Purcell R. H., 1992: Characterization of a prototype strain of hepatitis E virus. Proc. Natl Acad. Sci. U S A 89, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsarev, S. A. , Tsareva T. S., Emerson S. U., Govindarajan S., Shapiro M., Gerin J. L., and Purcell R. H., 1997: Recombinant vaccine against hepatitis E: dose response and protection against heterologous challenge. Vaccine 15, 1834–1838. [DOI] [PubMed] [Google Scholar]
- Viswanathan, R. , 1957: Epidemiology. Indian J. Med. Res. 45, 1–29. [PubMed] [Google Scholar]
- Vitral, C. L. , Pinto M. A., Lewis‐Ximenez L. L., Khudyakov Y. E., dos Santos D. R., and Gaspar A. M., 2005: Serological evidence of hepatitis E virus infection in different animal species from the Southeast of Brazil. Mem. Inst. Oswaldo Cruz 100, 117–122. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , and Ma X., 2010: Detection and sequences analysis of sheep hepatitis E virus RNA in Xinjiang autonomous region. Wei Sheng Wu Xue Bao 50, 937–941. [PubMed] [Google Scholar]
- Wang, Y. , Ling R., Erker J. C., Zhang H., Li H., Desai S., Mushahwar I. K., and Harrison T. J., 1999: A divergent genotype of hepatitis E virus in Chinese patients with acute hepatitis. J. Gen. Virol. 80, 169–177. [DOI] [PubMed] [Google Scholar]
- Wang, Y. C. , Zhang H. Y., Xia N. S., Peng G., Lan H. Y., Zhuang H., Zhu Y. H., Li S. W., Tian K. G., Gu W. J., Lin J. X., Wu X., Li H. M., and Harrison T. J., 2002: Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J. Med. Virol. 67, 516–521. [DOI] [PubMed] [Google Scholar]
- Wenzel, J. J. , Preiss J., Schemmerer M., Huber B., Plentz A., and Jilg W., 2011: Detection of hepatitis E virus (HEV) from porcine livers in Southeastern Germany and high sequence homology to human HEV isolates. J. Clin. Virol. 52, 50–54. [DOI] [PubMed] [Google Scholar]
- WHO , 2013: Hepatitis E. Available at: http://www.who.int/mediacentre/factsheets/fs280/en/ (accessed 23 January 2014).
- Wibawa, I. D. , Muljono D. H., Mulyanto, Suryadarma I. G., Tsuda F., Takahashi M., Nishizawa T., and Okamoto H., 2004: Prevalence of antibodies to hepatitis E virus among apparently healthy humans and pigs in Bali, Indonesia: Identification of a pig infected with a genotype 4 hepatitis E virus. J. Med. Virol. 73, 38–44. [DOI] [PubMed] [Google Scholar]
- Wichmann, O. , Schimanski S., Koch J., Kohler M., Rothe C., Plentz A., Jilg W., and Stark K., 2008: Phylogenetic and case‐control study on hepatitis E virus infection in Germany. J. Infect. Dis. 198, 1732–1741. [DOI] [PubMed] [Google Scholar]
- Widén, F. , Sundqvist L., Matyi‐Toth A., Metreveli G., Belák S., Hallgren G., and Norder H., 2011: Molecular epidemiology of hepatitis E virus in humans, pigs and wild boars in Sweden. Epidemiol. Infect. 139, 361–371. [DOI] [PubMed] [Google Scholar]
- Woo, P. C. , Lau S. K., Teng J. L., Tsang A. K., Joseph M., Wong E. Y., Tang Y., Sivakumar S., Xie J., Bai R., Wernery R., Wernery U., and Yuen K. Y., 2014: New hepatitis E virus genotype in camels, the Middle East. Emerg. Infect. Dis. 20, 1044–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Pan Y., Baloch A. R., Tian L., Wang M., Na W., Ding L., and Zeng Q., 2014: Hepatitis e virus genotype 4 in yak, northwestern china. Emerg. Infect. Dis. 20, 2182–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, S. , Purcell R. H., and Emerson S. U., 1994: A new Chinese isolate of hepatitis E virus: comparison with strains recovered from different geographical regions. Virus Genes 9, 23–32. [DOI] [PubMed] [Google Scholar]
- Yu, C. , Zimmerman C., Stone R., Engle R. E., Elkins W., Nardone G. A., Emerson S. U., and Purcell R. H., 2007: Using improved technology for filter paper‐based blood collection to survey wild Sika deer for antibodies to hepatitis E virus. J. Virol. Methods 142, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Emerson S. U., Nguyen H., Engle R. E., Govindarajan S., Gerin J. L., and Purcell R. H., 2001: Immunogenicity and protective efficacy of a vaccine prepared from 53 kDa truncated hepatitis E virus capsid protein expressed in insect cells. Vaccine 20, 853–857. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Shen Q., Mou J., Gong G., Yang Z., Cui L., Zhu J., Ju G., and Hua X., 2008a: Hepatitis E virus infection among domestic animals in eastern China. Zoonoses Public Health 55, 291–298. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Shen Q., Mou J., Yang Z. B., Yuan C. L., Cui L., Zhu J. G., Hua X. G., Xu C. M., and Hu J., 2008b: Cross‐species infection of hepatitis E virus in a zoo‐like location, including birds. Epidemiol. Infect. 136, 1020–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Ma Z., Harrison T. J., Feng R., Zhang C., Qiao Z., Fan J., Ma H., Li M., Song A., and Wang Y., 2009: A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J. Med. Virol. 81, 1371–1379. [DOI] [PubMed] [Google Scholar]
- Zhao, Q. , Sun Y. N., Hu S. B., Wang X. J., Xiao Y. H., Hsu W. H., Xiao S. Q., Wang C. B., Mu Y., Hiscox J. A., and Zhou E. M., 2013: Characterization of antigenic domains and epitopes in the ORF3 protein of a Chinese isolate of avian hepatitis E virus. Vet. Microbiol. 167, 242–249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Correction added on 20 June 2015, after first online publication: Legend of Table S1 has been corrected.]
Table S1. Detection of HEV or HEV‐related viruses in animals.


