Summary
Equine coronaviruses (ECoV) are the only coronavirus known to infect horses. So far, data on ECoV infection in horses remain limited to the USA, France and Japan and its geographic distribution is not well understood. We carried out RT‐PCR on 306 nasal and 315 rectal swabs and tested 243 sera for antibodies to detect coronavirus infections in apparently healthy horses in Saudi Arabia and Oman. We document evidence of infection with ECoV and HKU23 coronavirus by RT‐PCR. There was no conclusive evidence of Middle East respiratory syndrome coronavirus infection in horses. Serological data suggest that lineage A betacoronavirus infections are commonly infecting horses in Saudi Arabia and Oman but antibody cross‐reactivities between these viruses do not permit us to use serological data alone to identify which coronaviruses are causing these infections.
Keywords: cross‐neutralization, equine coronavirus, HKU23, middle east respiratory syndrome coronavirus, polymerase chain reaction, serology
1. INTRODUCTION
Coronaviruses infect a wide range of mammals and birds, exhibit a marked tropism for epithelial cells of the respiratory and enteric tracts and cause a remarkably diverse spectrum of diseases in humans and livestock. They typically have a restricted host range, infecting only their natural host and closely related animal species but do have the capacity to cross the species barrier to infect new hosts. Equine coronavirus (ECoV) is the only coronavirus known to infect or cause disease in horses (Balasuriya, 2013). It belongs to the genus Betacoronavirus lineage A, as does human coronavirus OC43, bovine coronavirus and porcine hemagglutinating encephalomyelitis virus. Equine coronavirus was first isolated from faeces of a diarrhoeic foal in 1999 (ECoV‐NC99) in North Carolina, USA (Guy, Breslin, Breuhaus, Vivrette, & Smith, 2000), and was initially believed to only affect foals. Since 2010, there have been several reports of ECoV‐associated respiratory and enteric infections in adult horses in Japan, Europe and the United States, but its global distribution is still poorly defined (Kooijman, Mapes, & Pusterla, 2016; Miszczak et al., 2014; Oue, Morita, Kondo, & Nemoto, 2013; Pusterla, Holzenkaempfer, Mapes, & Kass, 2015).
Middle East respiratory syndrome coronavirus (MERS‐CoV) is a betacoronavirus within lineage C first identified in humans in 2012 and continues to pose a threat to global health (WHO, 2016 May 27). The evidence points to dromedary camels as a natural host and a major source for human MERS‐CoV infections (Alexandersen, Kobinger, Soule, & Wernery, 2014; Chu et al., 2015; Gutierrez, Tejedor‐Junco, Gonzalez, Lattwein, & Renneker, 2015; Memish et al., 2014; Perera et al., 2013; Reusken et al., 2014). There is no convincing evidence of MERS‐CoV infections in other domestic livestock species so far (Adney et al., 2016; Alexandersen et al., 2014; Hemida et al., 2013; Meyer et al., 2015; Perera et al., 2013). Sequence similarity comparisons of dipeptidyl peptidase‐4 (DPP4 [CD26]), the functional receptor for MERS‐CoV, have revealed that equine DPP4 is phylogentically closely related to human DPP4 and the binding affinity of MERS‐CoV spike S1 domain for equine DPP4 is similar to that of human and camel DPP4, raising the possibility that MERS‐CoV infections may occur among horses (Barlan et al., 2014). Serological studies of horses originating from Spain and the United Arab Emirates gave negative results for MERS‐CoV (Alexandersen et al., 2014; Meyer et al., 2015), but it is still relevant to investigate for evidence of MERS‐CoV in horses in areas endemic for MERS‐CoV.
We have tested for coronavirus infections in apparently healthy horses in Saudi Arabia and Oman by testing nasal and rectal swabs by RT‐PCR assays for MERS‐coronavirus and using a pan‐coronavirus RT‐PCR with potential to detect all coronaviruses, hitherto known and unknown. Following the detection of ECoV and HKU23 coronavirus (HKU23) by the pan‐coronavirus RT‐PCR, we tested the swab specimens with specific RT‐PCR assays for ECoV and HKU23 and tested the sera with serological assays to detect ECoV, bovine CoV (BCoV) (closely related to HKU23), as well as MERS‐CoV.
2. MATERIALS AND METHODS
2.1. Specimen collection
Serum samples (n = 243), nasal swabs (n = 306) and rectal swabs (n = 315) were collected from apparently healthy adult (>1 years of age) horses from East and Central provinces of the Kingdom of Saudi Arabia and Oman. Many horses had all three samples collected. These specimens were collected during the period February 2014 to January 2015. The numbers of specimens collected from different geographic regions of Saudi Arabia and Oman are indicated in Table 1. The animal management practices and exposures to other domestic animal species such as dromedaries are indicated. Farming activities include activities related to date palm plantations as well as other farming activities. Racing activities refer to horse racing within Saudi Arabia rather than participation in international races.
Table 1.
Geographic location of specimens collected and animal management practices
| Region | City | Provider code | Animal management practices | Contact with other animals | Animals in contact with | Nasal swabs | Rectal swabs | Sera | Virus RNA detected, if any |
|---|---|---|---|---|---|---|---|---|---|
| Saudi Arabia (Central) | Riyadh | 1 | Breeding | Occasionally | Camels, sheep, goat, chickens | 14 | 14 | 14 | ECoV (n = 5) |
| 2 | Farming, Breeding | Occasionally | Camels, sheep, goat | 0 | 0 | 33 | |||
| Saudi Arabia (East) | Dammam | 3 | Farming, Breeding | Frequent | Camels, sheep, goat, chickens | 103 | 103 | 19 | HKU23 |
| 4 | Breeding | Frequent | Camels, sheep, goat | 6 | 6 | 6 | |||
| Saudi Arabia (East) | Al‐Hasa | 5 | Farming, Breeding | Occasionally | Camels, sheep, goat | 39 | 39 | 39 | |
| 6 | Farming | Frequently | Camels, sheep, goat, chickens | 27 | 27 | 0 | |||
| 7 | Farming | Frequently | Camels, sheep, goat, chickens, dogs | 26 | 26 | 15 | |||
| 8 | Farming, Breeding | Frequent | Camels, sheep, goat, chickens | 22 | 22 | 12 | |||
| Saudi Arabia (Central) | Al‐Qassem | 9 | Racing | Rare | Camels, sheep, goat | 20 | 19 | 19 | |
| Saudi Arabia (East) | Qateef | 10 | Farming | Frequent | Camels, sheep, goat, chickens | 41 | 41 | 24 | HKU23 |
| 11 | Farming | Frequent | Camels, sheep, goat, chickens | 0 | 0 | 13 | |||
| Saudi Arabia (East) | Aljobail | 12 | Farming | Frequent | Camels, sheep, goat, chickens | 0 | 0 | 5 | |
| Oman | 13 | Racing, breeding | Frequent | Camels, dogs | 8 | 18 | 44 | ||
| Total | 306 | 315 | 243 | ||||||
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.2. Cells and viruses
Vero cells (ATCC CCL‐81) were used to culture and propagate MERS‐CoV and the HRT‐18G cell line (ATCC, CRL‐11663, American Type Culture Collection, Manassas, VA, USA) for culture of ECoV and BCoV. The culture medium for both the cell lines was DMEM (Cat:10569; Life Technologies, Carlsbad, CA, USA) supplemented with 10% foetal calf serum (Cat:16140; Life Technologies).
The prototype viruses used were MERS‐CoV (EMC/2012 obtained from Dr R Fouchier, Erasmus MC, Rotterdam), the ECoV‐NC99 strain (obtained from Dr. Udeni B.R. Balasuriya, Maxwell H. Gluck Equine Research Center, University of Kentucky, USA) (Guy et al., 2000; Zhang et al., 2007) and the bovine coronavirus (ATCC BRCV‐OK‐0514‐2).
2.3. RT‐PCR testing
Viral RNA was extracted from nasal and rectal swabs using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany) following the manufacturer's instructions. RNA extracts were tested for evidence of conserved coronavirus nucleic acid genetic sequences using previously reported RT‐PCR assays (Chu et al., 2014), RTqPCR assay for MERS‐CoV upE gene (Corman et al., 2012), RTqPCR assay for ECoV (Miszczak et al., 2014), and a RTqPCR assay for HKU23 reported below. RNA was reverse transcribed in a 20 μl reaction mixture containing 1× first‐strand buffer, 5 mM DTT, 0.5 mM deoxynucleotide triphosphates (dNTP), 2.5 ng/μl random hexamers and 200 units of SuperScript III (Life Technologies).
The pan‐coronavirus nested PCR targeted the RNA‐dependent RNA polymerase (RdRp) gene of coronaviruses developed by us as previously described (Chu et al., 2014). Using the cDNA synthesized as described above, first round PCR was carried out using forward primer 5′‐GGKTGGGAYTAYCCKAARTG‐3′ (position 15,287 of ECoV strain NC99; GenBank accession number EF446615) and reverse primer 5′‐TGYTGTSWRCARAAYTCRTG‐3′ (position 15,869 of ECoV strain NC99) and Phusion High Fidelity PCR Master Mix Kit (Thermo Scientific). A 25 μl reaction mixture contained 12.5 μl of 2× phusion master mix, 1.5 μl (10 pmol) forward primer, 1.5 μl (10 pmol) reverse primer, 4.5 μl ddH2O and 5 μl of cDNA. The PCR cycler conditions for the amplification were 98°C for 2 min, 40 cycles of amplification (98°C for 15 s, 48°C for 15 s, 72°C for 30 s), then 72°C for 2 min. An aliquot of 0.5 μl of the first PCR product was then further amplified in a second round PCR prepared as first around PCR but using a new set of primers (forward primer 5′‐GGTTGGGACTATCCTAAGTGTGA‐3′ (position 15,287 of ECoV strain NC99), reverse primer 5′‐CCATCATCAGATAGAATCATCAT‐3′ (position 15,721 of ECoV strain NC99) which amplify a final PCR product of 440 bp. The final PCR amplicons (440 bp) were gel purified using QIAquick gel purification kit (QIAGEN) and analysed by DNA sequencing using the primers used for the second round PCR. To amplify an extended RdRp gene sequence of both equine CoV and HKU23‐like viruses, PCR was prepared as above with forward primer 5′‐TACGCAGGATGGTAATGCTG‐3′ (position 14,767 of ECoV strain NC99) and reverse primer 5′‐TTGTGCGCATTCATTCGCAAG‐3′ (position 15,404 of ECoV strain NC99) and 5 μl of cDNA as template for the reaction. PCR products with expected size of 658 bp were purified for DNA sequencing using the forward and reverse primers.
We developed a two‐step real‐time quantitative RT‐PCR (RT‐qPCR) assay for the detecting HKU23 targeting N gene of the virus. Virus RNA was reverse transcribed as detailed above. Real‐time PCR was performed using Power SYBR Green Master Mix (Life Technologies) in a 20 μl reaction containing 5 pmol forward primer 5′‐CTGCCACGATGGTATTTTTAC‐3′ (position 29,758 of HKU23 strain 362F) and 5 pmol reverse primer 5′‐GGTATTGACATCAGCCTGGT‐3′ (position 29,852 of HKU23 strain 362F) with 40 cycles of amplification (95°C for 15 s, 62°C for 40 s) in a 20 μl volume followed by melting curve analysis. Positive result was interpreted by a detectable Ct value >40 considered together with a amplicon melting temperature within the range of 81.64°C to 82.64°C.
The details of all RT‐PCR and real‐time PCR assays used in this study are summarized in Table S1.
2.4. PCR amplicon sequencing and analysis
The primers used for sequencing were those used to generate the PCR products. DNA amplicons were purified and sequenced in both forward and reverse directions with the PCR primers using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and a 96‐capillary 3730xl DNA Analyzer (Applied Biosystems) according to manufacturer's instructions. Sequences were assembled using BioEdit 7.0 and MUSCLE for multiple sequence alignment. Phylogeny analysis was performed by maximum likelihood method using PhyML with GTR nucleotide substitution model and SH‐like approximate likelihood ratio test for estimating branch support.
2.5. Serology
All sera were tested using previously validated MERS‐CoV spike pseudoparticle neutralization test (ppNT) (Hemida et al., 2014; Perera et al., 2013), and the positive sera were confirmed using microneutralization (MN) test and plaque reduction neutralization tests (PRNT) using live MERS‐CoV strain EMC in a Biosafety Level 3 containment laboratory (Park et al., 2015). Selected sera were tested for antibodies to ECoV and BCoVs (Perera et al., 2013). The North American ECoV‐NC99 strain was propagated in HRT‐18G cells to make virus stocks for MN assay. ECoV MN assays were carried out by mixing 100 μl of serial 2‐fold dilutions (1:10 to 1:640) of heat‐inactivated sera (56°C for 30 min) with equal volumes of 200 tissue culture infectious doses (TCID50) of ECoV in 96‐well microtitre plates (Cat:92096, TPP tissue culture plates, Switzerland). Plates were incubated for 1 hr at 37°C. The virus–serum mixture was then added in quadruplicate to HRT‐18G cell monolayers in 96‐well microtitre plates followed by 1 hr adsorption at 37°C. The plates were incubated at 37°C in 5% CO2 in a humidified incubator for 5 days, when virus back titrations were inspected to confirm that the virus dose used was within the expected range. The antibody titre was the highest serum dilution that resulted in complete neutralization of cytopathic effect in ≥2 of the four wells replicate wells at each serum dilution. Antibody titre refers to the dilution of the serum in the serum dilution–virus mixture. All antibody titres are denoted as the reciprocal of the titre.
BCoV is ubiquitous in cattle worldwide and is genetically and antigenically related with HKU23, a coronavirus endemic in dromedaries (Woo et al., 2014, 2016). Antibody to BCoV was tested using MN assay as described previously (Park et al., 2015; Perera et al., 2013). Serology for BCoV and ECoV was carried out with culture medium without foetal calf serum. Known positive and negative control sera were included in all serology assays.
2.6. Reference sera used in investigation of cross‐reactivity in serological assays
Cross‐reactive responses in the MERS‐CoV ppNT, BCoV MN and ECoV MN assays were assessed using a panel of previously characterized immune or hyperimmune reference antisera generated against alpha, beta and gamma coronaviruses. These sera were either obtained from BEI Resources (animal CoV reagents supplied to BEI by Dr Linda Saif http://www.beiresources.org/About/BEIResources.aspx) or kindly provided by Dr. Linda Saif or Dr Stanley Perlman. Equine sera known to be seropositive to ECoV were provided by Dr. Nicola Pusterla, Department of Veterinary Medicine and Epidemiology, School of Veterinary Medicine, University of California, Davis, CA, USA. The homologous antibody titres to the immunized virus were also obtained, where available, from the providers of these respective antisera. The serum panel for alpha coronavirus included sera generated against porcine respiratory coronavirus, feline infectious peritonitis virus, canine enteric coronavirus and porcine transmissible gastroenteritis virus. Sera produced against mouse hepatitis virus strains JHM and A59, SARS coronavirus, BCoV, ECoV represented the betacoronavirus group. Furthermore, an immune serum for infectious bronchitis virus was used as representative for the gammacoronavirus group. All these reference sera, as well as additional ECoV immune sera and MERS‐CoV‐negative camel sera, were tested in ppNT or MN assays in parallel against MERS‐CoV, BCoV, as well as ECoV, to assess possible cross‐reactive antibody responses among these three coronaviruses. Some of these reference sera, with the exception of horse and camel sera, had been previously tested against MERS‐CoV and BCoV, and the results were reported by us (Hemida et al., 2014).
3. RESULTS AND DISCUSSION
All nasal and rectal swabs were tested using the pan‐coronavirus nested RT‐PCR assay, ECoV and MERS‐CoV upE gene RT‐qPCR assays. All swabs were negative in the MERS‐CoV RTqPCR assay. Five rectal swabs (5/315) were positive for coronavirus RNA by the pan‐coronavirus assay (Tables 1 and 2). Sequencing of the 440 base pair nucleotide fragment from the RdRp gene confirmed four of these to be ECoV and the other to be dromedary camel HKU23‐like virus designated as equine HKU23 CoV SA1354. Additional sequence was obtained from these specimens to generate a nucleotide sequence of 933 nucleotides (position 14,780 to 15,712 of ECoV strain NC99) which was used for analysis of nucleotide differences in comparison with other coronaviruses (GenBank accession numbers KY458228‐KY458232) (Table 3) and phylogenetic analysis which confirmed the identification of the coronaviruses to be ECoV and HKU23, respectively (Figure 1). This result led to the testing of 306 nasal and 315 rectal swab specimens by specific RTqPCR assays for ECoV and RTqPCR assay for HKU23. Testing by the ECoV specific RTqPCR led to the detection of five rectal swabs specimens, four of which had been detected by the pan‐coronavirus RT‐PCR assay as well as an additional positive rectal swab (Ct value 36.3) from a horse in the Riyadh area (Table 2). Using the HKU23‐specific RTqPCR, two positives were detected including SA1354 identified as HKU23 by sequencing the pan‐coronavirus RT‐PCR assay amplicon and an additional positive result with a high Ct value of 37.6 from a nasal swab sample. None of the other nasal swabs had evidence of coronavirus RNA. Compared to the pan‐coronavirus assay, the specific ECoV‐ and HKU23‐specific assays detected one additional specimen that was weakly positive for each of these viruses (Table 2).
Table 2.
RT‐qPCR‐positive specimens
| Swab type | Location and provider code | Pan CoV nested RT‐PCR result with sequencing of PCR product | MERS‐CoV UpE_RT‐qPCR | EqCoV_RT‐qPCR_Ct | HKU23 RT‐qPCR Ct value | HKU23 RT‐qPCR Tma |
|---|---|---|---|---|---|---|
| Nasal | Al‐Quassem (10) | Negative | Negative | Negative | 37.46 | 82.26 |
| Rectal | Dammam (3) | HKU23 | Negative | Negative | 34.67 | 82.25 |
| Rectal | Riyadh (1) | Equine CoV | Negative | 35.99 | Negative | |
| Rectal | Riyadh (1) | Equine CoV | Negative | 29.38 | Negative | |
| Rectal | Riyadh (1) | Equine CoV | Negative | 30.63 | Negative | |
| Rectal | Riyadh (1) | Negative | Negative | 36.32 | Negative | |
| Rectal | Riyadh (1) | Equine CoV | Negative | 34.87 | Negative |
aTm of camel HKU23 is 82.138 degree C. Screening horse samples for HKU23 RTqPCR allowed Tm range 81.638 to 82.638 degree C.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 3.
Jukes‐Cantor corrected distance nucleotide difference (%) comparing 919 nucleotides of RdRp gene of MERS‐CoV, BCoV, HKU23 and EqCoV with the coronaviruses detected from equine rectal swabs. Viruses detected from this study were bolded
| SA1360 Equine CoV | SA1362 Equine CoV | SA1364 Equine CoV | SA1382 Equine CoV | Equine CoV NC99 | SA1354 Camel HKU23‐like | Camel CoV HKU23‐265F | Camel CoV HKU23‐Ry123 | Bovine CoV BCoV‐ENT | Human CoV OC43 5617‐07 | Human CoV MERS | Bat CoV HKU4 | Bat CoV HKU5 | Mouse hepatitis virus‐JHM | Human CoV HKU1 | Human CoV SARS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SA1360 Equine CoV | – | |||||||||||||||
| SA1362 Equine CoV | 0.2 | – | ||||||||||||||
| SA1364 Equine CoV | 0.1 | 0.1 | – | |||||||||||||
| SA1382 Equine CoV | 0.1 | 0.1 | 0.0 | – | ||||||||||||
| Equine CoV NC99 | 0.4 | 0.4 | 0.3 | 0.3 | – | |||||||||||
| SA1354 Camel HKU23‐like | 2.1 | 2.1 | 2.0 | 2.0 | 1.9 | – | ||||||||||
| Camel CoV HKU23‐265F | 2.3 | 2.3 | 2.2 | 2.2 | 2.1 | 0.2 | – | |||||||||
| Camel CoV HKU23‐Ry123 | 2.1 | 2.1 | 2.0 | 2.0 | 1.9 | 0.0 | 0.2 | – | ||||||||
| Bovine CoV BCoV‐ENT | 1.7 | 1.7 | 1.5 | 1.5 | 1.4 | 1.8 | 1.8 | 1.8 | – | |||||||
| Human CoV OC43 5617‐07 | 3.6 | 3.6 | 3.5 | 3.5 | 3.3 | 3.5 | 3.2 | 3.5 | 2.8 | – | ||||||
| Human CoV MERS | 43.2 | 43.2 | 43.0 | 43.0 | 42.5 | 42.3 | 42.3 | 42.3 | 42.9 | 42.1 | – | |||||
| Bat CoV HKU4 | 39.5 | 39.5 | 39.3 | 39.3 | 39.1 | 39.6 | 39.5 | 39.6 | 38.9 | 39.6 | 21.7 | – | ||||
| Bat CoV HKU5 | 40.8 | 40.8 | 40.6 | 40.6 | 40.4 | 41.1 | 41.1 | 41.1 | 41.1 | 40.4 | 2.0 | 20.7 | – | |||
| Mouse hepatitis virus‐JHM | 14.3 | 14.5 | 14.3 | 14.3 | 14.0 | 14.2 | 14.0 | 14.2 | 13.8 | 13.4 | 44.2 | 42.1 | 41.5 | – | ||
| Human CoV HKU1 | 15.9 | 16.1 | 15.9 | 15.9 | 15.8 | 15.7 | 15.4 | 15.7 | 15.3 | 14.6 | 42.7 | 37.6 | 40.8 | 16.6 | – | |
| Human CoV SARS | 43.6 | 43.4 | 43.4 | 43.4 | 43.6 | 43.6 | 43.4 | 43.6 | 43.8 | 43.6 | 42.1 | 41.5 | 43.2 | 43.0 | 41.5 | – |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 1.
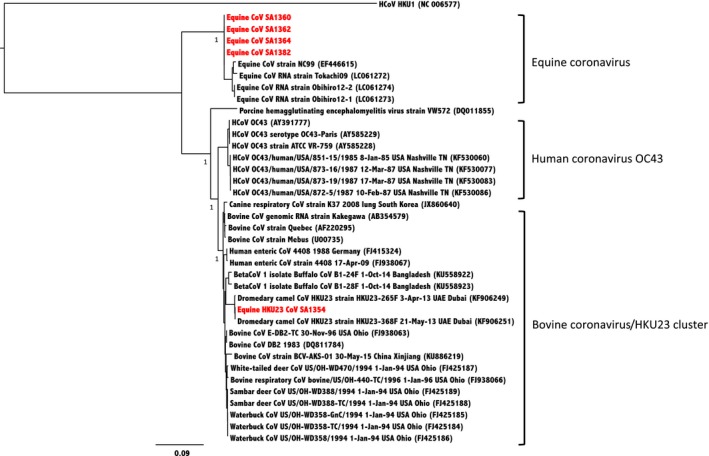
Phylogenetic analysis of equine coronavirus, human coronavirus OC43 and bovine coronavirus/dromedary camel coronavirus HKU23 cluster. The maximum likelihood phylogenetic tree rooted to HKU1 as an outgroup was generated using virus RdRp gene alignment of 934 nt by PhyML with approximate likelihood ratio test (aLRT) SH‐like branch support. Sequences generated from this study are highlighted in red. GenBank accession numbers of are indicated in brackets. Colour figure can be viewed at http://www.wileyonlinelibrary.com/]
The five pan‐coronavirus PCR CoV‐positive specimens had Ct ranging from 29.4 to 35.9 in either the ECoV or HKU23 specific assays. The pan‐coronavirus RT‐PCR was not sensitive at detecting specimens with Ct higher than 36 in the ECoV or HKU23 assays. However, the results do demonstrate the importance of using the pan‐coronavirus assay when investigating coronavirus activity in animal species in new geographic areas. Our initial focus was to explore for evidence of MERS‐CoV infections in horses in Saudi Arabia and Oman. None of the rectal swabs were positive for MERS‐CoV RNA, but the application of the pan‐coronavirus RT‐PCR to these specimens revealed the presence of two other coronaviruses infecting horses in this region, that is ECoV and HKU23.
All seven horses from whom ECoV or HKU23 were detected were apparently healthy. The specimens positive for ECoV RNA all came from different horses at a horse breeding facility in Riyadh. The two horses positive for HKU23 RNA were from Dammam and Qateef, Saudi Arabia, where horses were used for farming, and in Dammam, also for breeding (Tables 1 and 2). In both cases, these horses frequently came into contact with camels and other domestic livestock. HKU23 coronavirus is known to be endemic in camels (Woo et al., 2014).
We investigated for serologic evidence of MERS‐CoV infections in the 243 horse sera using ppNT screening followed by endpoint titrations. Any sera positive at a screening reciprocal antibody titre of 20 in ppNT were confirmed using MN assay and PRNT assay using live MERS‐CoV strain EMC. Possible serologic cross‐reactions with ECoV and BCoV were evaluated in MN assays which are the most specific serological assays available. Of the 243 sera tested, only five (2%) had detectable MERS‐CoV antibody in the MERS‐ppNT assay at a screening titre of 20; four of these were from Saudi Arabia, and one was from Oman. The endpoint MERS‐ppNT titres of these five sera ranged from 40 to 160 but only serum 9‐2 which had the highest MERS‐ppNT titre was positive in the MERS‐CoV MN test (titre 20) (Table 4). In the confirmatory PRNT which is considered the “gold standard” serological test for MERS‐CoV, horse serum 9‐2 reduced plaque numbers by 90% (PRNT90) up to a titre of 40. This serum was negative in ECoV and BCoV MN assays (Table 4). This serum was from a racing horse stable in Al‐Qassem and may represent a rare example of transmission of MERS‐CoV to horses or cross‐reaction with another yet undocumented coronavirus infecting horses. This horse was not known to frequently come into contact with camels. Three of the other sera (serum ID nos 5.35; 5.41; 2.12) positive in the MERS‐CoV ppNT assay had detectable ECoV MN antibody and one of these also had detectable BCoV MN antibody (Table 4). Thus, these may well represent evidence of cross‐reactive responses.
Table 4.
MERS‐CoV serology results
| Horse ID | Location and provider code | Reciprocal antibody titre | |||||
|---|---|---|---|---|---|---|---|
| MERS‐ppNT | MERS MN | MERS PRNT90 | MERS PRNT50 | BCoV MN | ECoV MN | ||
| 5‐36 | Al‐Hasa (5) | 40 | <20 | <20 | 20 | <20 | 80 |
| 5‐41 | Al‐Hasa (5) | 40 | <20 | <20 | 20 | <20 | 20 |
| 9‐2 | Al‐Qassem (9) | 160 | 20 | 40 | 80 | <20 | <20 |
| 13‐12 | Oman (13) | 40 | <20 | <20 | 20 | <20 | <20 |
| 2‐12 | Riyadh (2) | 40 | <20 | <20 | 20 | 20 | 40 |
ppNT, pseudotype neutralization test; MN, microneutralization test; PRNT, plaque reduction neutralization test.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The extent of the serological cross‐reactivity of MERS‐CoV, ECoV and BCoV was systematically investigated. We did not have an isolate of HKU23 virus available for serological testing and BCoV was used as a virus that is genetically closely related to HKU23 with pairwise amino acid similarity of 94.1% in the spike protein gene, the determinant of antigenic cross‐reaction in neutralization tests (Woo et al., 2014). Horse sera obtained from the USA reported to be strongly reactive by ELISA (optical density ranging from 1.458 to 1.939) for ECoV showed high titre (>1,280) for ECoV in MN assay as expected, but also had high antibody titres (ranging from 160 to 1,280) in BCoV MN tests (Table 5). As these horse sera were from horses in the “field,” they may well have been exposed to multiple coronaviruses. Thus, these results may reflect cross‐reactivity or co‐infection of US horses with ECoV‐ and BCoV‐like viruses. Two of these sera showed low titres (20) in the MERS‐CoV ppNT assay but were negative in the more stringent MERS‐CoV MN assay. As MERS‐CoV is not circulating in the USA, these results suggest cross‐reactivity between MERS‐CoV and ECoV or related CoVs may occur, albeit at low titre. Similarly, a BCoV immune serum from an experimentally infected gnotobiotic calf showed detectable, but 16‐fold reduced antibody titre with ECoV but no cross‐reaction with MERS‐CoV. Spike proteins of equine coronavirus and bovine coronavirus have an overall amino acid similarity of 80.8% and 73.1% similarity of the S1 region which contains the receptor‐binding domain and neutralization epitopes. This probably explains the large difference in neutralization titres observed in Table 5. A dromedary serum from Kazakhstan, a region known to be free of MERS‐CoV activity (Miguel et al., 2016), had detectable antibody to BCoV (probably reflecting HKU23 or BCoV infection) and also to ECoV. We cannot conclude whether this reflects cross‐reactivity or infection with BCoV, HKU23 and/or ECoV. None of the beta coronavirus immune sera to SARS‐CoV or mouse hepatitis virus, nor immune sera to alpha or gamma coronaviruses gave cross‐reactions in BCoV or ECoV MN assays (Table 5). As had been previously reported, none of these dromedary sera cross‐reacted with MERS‐CoV (Hemida et al., 2014).
Table 5.
Cross‐neutralization titres (denoted as reciprocal titres) for Middle East respiratory coronavirus (MERS‐CoV), bovine coronavirus (BCoV) and equine coronavirus (ECoV) in hyperimmune or naturally infected sera known to be positive for different coronaviruses
| Genus | Sera panel—Catalogue numbers provided for sera obtained from BEI Resource | Homologous antibody titre by ELISA unless specified | MERS‐ppNT | MERS MN | BCoV MN | ECoV MN |
|---|---|---|---|---|---|---|
| Alpha coronavirus | NR2727 – Guina pig antiserum to canine coronavirus | 4,094b | <20 | <20 | <20 | <20 |
| NR460 – pig antiserum to porcine respiratory coronavirus | 1,200a | <20 | <20 | <20 | <20 | |
| NR2518 – Guina pig antisera feline infectious peritonitis virus | 2,000a | <20 | <20 | <20 | <20 | |
| NR458 – pig antisera for porcine transmissible gastroenteritis virus | 1,400a | <20 | <20 | <20 | <20 | |
| Beta coronavirus | ECoV‐positive sera 459 (horse, USA) | qPCR negative; ELISA OD 1.459d | <20 | <20 | 160 | >1,280 |
| ECoV‐positive sera 463 (horse, USA) | qPCR positive; ELISA OD 1.525d | <20 | <20 | 1,280 | >1,280 | |
| ECoV‐positive sera 472 (horse, USA) | qPCR negative; ELISA OD 1.458d | 20 | <20 | 640 | >1,280 | |
| ECoV‐positive sera 486 (horse, USA) | qPCR negative; ELISA OD 1.939d | 20 | <20 | 1,280 | >1,280 | |
| ECoV‐negative sera 2 (horse, Hong Kong) | Not relevant | <20 | <20 | <20 | <20 | |
| BCoV‐positive camel serum #740293e | Not relevant | <20 | ND | 640 | 80 | |
| BCoV‐positive camel serum #467468e | Not relevant | <20 | ND | 80 | <20 | |
| C99‐10 – BCoV antisera from guinea pig | 20,480b | <20 | <20 | 320 | <20 | |
| Bcov antisera from germ free bovine calf | 580b – neutralisation titre | <20 | <20 | 1,280 | 80 | |
| NR456 – BCoV antisera from gnotobiotic calf | 10,000a | <20 | <20 | 80 | <20 | |
| NRC772 – Rabbit antisera SARS S protein high titre | 640 | <20 | <20 | <20 | <20 | |
| Mouse hepatitis virus(JHM strain) hyperimmunized mouse dam 1 | 1,778c – neutralisation titre | <20 | <20 | <20 | <20 | |
| Mouse hepatitis virus(JHM strain) hyperimmunized mouse dam 2 | 363c – neutralisation titre | <20 | <20 | <20 | <20 | |
| Mouse hepatitis virus(A59 strain)‐infected mice | 1,000c – neutralization titre | <20 | <20 | <20 | <20 | |
| NRC774 – antisera for SARS coronavirus sero titre | <10 | <20 | <20 | <20 | <20 | |
| NRC769 – Rabbit antisera to SARS S protein sero titre | <10 | <20 | <20 | <20 | <20 | |
| Gamma coronavirus | NR2515‐Guina pig antiserum to infectious bronchitis virus | 50,000a | <20 | <20 | <20 | <20 |
ppNT, Pseudoparticle neutralization test; MN, micro neutralization test; PRNT, plaque reduction neutralization test; BCoV, bovine coronavirus; ECoV, equine coronavirus; OD, optical density.
Equine antisera against ECoV were kindly provided by Professor Nicola Pusterla, Department of Veterinary Medicine and Epidemiology, School of Veterinary Medicine, University of California, Davis, CA, USA.
Homologous titres or OD results obtained from BEI resources, Linda Saif, Stanly Perlman and Udeni Balasuriya represented as a, b, c, d respectively. eField collected dromedary sera from Kazakhstan, a region known to be free of MERS‐CoV infection.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Fifty‐four horse sera from Saudi Arabia and Oman were selected to represent different collection locations, excluding those five sera previously reported to be MERS‐CoV ppNT positive (Table 6). These sera were seronegative for MERS‐CoV (as expected); 40 (74%) of them had detectable MN antibody to ECoV (titres ranging from <20 to 640; median 40), and 18 (33.3%) had MN antibodies to BCoV (titres ranging from <20 to 160; median <20). The reciprocal geometric mean antibody titres (assigning sera with titres <20 the value of 10 for computational purposes) for ECoV and BCoV were 44 and 25.5, respectively. There were twenty‐four sera with undetectable antibody to BCoV with detectable ECoV antibody titres ranging from 20 to 640. The titres to ECoV were higher than or equal to titres to BCoV, with two exceptions where the titre to BCoV (20) was marginally higher than that with ECoV (Table 6). These results are compatible with the RT‐PCR detection of ECoV in rectal swabs in horses and probably suggest that ECoV or a closely related virus is commonly infecting horses in many regions of the Arabian Peninsula. Two of the five horses in whom ECoV RNA was detected by RTqPCR had low ECoV MN antibody titres (20) while the other three had titres ranging from 160 to 640 (Table 6). This may reflect virus shedding persists into convalescence or that re‐infection of previously seropositive horses can occur.
Table 6.
Seropositivity to BCoV and ECoV in horses from different geographic locations
| Location | Provider code and management system | Sample ID | Reciprocal BCoV MN titre | Reciprocal ECoV MN Titre | PCR results |
|---|---|---|---|---|---|
| Riyadh | 1 (Breeding) | 1‐1a | <20 | 20 | ECoV PCR positive |
| 1‐2a | <20 | 20 | ECoV PCR positive | ||
| v1‐3a | 20 | 160 | ECoV PCR positive | ||
| 1‐4 | <20 | 80 | |||
| 1‐5 | <20 | 160 | |||
| 1‐6 | <20 | 40 | |||
| 1‐8a | <20 | 640 | ECoV PCR positive | ||
| 1‐12a | 80 | 320 | ECoV PCR positive | ||
| 2 (Farming, breeding) | 2‐13 | 40 | 160 | ||
| 2‐14 | <20 | 80 | |||
| Dammam | 3 (Farming, breeding) | 3‐11 | <20 | <20 | |
| 3‐12 | <20 | 40 | |||
| 3‐41a | <20 | <20 | HKU23 PCR positive | ||
| Al‐Hasa | 5 (Breeding) | 5‐56 | <20 | 40 | |
| 5‐57 | 160 | 320 | |||
| 5‐58 | 40 | 80 | |||
| 5‐59 | 80 | 160 | |||
| 8 (Farming, breeding) | 8‐2 | 20 | 80 | ||
| 8‐3 | 1:80 | 40 | |||
| 8‐4 | <20 | 640 | |||
| 8‐5 | <20 | <20 | |||
| 8‐6 | <20 | 20 | |||
| 7 (Farming) | 7‐1 | <20 | 20 | ||
| 7‐2 | 20 | <20 | |||
| 7‐3 | <20 | <20 | |||
| 7‐4 | <20 | 20 | |||
| 7‐1 | <20 | <20 | |||
| 7‐2 | <20 | <20 | |||
| Al‐Qassem | 9 (Racing) | 9‐A3 | 20 | <20 | |
| 9‐A4 | <20 | 40 | |||
| 9‐A5 | <20 | <20 | |||
| 9‐B1 | <20 | <20 | |||
| 9‐C1 | 20 | 20 | |||
| 9‐C2 | 80 | 640 | |||
| 9‐E1 | <20 | <20 | |||
| 9‐E2 | <20 | <20 | |||
| Qateef | 10 (Farming) | 10‐13 | <20 | 80 | |
| 10‐14 | <20 | 20 | |||
| 10‐15 | <20 | 80 | |||
| 10‐16 | 20 | 160 | |||
| 11 (Farming) | 11‐8 | 80 | 40 | ||
| 11‐9 | <20 | 320 | |||
| 11‐11 | <20 | <20 | |||
| 11‐12 | <20 | <20 | |||
| 11‐13 | <20 | 80 | |||
| Aljobail | 12 (Farming) | 12‐1 | 40 | 40 | |
| Oman | 13 (Racing, breeding) | 13‐1 | <20 | 20 | |
| 13‐2 | <20 | 80 | |||
| 13‐3 | <20 | 40 | |||
| 13‐4 | 20 | 160 | |||
| 13‐5 | 20 | 40 | |||
| 13‐6 | 20 | 160 | |||
| 13‐10 | 20 | 40 | |||
| 13‐11 | <20 | 80 |
aHorses found to be coronavirus RNA positive in swabs by RT‐PCR or RT‐qPCR.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The RTqPCR detection of two HKU23‐like viruses indicates that HKU23 which is common in dromedaries in the region is also infecting horses. In the absence of an HKU23 isolate for use in neutralization tests and given the potential serological cross‐reactivity between these three viruses, it is difficult to conclude how commonly equines are infected with HKU23. The antibody titres to BCoV, which is genetically closely related to HKU23 in the spike protein gene, may suggest that BCoV‐ or HKU23‐like virus infection of horses does occur in Saudi Arabia.
The serological evidence of one of the 243 sera with detectable MERS‐CoV antibody titres in the PRNT90 test raises the question whether MERS‐CoV may occasionally cross species to horses. This serum did not have any detectable antibody to ECoV or BCoV. However, it is not possible to exclude the possibility that this observation reflects cross‐reaction with ECoV, BCoV or other yet unknown coronaviruses. Confirmation of natural infection of horses with MERS‐CoV can only come from detection of MERS‐CoV RNA in horse specimens. Recent experimental infection of horses with MERS‐CoV did not result in virus replication or seroconversion (Adney et al., 2016).
In conclusion, the serological data suggest that lineage A betacoronaviruses are commonly infecting horses in Saudi Arabia and Oman but antibody cross‐reactions between these viruses do not permit us to use serological data alone to identify which coronaviruses are causing these infections. RT‐PCR detection of ECoV and HKU23 in equine swabs confirms the circulation of these two viruses in horses in Saudi Arabia. Cocirculation of related viruses may provide potential for recombination, a potential means of generating genetic diversity and facilitating host jumps in coronaviruses. This is the first report of HKU23 being detected in horses and the first detection of ECoV in Asia outside of Japan.
CONFLICT OF INTEREST
We declare that we have no conflict of interests.
Supporting information
ACKNOWLEDGEMENTS
Equine antisera against ECoV were kindly provided by Professor Nicola Pusterla, Department of Veterinary Medicine and Epidemiology, School of Veterinary Medicine, University of California, Davis, CA, USA. Reference coronavirus immune sera were provided by BEI Resources (http://www.beiresources.org/About/BEIResources.aspx) or kindly provided by Dr Linda Saif or Dr Stanley Perlman. This work was supported by research grants from the US National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract no. HHSN272201500006C), a commissioned grant from the Health and Medical Research Fund, Food and Health Bureau, Government of the Hong Kong Special Administrative Region and a grant from the King Abdul‐Aziz city of Science and Technology (KACST), Grant No (ARP‐34‐117).
Hemida MG, Chu DKW, Perera RAPM, et al. Coronavirus infections in horses in Saudi Arabia and Oman. Transbound Emerg Dis. 2017;64:2093–2103. 10.1111/tbed.12630
REFERENCE
- Adney, D. R. , Brown, V. R. , Porter, S. M. , Bielefeldt‐Ohmann, H. , Hartwig, A. E. , & Bowen, R. A. (2016). Inoculation of goats, sheep, and horses with MERS‐CoV does not result in productive viral shedding. Viruses, 8(8), 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersen, S. , Kobinger, G. P. , Soule, G. , & Wernery, U. (2014). Middle East respiratory syndrome coronavirus antibody reactors among camels in Dubai, United Arab Emirates, in 2005. Transboundary and Emerging Diseases, 61, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasuriya, U. B. R. (2013). Coronaviridae In McVey D. S., Kennedy M. & Chengappa M. M. (Eds.), Veterinary microbiology, 3rd edn (pp. 456–473). Ames, IA: Wiley‐Blackwell. [Google Scholar]
- Barlan, A. , Zhao, J. , Sarkar, M. K. , Li, K. , McCray, P. B. Jr , Perlman, S. , & Gallagher, T. (2014). Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. Journal of Virology, 88, 4953–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, D. K. , Oladipo, J. O. , Perera, R. A. , Kuranga, S. A. , Chan, S. M. , Poon, L. L. , & Peiris, M. (2015). Middle East respiratory syndrome coronavirus (MERS‐CoV) in dromedary camels in Nigeria, 2015. Euro Surveillance, 20(49), doi: 10.2807/1560-7917.ES.2015.20.49.30086. [DOI] [PubMed] [Google Scholar]
- Chu, D. K. , Poon, L. L. , Gomaa, M. M. , Shehata, M. M. , Perera, R. A. , Abu Zeid, D. , … Kayali, G. (2014). MERS coronaviruses in dromedary camels, Egypt. Emerging Infectious Diseases, 20, 1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M. , Muller, M. A. , Costabel, U. , Timm, J. , Binger, T. , Meyer, B. , … Drosten, C. (2012). Assays for laboratory confirmation of novel human coronavirus (hCoV‐EMC) infections. Euro Surveillance, 17(49), 20334. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. , Tejedor‐Junco, M. T. , Gonzalez, M. , Lattwein, E. , & Renneker, S. (2015). Presence of antibodies but no evidence for circulation of MERS‐CoV in dromedaries on the Canary Islands, 2015. Euro Surveillance, 20(37), doi: 10.2807/1560-7917.ES.2015.20.37.30019. [DOI] [PubMed] [Google Scholar]
- Guy, J. S. , Breslin, J. J. , Breuhaus, B. , Vivrette, S. , & Smith, L. G. (2000). Characterization of a coronavirus isolated from a diarrheic foal. Journal of Clinical Microbiology, 38, 4523–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida, M. G. , Perera, R. A. , Al Jassim, R. A. , Kayali, G. , Siu, L. Y. , Wang, P. , … Peiris, M. (2014). Seroepidemiology of Middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Euro Surveillance, 19(23), pii: 20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida, M. G. , Perera, R. A. , Wang, P. , Alhammadi, M. A. , Siu, L. Y. , Li, M. , Poon, L. L. , Saif, L. , Alnaeem, A. , & Peiris, M. (2013). Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveillance, 18(50), 20659. [DOI] [PubMed] [Google Scholar]
- Kooijman, L. J. , Mapes, S. M. , & Pusterla, N. (2016). Development of an equine coronavirus‐specific enzyme‐linked immunosorbent assay to determine serologic responses in naturally infected horses. Journal of Veterinary Diagnostic Investigation, 28, 414–418. [DOI] [PubMed] [Google Scholar]
- Memish, Z. A. , Cotten, M. , Meyer, B. , Watson, S. J. , Alsahafi, A. J. , Al Rabeeah, A. A. , … Drosten, C. (2014). Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerging Infectious Diseases, 20, 1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B. , Garcia‐Bocanegra, I. , Wernery, U. , Wernery, R. , Sieberg, A. , Muller, M. A. , … Eckerle, I. (2015). Serologic assessment of possibility for MERS‐CoV infection in equids. Emerging Infectious Diseases, 21, 181–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel, E. , Perera, R. A. , Baubekova, A. , Chevalier, V. , Faye, B. , Akhmetsadykov, N. , … Peiris, M. (2016). Absence of middle east respiratory syndrome coronavirus in camelids, Kazakhstan, 2015. Emerging Infectious Diseases, 22(3), 555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miszczak, F. , Tesson, V. , Kin, N. , Dina, J. , Balasuriya, U. B. , Pronost, S. , & Vabret, A. (2014). First detection of equine coronavirus (ECoV) in Europe. Veterinary Microbiology, 171, 206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oue, Y. , Morita, Y. , Kondo, T. , & Nemoto, M. (2013). Epidemic of equine coronavirus at Obihiro Racecourse, Hokkaido, Japan in 2012. Journal of Veterinary Medical Science, 75, 1261–1265. [DOI] [PubMed] [Google Scholar]
- Park, W. B. , Perera, R. A. , Choe, P. G. , Lau, E. H. , Choi, S. J. , Chun, J. Y. , … Oh, M. D. (2015). Kinetics of serologic responses to MERS coronavirus infection in humans, South Korea. Emerging Infectious Diseases, 21, 2186–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, R. A. , Wang, P. , Gomaa, M. R. , El‐Shesheny, R. , Kandeil, A. , Bagato, O. , … Kayali, G. (2013). Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveillance, 18(36), 20574. [DOI] [PubMed] [Google Scholar]
- Pusterla, N. , Holzenkaempfer, N. , Mapes, S. , & Kass, P. (2015). Prevalence of equine coronavirus in nasal secretions from horses with fever and upper respiratory tract infection. Veterinary Record, 177, 289. [DOI] [PubMed] [Google Scholar]
- Reusken, C. B. , Messadi, L. , Feyisa, A. , Ularamu, H. , Godeke, G. J. , … Koopmans, M. P. (2014). Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerging Infectious Diseases, 20, 1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2016). Middle East respiratory syndrome coronavirus (MERS‐CoV). Retrieved from http://www.who.int/emergencies/mers-cov/en/ (accessed May 27, 2016).
- Woo, P. C. , Lau, S. K. , Fan, R. Y. , Lau, C. C. , Wong, E. Y. , Joseph, S. , … Yuen, K. Y. (2016). Isolation and characterization of dromedary camel coronavirus UAE‐HKU23 from dromedaries of the middle east: Minimal serological cross‐reactivity between MERS coronavirus and dromedary camel coronavirus UAE‐HKU23. International Journal of Molecular Sciences, 17(5), pii: E691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P. C. , Lau, S. K. , Wernery, U. , Wong, E. Y. , Tsang, A. K. , Johnson, B. , … Yuen, K. Y. (2014). Novel betacoronavirus in dromedaries of the Middle East, 2013. Emerging Infectious Diseases, 20, 560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Guy, J. S. , Snijder, E. J. , Denniston, D. A. , Timoney, P. J. , & Balasuriya, U. B. (2007). Genomic characterization of equine coronavirus. Virology, 369, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


