Abstract
Background
Epidemiologic studies of human adenovirus (HAdV) in allogeneic hematopoietic cell transplantation (HCT) recipients have been conducted mostly in European countries where HAdV 2 (species C) has been most prevalent in the community. The main objective of this study was to investigate the epidemiology and the characteristics of HAdV infection in Korean allogeneic HCT recipients (<19 years).
Methods
In a prospective study from April 2012 to September 2015, HAdV in blood, urine, and stool specimens were monitored weekly from transplantation to day 100 or after if clinically suspected. HAdV infection was defined as positive HAdV PCR result in any specimens regardless of symptoms.
Results
A total of 1734 specimens were collected from 57 consecutively enrolled recipients. The cumulative incidence of HAdV infection at day 100, and 1 year was 10%, and 20%, and the incidence of viremia was 2% and 6%, respectively. The median onset time from HCT to viremia was 221 days (range, 7‐596 days). All viremia cases were caused by only HAdV 3 (species B), whereas several types were detected in stool. Among patients with HAdV infection, lower absolute lymphocyte counts and extensive chronic graft‐vs‐host disease were associated with viremia (P = .028 and P = .006, respectively).
Conclusions
Compared to western studies, this study demonstrated a lower incidence and delayed onset of HAdV infections and HAdV 3 was most prevalent in Korea.
Keywords: allogeneic hematopoietic cell transplantation, children, epidemiology, human adenovirus, virus type
1. INTRODUCTION
Human adenovirus (HAdV) viremia is known to be common in allogeneic hematopoietic cell transplantation (HCT) recipients, particularly in children. The incidence of HAdV viremia following allogeneic HCT has been reported as 6%‐50% in children, and 3%‐15% in adults. Although variable, most studies reported that HAdV infection including viremia usually occurred within the first 100‐180 days after transplantation.1, 2, 3, 4, 5, 6, 7, 8 Endogenous reactivation of latent virus was regarded as the major mode of infection; however, de novo infection in nosocomial and community settings could also occur.6, 9 HAdV‐attributable mortality is approximately 1%; however, in disseminated cases, the rate may increase up to 80%‐100%.6, 7, 10 The major risk factor for severe invasive HAdV disease or dissemination is T‐cell depletion, including ex vivo T‐cell depletion and use of alemtuzumab. Younger age, unrelated donor graft, cord blood, prolonged immunosuppressive treatment, and an absolute lymphocyte count (ALC) <300/μL are additional risk factors.2, 6, 7, 11, 12 With this background, the 4th European Society of Infectious Diseases in Leukemia (ECIL‐4) recommended routine HAdV monitoring using quantitative polymerase chain reaction (PCR) in peripheral blood, especially in high‐risk patients.7
However, the correlation between HAdV species and clinical features in immunocompromised population has not been established yet. HAdVs cause a variety of clinical manifestations depending on virus type and tissue tropism in immunocompetent host. For example, species B:1, C, and E can cause respiratory illness, whereas species F and G can mainly cause gastroenteritis. Species B:2 (eg HAdV 11, 34, and 35) are responsible for urinary tract infection, and species B and D are known to be associated with keratoconjunctivitis.13, 14 Serious complications often result from lower respiratory tract illness, particularly in association with HAdV 3, 7, and 21 (all 3 species B).15, 16
We hypothesized HAdV types could also affect the clinical features including the severity and/or dissemination in allogeneic HCT recipients. The main objectives of this study were to investigate the epidemiology of HAdV infections and the clinical correlations of HAdV types among Korean pediatric allogeneic HCT recipients. In addition, we explored potential risk factors for HAdV infection and/or viremia and HAdV‐attributable mortality.
2. MATERIALS AND METHODS
2.1. Patients and collection of samples
This study was conducted at Samsung Medical Center (SMC), Seoul, Korea, from 1 April 2012 to 30 September 2015. All consecutive pediatric recipients aged less than 19 years who received allogeneic HCT at the pediatric cancer center and agreed to participate were included. Informed consent and assent were obtained at the time of enrollment. This study was approved by the institutional review board (IRB) of SMC (IRB #2011‐11‐066‐010).
Recipients were screened for respiratory virus infection before the admission for HCT in our center. In enrolled patients, blood, urine, and stool specimens were collected prospectively from the infusion date of transplantation to day 100. About 1 mL of EDTA whole blood was collected weekly from the day of transplantation (day 0, day 7, day 14, etc.). Urine and stool samples were collected weekly as well as blood samples. These collected specimens were stored at −80°C until DNA extraction. After day 100, urine and stool specimens were further tested whenever clinically suspected. If HAdV was detected, we performed additional quantitative real‐time PCR using blood specimens to detect HAdV viremia. Respiratory specimens were tested using multiplex respiratory virus PCR (LG Life Science, Korea) if respiratory symptoms were present as a part of routine care practice.
2.2. DNA purification, amplification, and HAdV type analysis
DNA was extracted from 200 μL aliquots of clinical samples using the QIAamp Blood or Stool Mini Kit (QIAGEN, Hilden, Germany). Eluted DNA was stored at −20°C until PCR amplification. Quantitative real‐time PCR was used to detect HAdV in blood specimens using a commercial Adenovirus R‐Gene® kit (Argene, Varhilles, France).17 The lower limit for quantitation was 200 copies/mL. Qualitative conventional PCR was performed to detect HAdV in urine and stool specimens using a premix kit (Bioneer, Daejeon, Korea) following the manufacturer's protocol.
Human adenovirus type was determined by sequencing hyper‐variable region 1‐6 of the hexon gene. Nested PCR was performed to secure sufficient HAdV DNA for sequencing, using a previously described method with minor modifications (Method S1).18 Sequencing was performed in both directions using the 2nd round PCR primers. Sequencher™ 4.9 software (Gene Codes, Ann Arbor, MI, USA) and Basic Local Alignment Search Tool (BLAST) software (http://blast.ncbi.nlm.nih.gov/) were used to identify HAdV types. Phylogenetic and molecular analyses were conducted using MEGA 6.06. Reference HAdV strains are shown in Table S1.
2.3. Collection of clinical information
Epidemiologic data and patient characteristics including age at HCT, sex, underlying disease, type of donor, source of graft, use of total body irradiation (TBI), use of anti‐thymoglobulin (ATG), and conditioning regimen were collected from medical records at the time of enrollment. We performed weekly monitoring of the clinical data including symptoms, laboratory data, the severity of graft‐vs‐host disease (GVHD), the maximal dose of systemic corticosteroids, co‐infection complications until post‐transplant day 100, and mortality through medical chart review. In patients with HAdV infection, clinical data including details of co‐infections and the treatment was also reviewed. Co‐infection was defined as any infection caused by other pathogens in addition to HAdV before and after 7 days of HAdV‐positive period. We used previously proposed definitions of HAdV infection and disease.7 If 2 or more types of HAdV infection occurred simultaneously or sequentially, the terminology “multiple HAdV infections” was used. We defined HAdV infection as the identification of adenovirus from any surveillance sample regardless of symptoms. HAdV viremia was defined as positive HAdV PCR result in peripheral blood and local HAdV infection such as colitis, cystitis, and respiratory tract infections was defined as positive HAdV PCR result or antigen detection in corresponding body fluids other than peripheral blood. Probable disease was defined as HAdV infection plus corresponding symptoms and signs without histological confirmation. Proven disease was defined as HAdV infection plus corresponding symptoms and signs with histological confirmation.7
2.4. Conditioning regimens, prophylaxis of GVHD, and prophylaxis of infectious diseases
Conditioning regimens were determined by primary disease, age, and status according to the study protocols. High dose‐TBI (≥total of 10 Gy) and busulfan (16 mg/kg for oral doses or 12.8 mg/kg for intravenous doses) were usually employed as myeloablative (MA) conditioning regimens, depending on the specific protocol. All patients with relapsed neuroblastoma were treated with reduced‐intensity (RI) conditioning regimens. In vivo T‐cell depletion using alemtuzumab and ex vivo T‐cell depletion were not performed at our center. Calcineurin inhibitor alone or with other immunosuppressants (mycophenolate mofetil, corticosteroids, and methotrexate) was/were started on day −1 for GVHD prophylaxis. Cord blood recipients received cyclosporin A (CsA) plus mycophenolate mofetil. The diagnoses of acute and chronic GVHD were based on standard clinical criteria and biopsies were performed when possible.19 Acute GVHD of grade II or above was treated with systemic corticosteroids. Recipients received antiviral prophylaxis (acyclovir) from day −1 to day 28 and antifungal prophylaxis (micafungin) from day −1 to neutrophil engraftment, and Pneumocystis jirovecii prophylaxis (trimethoprim/sulfamethoxazole) from neutrophil engraftment until cessation of the immunosuppressive agents.20
2.5. Statistical analysis
Demographic and clinical data were compared using Fisher's exact test or chi‐square test for categorical variables and the Mann‐Whitney U test for continuous variables, as appropriate. Categorical variables were presented as frequencies (percentages) and continuous variables were summarized as medians (ranges). Logistic regression analysis was performed to evaluate the risk factors for HAdV infection or viremia. The relationships between potential risk factors and outcomes were tested by Cox proportional‐hazards regression. Variables with P < .1 in the univariate analysis were included in the multivariate analysis (model I). We also included other variables: age at HCT, sex, relapsed status of underlying disease (relapsed or not), and severe acute GVHD (grade ≥3) in additional multivariate analysis (model II). A phylogenetic tree was generated using the neighbor‐joining (NJ) method. The accuracy of the phylogenetic tree was tested by 1000 bootstrap replications. The cumulative incidences of HAdV infection or HAdV viremia and the relationships between overall survival and HAdV infection were estimated using the Kaplan‐Meier method and assessed using the log‐rank test. Two‐sided P of less than .05 was considered statistically significant. All statistical analyses were performed using SPSS version 19 (SPSS, Chicago, IL, USA) and GraphPad Prism version 7.03 (GraphPad Software, La Jolla, CA, USA).
3. RESULTS
3.1. Patient characteristics
Between April 2012 and September 2015, a total of 93 pediatric recipients received 97 allogeneic HCTs at SMC. Among them, 57 recipients (61%) with 58 HCT procedures were enrolled (Figure S1). Median age at HCT was 5.2 years (range, 0.6‐18.1 years) and 36 recipients (62%) were male. Median follow‐up was 2.1 years (range 0.2‐4.1 years). None of our patients received ex vivo T‐cell depletion. Other detailed characteristics are listed in Table 1.
Table 1.
Patient characteristics (n = 57)
| HAdV infection | No HAdV infection | P | |||
|---|---|---|---|---|---|
| N = 11 | % or range | N = 46 | % or range | ||
| Median age at HCT, years | 5.2 | 0.7‐17.5 | 5.5 | 0.6‐18.1 | .8 |
| Sex, male | 6 | 17 | 30 | 84 | .5 |
| Diagnosis | .2 | ||||
| Acute leukemia (AML/ALL) | 5 | 29 | 12 | 71 | |
| Aplastic anemia | 2 | 20 | 8 | 80 | |
| Neuroblastoma (all relapsed) | 3 | 27 | 8 | 73 | |
| Primary immunodeficiency disease | 1 | 20 | 4 | 80 | |
| Othersa | 0 | 0 | 14 | 100 | |
| Relapsed statusb | 6 | 29 | 15 | 71 | .3 |
| Donor type | .2 | ||||
| MRD (sibling) | 3 | 50 | 3 | 50 | |
| MUD | 6 | 15 | 33 | 85 | |
| MMRD (haploidentical) | 2 | 17 | 10 | 83 | |
| Donor cell source | .7 | ||||
| Peripheral blood | 9 | 22 | 32 | 78 | |
| Cord blood | 2 | 13 | 14 | 88 | |
| CD34 cell dose (x106 cells/kg) | |||||
| Peripheral blood | 9.2 | 1.6‐30.1 | 5.3 | 1.6‐16.6 | .1 |
| Cord blood | 0.3 | 0.2‐0.4 | 0.1 | 0.02‐1.6 | .6 |
| Conditioning intensity | .7 | ||||
| Myeloablative | 8 | 18 | 36 | 82 | |
| Reduced | 3 | 23 | 10 | 77 | |
| Conditioning regimen | .8 | ||||
| TBI‐based | 4 | 27 | 11 | 73 | |
| Busulfan‐based | 4 | 17 | 20 | 83 | |
| Others | 3 | 17 | 15 | 83 | |
| Use of ATG | 6 | 17 | 29 | 83 | .7 |
| Recipients’ pre‐HCT surveillance | |||||
| Respiratory virus (by PCR)c | 10 | 19 | 43 | 81 | 1.0 |
| CMV seropositivity | 10 | 19 | 44 | 81 | .5 |
| HSV seropositivity | 9 | 20 | 35 | 80 | 1.0 |
| Median neutrophil engraftment, days | 12 | 9‐19 | 12 | 8‐25 | .7 |
| Median dose of max. steroid until day 100, (mg/kg/d) | 2 | 0‐2 | 2 | 0‐10 | .4 |
| Infectious complication until day 100 | |||||
| CMV Antigenemia or diseased | 8 | 17 | 38 | 83 | .4 |
| EBV viremia | 3 | 18 | 14 | 82 | 1.0 |
| BKV cystitis | 7 | 19 | 30 | 81 | 1.0 |
| Other respiratory virus infection | 2 | 17 | 10 | 83 | 1.0 |
| Bacteraemia | 1 | 17 | 5 | 83 | 1.0 |
| Invasive fungal infectione | 1 | 20 | 4 | 80 | 1.0 |
| Acute GVHD ≥3 | 2 | 15 | 11 | 85 | 1.0 |
| Chronic GVHD | 5 | 22 | 18 | 78 | 1.0 |
| Extensive | 3 | 23 | 8 | 77 | 1.0 |
| N/A | – | – | |||
HAdV indicates human adenovirus; HCT, hematopoietic cell transplantation; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; MRD, matched related donor; MUD, matched unrelated donor; MMRD, mismatched related donor; TBI, total body irradiation; ATG, antithymocyte globulin; PCR, polymerase chain reaction; CMV, cytomegalovirus; HSV, herpes simplex virus; EBV, Epstein–Barr virus; BKV, BK virus; GVHD, graft‐vs‐host disease; N/A, not applicable.
Included 4 cases of juvenile myelomonocytic leukemia (JMML), 2 cases of hemophagocytic lymphohistiocytosis (HLH), 2 cases of myelodysplastic syndromes (MDS), 2 cases of sarcoma, 1 case of Fanconi anemia, 1 case of non‐Hodgkin lymphoma, 1 case of pure red cell aplasia, and 1 case of retinoblastoma.
The relapsed state was defined as a return of a malignant disease or the signs and symptoms of a disease after a period of improvement.
Among 53 recipients who had been tested pre‐HCT by multiplex respiratory virus PCR, 5 cases had respiratory virus detected (all rhinovirus).
All 47 recipients were treated with ganciclovir. Forty‐four patients had CMV antigenemia only and 3 recipients had CMV disease.
All 5 patients had invasive pulmonary aspergillosis (IPA) infection. Four cases were probable and 1 case was possible IPA.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.2. Detection rate of HAdV and the incidences of HAdV infection/viremia
A total of 819 blood samples, 452 stool samples, and 463 urine samples were tested, and HAdV was detected in 16 samples from 12 recipients (Table 2). The cumulative incidence of HAdV infection at day 100, 1 year, and 2 years were 10% (95% confidence interval [CI], 6.4%‐14.4%), 20% (95% CI, 14.9%‐25.9%) and 23% (95% CI, 17.0%‐28.8%), respectively (Figure 1). Four recipients developed HAdV viremia and the cumulative incidences at day 100, 1, and 2 years were 2% (95% CI, 0.0‐3.4%), 6% (95% CI, 3.2%‐8.2%) and 8% (95% CI, 4.3%‐12.3%), respectively (Figure 1). Median time from HCT to first HAdV infection was 113 days (range, 7‐596 days) and to onset of HAdV viremia was 221 days (range, 7‐596 days). More than half of HAdV infections (58%, 7/12) occurred after day 100 from HCT. All of HAdV‐positive respiratory specimens (n = 5) were detected after day 100 (Table 3).
Table 2.
Human adenovirus (HAdV) typing and positivity according to specimen
| Specimen | Total number of samples | Positive number of samples | Positivity (%) | Type with speciesa (number of samples)b |
|---|---|---|---|---|
| Blood | 819 | 6 | 0.7 | B3 (4) |
| Urine | 463 | 1 | 0.2 | B35 (1) |
| Stool | 452 | 9 | 2.0 | C1 (3), B3 (1), C2 (1), A31 (1), F41 (1), ND (1)c |
ND indicates not detected.
Types are written as HAdV X (species) Y (type).
Among 16 HAdV‐positive samples, 15 samples were available for type sequencing and 12 representative samples were used.
HAdV was detected in the conventional PCR, but the stored sample was not amplified during the subsequent nested PCR for typing.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 1.
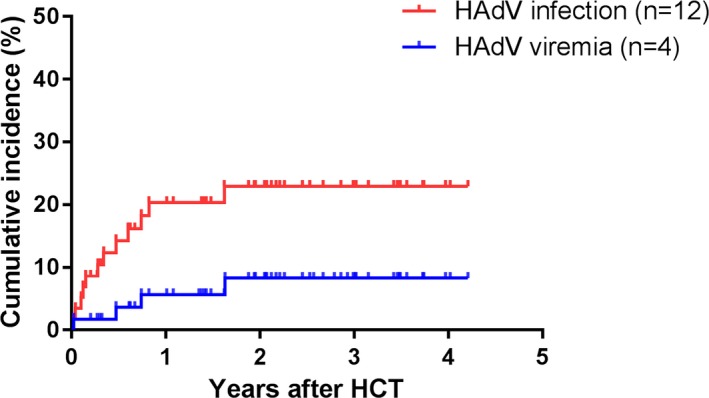
Cumulative incidence of human adenovirus (HAdV) infection and HAdV viremia in pediatric recipients with allogeneic hematopoietic cell transplantation. The cumulative incidence rates of HAdV infection and viremia at 1 year after HCT were 20% and 6% and 2 years after HCT were 23% and 8%, respectively. Median time to first HAdV infection after HCT was 113 days (range, 7‐596 days), whereas median time to first HAdV viremia was 221 days (range, 7‐596 days). HCT, hematopoietic cell transplantation
Table 3.
Detailed characteristics and outcomes among recipients with human adenovirus (HAdV) infection (n = 12)
| Case No. | Age/Sex | UD | Donor/Source | Max acute GVHD | Max chronic GVHD | Relapseda | Co‐infectionsb | HAdV infection‐associated characteristics | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Onset (day) | Specimen (type with species)c | ALC (/μL) | Max copies (/mL) | Symptom/Disease status | Antiviral treatment | Outcome | ||||||||
| Viremia, n = 4 | ||||||||||||||
| 016 | 17/F | AA | MRD/PB | 2 | E | No | CMV, BKV | 270, 278 | B (B3), S (C1) | 250 | 1800 | Melena | No | Resolved |
| 031 | 9/M | ALL | MUD/PB | 1 | E | Yes | CMV, BKV, RSV, NoV, IFI, | 591, 596 | R, B (B3) | 90 | 191 000 | Pneumonia | Yes | Deathe |
| 038‐2d | 3/F | AML | MRD/PB | 2 | 0 | Yes | CMV, IFI | 7, 16 | B (B3), U | 330 | 9300 | Hematuria | No | Resolved |
| 047 | 6/F | NBL | MMRD/PB | 2 | E | Yes | CMV, BKV, RSV, IFI | 172, 172, 252 | B (B3), R, S (B3) | 270 | 2000 | Pneumonia | Yes | Deathf |
| Non‐viremia, n = 8 | ||||||||||||||
| 002 | 20 mo/M | CGD | MUD/PB | 3 | 0 | No | CMV, EBV | 42 | U (B35) | 690 | N/A | Hematuria | No | Resolved |
| 018 | 10/M | AA | MUD/CB | 1 | 0 | No | CMV, EBV, PIV | 219 | R | 3610 | N/A | Sinusitis | No | Resolved |
| 028 | 8 mo/M | ALL | MRD/PB | 1 | L | Yes | No | 102 | S (C2) | 1620 | N/A | Diarrheal | No | Resolved |
| 038‡ | 2/F | AML | MRD/PB | 1 | 0 | No | CMV, RSV | 53 | S | 380 | N/A | Diarrheal | No | Resolved |
| 039 | 2/F | AML | MUD/PB | 0 | 0 | No | No | 15 | S (F41) | 210 | N/A | No | No | Resolved |
| 042 | 7/M | NBL | MUD/PB | 1 | L | Yes | BKV, HCoRV | 124 | R | 1390 | N/A | Nog | No | Resolved |
| 051 | 5/M | NBL | MMRD/PB | 2 | 0 | Yes | No | 301 | R | 730 | N/A | URTI | Yes | Resolved |
| 059 | 5/F | AML | MUD/CB | 3 | 0 | Yes | CMV, BKV | 50, 56 | S (A31), S (C1) | 1250 | N/A | Diarrheal | No | Resolved |
HAdV indicates human adenovirus; UD, underlying disease; GVHD, graft‐vs‐host disease; ALC, absolute lymphocyte count; AML, acute myeloid leukemia; NBL, neuroblastoma; AA, aplastic anemia; ALL, acute lymphoblastic leukemia; CGD, chronic granulomatous disease; MRD, matched related donor; MUD, matched unrelated donor; MMRD, mismatched related donor; PB, peripheral blood; CB, cord blood; E, extensive; L, localized; CMV, cytomegalovirus; IFI, invasive fungal infection; BKV, BK virus; RSV, respiratory syncytial virus; NoV, norovirus; EBV, Epstein–Barr virus; HCoRV, human coronavirus; PIV, parainfluenza virus; B, blood; U, urine, R, respiratory; S, stool; N/A, not applicable; URTI, upper respiratory tract infection.
The relapsed state is defined as a return of a malignant disease or the signs and symptoms of a disease after a period of improvement.
Co‐infection was defined as any infection caused by other pathogens in addition to HAdV before and after 7 days of HAdV‐positive period.
Cases without virus‐type information are presented as only sample types. Types are written as HAdV X (species) Y (type).
This recipient had received allogeneic hematopoietic cell transplantation (HCT) twice. The case No. 038 indicates the HAdV infection after 1st HCT, and the case No. 038‐2 indicates another HAdV infection after the 2nd HCT.
This recipient died due to progression of severe pneumonia with underlying poorly controlled extensive chronic GVHD. He also had RSV, CMV with antigenemia (maximum titer: 74/200 000 WBCs) and HAdV infections. RSV and HAdV were detected in respiratory specimens, but only HAdV was detected in blood specimens with very high titers (19 000 copies/mL) at the time of death. CMV was well‐controlled by antiviral management (0/200 000 WBCs at the time of death).
This recipient died due to progression of severe pneumonia with underlying poorly controlled extensive chronic GVHD. RSV and HAdV were detected in respiratory specimens, ut HAdV was the only pathogen detected in blood specimen at the time of death. Further study on HAdV titer in this patient could not be performed because research sample collections during the period of disease progression were not available owing to an outbreak of Middle‐East respiratory syndrome coronavirus (MERS‐CoV) at our center in 2015.31
This recipient was admitted to the hospital due to anemia and developed fever during red blood cell transfusion. Respiratory multiplex PCR was performed as a routine care practice.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.3. Characteristics of HAdV–infected patients
Among the 12 HAdV‐infected patients, 7 (58%, 7/12) experienced a relapse of their underlying diseases. One patient (recipient 038) had 2 separate episodes of HAdV infection; this patient developed HAdV diarrhea without viremia at day 53 after the 1st HCT, and developed another episode of HAdV infection (HAdV detected in urine and blood) at day 7 and 16 after the 2nd HCT which was performed more than 1 year after the 1st HCT. Ten of 12 HAdV infections were probable HAdV diseases and 2 were asymptomatic infections. Enteritis with diarrhea or melena (n = 4), and respiratory illness (n = 4) were the most common clinical manifestations, followed by hematuria (n = 2) and asymptomatic condition (n = 2) (Table 3).
Among 12 patients with HAdV infection, 3 of 4 patients with viremia (75%, 3/4) received intensive immunosuppressive agents due to extensive chronic GVHD, whereas patients without viremia did not experience extensive chronic GVHD (P = .006). Two patients with viremia had disseminated HAdV infection at the time of death. These 2 deceased patients had suffered from the progression of severe HAdV pneumonia, persistent HAdV viremia, and poorly controlled extensive chronic GVHD (Table 3). The ALCs of patients with viremia were significantly lower than those of patients without viremia (median, 260/μL; range, 90‐330/μL vs 990/μL; range, 210‐3610/μL) (P = .028).
3.4. HAdV typing by sequencing analysis
Among the total of 16 HAdV‐detected samples, 15 samples were available for type sequencing and 12 representative samples were studied (Table 2). Among them, HAdV 3 was the most commonly detected type (n = 5), followed by HAdV 1 (n = 3). There was only one case for HAdV 2, 31, 35, and 41, respectively (Table 3). A phylogenetic tree for those 12 HAdV is shown in Figure S2A.
In typing analysis, HAdV 3 was the only type detected in blood, while various types were detected in stool (HAdV 1, 2, 3, 31 and 41) (P = .029). One urine specimen was typed as HAdV 35 (Table 2). Of note, different types of HAdV were detected sequentially in the same recipient in 2 cases (Figure 2). In one patient (recipient 016), HAdV 3 was detected in blood (day 270) and HAdV 1 was detected in stool (day 278). In another patient (recipient 059), HAdV 31 was detected on day 50 and HAdV 1 was detected on day 56 in stool samples (Figure S2B).
Figure 2.
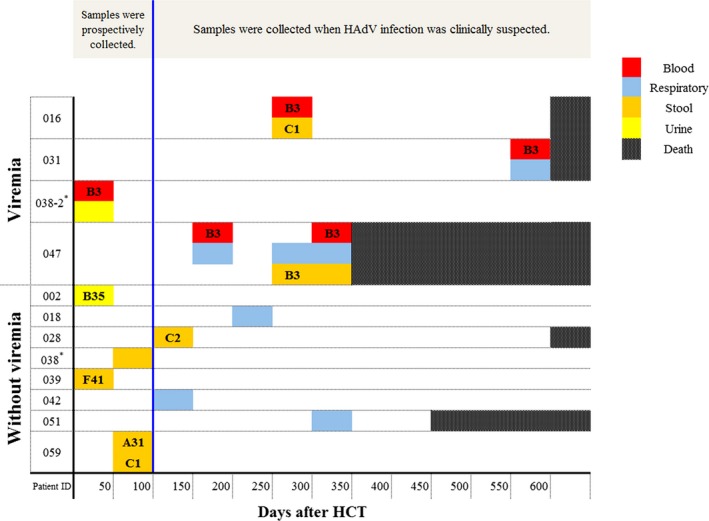
Distribution of human adenovirus (HAdV) positive specimens in 12 patients. In the typing analysis, HAdV 3 was the only type detected in all HAdV positive blood specimens, while various types were detected in stool specimens (HAdV 1, 2, 3, 31, and 41). Of note, different types were detected sequentially in recipient 016 and recipient 059. *This recipient had received allogeneic HCT twice. The case No. 038 indicates the HAdV infection after1st HCT, and the case No. 038‐2 indicates the HAdV infection after 2nd HCT. HCT, hematopoietic cell transplantation
3.5. Risk factors for HAdV infection and viremia
The univariate analysis did not identify potential risk factors for HAdV infection (Table S2). In cases of HAdV viremia (n = 3), extensive chronic GVHD was identified as a risk factor by univariate analysis (P = .013); however, we were unable to apply multivariate analysis due to the small number of cases (Table S3).
3.6. Overall survival and HAdV‐related mortality
In the univariate analysis, category of underlying disease, relapsed state of underlying disease, type of donor, intensity of conditioning regimen, use of ATG, extensive chronic GVHD, and existence of HAdV viremia were identified as potential risk factors for mortality (P < .1). HAdV viremia appeared to be a significant risk factor for mortality in univariate analysis (P = .045), but was not significant in multivariate analysis (model II) (hazard ratio [HR] 1.39, 95% CI 0.25‐7.56, P = .70) (Table S4). Relapsed state of underlying disease (HR 6.73, 95% CI 2.04‐22.19, P = .002) and extensive chronic GVHD (HR 5.49, 95% CI 1.35‐22.25, P = .017) were identified as significant risk factors for mortality in multivariate analysis (model II) (Table S4). Overall survival was not significantly different among recipients with HAdV infection (45%) vs recipients without HAdV infection (65%) (P = .72) (Figure 3).
Figure 3.
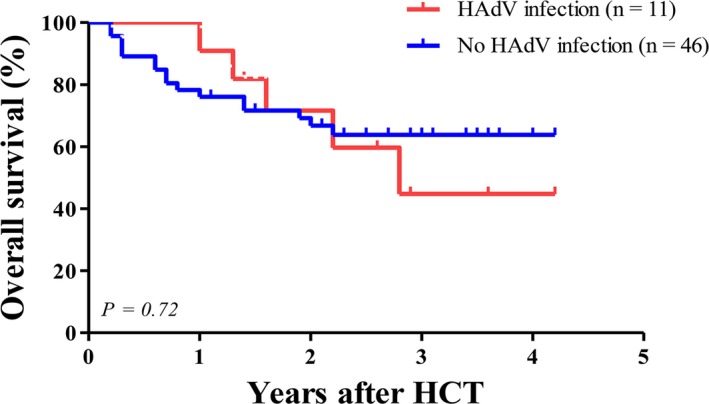
Overall survival after allogeneic hematopoietic cell transplantation between recipients with human adenovirus (HAdV) infection and those without HAdV infection. The Kaplan‐Meier curve showed no significant difference between recipients with HAdV infection and those without HAdV infection (45% in recipients with HAdV viremia vs 65% in recipients without HAdV viremia, P = .72 by log‐rank test)
4. DISCUSSION
We have described the epidemiology of HAdV infection in Korean allogeneic HCT recipients and identified the HAdV types. We observed only HAdV 3 in viremia patients, and multiple different HAdV types in other sites.
The results of this study highlight 2 epidemiologic findings that are different from those of previous studies. First, the cumulative incidence of HAdV viremia was lower than those of previous studies. In this study, the cumulative incidences of HAdV viremia at day 100 and 1 year were 2% and 6%, whereas the rates reported in previous studies were 6%‐50% in pediatric patients.7, 8, 19, 20 One possible explanation is the use of alemtuzumab or ex vivo T‐cell depletion, which we do not apply at our center. T‐cell depletion is used to reduce GVHD at certain centers, but is also a major risk factor for HAdV infection in HCT recipients because it is associated with significant immunosuppression during the early post‐transplant period.21 Therefore, the use of less intensive T‐cell depletion strategies at our center may explain the lower incidence of HAdV viremia, especially during the early post‐transplant period.6, 20, 22 Second, the onset of HAdV infection appeared to be delayed in our study compared to the results of previous studies. Previous studies including recipients who received alemtuzumab‐using T‐cell depletion or ex vivo T‐cell depletion reported that the majority of HAdV infections occurred before day 100 and were associated with high morbidity and mortality.2, 3, 19, 23, 24 Feuchtinger et al2 conducted a prospective study of 21 children and showed that 90% of HAdV infections with viremia occurred within 3 months after HCT. Lion et al24 also reported that the median onset days of HAdV detection after HCT was 18 days (range, day −7 to > day 100). In contrast, seven of 12 HAdV infections (58%) and 3 of 4 HAdV viremia cases (75%) occurred after post‐HCT day 100 in our study. The monitoring period should be applied according to the patient's immune status.7 Therefore, it may be necessary to expand regular HAdV monitoring in patients with delayed immune reconstitution.
Predominant HAdV types in allogeneic HCT recipients may differ according to country and/or geographic region, as well as those in general populations. In this study, HAdV 3 (n = 5) was identified as the most prevalent type, followed by HAdV 2 (n = 3). In contrast, previous studies that were mostly conducted in European countries reported that species C (HAdV types 2, 1, and 5 in order of frequency) and species A (HAdV 31) were the most prevalent species among pediatric HCT recipients.3, 8, 19 In community infection settings, HAdV 2, 1, and 3 were identified as the most prevalent genotypes in the UK and Croatia, whereas HAdV 3 (37%), 7 (23%), and 2 (11%) were identified as the prevalent types in Korea.13, 25 The results of type analyses in East Asian countries such as Taiwan and Japan also showed that species B (especially HAdV 3) was most prevalent in the community.13, 14, 26 HAdV C is well known for persistent infection in mucosal lymphocytes and tissues. Hence, HAdV C may be more likely to be in latent state before transplantation which can be linked to early post‐HCT reactivation. Difference in the prevalence of specific virus types in the community might be one of possible explanations why the incidence of HAdV infection in our study is lower than those of previous studies in European countries.27, 28
The high HAdV loads in blood have known to be an important risk factor for the disease severity and mortality. Therefore, the ECIL‐4 guidelines recommended quantitative PCR monitoring of HAdV in peripheral blood.7, 8 In addition to viral loads, we also questioned whether the severity of clinical manifestation or the rate of viremia would differ according to virus type, because all viremia cases were caused by HAdV 3 (n = 4) and 2 patients died of conditions related to disseminated HAdV infection. Taniguchi et al23 reported that disseminated HAdV infection occurred exclusively in species B virus‐infected recipients (HAdV 11 in 5 recipients, 34 and 35 in one recipient, respectively) among 110 Japanese adult recipients with haploidentical HCT. Feghoul et al3 reported surveillance data in 72 French pediatric allogeneic HCT recipients, and suggested that species C was not highly virulent, despite its prevalence. In contrast, Lion et al19, 24 reported that most fatal cases were associated with species C among 132 Austrian pediatric allogeneic HCT recipients. The relationship between HAdV types and clinical severity and/or dissemination remains uncertain, based on current data. Hence, further studies are needed.
We observed interesting findings in virus‐type analyses. First, different types of HAdV were detected in the same recipient within a short time interval (recipient 059) (Figure S2B). Second, the types of HAdV detected in stool were different from the types detected in blood in certain patients (recipient 016). Kroes et al reported similar results that data for 83 pediatric HCT recipients. Among 33 HAdV‐detected recipients, multiple HAdV types were detected in 12 recipients (36%).6, 29, 30 These findings support the hypothesis of endogenous reactivation from latent state in tissue, rather than consecutive HAdV infections from exogenous sources like other individuals or environment.27
Our findings need to be interpreted with caution. First, the refusal to participate and drop rate during the study period were high. Major reasons for refusal at enrollment and poor compliance included difficulty in collecting and delivering urine and stool specimens. Second, there is a possibility that we underestimated the incidence of HAdV infection and viremia due to the fact that routine surveillance was performed only until day 100 after HCT, because we found that significant numbers of HAdV infections occurred post‐HCT day 100. Lastly, the size of our study sample was relatively small to demonstrate definite risk factors for HAdV infection or viremia.
In summary, we observed lower incidence rate and more delayed onset pattern of HAdV infections than those of previous studies, which might be influenced by extent of T‐cell depletion and/or local HAdV epidemiology. HAdV 3 was the most common type, followed by HAdV 1, whereas HAdV 2 had been reported as the most prevalent type in western countries. Although we were unable to identify potential risk factors for HAdV infection or viremia, we found that HAdV‐infected recipients with viremia were more likely to have extensive chronic GVHD and lower lymphocyte counts than those without viremia.
In conclusion, this study demonstrated the distinct epidemiology of HAdV in Korean recipients. Watchful monitoring for HAdV viremia is recommended in HAdV‐infected HCT recipients, particularly in those with severe lymphopenia or extensive chronic GVHD. Additional studies are necessary to clarify the relationships between HAdV type and clinical severity or rate of viremia.
CONFLICTS OF INTEREST
The authors of this report have no conflicts of interest to disclose.
AUTHORS’ CONTRIBUTION
Ji‐Man Kang and Yae‐Jean Kim designed the study; Ji‐Man Kang, Ki‐Sup Park, and Jong Min Kim collected samples and performed experiments; Ji‐Man Kang and Jong Min Kim coordinated clinical data collection and did the statistical analyses; Ji‐Man Kang and Hee Jae Huh analyzed the sequencing data; Keon Hee Yoo, Ki Woong Sung Hong‐Hoe Koo, Chang‐Seok Ki, Nam Yong Lee and Yae‐Jean Kim supervised data collection and analysis, and reviewed the manuscript; and Ji‐Man Kang and Yae‐Jean Kim wrote the paper. All authors approved the final submitted manuscript.
Supporting information
Kang J‐M, Park K‐S, Kim JM, et al. Prospective monitoring of adenovirus infection and type analysis after allogeneic hematopoietic cell transplantation: A single‐center study in Korea. Transpl Infect Dis. 2018;20:e12885 10.1111/tid.12885
Funding information
This study was supported by an intramural fund of Samsung Medical Center.
REFERENCES
- 1. Teramura T, Naya M, Yoshihara T, Kanoh G, Morimoto A, Imashuku S. Adenoviral infection in hematopoietic stem cell transplantation: early diagnosis with quantitative detection of the viral genome in serum and urine. Bone Marrow Transplant. 2004;33:87‐92. [DOI] [PubMed] [Google Scholar]
- 2. Feuchtinger T, Lucke J, Hamprecht K, et al. Detection of adenovirus‐specific T cells in children with adenovirus infection after allogeneic stem cell transplantation. Br J Haematol. 2005;128:503‐509. [DOI] [PubMed] [Google Scholar]
- 3. Feghoul L, Chevret S, Cuinet A, et al. Adenovirus infection and disease in paediatric haematopoietic stem cell transplant patients: clues for antiviral pre‐emptive treatment. Clin Microbiol Infect. 2015;21:701‐709. [DOI] [PubMed] [Google Scholar]
- 4. Ohrmalm L, Lindblom A, Omar H, et al. Evaluation of a surveillance strategy for early detection of adenovirus by PCR of peripheral blood in hematopoietic SCT recipients: incidence and outcome. Bone Marrow Transplant. 2011;46:267‐272. [DOI] [PubMed] [Google Scholar]
- 5. Chakrabarti S, Mautner V, Osman H, et al. Adenovirus infections following allogeneic stem cell transplantation: incidence and outcome in relation to graft manipulation, immunosuppression, and immune recovery. Blood. 2002;100:1619‐1627. [DOI] [PubMed] [Google Scholar]
- 6. Lion T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev. 2014;27:441‐462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matthes‐Martin S, Feuchtinger T, Shaw PJ, et al. European guidelines for diagnosis and treatment of adenovirus infection in leukemia and stem cell transplantation: summary of ECIL‐4 (2011). Transpl Infect Dis. 2012;14:555‐563. [DOI] [PubMed] [Google Scholar]
- 8. Mynarek M, Ganzenmueller T, Mueller‐Heine A, et al. Patient, virus, and treatment‐related risk factors in pediatric adenovirus infection after stem cell transplantation: results of a routine monitoring program. Biol Blood Marrow Transplant. 2014;20:250‐256. [DOI] [PubMed] [Google Scholar]
- 9. Swartling L, Allard A, Torlen J, Ljungman P, Mattsson J, Sparrelid E. Prolonged outbreak of adenovirus A31 in allogeneic stem cell transplant recipients. Transpl Infect Dis. 2015;17:785‐794. [DOI] [PubMed] [Google Scholar]
- 10. Ganzenmueller T, Buchholz S, Harste G, Dammann E, Trenschel R, Heim A. High lethality of human adenovirus disease in adult allogeneic stem cell transplant recipients with high adenoviral blood load. J Clin Virol. 2011;52:55‐59. [DOI] [PubMed] [Google Scholar]
- 11. Lindemans CA, Leen AM, Boelens JJ. How I treat adenovirus in hematopoietic stem cell transplant recipients. Blood. 2010;116:5476‐5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yilmaz M, Chemaly RF, Han XY, et al. Adenoviral infections in adult allogeneic hematopoietic SCT recipients: a single center experience. Bone Marrow Transplant. 2013;48:1218‐1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lynch JP 3rd, Kajon AE. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin Respir Crit Care Med. 2016;37:586‐602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med. 1953;84:570‐573. [DOI] [PubMed] [Google Scholar]
- 15. Hage E, Huzly D, Ganzenmueller T, Beck R, Schulz TF, Heim A. A human adenovirus species B subtype 21a associated with severe pneumonia. J Infect. 2014;69:490‐499. [DOI] [PubMed] [Google Scholar]
- 16. Kim YJ, Hong JY, Lee HJ, et al. Genome type analysis of adenovirus types 3 and 7 isolated during successive outbreaks of lower respiratory tract infections in children. J Clin Microbiol. 2003;41:4594‐4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeulin H, Salmon A, Bordigoni P, Venard V. Comparison of in‐house real‐time quantitative PCR to the Adenovirus R‐Gene kit for determination of adenovirus load in clinical samples. J Clin Microbiol. 2010;48:3132‐3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol. 2006;151:1587‐1602. [DOI] [PubMed] [Google Scholar]
- 19. Lion T, Kosulin K, Landlinger C, et al. Monitoring of adenovirus load in stool by real‐time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia. 2010;24:706‐714. [DOI] [PubMed] [Google Scholar]
- 20. Watson T, MacDonald D, Song X, et al. Risk factors for molecular detection of adenovirus in pediatric hematopoietic stem cell transplantation recipients. Biol Blood Marrow Transplant. 2012;18:1227‐1234. [DOI] [PubMed] [Google Scholar]
- 21. Booth C, Lawson S, Veys P. The current role of T cell depletion in paediatric stem cell transplantation. Br J Haematol. 2013;162:177‐190. [DOI] [PubMed] [Google Scholar]
- 22. Hubmann M, Fritsch S, Zoellner AK, et al. Occurrence, risk factors and outcome of adenovirus infection in adult recipients of allogeneic hematopoietic stem cell transplantation. J Clin Virol. 2016;82:33‐40. [DOI] [PubMed] [Google Scholar]
- 23. Taniguchi K, Yoshihara S, Tamaki H, et al. Incidence and treatment strategy for disseminated adenovirus disease after haploidentical stem cell transplantation. Ann Hematol. 2012;91:1305‐1312. [DOI] [PubMed] [Google Scholar]
- 24. Lion T, Baumgartinger R, Watzinger F, et al. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood. 2003;102:1114‐1120. [DOI] [PubMed] [Google Scholar]
- 25. Tabain I, Ljubin‐Sternak S, Cepin‐Bogovic J, Markovinovic L, Knezovic I, Mlinaric‐Galinovic G. Adenovirus respiratory infections in hospitalized children: clinical findings in relation to species and serotypes. Pediatr Infect Dis J. 2012;31:680‐684. [DOI] [PubMed] [Google Scholar]
- 26. Sriwanna P, Chieochansin T, Vuthitanachot C, Vuthitanachot V, Theamboonlers A, Poovorawan Y. Molecular characterization of human adenovirus infection in Thailand, 2009‐2012. Virol J. 2013;10:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garnett CT, Talekar G, Mahr JA, et al. Latent species C adenoviruses in human tonsil tissues. J Virol. 2009;83:2417‐2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heemskerk B, Lankester AC, van Vreeswijk T, et al. Immune reconstitution and clearance of human adenovirus viremia in pediatric stem‐cell recipients. J Infect Dis. 2005;191:520‐530. [DOI] [PubMed] [Google Scholar]
- 29. Kroes AC, de Klerk EP, Lankester AC, et al. Sequential emergence of multiple adenovirus serotypes after pediatric stem cell transplantation. J Clin Virol. 2007;38:341‐347. [DOI] [PubMed] [Google Scholar]
- 30. Suzuki HI, Asai T, Okada K, et al. Disseminated adenovirus disease by multiple adenovirus serotypes following allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2008;14:353‐355. [DOI] [PubMed] [Google Scholar]
- 31. Cho SY, Kang JM, Ha YE, et al. MERS‐CoV outbreak following a single patient exposure in an emergency room in South Korea: an epidemiological outbreak study. Lancet. 2016;388:994‐1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


