Summary
Porcine‐transmissible gastroenteritis virus (TGEV) is a pathogenic coronavirus responsible for high diarrhoea‐associated morbidity and mortality in suckling piglets. We analysed the TGEV ORF3 gene using nested polymerase chain reaction and identified an ORF3a deletion in three field strains of TGEV collected from piglets in China in 2015. Eight TGEV ORF3 sequences were obtained in this study. Phylogenetic tree analysis of ORF3 showed that the eight TGEV ORF3 genes all belonged to the Miller cluster. CH‐LNCT and CH‐MZL were closely correlated with Miller M6, while CH‐SH was correlated with Miller M60. These results thus indicate that the existence of Miller, as well as the Purdue cluster, in Chinese field strains of TGEV. Furthermore, we found the first evidence for a large deletion in ORF3 resulting in the loss of ORF3a, previously reported in porcine respiratory coronavirus, in three field strains (CH‐LNCT, CH‐MZL, and CH‐SH) of TGEV. The results of the present study thus provide important information regarding the underlying evolution mechanisms of coronaviruses.
Keywords: deletion, ORF3a, transmissible gastroenteritis virus
1. Introduction
Porcine diarrhoea can lead to weight loss and death in piglets. Coronaviruses (CoVs) are a major cause of porcine diarrhoea in pig herds worldwide, leading to important economic losses (Zuniga, Pascual‐Iglesias, Sanchez, Sola, & Enjuanes, 2016). Coronaviruses comprise four genera: alpha‐, beta‐ (lineages A–D), gamma‐, and delta‐CoVs. The CoV antigen responsible for causing diarrhoea in swine has been identified (Ma et al., 2015). Porcine‐transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhoea virus (PEDV), and a natural TGEV deletion mutant porcine respiratory virus (PRCV) are alpha‐CoVs. Porcine hemagglutinating encephalomyelitis virus is a beta‐CoV, and porcine deltacoronavirus is a delta‐CoV. Coronaviruses have high mutation rates resulting in changes in tissue tropism, transmission pathways, and host specificities (Ma et al., 2015). Transmissible gastroenteritis virus is a member of the Coronaviridae family responsible for severe diarrhoea in suckling piglets (about 2 weeks old). It is an enveloped virus with a positive‐sense RNA genome of 28.5 kb, that was first reported in the USA in 1946 (Doyle & Hutchings, 1946). Transmissible gastroenteritis virus is currently divided into two distinct genogroups: the Miller cluster and the Purdue cluster (Hu et al., 2015). Transmissible gastroenteritis virus occurs in suckling piglets in swine‐producing areas in China (Weiwei et al., 2014). The TGEV genome was first sequenced by the analysis of polymerase chain reaction (PCR) products in 1993 (Chen, Pocock, & Britton, 1993).
The TGEV genome encodes nine proteins, including four structural proteins (spike, membrane, nucleocapsid, and envelope) and five non‐structural proteins (polyprotein 1a, polyprotein 1ab, ORF3a, ORF3b, and 7). Porcine respiratory virus is an S gene deletion mutant of TGEV first identified in Belgium in 1984 (Pensaert, Callebaut, & Vergote, 1986). Porcine respiratory virus can replicate in respiratory tissues in infected piglets and demonstrates selective tropism with very little replication in intestinal tissues (Wang & Zhang, 2015). Porcine respiratory virus was identified as a TGEV mutant because of their antigenic and genetic relationships, though sequence analysis showed heterogeneity in the S, ORF3a, and ORF3b genes between TGEV and PRCV (Vaughn, Halbur, & Paul, 1995). Sequence analysis of TGEV strains also showed that large deletions or insertions in the ORF3 gene region (Kwon, Saif, & Jackwood, 1998). Given the potential involvement of the TGEV ORF3 gene in virus virulence or tropism, it is necessary to establish a method for its characterization.
Reverse transcription PCR (RT‐PCR) was used to detect TGEV in 1991 (Page, Mawditt, & Britton, 1991), while nested PCR (nPCR) has been used to detect nucleic acids in members of the family Coronaviridae including PEDV (Chen et al., 2013), feline infectious peritonitis virus (Gamble, Lobbiani, Gramegna, Moore, & Colucci, 1997), canine coronavirus (Pratelli, Tempesta, Greco, Martella, & Buonavoglia, 1999), and TGEV (Rodriguez et al., 2012). Multiplex nPCR has also been used to differentiate between PEDV and TGEV infections (Jung, Kim, Kim, Kim, & Chae, 2003). In this study, we developed an nPCR‐based diagnostic field test for the rapid detection of TGEV infection in farmed pigs. Furthermore, we detected TGEV field strains belonging to Miller cluster in China, and also provided the first evidence for a large deletion in ORF3 resulting in the loss of ORF3a deletion in China.
2. Materials and Methods
2.1. Detection of viral RNA and sequencing
Samples or contents of the small intestine were collected from piglets with watery diarrhoea and dehydration at 44 pig farms in China in 2015. Samples were diluted with phosphate‐buffered saline to make 10% (v/v) suspensions. The suspensions were vortexed for 1 min and clarified by centrifugation for 10 min at 5,000 g and the supernatants were collected for RT‐nPCR. A volume of 30 μl of viral RNA was extracted from 140 μl of supernatants using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany), and first‐strand cDNA was synthesized with M‐MLV reverse transcriptase (Promega, Madison, WI, USA) using Oligo d(T)18 (TaKaRa, Dalian, China), according to the manufacturers’ instructions. Two primer pairs, TGE‐ORF3‐F1 and TGE‐ORF3‐F2, and TGE‐ORF3‐F1n and TGE‐ORF3‐F2n (Table S1), were designed and synthesized for RT‐nPCR according to the corresponding sequence of TGEV strain H16 (GenBank No. FJ755618). First PCR was performed in a total volume of 25 μl containing 10 ng of cDNA template, 1× EmeraldAmp PCR Master Mix (TaKaRa), and a 0.5 μM concentration of each primer. After initial denaturation at 95°C for 5 min, amplification was performed for 30 cycles, each consisting of denaturation at 95°C for 30 s, annealing at 50°C for 45 s, and elongation at 72°C for 60 s. Second PCR was performed in a total volume of 25 μl containing 1 μl of product of the first PCR, 1× EmeraldAmp PCR Master Mix (TaKaRa), and a 0.5 μM concentration of each primer. After initial denaturation at 95°C for 5 min, amplification was performed for 30 cycles as the described for the first PCR. The outer span, including primers TGE‐ORF3‐F1 and TGE‐ORF3‐F2, was 1602 base pairs (bp), and the inner span, including primers TGE‐ORF3‐F1n and TGE‐ORF3‐F2n, was 1417 bp. Fragments containing the full‐length TGEV ORF3 gene were amplified from samples using these primers. Furthermore, a pair of primers designated TGEV‐d‐F1 and TGEV‐d‐F2 was created to allow the rapid detection of TGEV field strains. PCR products were excised from 1.0% agarose gels, purified using an AxyPrep™ DNA Gel Extraction Kit (Axygen Scientific, Inc., CA, USA), cloned into the pMD18‐T, and introduced into Escherichia coli DH5α competent cells (TaKaRa) by transformation, according to the manufacturer's instructions. Eight recombinant DNA clones were sequenced by Comate Bioscience Company Limited (Jilin, China) and aligned using the MUSCLE method in MEGA5 software. A Construct/Test Maximum Likelihood Tree was obtained in Phylogeny analyse (a function of the MEGA software). Virus sequences used in this study are shown in Table S2.
3. Results and Discussion
Eight TGEV ORF3 sequences were obtained from eight farms in this study (Figure 1). The overall detection rate of TGEV in the samples by RT‐nPCR was 8/44 (18.2%). All the sequences in this study have been deposited in the GenBank database. The field strains and their accession numbers are shown in Table S3. The identities among the sequences ranged from 98.6% to 99.9%. Phylogenetic tree analysis of ORF3 showed that the eight TGEV ORF3 genes all belonged to the Miller cluster (Figure 1). CH‐SY and CH‐ZB were closely correlated with Miller M6, which is a virulent TGEV strain, while CH‐SH was correlated with Miller M60, which is an attenuated TGEV strain. Previous reports showed that the Purdue cluster also occurred in China (Weiwei et al., 2014). The current results thus indicate that the Miller, as well as the Purdue cluster, exists in Chinese field strains of TGEV. A large deletion (81 nucleotides [nt]) in ORF3 was found in three strains (CH‐LNCT, CH‐MZL, and CH‐SH) (Figure 2a). This may indicate that the ORF3‐deleted and non‐deleted TGEV field strains were both detected at farm in China.
Figure 1.
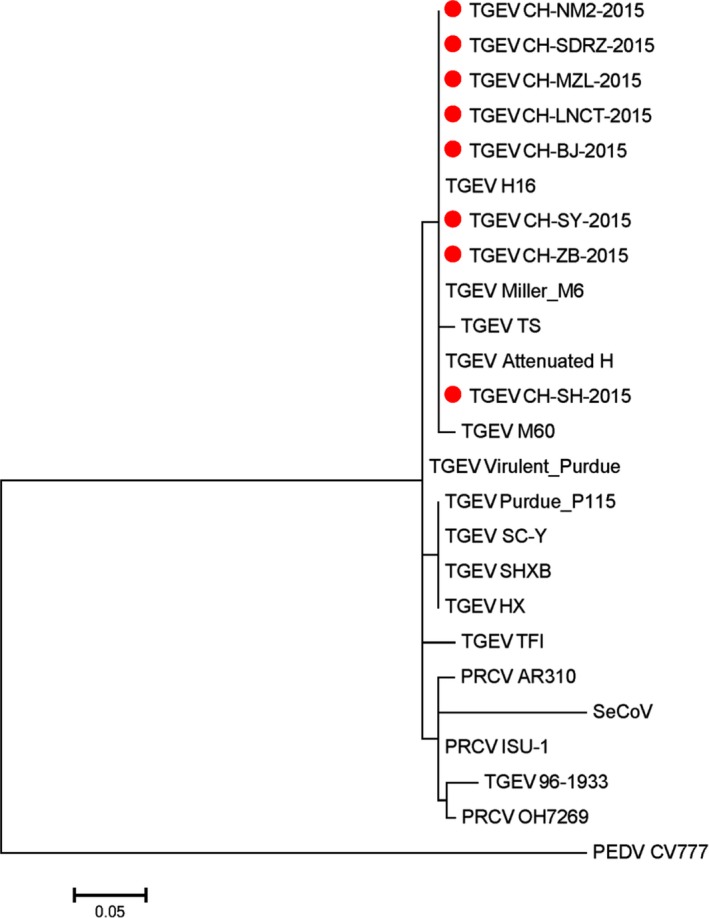
Phylogenetic tree based on the ORF3 gene sequence of TGEV virus strain. Sequences were analysed with a Construct/Test Maximum Likelihood Tree using the MUSCLE method in MEGA5.  Represents TGEV virus field strains in China in 2015. Scale bar indicates nucleotide substitutions. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Represents TGEV virus field strains in China in 2015. Scale bar indicates nucleotide substitutions. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Figure 2.
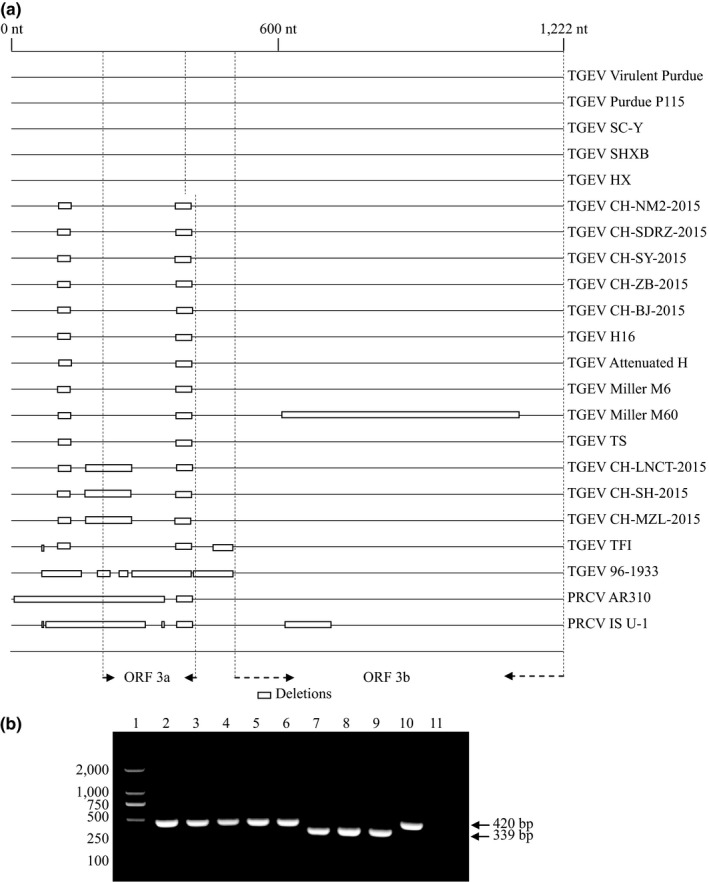
Deletions in ORF3 (3a and 3b) in TGEV field strains and PCR confirmation. (a) Deletions in ORF3 (3a and 3b) in TGEV field strains and other TGEV and PRCV strains. (b) PCR results of TGEV field strains with and without ORF3a deletion. Lane 1: DNA marker; lane 2: CH‐SDRZ; lane 3: CH‐SY; lane 4: CH‐ZB; lane 5: CH‐BJ; lane 6: CH‐NM2; lane 7: CH‐LNCT; lane 8: CH‐SH; lane 9: CH‐MZL; lane 10: Positive control (attenuated H); lane 11: negative control
We used the TGEV‐d‐F1 and TGEV‐d‐F2 primers to allow the rapid detection of TGEV strains with the 81‐nt deletion. The PCR products of TGE‐ORF3‐F1 and TGE‐ORF3‐F2 were used as a template. The PCR products of CH‐SDRZ, CH‐SY, CH‐ZB, and CH‐BJ were 420 bp indicating no deletion, while those of CH‐LNCT, CH‐SH, CH‐MZL were 339 bp, indicating the presence of the 81‐nt deletion (Figure 2b). This result validated the previous sequencing results. Transmissible gastroenteritis virus is difficult to grow in cell culture, and virus lacking 3a replicates particularly poorly in cell cultures (Paul, Vaughn, & Halbur, 1997). We tried to isolate the ORF3a‐deleted TGEV virus from samples prepared as above, but unfortunately were unable to do so (data not shown).
Previous reports detected a series of large deletions downstream of the TGEV S gene, resulting in loss of the transcription signal and the initiation codon of ORF3a. (McGoldrick, Lowings, & Paton, 1999). Similarly, a small plaque variant and two non‐pathogenic cell culture‐adapted strains of TGEV have deletions or insertions that alter ORF3 (Zhang et al., 2007). These results suggest that the ORF3 region of the TGEV genome is an important determinant for regulating viral virulence (Zuniga et al., 2016). Transmissible gastroenteritis virus strain 96‐1933 is a virulent variant isolated in England in 1996 (Jones & Paton, 1996), including an intact S gene, but with a deletion in ORF3a. There have been no other relevant reports of ORF3a‐deleted TGEV until the current study, which demonstrated an 81‐nt deletion in ORF3a. However, the locations of the current and 96‐1933 deletions differed; gene loss in 96‐1933 was due to a discontinuity in ORF3a, while the deletion of TGEV ORF3a in the current study was continuous, which has not previously been reported in TGEV.
Some TGEV viruses belonging to the Miller group have been found in China (attenuated H and TS) (Li et al., 2010; Wang et al., 2010), while Purdue strains, such as TGEV HX and SHXB strains, are also prevalent in China (Hu et al., 2015; Weiwei et al., 2014), thus providing increased opportunities for reorganization. The leader RNA‐binding site and ORF3a start site were included in the deleted region identified in TGEV the current study (Figure 3), and in the sequences of PRCV‐ISU‐1 (Lai, Welter, & Welter, 1995; Zhang et al., 2007). Previous studies on the genetic structures of PRCV and TGEV revealed two unique characteristics. First, the PRCV S gene lacks nt 621–681, resulting in a smaller S glycoprotein in PRCV than TGEV. Second, TGEV and PRCV differ in the ORF3 region; the leader RNA‐binding site (CTAAAC) in PRCV is altered or partially deleted before the ORF3a gene (Wang & Zhang, 2015).
Figure 3.
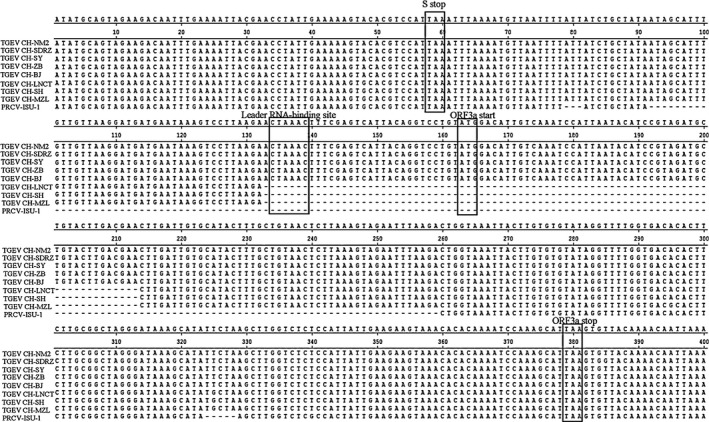
Deleted regions of leader RNA‐binding site and ORF3a start site in TGEV virus field strains in China in 2015, and in PRCV strain ISU‐1.
A deletion occurring at a common break point was previously found in non‐structural protein 3b of the naturally attenuated PRCV‐ISU‐1 and TGEV Miller M60 (Zhang et al., 2007). Sequence comparison showed that TGEV Miller strains had a closer relationship with PRCV‐ISU‐1 compared with Purdue strains. In this study, eight ORF3 sequences belonging to Miller cluster were identified in field strains of TGEV. Furthermore, a novel type of deletion within ORF3a was found in three TGEV field strains, suggesting that some of the differences among ORF3a sequences maybe with regard to PRCV. However, we have not yet succeeded in isolating ORF3a‐deleted TGEV, and have therefore been unable to evaluate its pathogenic mechanisms in pigs to date. However, further studies are planned to isolate this virus.
Chimeric viruses termed swine enteric coronaviruses (SeCoVs) were recently found in Central Eastern Europe (Belsham et al., 2016) and Italy (Boniotti et al., 2016) in 2016. We examined the relationship between ORF3 in SeCoVs and the ORF3 detected in this study by comparing the TGEV ORF3 sequences with the ORF3 gene of SeCoVs. As shown in Figure 1, the TGEV ORF3 sequences were not closely related to the ORF3 gene of SeCoVs, which was in turn closely related to PRCV AR310 and ISU‐1.
4. Conclusions
In summary, we characterized eight ORF3 genes in TGEV field strains from China. We also provided novel evidence for the existence of Miller cluster TGEV field strains in Chinese pig populations, indicating the complexity of TGEV field strains in China. Interestingly, we also identify a novel deletion in within ORF3a in three field strains of TGEV, similar to the deletion found in PRCV, thus reflecting the relationship between the TGEV and PRCV. The detected‐ORF3a deletion in the CH‐LNCT, CH‐MZL, and CH‐SH strains provides the basis for further evaluation pathogenicity and replication of TGEV, and further studies are needed to determine the role of TGEV ORF3a‐deletion field strains in porcine diarrhoea using an animal model. The results of the present study thus provide important information regarding the evolution of coronaviruses.
Supporting information
Acknowledgements
This study was funded by the National Key Technology Support Program (2015BAD12B02) and the National Natural Science Foundation of China (grant Nos. 31572541, and 31502092).
Zhang X, Zhu Y, Zhu X, et al. ORF3a deletion in field strains of porcine‐transmissible gastroenteritis virus in China: A hint of association with porcine respiratory coronavirus. Transbound Emerg Dis. 2017;64:698–702. 10.1111/tbed.12634
References
- Belsham, G. J. , Rasmussen, T. B. , Normann, P. , Vaclavek, P. , Strandbygaard, B. , & Botner, A. (2016). Characterization of a novel chimeric swine enteric coronavirus from diseased pigs in central eastern Europe in 2016. Transboundary and Emerging Diseases, 63, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti, M. B. , Papetti, A. , Lavazza, A. , Alborali, G. , Sozzi, E. , Chiapponi, C. , … Marthaler, D. (2016). Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerging Infectious Diseases, 22, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Liu, X. , Shi, D. , Shi, H. , Zhang, X. , Li, C. , … Feng, L. (2013). Detection and molecular diversity of spike gene of porcine epidemic diarrhea virus in China. Viruses, 5, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. M. , Pocock, D. H. , & Britton, P. (1993). Genomic organisation of a virulent Taiwanese strain of transmissible gastroenteritis virus. Advances in Experimental Medicine and Biology, 342, 23–28. [DOI] [PubMed] [Google Scholar]
- Doyle, L. P. , & Hutchings, L. M. (1946). A transmissible gastroenteritis in pigs. Journal of the American Veterinary Medical Association, 108, 257–259. [PubMed] [Google Scholar]
- Gamble, D. A. , Lobbiani, A. , Gramegna, M. , Moore, L. E. , & Colucci, G. (1997). Development of a nested PCR assay for detection of feline infectious peritonitis virus in clinical specimens. Journal of Clinical Microbiology, 35, 673–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. , Li, N. , Tian, Z. , Yin, X. , Qu, L. , & Qu, J. (2015). Molecular characterization and phylogenetic analysis of transmissible gastroenteritis virus HX strain isolated from China. BMC Veterinary Research, 11, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. O. , & Paton, D. J. (1996). Classical transmissible gastroenteritis returns. Veterinary Record, 138, 166–167. [PubMed] [Google Scholar]
- Jung, K. , Kim, J. , Kim, O. , Kim, B. , & Chae, C. (2003). Differentiation between porcine epidemic diarrhea virus and transmissible gastroenteritis virus in formalin‐fixed paraffin‐embedded tissues by multiplex RT‐nested PCR and comparison with in situ hybridization. Journal of Virological Methods, 108, 41–47. [DOI] [PubMed] [Google Scholar]
- Kwon, H. M. , Saif, L. J. , & Jackwood, D. J. (1998). Field isolates of transmissible gastroenteritis virus differ at the molecular level from the Miller and Purdue virulent and attenuated strains and from porcine respiratory coronaviruses. Journal of Veterinary Medical Science, 60, 589–597. [DOI] [PubMed] [Google Scholar]
- Lai, C. H. , Welter, M. W. , & Welter, L. M. (1995). The use of arms PCR and RFLP analysis in identifying genetic profiles of virulent, attenuated or vaccine strains of TGEV and PRCV. Advances in Experimental Medicine and Biology, 380, 243–250. [DOI] [PubMed] [Google Scholar]
- Li, J. Q. , Cheng, J. , Lan, X. , Li, X. R. , Li, W. , Yin, X. P. , … Liu, J. X. (2010). Complete genomic sequence of transmissible gastroenteritis virus TS and 3’ end sequence characterization following cell culture. Virologica Sinica, 25, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Zhang, Y. , Liang, X. , Lou, F. , Oglesbee, M. , Krakowka, S. , & Li, J. (2015). Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio, 6, e00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick, A. , Lowings, J. P. , & Paton, D. J. (1999). Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Archives of Virology, 144, 763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, K. W. , Mawditt, K. L. , & Britton, P. (1991). Sequence comparison of the 5′ end of mRNA 3 from transmissible gastroenteritis virus and porcine respiratory coronavirus. Journal of General Virology, 72, 579–587. [DOI] [PubMed] [Google Scholar]
- Paul, P. S. , Vaughn, E. M. , & Halbur, P. G. (1997). Pathogenicity and sequence analysis studies suggest potential role of gene 3 in virulence of swine enteric and respiratory coronaviruses. Advances in Experimental Medicine and Biology, 412, 317–321. [DOI] [PubMed] [Google Scholar]
- Pensaert, M. , Callebaut, P. , & Vergote, J. (1986). Isolation of a porcine respiratory, non‐enteric coronavirus related to transmissible gastroenteritis. Veterinary Quarterly, 8, 257–261. [DOI] [PubMed] [Google Scholar]
- Pratelli, A. , Tempesta, M. , Greco, G. , Martella, V. , & Buonavoglia, C. (1999). Development of a nested PCR assay for the detection of canine coronavirus. Journal of Virological Methods, 80, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, E. , Betancourt, A. , Relova, D. , Lee, C. , Yoo, D. , & Barrera, M. (2012). Development of a nested polymerase chain reaction test for the diagnosis of transmissible gastroenteritis of pigs. Revue Scientifique et Technique, 31, 1033–1044. [PubMed] [Google Scholar]
- Vaughn, E. M. , Halbur, P. G. , & Paul, P. S. (1995). Sequence comparison of porcine respiratory coronavirus isolates reveals heterogeneity in the S, 3, and 3‐1 genes. Journal of Virology, 69, 3176–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Chen, J. , Shi, H. , Qiu, H. , Xue, F. , Liu, C. , … Feng, L. (2010). Molecular characterization of a Chinese vaccine strain of transmissible gastroenteritis virus: Mutations that may contribute to attenuation. Virus Genes, 40, 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , & Zhang, Y. (2015). Genomic characterization of a new PRCV variant, United States, 2014. Transboundary and Emerging Diseases, 6, 1–3. [DOI] [PubMed] [Google Scholar]
- Weiwei, H. , Qinghua, Y. , Liqi, Z. , Haofei, L. , Shanshan, Z. , Qi, G. , … Qian, Y. (2014). Complete genomic sequence of the coronavirus transmissible gastroenteritis virus SHXB isolated in China. Archives of Virology, 159, 2295–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Hasoksuz, M. , Spiro, D. , Halpin, R. , Wang, S. , Stollar, S. , … Saif, L. (2007). Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology, 358, 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga, S. , Pascual‐Iglesias, A. , Sanchez, C. M. , Sola, I. , & Enjuanes, L. (2016). Virulence factors in porcine coronaviruses and vaccine design. Virus Research, 226, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


