Summary
Group A rotaviruses (RVAs) are a major cause of serious intestinal disease in piglets. In this study, a novel pig strain was identified in a stool sample from China. The strain was designated RVA/Pig/China/LNCY/2016/G3P[13] and had a G3‐P[13]‐I5‐R1‐C1‐M1‐A8‐N1‐T1‐E1‐H1 genome. The viral protein 7 (VP7) and non‐structural protein 4 (NSP4) genes of RVA/Pig/China/LNCY/2016/G3P[13] were closely related to cogent genes of human RVAs, suggesting that a reassortment between pig and human strains had occurred. Recombination analysis showed that RVA/Pig/China/LNCY/2016/G3P[13] is a natural recombinant strain between the P[23] and P[7] RVA strains, and crossover points for recombination were found at nucleotides (nt) 456 and 804 of the VP4 gene. Elucidating the biological characteristics of porcine rotavirus (PoRV) will be helpful for further analyses of the epidemic characteristics of this virus. The results of this study provide valuable information for RVA recombination and evolution and will facilitate future investigations into the molecular pathogenesis of RVAs.
Keywords: porcine group A rotavirus, reassortant, recombinant, VP4
1. INTRODUCTION
Rotaviruses (RVs) are a major cause of intestinal disease in animals, including weaned piglets and young children (Vlasova, Amimo, & Saif, 2017). RVs form a genus of the Reoviridae family and contain a genome consisting of 11 double‐stranded RNA segments. The RV genome encodes six structural viral proteins (VPs; VP1‐VP4, VP6 and VP7) and five or six non‐structural proteins (NSP1‐NSP5/6) (Ramig, 1997). The RV VP6 protein has been used to differentiate RV species by classifying them into 10 groups from group A rotavirus (RVA) to group J rotavirus (RVJ) (Banyai et al., 2017; Matthijnssens et al., 2012; Mihalov‐Kovacs et al., 2015). Continuous RV classification is being performed by the Rega Instituut, KU Leuven (Belgium) (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/7th-RCWG-meeting). The most common group infecting humans and animals is RVA, which causes acute dehydrating diarrhoea (Vlasova et al., 2017).
To facilitate the classification of novel RVA strains, a Rotavirus Classification Working Group (RCWG) was established to develop a classification system for RVs (Matthijnssens, Ciarlet, Rahman et al., 2008b). Due to the high genetic diversity of RVA strains, the dual typing system (G and P genotypes) was extended to a whole‐genome sequence classification system with all 11 gene segments (Matthijnssens, Ciarlet, Heiman et al., 2008a). The classification and nomenclature of RVA genes is Gx‐P[x]‐Ix‐Rx‐Cx‐Mx‐Ax‐Nx‐Tx‐Ex‐Hx, where x indicates the corresponding number of genotypes (Matthijnssens et al., 2011). To date, 32 G, 47 P, 24 I, 18 R, 17 C, 17 M, 28 A, 18 N, 19 T, 24 E and 19 H genotypes have been identified among RVAs from humans and animals (Esona et al., 2017), as described at the 7th RCWG meeting (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/7th-RCWG-meeting) at Rega Instituut, KU Leuven (Belgium). In pigs, 12 G genotypes (G1 to G6, G8 to G12 and G26) and 16 P genotypes (P[1] to P[8], P[13], P[19], P[23], P[26], P[27], P[28], P[32] and P[34]) of RVA have been identified, and G3, G4, G5, G9 and G11 usually occur in combination with P[5], P[6], P[7], P[13] and P[28], respectively (Vlasova et al., 2017).
Infections are confirmed in piglets by the association of RVAs with diarrhoea worldwide. An analysis of molecular characterization data for RVAs in Kenya showed that 18.5% of pigs were positive for RVA (Amimo, Otieno, Okoth, Onono, & Bett, 2017). RVA is an important pathogen for diarrhoea in Belgian suckling pigs (Amimo et al., 2016). Epidemiological surveillance of porcine rotavirus (PoRV) conducted in Ohio, USA, showed that 9.4% of pigs were positive for RVA (Amimo, Vlasova, & Saif, 2013). Genetic diversity analyses showed that different genotype combinations of PoRV strains were present in Thailand (Saikruang et al., 2013). Previous reports have shown that diarrhoea caused by PoRV can also be found in China (Shi et al., 2012).
To study the biological characteristics of PoRV, further investigation of PoRV diversity is necessary. In this study, the whole genome of an RVA strain belonging to the G3P[13] genotype was sequenced. The genome sequence identified an emerging reassortant G3P[13] PoRV strain from China with natural crossover points in the VP4 gene. The results will be helpful for further analysis of the epidemic characteristics of PoRV. The results provide valuable information on RVA recombination and evolution and facilitate future investigations into the molecular pathogenesis of RVA.
2. MATERIALS AND METHODS
2.1. Specimen collection
In 2016, an intestinal tissue sample from a pig with diarrhoea was collected from a pig farm in Liaoning Chaoyang (LNCY), China. The intestinal contents were collected and mixed with PBS at a 1:10 ratio. The suspension was centrifuged at 8000× g and 4°C for 10 min, and the supernatant was stored at −20°C.
2.2. Viruses
The positive control viruses PoRV strain RVA/Pig/China/NMTL (Accession No. JF781163) (Shi et al., 2012), porcine epidemic diarrhoea virus (PEDV) strain CV777 (Accession No. AF353511) and transmissible gastroenteritis virus (TGEV) strain AHHF (Accession No. KX499468) (Zhang, Zhu, Zhu, Shi et al., 2017) were maintained in our laboratory.
2.3. Reverse transcription (RT)‐PCR and sequencing
Total RNA was extracted from the supernatant of the intestinal contents using the QIAamp Viral RNA Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. RT‐PCR was performed using the PrimeScript™ One Step RT‐PCR Kit Ver. 2 (TaKaRa, Dalian, China) according to the manufacturer's instructions. Primers are designed as shown in Table S1.
2.4. Sequence analysis
Sequences of the PoRV reference strains were obtained from GenBank (Tables S2 and S3). The nt sequences were analysed with DNASTAR software (DNAstar Inc., Madison, WI, USA). Phylogenetic trees were constructed with MEGA5.2 software using the maximum likelihood method.
2.5. 3D model of the VP4 protein
Using PyMOL software, the spatial distribution of the VP4 protein (70‐210 aa) was analysed in a 3D model using the SWISS‐MODEL server (Biasini et al., 2014).
2.6. Recombination analysis
Sequence recombination was analysed using RDP4 software (Martin, Murrell, Golden, Khoosal, & Muhire, 2015). The RDP, GENECONV, Chimaera, MaxChi, Bootscan, SiScan and 3Seq methods were employed with respective default parameters.
3. RESULTS
3.1. Genome sequence of the PoRV RVA/Pig/China/LNCY/2016/G3P[13] strain
To determine the causative agent of diarrhoea in the sample from the LNCY pig farm in China, total RNA was extracted from the small intestine contents and subjected to RT‐PCR. The samples were positive for PoRV but negative for TGEV and PEDV (Figure S1). The complete open reading frame (ORF) sequences for all 11 genome segments of strain RVA/Pig/China/LNCY/2016/G3P[13] were obtained using the primers as shown in Table S1. The accession numbers for the 11 segments deposited in GenBank are MF462321, MF462322, MF462323, MF462324, MF462325, MF462326, MF462316, MF462317, MF462318, MF462319 and MF462320 for VP1, VP2, VP3, VP4, VP6, VP7, NSP1, NSP2, NSP3, NSP4 and NSP5, respectively. The individual genotypes for VP7‐VP4‐VP6‐VP1‐VP2‐VP3‐NSP1‐NSP2‐NSP3‐NSP4‐NSP5 of RVA/Pig/China/LNCY/2016/G3P[13] were assigned as G3‐P[13]‐I5‐R1‐C1‐M1‐A8‐N1‐T1‐E1‐H1 using the online RotaC2.0 automated genotyping tool (Maes, Matthijnssens, Rahman, & Van Ranst, 2009). The VP7 and NSP4 genes of RVA/Pig/China/LNCY/2016/G3P[13] were closely related to cogent genes of human origin viruses (Table 1).
Table 1.
Nucleotide and amino acid identities of genome segments of strains RVA/Pig/China/LNCY/2016/G3P[13] compared with the closest strains from GenBank database
| Gene | Closely related strains | Nucleotide (%) | Amino acid (%) | Genotype | Accession No. |
|---|---|---|---|---|---|
| VP1 | RVA/Pig‐tc/USA/Gottfried/1983/G4[P6] | 95.2 | 99.2 | R1 | KR052746 |
| VP2 | RVA/Pig‐tc/USA/OSU/1977/G5[P7] | 93.0 | 98.8 | C1 | GU199515 |
| VP3 | RVA/Pig‐tc/MEX/YM/1983/G11[P7] | 87.5 | 95.0 | M1 | AY300922 |
| VP4 | RVA/Pig‐wt/VNM/14150_54/VP4 | 95.7 | 97.3 | P[13] | KX363348 |
| VP6 | RVA/Pig‐tc/MEX/YM/1983/G11[P7] | 90.2 | 96.7 | I5 | X69487 |
| VP7 | RVA/Human‐tc/USA/P/1974/G3[P8] | 86.0 | 94.2 | G3 | EF672602 |
| NSP1 | RVA/Pig‐tc/USA/Gottfried/1983/G4[P6] | 82.9 | 84.2 | A8 | U08431 |
| NSP2 | RVA/Pig‐tc/MEX/YM/1983/G11[P7] | 95.0 | 96.9 | N1 | GU199517 |
| NSP3 | RVA/Pig‐tc/MEX/YM/1983/G11[P7] | 88.7 | 96.5 | T1 | GU199518 |
| NSP4 | RVA/Human‐tc/USA/P/1974/G3[P8] | 94.7 | 96.0 | E1 | EF672603 |
| NSP5 | RVA/Pig‐tc/MEX/YM/1983/G11[P7] | 96.5 | 98.0 | H1 | X69486 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.2. Sequence analysis of the VP7 gene
The VP7 gene contains 981 nt encoding 326 amino acids (aa). The VP7 type was determined for the PoRV strains. A phylogenetic tree was constructed using a whole VP7 gene sequence (981 nt) with selected G genotype sequences from GenBank. RVA/Pig/China/LNCY/2016/G3P[13] demonstrated the closest relationship with RVA/Human‐tc/USA/P/1974/G3[P8] (EF672602) (Figure S2A). An analysis of PoRV G3 strains identified as RVA/Pig/China/LNCY/2016/G3P[13] and the PoRV VP7 gene sequences available in GenBank showed that the nt identity rates varied from 79.3% to 88.0%. In a previous study, G3 genotypes were shown to be genetically diverse and clustered into two distinct lineages (G3a and G3b) (Mino et al., 2013). The RVA/Pig/China/LNCY/2016/G3P[13] strain found in this study belonged to G3a (Figure 1). Furthermore, a close relationship was found for RVAs (Maes et al., 2009) between RVA/Pig/China/LNCY/2016/G3P[13] and a number of RVA G3 strains using the RotaC2.0 automated genotyping tool. The RotaC results showed that the sequence was genotype G3 (glycolated) and that the query sequence was most similar to G3‐RVA/Human‐tc/USA/P/1974/G3[P8] (86.0%) (Figure S2B).
Figure 1.
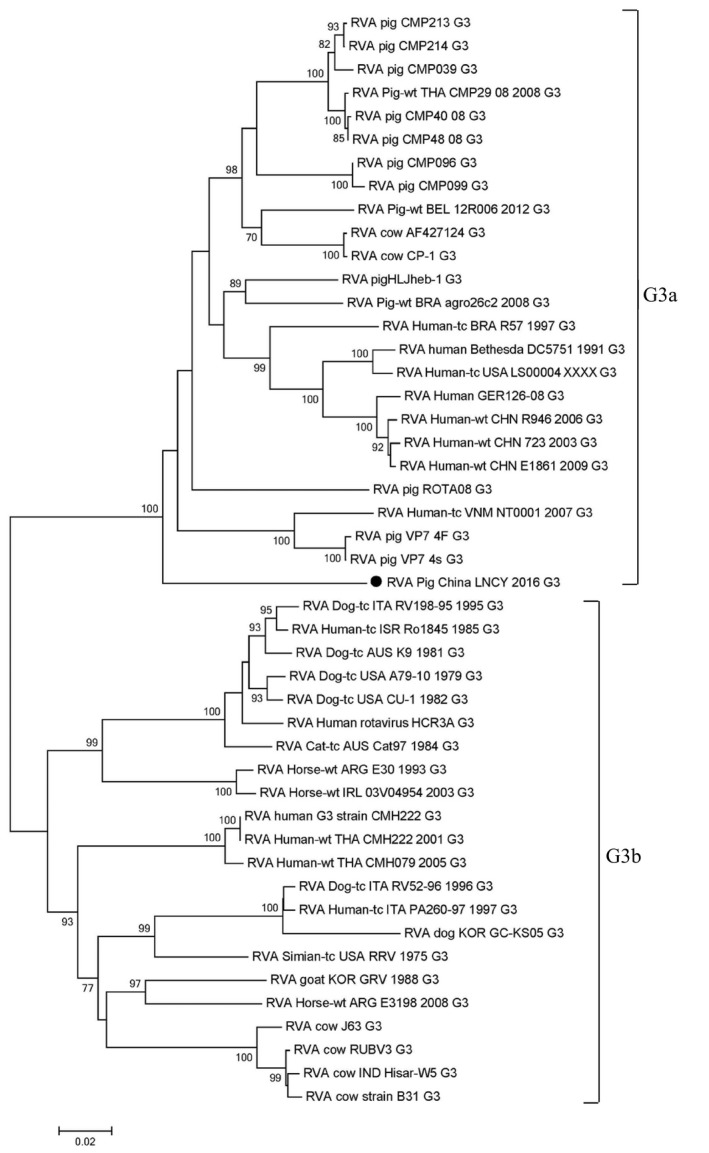
Phylogenetic analysis of RVA/Pig/China/LNCY/2016/G3P[13] based on the VP7 sequence in the G3 genotype. Phylogenetic trees were constructed using the Construct/Test Neighbour‐Joining method with MEGA 5.2 software, and tree reliability was evaluated using the bootstrap method with 1,000 replications. Only bootstrap values >70% at branch points are shown
3.3. Sequence analysis of the VP4 gene
The VP4 gene contains 2337 nt encoding 778 aa. The VP4 type was determined for the PoRV strains. A phylogenetic tree was constructed using the whole VP4 gene sequence (2337 nt) with selected P genotype sequences from GenBank. RVA/Pig/China/LNCY/2016/G3P[13] demonstrated the closest relationship with RVA/Pig‐wt/VNM/14150_54/VP4 (KX363348) (Figure S3A). In a previous study, P[13] genotypes were shown to be genetically diverse and clustered into three distinct lineages (an India/Thailand lineage, a Japanese lineage and a new USA lineage) (Amimo et al., 2013). In this study, a phylogenetic tree was constructed using whole VP4 gene sequences (2337 nt) with selected P[13] genotype sequences from GenBank. Analysis of PoRV P[13] identified as RVA/Pig/China/LNCY/2016/G3P[13] and the PoRV VP4 gene sequences available in GenBank showed that the nt identity rates varied from 82.4% to 95.7%. We clustered the P[13] sequences into four lineages (lineage I, lineage II, lineage III and lineage IV) and found that RVA/Pig/China/LNCY/2016/G3P[13] belonged to lineage IV (Figure 2). Furthermore, a close relationship was found for group A rotaviruses (Maes et al., 2009) between RVA/Pig/China/LNCY/2016/G3P[13] and a number of RVA P[13] strains using the RotaC2.0 automated genotyping tool. The RotaC results showed that the sequence was genotype P[13] (protease‐sensitive), and the query sequence was most similar to PoRV strain A46 (AY050274) with the highest similarity (83.5%); this strain was collected in 2001 in the USA (Figure S3B).
Figure 2.
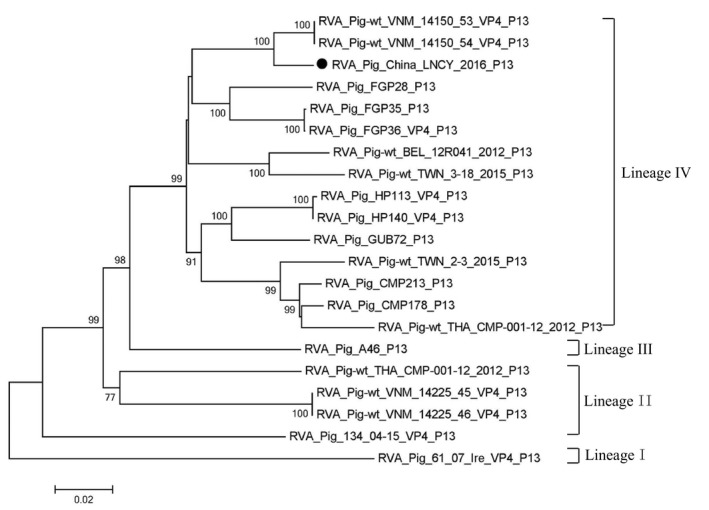
Phylogenetic analysis of RVA/Pig/China/LNCY/2016/G3P[13] based on the VP4 sequence in the P[13] genotype. Phylogenetic trees were constructed using the Construct/Test Neighbour‐Joining Tree method with MEGA 5.2 software, and tree reliability was evaluated using the bootstrap method with 1,000 replications. Only bootstrap values >70% at branch points are shown
3.4. Sequence analysis of VP6, VP1, VP2 and VP3
Using the RotaC2.0 automated genotyping tool for RVAs (Maes et al., 2009), the VP6, VP1, VP2 and VP3 sequences were compared between RVA/Pig/China/LNCY/2016/G3P[13] and a number of RVAs. The VP1 sequence was genotype R1 (RNA‐dependent RNA polymerase); the query sequence was most similar to R1‐RVA/Pig‐tc/USA/Gottfried/1983/G4[P6] (95.2%) (Figure S4A). The VP2 sequence was genotype C1 (core shell protein); the query VP2 sequence was most similar to C1‐RVA/Pig‐tc/USA/OSU/1977/G5[P7] (93.0%) (Figure S4B). The VP3 sequence was genotype M1 (methyltransferase); the query VP3 sequence was most similar to M1‐RVA/Pig‐tc/MEX/YM/1983/G11[P7] (87.5%) (Figure S4C). The VP6 sequence was genotype I5 (intermediate capsid); the query VP6 sequence was most similar to I5‐RVA/Pig‐tc/MEX/YM/1983/G11[P7] (90.2%) (Figure S4D).
3.5. Sequence analysis of NSP1, NSP2, NSP3, NSP4 and NSP5
Using the RotaC2.0 automated genotyping tool for RVAs (Maes et al., 2009), the NSP1, NSP2, NSP3, NSP4 and NSP5 sequences were compared between RVA/Pig/China/LNCY/2016/G3P[13] and a number of RVAs. The NSP1 sequence was genotype A8 (interferon antagonist); the query NSP1 sequence was most similar to A8‐RVA/Pig‐tc/USA/Gottfried/1983/G4[P6] (82.9%) (Figure S5A). The NSP2 sequence was genotype N1 (NTPase); the query sequence was most similar to N1‐RVA/Pig‐tc/MEX/YM/1983/G11[P7] (95.0%) (Figure S5B). The NSP3 sequence was genotype T1 (translation enhancer); the query NSP3 sequence was most similar to T1‐RVA/Pig‐tc/MEX/YM/1983/G11[P7] (88.7%) (Figure S5C). The NSP4 sequence was genotype E1 (enterotoxin); the query NSP4 sequence was most similar to E1‐RVA/Human‐tc/USA/P/1974/G3[P8] (94.7%) (Figure S5D). The NSP5 sequence was genotype H1 (phosphoprotein); the query NSP5 sequence was most similar to H1‐RVA/Pig‐tc/MEX/YM/1983/G11[P7] (96.5%) (Figure S5E).
3.6. Sequence divergence of VP7 and VP4 in strains from China
The structures of the neutralizing epitopes have been identified on the VP7 (7‐1 and 7‐2) (Aoki et al., 2009), VP5* and VP8* proteins (8‐1, 8‐2, 8‐3 and 8‐4) (Dormitzer, Nason, Prasad, & Harrison, 2004; Naseer, Jarvis, Ciarlet, & Marthaler, 2017). The two neutralizing epitope regions of the RVA/Pig/China/LNCY/2016/G3P[13] VP7 protein were compared with those of all PoRV strains from China obtained from GenBank and the three rotavirus vaccine RCE strains (Gottfried G4P[6], OSU G5P[7] and A2 G9P[6]) available in the ProSystems RCE (Merck). A total of six aa positions (91T, 97A, 99W, 211T, 238L and 291K) in neutralizing epitope in 7‐1 and three aa positions (217E, 223E and 264V) in neutralizing epitope in 7‐2 were conserved (Figure 3a). Six aa (94, 96, 98, 213, 147 and 190) in the 7‐1 and 7‐2 aa sequences differed between RVA/Pig/China/LNCY/2016/G3P[13] and the RCE strain A2‐G9P[6]‐USA. Nine aa (87, 94, 98, 100, 213, 147, 148, 190 and 221) in the 7‐1 and 7‐2 aa sequences differed between RVA/Pig/China/LNCY/2016/G3P[13] and the RCE strain OSU‐G5P[7]‐USA. Six aa (87, 98, 147, 148, 221 and 242) differed in the 7‐1 and 7‐2 aa sequences between RVA/Pig/China/LNCY/2016/G3P[13] and the RCE strain Gottfried G4P[6]‐USA. Furthermore, in RVA/Pig/China/LNCY/2016/G3P[13], the two aa positions located at 98S in 7‐1 and 147A in 7‐2 were not found in any of the RCE strains.
Figure 3.
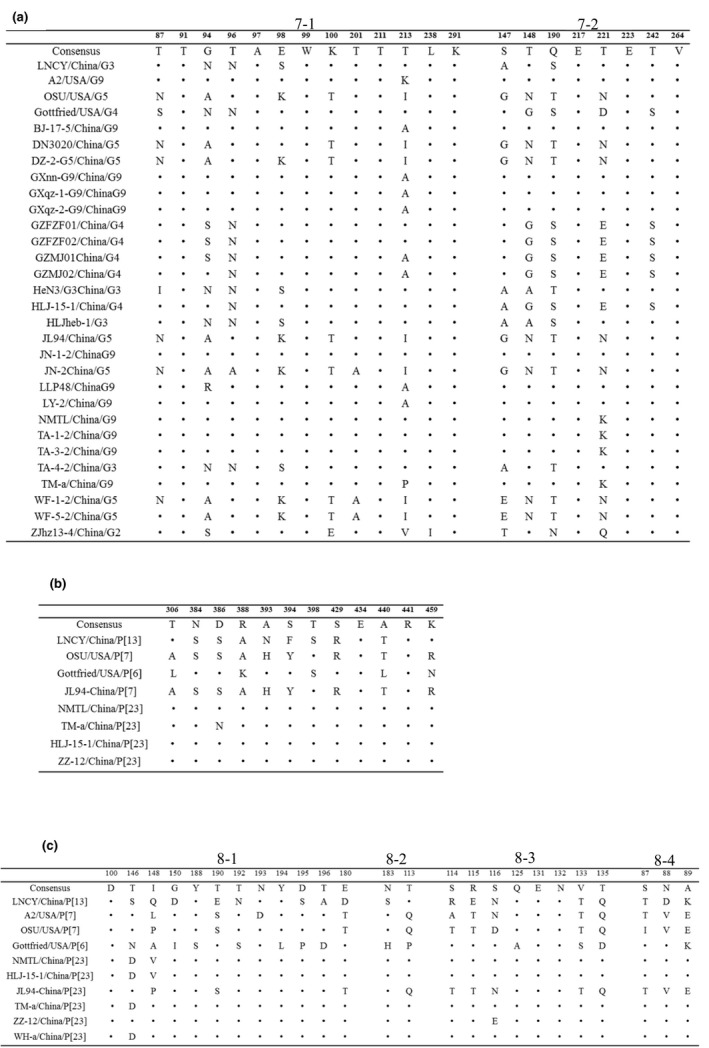
Neutralizing epitopes on the VP7 and VP4 (VP5* and VP8*) proteins in Chinese strains. (a) Neutralizing epitopes on the VP7 protein. (b) Neutralizing epitopes on the VP5* protein. (c) Neutralizing epitopes on the VP8* protein. Dots represent the same residues
Trypsin cleavage of VP4 generates two fragments (VP5* and VP8*) (Dunn et al., 1995; Patton, Hua, & Mansell, 1993; Ruggeri & Greenberg, 1991). Only a partial sequence of the VP4 of porcine rotavirus strain A2 was found (AB180977, 783 nt). Therefore, we only compared the VP5* neutralizing epitope region of RVA/Pig/China/LNCY/2016/G3P[13] with all PoRV strains from China obtained from GenBank and the RCE strains (Gottfried G4P[6], OSU G5P[7]) available in the ProSystems RCE (Merck). The results show that two aa positions (434E and 441R) in the neutralizing epitope in VP5* were conserved (Figure 3b). Five aa (306, 393, 394, 398 and 459) in the VP5* neutralizing epitope differed between RVA/Pig/China/LNCY/2016/G3P[13] and the RCE strain OSU‐G5P[7]‐USA (Figure 3b). Nine aa (306, 384, 386, 388, 393, 394, 429, 440 and 459) in the VP5* neutralizing epitope differed between RVA/Pig/China/LNCY/2016/G3P[13] and the RCE strain Gottfried G4P[6]‐USA (Figure 3b). Only two aa positions were conserved in the neutralizing epitope in VP8*: one aa (100D) in 8‐1 and the other aa (131E and 132N) in 8‐3 (Figure 3c).
3.7. A six‐nt insertion impacts the spatial structure of the VP4 protein
We identified an additional six nt (nt 559‐561 and nt 568‐570) in the VP8* gene of RVA/Pig/China/LNCY/2016/G3P[13] compared with the corresponding genes of other Chinese strains (Figure 4a). No other Chinese strains were previously reported to contain six additional nt in the VP8* protein (Figure 4a). However, the presence of six‐nt gains at the same positions was found in strains from other countries, including Thailand, India, Japan, Australia, and the USA (Figure 4a). The additional six nt resulted in four aa changes (aa 184, 185, 186 and 190) and the loss of two aa from the VP8* protein (aa 188 and 189) (Figure 4a). Further analysis of the influence of these aa changes on the spatial structure of the VP8* protein was performed. The VP8* protein sequences (70‐210 aa) of RVA/Pig/China/LNCY/2016/G3P[13] and RVA/Pig/China/NMTL were searched against the SWISS‐MODEL template library. For the best homology modelling with VP8* protein sequences (70‐210 aa) of RVA/Pig/China/LNCY/2016/G3P[13] while searching against templates, the solution structure of the rotavirus spike protein (SMTL id 2p3j.1) (Kraschnefski et al., 2009) was selected for model construction. Based on the results, the spatial structure of the VP8* protein of RVA/Pig/China/LNCY/2016/G3P[13] was different from that of RVA/Pig/China/NMTL. The RVA/Pig/China/NMTL strain forms a longer β‐sheet in the aa region of 184TTGY187 than in the aa region of 184APQEFG190 in RVA/Pig/China/LNCY/2016/G3P[13] (Figure 4b).
Figure 4.
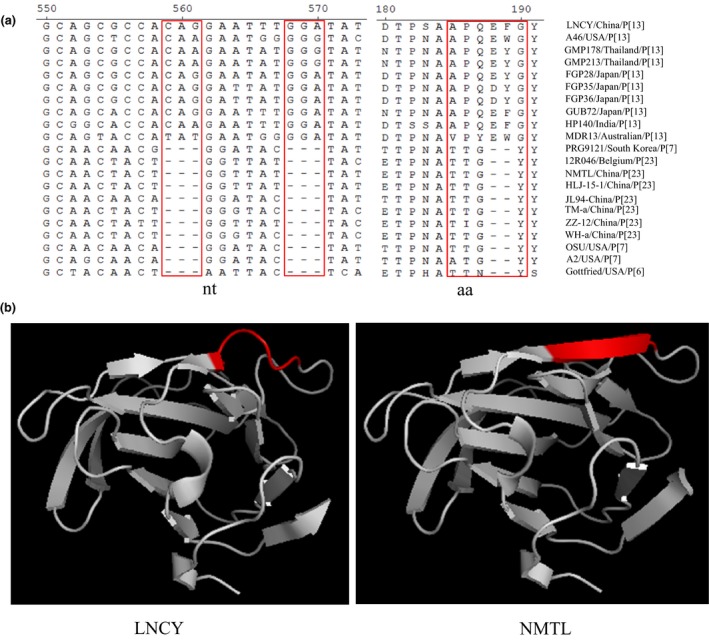
A two‐aa deletion in the VP8* protein. (a) six‐nt deletions and a two‐aa deletion in the VP8 protein of a Chinese strain. (b) Specific structures of the VP8* protein (70‐210 aa) from RVA/Pig/China/LNCY/2016/G3P[13] and RVA/Pig/China/NMTL. LNCY represents RVA/Pig/China/LNCY/2016/G3P[13], and NMTL represents RVA/Pig/China/NMTL
3.8. Recombination of RVA/Pig/China/LNCY/2016/G3P[13]
To further analyse the association between VP4 from RVA/Pig/China/LNCY/2016/G3P[13] and the existing isolates, a genetic analysis was completed between RVA/Pig/China/LNCY/2016/G3P[13] and the existing RVA isolates using RDP4 software (Martin et al., 2015). The VP4 sequence of RVA/Pig/China/LNCY/2016/G3P[13] is shown in Figure 5a. Crossover points for a potential recombination zone were found at nt 456‐804 of the VP4 gene (Figure 5b), but no gene recombination was detected in the other viral proteins. The major parental strain for the recombination located in the VP4 gene of RVA/Pig/China/LNCY/2016/G3P[13] was RVA/Pig/PRG9121/2006/G9P[7] (JF796737) (Kim et al., 2012), and the minor parental strain was RVA/Pig‐wt/BEL/12R046/2012/G9P[23] (KM820720) (Theuns et al., 2015) (Figure 4b). These results indicate that P[13] of RVA/Pig/China/LNCY/2016/G3P[13] was the result of recombination of P[23] and P[7] RVA genes.
Figure 5.
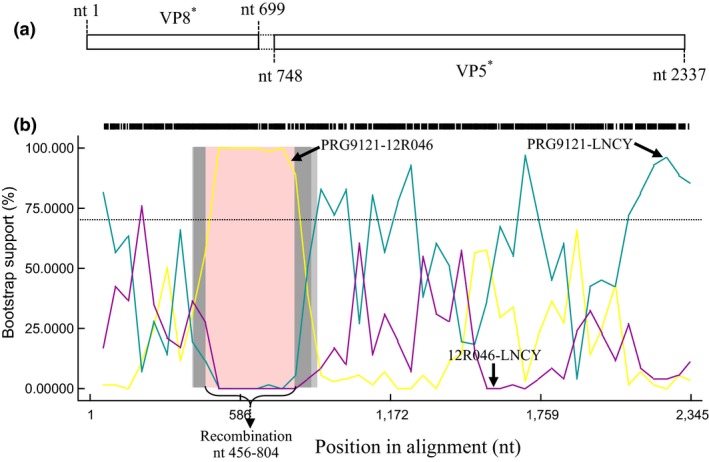
Recombination analysis of RVA/Pig/China/LNCY/2016/G3P[13] and other PoRV strains in the VP4 gene. (a) VP4 gene organization of PoRV. (b) Recombination analysis of the VP4 gene in RVA/Pig/China/LNCY/2016/G3P[13], RVA/Pig/PRG9121/2006/G9P[7] and RVA/Pig‐wt/BEL/12R046/2012/G9P[23]. LNCY represents RVA/Pig/China/LNCY/2016/G3P[13] (MF462324), PRG9121 represents RVA/Pig/PRG9121/2006/G9P[7] (JF796737), and 12R046 represents RVA/Pig‐wt/BEL/12R046/2012/G9P[23] (KM820720). Recombination crossover points are located at nt 456‐804
4. DISCUSSION
Many types of viruses cause diarrhoea in animals, including TGEV (Zhang, Zhu, Zhu, Chen et al., 2017), swine enteric coronaviruses (SeCoVs) (Belsham et al., 2016), PEDV (Lohse et al., 2016; Tian et al., 2014; Zhang, Tian et al., 2017), porcine deltacoronavirus (Lee et al., 2016; Mai et al., 2017; Saeng‐Chuto et al., 2017) and rotaviruses (Kim et al., 2016). Rotaviruses cause acute dehydrating diarrhoea in various hosts, including children and young animals (Jiang, Liu, & Tan, 2017). Further information on rotavirus diversity is necessary to improve our understanding of the biological characteristics and evolution of rotaviruses. In 2016, intestinal tissue was collected from a piglet with LNCY‐induced diarrhoea accompanied by vomiting at a pig farm in China. The entire RVA/Pig/China/LNCY/2016/G3P[13] genome was successfully sequenced, and evidence of a recombination event was identified.
Reassortment events between humans and animal strains have been found previously (Banyai et al., 2009; Doan et al., 2013; Esona et al., 2009, 2017; Ghosh et al., 2010; He et al., 2013, 2017; Jere, Mlera, O'Neill, Peenze, & van Dijk, 2012; Malik et al., 2016; Masuda et al., 2014; Matthijnssens et al., 2009; Mullick et al., 2013; Steyer, Sagadin, Kolenc, & Poljsak‐Prijatelj, 2013). Of the 11 segments of the RVA/Pig/China/LNCY/2016/G3P[13] strain, nine gene segments (VP1, VP2, VP3, VP4, VP6, NSP1, NSP2, NSP3 and NPS5) possessed sequences nearly identical with those of the porcine strains, suggesting that RVA/Pig/China/LNCY/2016/G3P[13] was possibly of porcine origin. However, the VP7 and NSP4 genes of this porcine strain had human origins because the VP7 and NSP4 genes had the closest relationships with G3‐RVA/Human‐tc/USA/P/1974/G3[P8] and E1‐RVA/Human‐tc/USA/P/1974/G3[P8], respectively. These results indicated that RVA/Pig/China/LNCY/2016/G3P[13] might be a reassortant between porcine and human strains.
Rotavirus VP7 forms soluble calcium‐dependent trimers (Dormitzer, Greenberg, & Harrison, 2000). Protection against RV infection can be mediated by neutralizing antibodies that target epitopes on both VP4 and VP7 (Taniguchi et al., 1991). The structures of the neutralizing epitopes on the VP7 protein have been identified. The epitopes on the exposed surface of the VP7 protein are clustered into two regions: 7‐1 (aa 87, 91, 94, 96, 97, 98, 99, 100, 201, 211, 213, 238 and 291) and 7‐2 (aa 147, 148, 190, 217, 221, 223, 242 and 264) (Aoki et al., 2009). The neutralizing epitope of aa 94 to 99 was previously elucidated in the RVA VP7 protein (Mackow et al., 1988). The RV surface is decorated with 60 spikes, each of which is composed of a VP4 dimer (a viral haemagglutinin) (Tihova, Dryden, Bellamy, Greenberg, & Yeager, 2001). VP4 is a trimer at its base, on the surface only a dimer is exposed while the third monomer is folded away or removed (Settembre, Chen, Dormitzer, Grigorieff, & Harrison, 2011). Trypsin cleavage of the RVs VP4 generates two fragments (VP8* and VP5*); VP8* binds sialic acid (SA), whereas VP5* contains an integrin‐binding motif and a hydrophobic region that facilitate cell membrane penetration and integrin‐binding (Dowling, Denisova, LaMonica, & Mackow, 2000; Pesavento, Crawford, Estes, & Prasad, 2006). VP8* of RVs mediates cell attachment by interacting with histo‐blood group antigens (HBGAs) (Hu et al., 2012; Sun et al., 2016). The structures of the neutralizing epitopes on the VP5* and VP8* proteins have been identified. The epitopes on VP8* are clustered into four regions (8‐1, 8‐2, 8‐3 and 8‐4) (Dormitzer et al., 2004; Naseer et al., 2017). Figure 3 shows minimal differences in the neutralizing epitopes G3, G4, G5 and G9 between strains from China and the RCE strains, suggesting that the RCE strains protect against these genotypes in China. However, the PoRV P genotypes currently available in GenBank mainly are P[23]. At present, relatively few P genotypes from China can be found in GenBank. More reports of the P genotype of PoRV are needed in China, which will provide information for the prevention of PoRV and the development of PoRV vaccines.
Recombination has been shown to be an important means of viral evolution (Simmonds, 2006). Recombination is considered a potential mechanism for antigenic diversity and a possible mechanism to escape from vaccine‐imposed selective pressure but may not contribute to the long‐term evolution of RVAs (Woods, 2015). Several reports have identified sequences with recombination in VP4 (Esona et al., 2017), VP6 (Jere, Mlera, Page, van Dijk, & O'Neill, 2011; Marthaler et al., 2014), VP7 (Martinez‐Laso et al., 2009; Parra, Bok, Martinez, & Gomez, 2004; Phan et al., 2007a, 2007b; Suzuki, Gojobori, & Nakagomi, 1998), NSP1 (Esona et al., 2017), NSP2 (Jere et al., 2011), NSP3 and NSP4 (Donker, Boniface, & Kirkwood, 2011) (Table S4). Intramolecular recombination could contribute to the development of new RVs phenotypes (Ramig, 1997). To date, no reports have demonstrated a recombinant PoRV in China. In this study, evidence for a recombination crossover event in the VP4 gene (nt 456‐804, aa 152‐268) was provided in a Chinese isolate. This result may indicate that PoRV infection in farms in China is becoming increasingly complex. In the future, studies of the genetic evolution of new PoRVs in China will be necessary.
In the current study, we identified aa differences in the neutralizing antigen epitopes of VP7, VP5* and VP8* (VP4 is cleaved into VP5* and VP8*) between Chinese PoRVs and the porcine RCE strains. Some of the same epitopes were found for the VP7 protein in the PoRV strains, including RVA/Pig/China/LNCY/2016/G3P[13], suggesting that the RCE strains would provide some protection against Chinese porcine genotypes. However, many neutralizing antigen epitope differences were identified on the VP4 protein between the Chinese PoRV strains and the RCE strains, suggesting the reduced effectiveness of the RCE strains. Rotavirus antigens and serotypes are diverse, suggesting that obtaining extensive protective immunity requires immunization or infection with multiple antigens (Nair et al., 2017). More information is needed to understand the diversity of field strains and to develop an effective polyvalent vaccine as a possible means of PoRV control in China.
VP8* is a cleaved product of the VP4 spike protein that contains a shallow groove domain responsible for binding to SAs on cellular glycans (Desselberger, 2014; Lopez & Arias, 2004) or interacting with HBGA for virus particle attachment (Jiang et al., 2017; Sun et al., 2016). In this study, we reported a six‐nt gain in the VP8* protein that resulted in a special structural change, which might affect attachment of the VP8* protein to host cells through interactions with SAs or HBGAs. This possibility will be a major focus of our future study.
Concurrent infection of a single host cell line by different RVs strains is a prerequisite for the occurrence of recombination events in nature (Esona et al., 2017). In developing countries, animal husbandry (i.e., cattle, sheep and pigs) is closely related to human populations, which provides potential possibilities for reassortment and recombination events. Monitoring newly emerging RVs in animals is important for the control of RVs infection in both humans and animals. The exact mechanism of recombination in rotaviruses is still unclear. Hypotheses have suggested that there may be a direct link between recombination and the formation of the secondary RNA structure, because these RNA structures induce stops, stalls or delays that favour strand invasion (Perez‐Losada, Arenas, Galan, Palero, & Gonzalez‐Candelas, 2015). The secondary structures in recombinant gene segments and in short stretches of AU‐rich sequence might be associated with recombination events in RVs (Esona et al., 2017). However, copy choice plays an important role in RNA virus recombination (Kirkegaard & Baltimore, 1986). Copy choice recombination might occur in RVs during plus‐ and minus‐strand RVs RNA synthesis (Desselberger, 1996). Rotavirus recombination most likely occurs during the genome transcription stage in the viral life cycle (Kojima, Taniguchi, Urasawa, & Urasawa, 1996). RVA/Pig/China/LNCY/2016/G3P[13] is a recombinant virus between the P[7] and P[23] genotypes. It will be important to investigate the pathogenicity of this virus in pigs in future work.
In summary, in this study, an example of a natural recombination event in China between the P[7] and P[23] genotypes was observed, and crossover points (nt 456‐804) for potential recombination were found in the VP4 gene. RVA/Pig/China/LNCY/2016/G3P[13] was a reassortant strain between pig and human strains. These results provide valuable information on RVs recombination and evolution and will facilitate future investigation of the molecular pathogenesis of porcine RVAs.
Supporting information
ACKNOWLEDGEMENTS
This study was supported by The National Key Research and Development Program of China (2015BAD12B02, 2016YFD0500103, 2017YFD0500104) and the National Natural Science Foundation of China (31572541, 31502092 and 31602072).
Jing Z, Zhang X, Shi H, et al. A G3P[13] porcine group A rotavirus emerging in China is a reassortant and a natural recombinant in the VP4 gene. Transbound Emerg Dis. 2018;65:e317–e328. 10.1111/tbed.12756
REFERENCES
- Amimo, J. O. , El Zowalaty, M. E. , Githae, D. , Wamalwa, M. , Djikeng, A. , & Nasrallah, G. K. (2016). Metagenomic analysis demonstrates the diversity of the fecal virome in asymptomatic pigs in East Africa. Archives of Virology, 161, 887–897. 10.1007/s00705-016-2819-6 [DOI] [PubMed] [Google Scholar]
- Amimo, J. O. , Otieno, T. F. , Okoth, E. , Onono, J. O. , & Bett, B. (2017). Risk factors for rotavirus infection in pigs in Busia and Teso subcounties. Western Kenya. Tropi. Anim. Health Pro., 49, 105–112. 10.1007/s11250-016-1164-9 [DOI] [PubMed] [Google Scholar]
- Amimo, J. O. , Vlasova, A. N. , & Saif, L. J. (2013). Detection and genetic diversity of porcine group A rotaviruses in historic (2004) and recent (2011 and 2012) swine fecal samples in Ohio: Predominance of the G9P[13] genotype in nursing piglets. Journal of Clinical Microbiology, 51, 1142–1151. 10.1128/JCM.03193-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, S. T. , Settembre, E. C. , Trask, S. D. , Greenberg, H. B. , Harrison, S. C. , & Dormitzer, P. R. (2009). Structure of rotavirus outer‐layer protein VP7 bound with a neutralizing Fab. Science, 324, 1444–1447. 10.1126/science.1170481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyai, K. , Esona, M. D. , Mijatovic, S. , Kerin, T. K. , Pedreira, C. , Mercado, J. , … Gentsch, J. R. (2009). Zoonotic bovine rotavirus strain in a diarrheic child. Nicaragua. J. Clin. Virol., 46, 391–393. 10.1016/j.jcv.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Banyai, K. , Kemenesi, G. , Budinski, I. , Foldes, F. , Zana, B. , Marton, S. , … Jakab, F. (2017). Candidate new rotavirus species in Schreiber's bats. Serbia. Infect. Genet. Evol., 48, 19–26. 10.1016/j.meegid.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham, G. J. , Rasmussen, T. B. , Normann, P. , Vaclavek, P. , Strandbygaard, B. , & Botner, A. (2016). Characterization of a novel chimeric swine enteric coronavirus from diseased pigs in central eastern Europe in 2016. Transbound. Emerg. Dis., 63, 595–601. 10.1111/tbed.12579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini, M. , Bienert, S. , Waterhouse, A. , Arnold, K. , Studer, G. , Schmidt, T. , … Schwede, T. (2014). SWISS‐MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 42, 252–258. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger, U. (1996). Genome rearrangements of rotaviruses. Advances in Virus Research, 46, 69–95. 10.1016/S0065-3527(08)60070-6 [DOI] [PubMed] [Google Scholar]
- Desselberger, U. (2014). Rotaviruses. Virus Research, 190, 75–96. 10.1016/j.virusres.2014.06.016 [DOI] [PubMed] [Google Scholar]
- Doan, Y. H. , Nakagomi, T. , Aboudy, Y. , Silberstein, I. , Behar‐Novat, E. , Nakagomi, O. , & Shulman, L. M. (2013). Identification by full‐genome analysis of a bovine rotavirus transmitted directly to and causing diarrhea in a human child. Journal of Clinical Microbiology, 51, 182–189. 10.1128/JCM.02062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donker, N. C. , Boniface, K. , & Kirkwood, C. D. (2011). Phylogenetic analysis of rotavirus A NSP2 gene sequences and evidence of intragenic recombination. Infect. Genet. Evol., 11, 1602–1607. 10.1016/j.meegid.2011.05.024 [DOI] [PubMed] [Google Scholar]
- Dormitzer, P. R. , Greenberg, H. B. , & Harrison, S. C. (2000). Purified recombinant rotavirus VP7 forms soluble, calcium‐dependent trimers. Virology, 277, 420–428. 10.1006/viro.2000.0625 [DOI] [PubMed] [Google Scholar]
- Dormitzer, P. R. , Nason, E. B. , Prasad, B. V. , & Harrison, S. C. (2004). Structural rearrangements in the membrane penetration protein of a non‐enveloped virus. Nature, 430, 1053–1058. 10.1038/nature02836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling, W. , Denisova, E. , LaMonica, R. , & Mackow, E. R. (2000). Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. Journal of Virology, 74, 6368–6376. 10.1128/JVI.74.14.6368-6376.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, S. J. , Fiore, L. , Werner, R. L. , Cross, T. L. , Broome, R. L. , Ruggeri, F. M. , & Greenberg, H. B. (1995). Immunogenicity, antigenicity, and protection efficacy of baculovirus expressed VP4 trypsin cleavage products, VP5(1)* and VP8* from rhesus rotavirus. Archives of Virology, 140, 1969–1978. 10.1007/BF01322686 [DOI] [PubMed] [Google Scholar]
- Esona, M. D. , Geyer, A. , Banyai, K. , Page, N. , Aminu, M. , Armah, G. E. , … Gentsch, J. R. (2009). Novel human rotavirus genotype G5P[7] from child with diarrhea. Cameroon. Emerg. Infect. Dis., 15, 83–86. 10.3201/eid1501.080899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona, M. D. , Roy, S. , Rungsrisuriyachai, K. , Sanchez, J. , Vasquez, L. , Gomez, V. , … Vazquez, M. (2017). Characterization of a triple‐recombinant, reassortant rotavirus strain from the Dominican Republic. Journal of General Virology, 98, 134–142. 10.1099/jgv.0.000688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S. , Alam, M. M. , Ahmed, M. U. , Talukdar, R. I. , Paul, S. K. , & Kobayashi, N. (2010). Complete genome constellation of a caprine group A rotavirus strain reveals common evolution with ruminant and human rotavirus strains. Journal of General Virology, 91, 2367–2373. 10.1099/vir.0.022244-0 [DOI] [PubMed] [Google Scholar]
- He, B. , Huang, X. , Zhang, F. , Tan, W. , Matthijnssens, J. , Qin, S. , … Tu, C. (2017). Group A rotaviruses in chinese bats: Genetic composition, serology, and evidence for bat‐to‐human transmission and reassortment. Journal of Virology, 91, 10.1128/jvi.02493-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B. , Yang, F. , Yang, W. , Zhang, Y. , Feng, Y. , Zhou, J. , … Tu, C. (2013). Characterization of a novel G3P[3] rotavirus isolated from a lesser horseshoe bat: A distant relative of feline/canine rotaviruses. Journal of Virology, 87, 12357–12366. 10.1128/JVI.02013-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, L. , Crawford, S. E. , Czako, R. , Cortes‐Penfield, N. W. , Smith, D. F. , Le Pendu, J. , … Prasad, B. V. (2012). Cell attachment protein VP8* of a human rotavirus specifically interacts with A‐type histo‐blood group antigen. Nature, 485, 256–259. 10.1038/nature10996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jere, K. C. , Mlera, L. , O'Neill, H. G. , Peenze, I. , & van Dijk, A. A. (2012). Whole genome sequence analyses of three African bovine rotaviruses reveal that they emerged through multiple reassortment events between rotaviruses from different mammalian species. Veterinary Microbiology, 159, 245–250. 10.1016/j.vetmic.2012.03.040 [DOI] [PubMed] [Google Scholar]
- Jere, K. C. , Mlera, L. , Page, N. A. , van Dijk, A. A. , & O'Neill, H. G. (2011). Whole genome analysis of multiple rotavirus strains from a single stool specimen using sequence‐independent amplification and 454(R) pyrosequencing reveals evidence of intergenotype genome segment recombination. Infect. Genet. Evol., 11, 2072–2082. 10.1016/j.meegid.2011.09.023 [DOI] [PubMed] [Google Scholar]
- Jiang, X. , Liu, Y. , & Tan, M. (2017). Histo‐blood group antigens as receptors for rotavirus, new understanding on rotavirus epidemiology and vaccine strategy. Emerg. Microbes Infect., 6, 10.1038/emi.2017.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. H. , Matthijnssens, J. , Kim, H. J. , Kwon, H. J. , Park, J. G. , Son, K. Y. , … Cho, K.O. (2012). Full‐length genomic analysis of porcine G9P[23] and G9P[7] rotavirus strains isolated from pigs with diarrhea in South Korea. Infect. Genet. Evol., 12, 1427–1435. 10.1016/j.meegid.2012.04.028 [DOI] [PubMed] [Google Scholar]
- Kim, H. K. , Yoon, S. W. , Kim, D. J. , Koo, B. S. , Noh, J. Y. , Kim, J. H. , … Jeong, D. G. (2016). Detection of Severe Acute Respiratory Syndrome‐Like, Middle East Respiratory Syndrome‐Like Bat Coronaviruses and Group H Rotavirus in Faeces of Korean Bats. Transbound. Emerg. Dis., 63, 365–372. 10.1111/tbed.2016.63.issue-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard, K. , & Baltimore, D. (1986). The mechanism of RNA recombination in poliovirus. Cell, 47, 433–443. 10.1016/0092-8674(86)90600-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, K. , Taniguchi, K. , Urasawa, T. , & Urasawa, S. (1996). Sequence analysis of normal and rearranged NSP5 genes from human rotavirus strains isolated in nature: Implications for the occurrence of the rearrangement at the step of plus strand synthesis. Virology, 224, 446–452. 10.1006/viro.1996.0551 [DOI] [PubMed] [Google Scholar]
- Kraschnefski, M. J. , Bugarcic, A. , Fleming, F. E. , Yu, X. , von Itzstein, M. , Coulson, B. S. , & Blanchard, H. (2009). Effects on sialic acid recognition of amino acid mutations in the carbohydrate‐binding cleft of the rotavirus spike protein. Glycobiology, 19, 194–200. 10.1093/glycob/cwn119 [DOI] [PubMed] [Google Scholar]
- Lee, J. H. , Chung, H. C. , Nguyen, V. G. , Moon, H. J. , Kim, H. K. , Park, S. J. , … Park, B. K. (2016). Detection and phylogenetic analysis of porcine deltacoronavirus in Korean swine farms, 2015. Transbound. Emerg. Dis., 63, 248–252. 10.1111/tbed.2016.63.issue-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse, L. , Krog, J. S. , Strandbygaard, B. , Rasmussen, T. B. , Kjaer, J. , Belsham, G. J. , & Botner, A. (2016). Experimental infection of young pigs with an early European strain of porcine epidemic diarrhoea virus and a recent US strain. Transbound. Emerg. Dis., 10, 10.1111/tbed.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, S. , & Arias, C. F. (2004). Multistep entry of rotavirus into cells: A Versaillesque dance. Trends in Microbiology, 12, 271–278. 10.1016/j.tim.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Mackow, E. R. , Shaw, R. D. , Matsui, S. M. , Vo, P. T. , Benfield, D. A. , & Greenberg, H. B. (1988). Characterization of homotypic and heterotypic VP7 neutralization sites of rhesus rotavirus. Virology, 165, 511–517. 10.1016/0042-6822(88)90595-8 [DOI] [PubMed] [Google Scholar]
- Maes, P. , Matthijnssens, J. , Rahman, M. , & Van Ranst, M. (2009). RotaC: A web‐based tool for the complete genome classification of group A rotaviruses. BMC Microbiology, 9, 238 10.1186/1471-2180-9-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai, K. , Feng, J. , Chen, G. , Li, D. , Zhou, L. , Bai, Y. , … Ma, J. (2017). The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong Province in Southern China. Transbound. Emerg. Dis., 27, 10.1111/tbed.12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, Y. S. , Kumar, N. , Sharma, K. , Saurabh, S. , Dhama, K. , Prasad, M. , … Singh, R. K. (2016). Multispecies reassortant bovine rotavirus strain carries a novel simian G3‐like VP7 genotype. Infect. Genet. Evol., 41, 63–72. [Epub ahead of print] 10.1016/j.meegid.2016.03.023 [DOI] [PubMed] [Google Scholar]
- Marthaler, D. , Suzuki, T. , Rossow, K. , Culhane, M. , Collins, J. , Goyal, S. , … Matthijnssens, J. (2014). VP6 genetic diversity, reassortment, intragenic recombination and classification of rotavirus B in American and Japanese pigs. Veterinary Microbiology, 172, 359–366. 10.1016/j.vetmic.2014.05.015 [DOI] [PubMed] [Google Scholar]
- Martin, D. P. , Murrell, B. , Golden, M. , Khoosal, A. , & Muhire, B. (2015). DP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol., 1, vev003 10.1093/ve/vev003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Laso, J. , Roman, A. , Rodriguez, M. , Cervera, I. , Head, J. , Rodriguez‐Avial, I. , & Picazo, J. J. (2009). Diversity of the G3 genes of human rotaviruses in isolates from Spain from 2004 to 2006: Cross‐species transmission and inter‐genotype recombination generates alleles. Journal of General Virology, 90, 935–943. 10.1099/vir.0.007807-0 [DOI] [PubMed] [Google Scholar]
- Masuda, T. , Nagai, M. , Yamasato, H. , Tsuchiaka, S. , Okazaki, S. , Katayama, Y. , … Mizutani, T. (2014). Identification of novel bovine group A rotavirus G15P[14] strain from epizootic diarrhea of adult cows by de novo sequencing using a next‐generation sequencer. Veterinary Microbiology, 171, 66–73. 10.1016/j.vetmic.2014.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens, J. , Ciarlet, M. , Heiman, E. , Arijs, I. , Delbeke, T. , McDonald, S. M. , … Van Ranst, M. (2008a). Full genome‐based classification of rotaviruses reveals a common origin between human Wa‐Like and porcine rotavirus strains and human DS‐1‐like and bovine rotavirus strains. Journal of Virology, 82, 3204–3219. 10.1128/JVI.02257-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens, J. , Ciarlet, M. , McDonald, S. M. , Attoui, H. , Banyai, K. , Brister, J. R. , … Van Ranst, M. (2011). Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Archives of Virology, 156, 1397–1413. 10.1007/s00705-011-1006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens, J. , Ciarlet, M. , Rahman, M. , Attoui, H. , Banyai, K. , Estes, M. K. , … Van Ranst, M. (2008b). Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Archives of Virology, 153, 1621–1629. 10.1007/s00705-008-0155-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens, J. , Otto, P. H. , Ciarlet, M. , Desselberger, U. , Van Ranst, M. , & Johne, R. (2012). VP6‐sequence‐based cutoff values as a criterion for rotavirus species demarcation. Archives of Virology, 157, 1177–1182. 10.1007/s00705-012-1273-3 [DOI] [PubMed] [Google Scholar]
- Matthijnssens, J. , Potgieter, C. A. , Ciarlet, M. , Parreno, V. , Martella, V. , Banyai, K. , … Van Ranst, M. (2009). Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? Journal of Virology, 83, 2917–2929. 10.1128/JVI.02246-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalov‐Kovacs, E. , Gellert, A. , Marton, S. , Farkas, S. L. , Feher, E. , Oldal, M. , … Banyai, K. (2015). Candidate new rotavirus species in sheltered dogs. Hungary. Emerg. Infect. Dis., 21, 660–663. 10.3201/eid2104.141370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino, S. , Matthijnssens, J. , Badaracco, A. , Garaicoechea, L. , Zeller, M. , Heylen, E. , … Parreno, V. (2013). Equine G3P[3] rotavirus strain E3198 related to simian RRV and feline/canine‐like rotaviruses based on complete genome analyses. Veterinary Microbiology, 161, 239–246. 10.1016/j.vetmic.2012.07.033 [DOI] [PubMed] [Google Scholar]
- Mullick, S. , Mukherjee, A. , Ghosh, S. , Pazhani, G. P. , Sur, D. , Manna, B. , … Chawla‐Sarkar, M. (2013). Genomic analysis of human rotavirus strains G6P[14] and G11P[25] isolated from Kolkata in 2009 reveals interspecies transmission and complex reassortment events. Infect. Genet. Evol., 14, 15–21. 10.1016/j.meegid.2012.11.010 [DOI] [PubMed] [Google Scholar]
- Nair, N. , Feng, N. , Blum, L. K. , Sanyal, M. , Ding, S. , Jiang, B. , … Greenberg, H. B. (2017). VP4‐ and VP7‐specific antibodies mediate heterotypic immunity to rotavirus in humans. Science Translational Medicine, 9, 1–12. 10.1126/scitranslmed.aam5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer, O. , Jarvis, M. C. , Ciarlet, M. , & Marthaler, D. G. (2017). Genotypic and epitope characteristics of group A porcine rotavirus strains circulating in Canada. Virology, 507, 53–63. 10.1016/j.virol.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Parra, G. I. , Bok, K. , Martinez, M. , & Gomez, J. A. (2004). Evidence of rotavirus intragenic recombination between two sublineages of the same genotype. Journal of General Virology, 85, 1713–1716. 10.1099/vir.0.79851-0 [DOI] [PubMed] [Google Scholar]
- Patton, J. T. , Hua, J. , & Mansell, E. A. (1993). Location of intrachain disulfide bonds in the VP5* and VP8* trypsin cleavage fragments of the rhesus rotavirus spike protein VP4. Journal of Virology, 67, 4848–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Losada, M. , Arenas, M. , Galan, J. C. , Palero, F. , & Gonzalez‐Candelas, F. (2015). Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol., 30, 296–307. 10.1016/j.meegid.2014.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento, J. B. , Crawford, S. E. , Estes, M. K. , & Prasad, B. V. (2006). Rotavirus proteins: Structure and assembly. Current Topics in Microbiology and Immunology, 309, 189–219. [DOI] [PubMed] [Google Scholar]
- Phan, T. G. , Okitsu, S. , Maneekarn, N. , & Ushijima, H. (2007a). Evidence of intragenic recombination in G1 rotavirus VP7 genes. Journal of Virology, 81, 10188–10194. 10.1128/JVI.00337-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T. G. , Okitsu, S. , Maneekarn, N. , & Ushijima, H. (2007b). Genetic heterogeneity, evolution and recombination in emerging G9 rotaviruses. Infect. Genet. Evol., 7, 656–663. 10.1016/j.meegid.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Ramig, R. F. (1997). Genetics of the rotaviruses. Annual Review of Microbiology, 51, 225–255. 10.1146/annurev.micro.51.1.225 [DOI] [PubMed] [Google Scholar]
- Ruggeri, F. M. , & Greenberg, H. B. (1991). Antibodies to the trypsin cleavage peptide VP8 neutralize rotavirus by inhibiting binding of virions to target cells in culture. Journal of Virology, 65, 2211–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeng‐Chuto, K. , Lorsirigool, A. , Temeeyasen, G. , Vui, D. T. , Stott, C. J. , Madapong, A. , … Nilubol, D. (2017). Different Lineage of Porcine Deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound. Emerg. Dis., 64, 3–10. 10.1111/tbed.2017.64.issue-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikruang, W. , Khamrin, P. , Chaimongkol, N. , Suantai, B. , Kongkaew, A. , Kongkaew, S. , … Maneekarn, N. (2013). Genetic diversity and novel combinations of G4P[19] and G9P[19] porcine rotavirus strains in Thailand. Veterinary Microbiology, 161, 255–262. 10.1016/j.vetmic.2012.07.036 [DOI] [PubMed] [Google Scholar]
- Settembre, E. C. , Chen, J. Z. , Dormitzer, P. R. , Grigorieff, N. , & Harrison, S. C. (2011). Atomic model of an infectious rotavirus particle. EMBO Journal, 30, 408–416. 10.1038/emboj.2010.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Chen, J. , Li, H. , Sun, D. , Wang, C. , & Feng, L. (2012). Molecular characterization of a rare G9P[23] porcine rotavirus isolate from China. Archives of Virology, 157, 1897–1903. 10.1007/s00705-012-1363-2 [DOI] [PubMed] [Google Scholar]
- Simmonds, P. (2006). Recombination and selection in the evolution of picornaviruses and other Mammalian positive‐stranded RNA viruses. Journal of Virology, 80, 11124–11140. 10.1128/JVI.01076-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyer, A. , Sagadin, M. , Kolenc, M. , & Poljsak‐Prijatelj, M. (2013). Whole genome sequence analysis of bovine G6P[11] rotavirus strain found in a child with gastroenteritis. Infect. Genet. Evol., 13, 89–95. 10.1016/j.meegid.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Sun, X. , Li, D. , Peng, R. , Guo, N. , Jin, M. , Zhou, Y. , … Duan, Z. J. (2016). Functional and structural characterization of P[19] rotavirus VP8* interaction with histo‐blood group antigens. Journal of Virology, 90, 9758–9765. 10.1128/JVI.01566-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y. , Gojobori, T. , & Nakagomi, O. (1998). Intragenic recombinations in rotaviruses. FEBS Letters, 427, 183–187. 10.1016/S0014-5793(98)00415-3 [DOI] [PubMed] [Google Scholar]
- Taniguchi, K. , Urasawa, T. , Kobayashi, N. , Ahmed, M. U. , Adachi, N. , Chiba, S. , & Urasawa, S. (1991). Antibody response to serotype‐specific and cross‐reactive neutralization epitopes on VP4 and VP7 after rotavirus infection or vaccination. Journal of Clinical Microbiology, 29, 483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns, S. , Heylen, E. , Zeller, M. , Roukaerts, I. D. , Desmarets, L. M. , Van Ranst, M. , … Matthijnssens, J. (2015). Complete genome characterization of recent and ancient Belgian pig group A rotaviruses and assessment of their evolutionary relationship with human rotaviruses. Journal of Virology, 89, 1043–1057. 10.1128/JVI.02513-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, P. F. , Jin, Y. L. , Xing, G. , Qv, L. L. , Huang, Y. W. , & Zhou, J. Y. (2014). Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerging Infectious Diseases, 20, 1735–1738. 10.3201/eid2010.140338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tihova, M. , Dryden, K. A. , Bellamy, A. R. , Greenberg, H. B. , & Yeager, M. (2001). Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: Implications for cell entry. Journal of Molecular Biology, 314, 985–992. 10.1006/jmbi.2000.5238 [DOI] [PubMed] [Google Scholar]
- Vlasova, A. N. , Amimo, J. O. , & Saif, L. J. (2017). Porcine rotaviruses: Epidemiology, immune responses and control strategies. Viruses, 9, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, R. J. (2015). Intrasegmental recombination does not contribute to the long‐term evolution of group A rotavirus. Infect. Genet. Evol., 32, 354–360. 10.1016/j.meegid.2015.03.035 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Tian, Y. , Lin, S. L. , Sun, S. F. , Chen, J. , Wang, G. S. , … Jiang, S. J. (2017). Two Distinct Genotypes of Porcine Epidemic Diarrhoea Virus in Vaccinated Pig Flocks in Shandong Province of China, 2012‐2015. Transbound. Emerg. Dis., 64, 1549–1556. 10.1111/tbed.12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Zhu, Y. , Zhu, X. , Chen, J. , Shi, H. , Shi, D. , … Feng, L. (2017). ORF3a deletion in field strains of porcine‐transmissible gastroenteritis virus in China: A hint of association with porcine respiratory coronavirus. Transbound. Emerg. Dis., 64, 698–702. 10.1111/tbed.12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Zhu, Y. , Zhu, X. , Shi, H. , Chen, J. , Shi, D. , … Feng, L. (2017). Identification of a natural recombinant transmissible gastroenteritis virus between purdue and miller clusters in China. Emerg. Microbes Infect., 6, E74 10.1038/emi.2017.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


