Summary
Bats are a unique group of mammals well suited to be hosts for emerging viruses. With current rates of deforestation and urbanization, redistribution of bat habitats to urban and suburban areas may bring bats into closer contact with livestock and humans. Common flying fox, Pteropus medius (previously known as Pteropus giganteus), forms large communal roosts on treetops, often in close proximity to human habitation in Sri Lanka. This report describes the detection of coronavirus RNA in P. medius bat guano collected in Peradeniya, Sri Lanka. These viruses had >97% nucleotide identity with coronaviruses detected in Cynopterus sphinx, Scotophilus heathii and S. kuhlii bats in Thailand. Pteropus medius is widespread in Asia and appears to excrete group D coronaviruses, which are hitherto confined to bats; however, these findings may have public health implications in the future.
Keywords: coronavirus, flying fox, Sri Lanka
1. INTRODUCTION
Bats contribute to maintain a balanced ecosystem through disseminating plant seeds or eating some types of insects. Some bat species are at risk and endangered. The displacement and redistribution of bats from their natural habitats expose susceptible human and animal populations to micro‐organisms (Han et al., 2015). Bats are a unique group of mammals that can fly long distances for daily foraging and aggregate in large colonies. Daily movement patterns, compact roosting structure, hibernation, periodic migration and long lifespan may make bats suitable hosts for emerging viruses (Han et al., 2015).
Coronaviruses (CoV) are a group of enveloped, single‐stranded (positive‐sense) RNA viruses that belong to the order Nidovirales, family Coronaviridae. They are divided into four major groups: alpha, beta, gamma and delta coronaviruses, only the alpha and beta coronaviruses being known to have infected humans (de Groot et al., 2013). The earliest known human coronaviruses (HCoV), HCoV‐229E and HCoV‐OC43 are causes of mild upper respiratory infections. The identification of the precursor of Severe Acute Respiratory Syndrome coronavirus (SARS‐CoV) in Rhinolophid bats led to more intensive investigations of coronaviruses in humans and in bats (Han et al., 2015). NL63 and HKU1 are newly discovered coronaviruses endemic in humans (Woo et al., 2005; Pyrc, Berkhout, & Hoek, 2007), and MERS‐coronavirus is a newly discovered zoonotic coronavirus (de Groot et al., 2013). Recent studies have shown that bat coronaviruses were the source of many human coronaviruses including SARS‐CoV, HCoV‐229E and NL63 (Han et al., 2015).
2. MATERIALS AND METHODS
A total of 51 treetops inhabited by fruit bats (Pteropus medius) were numbered on the first site visit, and these treetops were clustered in the Western corner of the Royal Botanical Gardens, bordering the River Mahaweli. A total of 50 bat guano samples were collected from randomly chosen treetops based on the original numbering. Of the 51 treetops numbered, 10 were chosen in the following order: 1st, 5th, 10th, 15th, 20th, 25th, 30th, 35th, 40th and 45th and sampled from February to March 2014. The ground areas under the selected trees were covered with clean polythene sheets. The accumulations of droppings were observed and identified immediately prior to collection to confirm the origin of the guano. About 1–2 g of each bat guano sample was collected into a vial with viral transport medium (VTM). The guano samples were transported to the Virology Laboratory, Faculty of Medicine, University of Peradeniya for screening with RT‐PCR work. The ethical clearance (EC No: 2012/EC/52) for the study was obtained from the Ethical Review Committee of the Faculty of Medicine, University of Peradeniya, Sri Lanka.
Viral RNA was extracted using QIAGEN QIAamp Viral RNA kit (QIAGEN, Hilden, Germany) as recommended by the manufacturer. The cDNA was generated from RNA using Superscript III reverse transcriptase (Invitrogen) with random hexamers. The cDNA was screened for the presence of coronavirus using a broad range hemi‐nested RT‐PCR targeting conserved regions of the RdRp gene for detecting known and unknown coronaviruses as previously described (Hemida et al., 2017). Positive and negative controls were included for each run. RT‐PCR products were analysed using agarose gel electrophoresis to identify a product size of 440 base pairs indicating the presence of known or unknown coronaviruses. DNA sequencing was carried out on four strongly positive RT‐PCR products at School of Public Health, University of Hong Kong, Hong Kong.
3. RESULTS AND DISCUSSION
The bats from which the guano samples taken were morphologically identified as Pteropus medius (previously known as P. giganteus) belonging to the family Pteropodedae. Eight (16%) of the 50 bat guano samples were positive for coronavirus; three of 10 collected on 1 February, four of 10 collected on 8 February, zero of 16 collected on 10 March and one of 14 collected on 19 March. Genome sequencing of the RT‐PCR products was carried out on four selected bat guano; BtCoV Peradeniya‐HKU CN10 collected on 1 February and BtCoV Peradeniya‐HKU CN12, CN14 and CN15 collected on 8 February. The virus sequences showed >97% nucleotide identity with lineage D beta coronaviruses previously identified in Thailand in a fruit bat species, Cynopterus sphinx and insectivorous bats, Scotophilus heathii and Scotophilus kuhlii (Wacharapluesadee et al., 2015) (Figure 1). These viruses had approximately 75% nucleotide identity to the HKU9 like bat coronaviruses, which have widespread geographic distribution.
Figure 1.
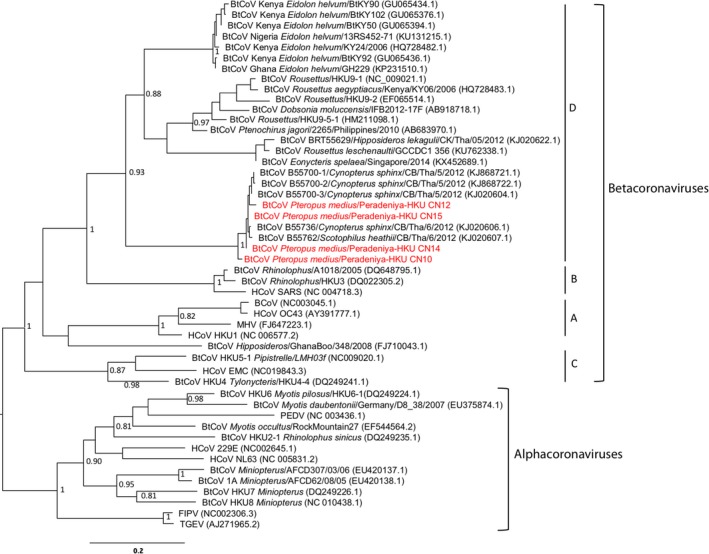
Phylogenetic analysis of coronavirus RdRp gene nucleotide sequences. The tree was constructed by maximum‐likelihood method using PhyML with an alignment of 339 nt. And the branch support was estimated by aLRT method, and values larger than 0.8 were indicated in the tree. The tree was mid‐point rooted at the branch between alpha and betacoronaviruses. Bat coronaviruses from Sri Lanka identified in this study were highlighted in red. The groups A‐D within the betacoronavirus lineage are indicated. GenBank accession number of sequences is shown in brackets [Colour figure can be viewed at http://wileyonlinelibrary.com]
Fruit bats, generally known as common flying foxes of genus Pteropus and family Pteropodidae, are a routine sight, in both rural and semi‐urbanized habitats, in Sri Lanka. They may roost in trees closer to human habitation. In 2002, there were 30 bat species documented in Sri Lanka (Yapa, Randeniya, & Ratnasooriya, 2002). Recently, a novel bat lyssavirus designated as Gannoruwa bat lyssavirus was discovered in Pteropus medius bats in the same geographic area of Peradeniya (Gunawardena et al., 2016). Over the last few years, identification of new coronaviruses in bats including the most recently reported lineage D betacoronavirus in cave nectar bats (Eonycteris spelaea) in Singapore (Mendenhall et al., 2017) led us to investigate coronaviruses in fruit bats in Sri Lanka. Detection of lineage D betacoronavirus in guano collected from Pteropus medius bats shows the presence of these viruses in bats, which roost in public places such as the Botanic gardens. Lineage D betacoronaviruses are strictly confined to bats up to now; however, they might evolve to become harmful to humans with public health implications. Pteropus medius are one of the largest species of bats, roost in large trees near waterways or rivers and fly distances of up to 150 Km daily for foraging (Newman, Field, Jong, & Epstein, 2011). They are widespread in South Asia and can be commonly found in peri‐domestic settings. In some cultures, these species are also hunted and consumed as food. New bats are born during February–May, and the samples collected in this study were from this period of new bat birthing and may explain the high rates of virus detection observed.
A 7‐week period of sampling has not enabled us to study the seasonality of the virus activity in the current study. The short fragment of the virus sequenced limits us from assigning a definitive taxonomic status to what appears to be a new coronavirus species and the public health implications of these findings need to be further explored.
CONFLICT OF INTEREST
The authors of the work have no conflict of interests to disclose.
ACKNOWLEDGEMENTS
This research was funded in part from University of Peradeniya RG/AF/13/38/M and the US National Institutes of Health (Contract No. HHSN272201500006C).
Kudagammana HDWS, Thevanesam V, Chu DKW, Eriyagama NB, Peiris JSM, Noordeen F. Coronaviruses in guano from Pteropus medius bats in Peradeniya, Sri Lanka. Transbound Emerg Dis. 2018;65:1122–1124. 10.1111/tbed.12851
REFERENCES
- de Groot, R. J. , Baker, S. C. , Baric, R. S. , Brown, C. S. , Drosten, C. , Enjuanes, L. , … Ziebuhr, J. (2013). Middle East respiratory syndrome coronavirus (MERS‐CoV): Announcement of the Coronavirus Study Group. Journal of Virology, 87, 7790–7792. 10.1128/JVI.01244-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena, P. S. , Marston, D. A. , Ellis, R. J. , Wise, E. L. , Karawita, A. C. , Breed, A. C. , … Fooks, A. R. (2016). Lyssavirus in Indian FlyingFoxes, Sri Lanka. Emerging Infectious Diseases, 22, 1456–1459. 10.3201/eid2208.151986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, H. J. , Wen, H. L. , Zhou, C. M. , Chen, F. F. , Luo, L. M. , Liu, J. W. , & Yu, X. J. (2015). Bats as reservoirs of severe emerging infectious diseases. Virus Research, 205, 1–6. 10.1016/j.virusres.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida, M. G. , Chu, D. K. , Perera, R. A. , Ko, R. L. , So, R. T. , & Ng, B. C. (2017). Coronavirus infections in horses in Saudi Arabia and Oman. Transboundary and Emerging Diseases, 64, 2093–2103. 10.1111/tbed.12630 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall, I. H. , Borthwick, S. , Neves, E. S. , Low, D. , Linster, M. , Liang, B. , … Smith, G. J. D. (2017). Identification of a lineage D betacoronavirus in cave nectar bats (Eonycteris spelaea) in Singapore and an overview of lineage D reservoir ecology in SE Asian bats. Transboundary and Emerging Diseases, 64, 1790–1800. 10.1111/tbed.12568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, S. H. , Field, H. E. , Jong, C. E. , & Epstein, J. H. (2011). Investigating the role of bats in emerging zoonoses: Balancing ecology, conservation and public health interests (1st Ed). Rome, Italy: FAO of the United Nations, Animal Production and Health Manual No 12: ISSN 1810‐1119. [Google Scholar]
- Pyrc, K. , Berkhout, B. , & Hoek, L. (2007). The novel human coronaviruses NL63 and HKU1. Journal of Virology, 81, 3051–3057. 10.1128/JVI.01466-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee, S. , Duengkae, P. , Rodpan, A. , Kaewpom, T. , Maneeorn, P. , Kanchanasaka, B. , … Hemachudha, T. (2015). Diversity of coronavirus in bats from Eastern Thailand. Virol Journal, 11, 12–57. 10.1186/s12985-015-0289-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P. C. , Lau, S. K. , Chu, C. M. , Chan, K. H. , Tsoi, H. W. , Huang, Y. , … Yuen, K. Y. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. Journal of Virology, 2005(79), 884–895. 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapa, W. B. , Randeniya, P. V. , & Ratnasooriya, W. D. (2002). Ecology and biology of bats in Sri Lanka. A survey on the distribution of bats in Sri Lanka. Final report, Colombo: NSF. URL: http://archive.cmb.ac.lk:8080/research/bitstream/70130/1081/1/YapaRathna_Review%20article%20on%20Sri%20Lankan%20Bats.pdf


