Summary
A novel circovirus called porcine circovirus type 3 (PCV3) was recently reported to exist in the USA. This circovirus is associated with porcine dermatitis, nephropathy syndrome and reproductive failure. This study reports on the first identification, widely epidemic, different phylogenetic clusters, potential role in sow reproductive failure and possible origins of PCV3 in China.
Keywords: China, complete genome, genetic characteristic, identification, porcine circovirus type 3
1. Introduction
Porcine circovirus (PCV) is a small, non‐enveloped, single‐stranded circular DNA virus belonging to the family Circoviridae, genus Circovirus (Mankertz et al., 2004) that has caused a wide spectrum of diseases in pigs and other animals (Bexton et al., 2015; Li et al., 2013). Two circovirus species have been reported in pigs: PCV type 1 (PCV1) (Tischer, Rasch, & Tochtermann, 1974) and PCV type 2 (PCV2) (Allan et al., 1998). PCV1 is non‐pathogenic to pigs (Tischer et al., 1986). PCV2 is the primary causative agent of PCV‐associated diseases, which cause significant economic losses to the swine industry worldwide (Rose, Opriessnig, Grasland, & Jestin, 2012). The capsid in the proteins of circoviruses is the major structural protein and antigenic characteristics that determine proteins (Nawagitgul et al., 2000).
Since 2010, new circoviruses from porcine samples were only initially characterized in some studies (Li et al., 2010; Zhang, Li, Deng, Kapusinszky, & Delwart, 2014). In 2016, a novel circovirus, called PCV type 3 (PCV3), was reported to exist in the USA (Palinski et al., 2016). PCV3 is associated with porcine dermatitis, nephropathy syndrome and reproductive failure (Palinski et al., 2016) and cardiac and multisystemic inflammation (Phan et al., 2016). In 2016, a total of 356 sows at three farms in the Liaoning and Jiangxi Provinces and Chongqin City suffered from reproductive failure and acute loss of neonatal piglets. General pig pathogens, such as porcine pseudorabies virus (PRV), PCV2, porcine reproductive and respiratory syndrome virus (PRRSV), porcine epidemic diarrhoea virus (PEDV), porcine transmissible gastroenteritis virus (TGEV), porcine rotavirus (RV) and classical swine fever virus (CSFV), were not detected in these cases. PCV3 was the only detected pathogen. The geographical distribution, prevalence, tissue tropism, role in diseases and origin of PCV3 in China are unknown. This study reports on the first identification, widely epidemic, wide tissue tropism, different phylogenetic clusters, potential role in sow reproductive failure and possible origins of PCV3 in China.
2. Materials and Methods
A total of 356 sows at three farms in the Liaoning and Jiangxi Provinces and Chongqin City have suffered from reproductive failure and acute loss of neonatal piglets since March 2016. The stillborn rate of the delivery sows ranged from 5.2% to 20.1% at these three farms, while sow mortality ranged from 5.4% to 10.5%. The general pig pathogens, such as PRV, PCV2, PRRSV, PEDV, TGEV, RV and HCV, were excluded by reverse transcription polymerase chain reaction (PCR) or simple PCR methods (Li, Li, Yan, Chen, & He, 2009; Yu et al., 2016; Zhang & He, 2010). PCV3 was the only detected pathogen in these cases. To better understand the infection status, epidemic status, geographical distribution, potential pathogenicity and genetic characteristics of PCV3 in China, a total of 222 samples (i.e., stillborn, tissues, semen and serum) were collected from 35 farms in 11 provinces or districts (i.e., Anhui, Chongqing, Fujian, Hebei, Henan, Hunan, Jiangsu, Jiangxi, Liaoning, Shenyang and Zhejiang) in China. These samples were subjected to pathogen detection in the Diagnostic Center of Animal Epidemic Diseases of Huazhong Agricultural University. The aforementioned samples were individually collected, stored, distilled in 2‐ or 5‐ml EP tubes or clear package bags and then transported utilizing ice boxes. The stillborn and tissues samples were homogenized and diluted 10‐fold with phosphate‐buffered saline (PBS; 0.1 M, pH 7.4). The semen and serum samples were diluted 10‐fold with PBS (0.1 M, pH 7.4). All the samples were frozen and thawed twice to release, further subjected to a vortex for 5 min, and centrifuged at 11,000 rpm (Eppendorf, Germany) for 8 min at 4°C. The supernatants were utilized for DNA and RNA extraction immediately or stored at −80°C refrigerator for further usage. A pair of primers was designed to detect PCV3 (Table 1). The detection limit of this method was 20.5 pg nuclear acid. The primers have no cross‐reactions with the PR, PCV2, TGEV, TGEV, RV, PRRSV, porcine parvovirus and deltacoronavirus temples stored in our laboratory. Two pairs of primers were utilized for the entire genome sequencing (Table 1).
Table 1.
List of primers used in this study
| Primer name | Nucleotide sequence | Primer location (nt)a | Product length (bp) | Purpose |
|---|---|---|---|---|
| PCV3‐1‐F | TTACTTAGAGAACGGACTTGTAACG | 1339‐1363 | 649 | Detection |
| PCV3‐1‐R | AAATGAGACACAGAGCTATATTCAG | 1987‐1965 | ||
| PCV3‐genome‐1‐F | TAGTATTACCCGGCACCTCGGAACC | 1‐25 | 1257 | Genome sequencing |
| PCV3‐genome‐1‐R | ACAGGTAAACGCCCTCGCATGTGGG | 1233‐1257 | ||
| PCV3‐genome‐2‐F | TTGCACTTGTGTACAATTATTGCG | 1120‐1143 | 1075 | Genome sequencing |
| PCV3‐genome‐2‐R | ATCTTCAGGACACTCGTAGCACCAC | 2170‐0‐194 |
Numbers correspond to positions within the strain PCV3/CN/Fujian‐5/2016.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The reaction conditions were as follows: pre‐denaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min and a final extension at 72°C for 10 min. The amplicons for the genome sequencing were gel‐purified, cloned into pMD®18‐T Vector (Takara, Japan) and sequenced in both directions at the GenScript company (Nanjing, China). The amplicons for detection were gel‐purified and sequenced. The genome sequences were assembled utilizing software MEGA v6.06 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013). The complete genome of the 9 PCV3 isolates and partial capsid gene (474 bp) of the 33 PCV3 isolates were deposited in the GenBank under the GenBank accession numbers from KY075986.1 to KY075994.1 and from KY075995.1 to KY076027.1, respectively (Appendix 1).
The genome sequences of the 26 coronaviruses (CVs) were downloaded from the GenBank (Figure 1). Multiple sequence alignments were generated with MEGA v6.06. The genetic identities of these isolates were analysed with the Lasergene package (DNAStar Inc., Madison, WI, USA). The phylogenetic trees of the genomes of nine CV strains collected from Fujian, Jiangxi, Henan and Chongqin, as well as the partial capsid genes of 33 PCVs collected from Anhui, Henan, Fujian, Jiangxi, Hubei, Liaoning and Chongqin, were constructed utilizing MEGA v6.06 with p‐distance‐based, neighbour‐joining method (bootstrap analysis with 1,000 replicates). A similarity plots analysis of nine Chinese isolates in this study, 1 USA PCV3 isolate, and 5 bat CVs (i.e., BtMr‐CV/GD2012 [KJ641711.1], BtPspp.‐CV/GD2012 [KJ641716.1], BtRs‐CV/HuB2013 [KJ641723.1], BtRa‐CV/JS2013 [KJ641724.1] and BtPa‐CV‐1/NX2013 [KJ641727.1]) was performed by the sliding window method as implemented in the SimPlot, v. 3.5.1 package (Lole et al., 1999). The similarity plot of these PCV3 strains was drawn, and the PCV3/CN/Fujian‐5/2016 was set as a query strain.
Figure 1.
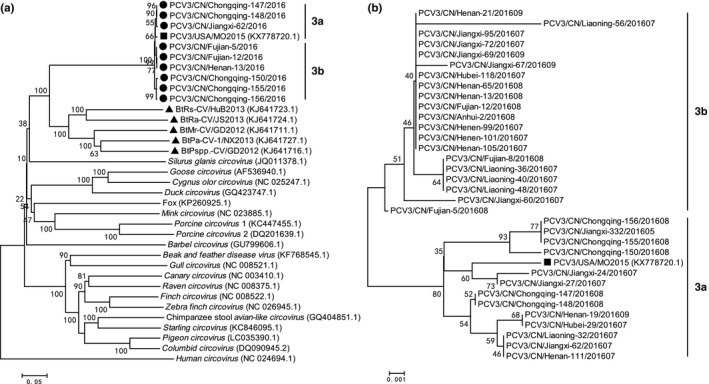
(a) Phylogenetic tree of the complete genome of different circovirus species. (b) Phylogenetic tree of partial capsid gene of different Chinese PCV3 isolates with a USA PCV3 isolates PCV3‐USA‐MO2015 (KX778720.1). The phylogenetic tree was constructed using the p‐distance‐based neighbour‐joining method in MEGA 6.06 software. Bootstrap values were calculated with 1,000 replicates. Black solid circles indicate the strains from China. Black solid square indicates the strain from USA. Black solid triangles indicate the bat circovirus strains from China. Scale bars indicate nucleotide substitutions per site.
3. Results and Discussion
The PCV3‐positive rate at the farm level was 68.6% (24/35). The positive rates of PCV2 and PCV3 in these samples were 62.2% (138/222) and 34.7% (77/222), respectively. The co‐infection rate of PCV3 with PCV2 was 15.8% (35/222). The co‐infection of PCV2 with other pathogens has been widely reported (Ellis, 2014). The co‐infection rate of PCV3 and PCV2 was 15.8% (35/222) in the above detection results. The emergence of PCV2 is believed to be an important reason for the failure of PCV2 vaccination in several farms (Reiner, Hofmeister, & Willems, 2015; Seo et al., 2014; Xiao, Halbur, & Opriessnig, 2012). Whether co‐infection with PCV3 also plays a role in these cases must be further researched. The singular infection rate of PCV3 in the samples was 18.9% (42/222). PCV3 can be detected in the samples collected from all 11 provinces or districts. Results show that PCV3 has spread widely in China. PCV3 can be detected in the samples: brain [73.5% (25/34); including 57.1% (8/14) in the brain of stillbirth samples], lung [4/7; including 66.7% (2/3) in the lungs of stillbirth samples], lymph node [36.6% (15/41)], tonsil [66.7% (4/6)], semen [8.5%(4/47)] and serum [28.7%(25/87)]. These results suggest that PCV3 can be detected from different porcine tissues. PCV3 detection in the stillbirth and semen samples suggests the risk of a vertical transmission of PCV3.
The nucleotide similarity of the complete genomes of 9 PCV3 isolates collected from four provinces or districts and partial capsid gene (474 bp) of 33 PCV3 isolates from seven provinces or districts were analysed and compared to identify the genetic characteristics of PCV3 in China. The genome and capsid nucleotide similarity of PCV3 isolates from China with the USA isolates [i.e., PCV3‐USA‐MO2015 (KX778720.1)] ranged from 99.0% to 99.1% and 98.4% to 98.9%, respectively. The genome and capsid nucleotide similarity of the PCV3 isolates from China ranged from 99.0% to 100% and 98.4 to 100%, respectively. The results show that the divergence level of the capsid was higher than that of the complete genome.
Chinese PCV3 isolates were clustered into the same cluster with the USA isolates (i.e., PCV3‐USA‐MO2015 [KX778720.1]) in the complete genome‐based phylogenetic tree analysis. These isolates also showed a close evolution relationship with Chinese Bat CVs (Figure 1a), such as BtMr‐CV/GD2012 (KJ641711.1), BtPspp.‐CV/GD2012 (KJ641716.1), BtRs‐CV/HuB2013 (KJ641723.1), BtRa‐CV/JS2013 (KJ641724.1) and BtPa‐CV‐1/NX2013 (KJ641727.1) (Figure 1a). In the complete genome and partial capsid gene‐based phylogenetic tree analysis in Figures 1a and b, the Chinese PCV3 isolates were divided into two clusters, designed as 3a and 3b (Figure 1). This process shows the divergence of the PCV3 isolates in China. The USA PCV3 isolates (i.e., PCV3‐USA‐MO2015 [KX778720.1]) belonged to cluster 3a. The sequence similarity of these CV isolates is plotted in Figure 2 to study the genetic relationship of the Chinese PCV3 isolates with the Chinese Bat CVs. Two recombination regions were identified between the bat CVs and PCV3 isolates in 264‐564nt and 714‐1148nt (Figure 2). These results suggest that PCV3 can be the recombination results of the bat CVs.
Figure 2.
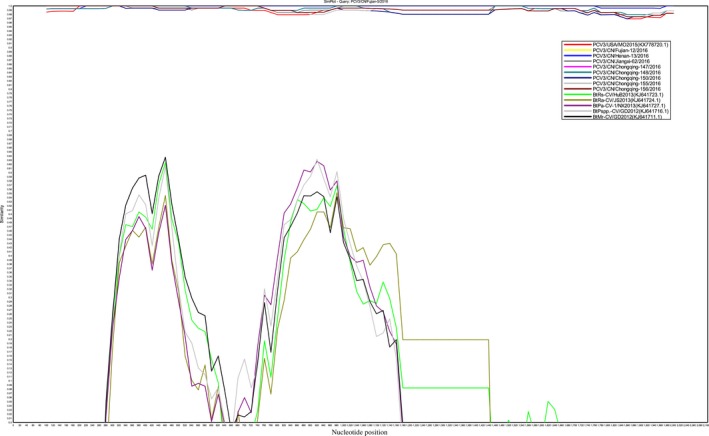
Similarity plot of the whole genome of 10 PCV3 isolates with 5 Bat circovirus isolates. The Chinese PCV3 isolates, PCV3/CN/Fujian‐5/2016, was set as query strain. The similarity plot was constructed using the two‐parameter (Kimura) distance model with a sliding window of 200 bp and step size of 20 bp. The vertical and horizontal axes indicate the nucleotide similarity per cent and nucleotide position (bp) in the graph, respectively.
PCV3 has been poorly understood because it was first identified in the USA in 2010 (Li et al., 2010). PCV3 has been identified as a pathogen agent associated with pig diseases (Palinski et al., 2016). However, the epidemic status and genetic characteristics of PCV3 in China are unclear. This paper reports on the first identification, widely epidemic, different phylogenetic clusters, potential role in sow reproductive failure and possible origins of PCV3 in China. We hypothesize that PCV3 is a potential zoonotic agent that can be vertically transmitted based on the above results. It can be detected from different tissues, such as brain, lung, lymph node and tonsil tissues. The above results are crucial to better understand PCV3 in China. Further studies are required to isolate PCV3, explore its biological and pathogenic characteristics and monitor its singular and co‐infection status.
Conflict of interest
All authors have declared no conflict of interest.
Acknowledgements
This work was supported by China Agriculture Research System (No. CARS‐36) and China Scholarship Council No. 201606760012 to F. Chen.
Appendix 1.
1.1.
| No. | Strain Name | GenBank accession no. | Collection nations | Collection date | Purpose |
|---|---|---|---|---|---|
| 1 | PCV3/CN/Fujian‐5/2016 | KY075986.1 | China:Fujian | 2016 | Complete genome analysis |
| 2 | PCV3/CN/Fujian‐12/2016 | KY075987.1 | China:Fujian | 2016 | Complete genome analysis |
| 3 | PCV3/CN/Henan‐13/2016 | KY075988.1 | China:Henan | 2016 | Complete genome analysis |
| 4 | PCV3/CN/Jiangxi‐62/2016 | KY075989.1 | China:Jiangxi | 2016 | Complete genome analysis |
| 5 | PCV3/CN/Chongqing‐147/2016 | KY075990.1 | China:Chongqing | 2016 | Complete genome analysis |
| 6 | PCV3/CN/Chongqing‐148/2016 | KY075991.1 | China:Chongqing | 2016 | Complete genome analysis |
| 7 | PCV3/CN/Chongqing‐150/2016 | KY075992.1 | China:Chongqing | 2016 | Complete genome analysis |
| 8 | PCV3/CN/Chongqing‐155/2016 | KY075993.1 | China:Chongqing | 2016 | Complete genome analysis |
| 9 | PCV3/CN/Chongqing‐156/2016 | KY075994.1 | China:Chongqing | 2016 | Complete genome analysis |
| 10 | PCV3/CN/Anhui‐2/201608 | KY075995.1 | China:Anhui | 2016 | Partial capsid gene analysis |
| 11 | PCV3/CN/Fujian‐5/201608 | KY075996.1 | China:Fujian | 2016 | Partial capsid gene analysis |
| 12 | PCV3/CN/Fujian‐8/201608 | KY075997.1 | China:Fujian | 2016 | Partial capsid gene analysis |
| 13 | PCV3/CN/Fujian‐12/201608 | KY075998.1 | China:Fujian | 2016 | Partial capsid gene analysis |
| 14 | PCV3/CN/Henan‐13/201608 | KY075999.1 | China:Henan | 2016 | Partial capsid gene analysis |
| 15 | PCV3/CN/Henan‐19/201609 | KY076000.1 | China:Henan | 2016 | Partial capsid gene analysis |
| 16 | PCV3/CN/Henan‐21/201609 | KY076001.1 | China:Henan | 2016 | Partial capsid gene analysis |
| 17 | PCV3/CN/Jiangxi‐24/201607 | KY076002.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
| 18 | PCV3/CN/Jiangxi‐27/201607 | KY076003.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
| 19 | PCV3/CN/Hubei‐29/201607 | KY076004.1 | China:Hubei | 2016 | Partial capsid gene analysis |
| 20 | PCV3/CN/Liaoning‐32/201607 | KY076005.1 | China:Liaoning | 2016 | Partial capsid gene analysis |
| 21 | PCV3/CN/Liaoning‐36/201607 | KY076006.1 | China:Liaoning | 2016 | Partial capsid gene analysis |
| 22 | PCV3/CN/Liaoning‐40/201607 | KY076007.1 | China:Liaoning | 2016 | Partial capsid gene analysis |
| 23 | PCV3/CN/Liaoning‐48/201607 | KY076008.1 | China:Liaoning | 2016 | Partial capsid gene analysis |
| 24 | PCV3/CN/Liaoning‐56/201607 | KY076009.1 | China:Liaoning | 2016 | Partial capsid gene analysis |
| 25 | PCV3/CN/Jiangxi‐60/201607 | KY076010.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
| 26 | PCV3/CN/Jiangxi‐62/201607 | KY076011.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
| 27 | PCV3/CN/Henan‐65/201608 | KY076012.1 | China:Henan | 2016 | Partial capsid gene analysis |
| 28 | PCV3/CN/Jiangxi‐67/201609 | KY076013.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
| 29 | PCV3/CN/Jiangxi‐69/201609 | KY076014.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
| 30 | PCV3/CN/Jiangxi‐72/201607 | KY076015.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
| 31 | PCV3/CN/Jiangxi‐95/201607 | KY076016.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
| 32 | PCV3/CN/Henen‐99/201607 | KY076017.1 | China:Henan | 2016 | Partial capsid gene analysis |
| 33 | PCV3/CN/Henen‐101/201607 | KY076018.1 | China:Henan | 2016 | Partial capsid gene analysis |
| 34 | PCV3/CN/Henan‐105/201607 | KY076019.1 | China:Henan | 2016 | Partial capsid gene analysis |
| 35 | PCV3/CN/Henan‐111/201607 | KY076020.1 | China:Henan | 2016 | Partial capsid gene analysis |
| 36 | PCV3/CN/Hubei‐118/201607 | KY076021.1 | China:Hubei | 2016 | Partial capsid gene analysis |
| 37 | PCV3/CN/Chongqing‐147/201608 | KY076022.1 | China:Chongqing | 2016 | Partial capsid gene analysis |
| 38 | PCV3/CN/Chongqing‐148/201608 | KY076023.1 | China:Chongqing | 2016 | Partial capsid gene analysis |
| 39 | PCV3/CN/Chongqing‐150/201608 | KY076024.1 | China:Chongqing | 2016 | Partial capsid gene analysis |
| 40 | PCV3/CN/Chongqing‐155/201608 | KY076025.1 | China:Chongqing | 2016 | Partial capsid gene analysis |
| 41 | PCV3/CN/Chongqing‐156/201608 | KY076026.1 | China:Chongqing | 2016 | Partial capsid gene analysis |
| 42 | PCV3/CN/Jiangxi‐332/201605 | KY076027.1 | China:Jiangxi | 2016 | Partial capsid gene analysis |
Ku X, Chen F, Li P, et al. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. 2017;64:703–708. 10.1111/tbed.12638
References
- Allan, G. M. , McNeilly, F. , Kennedy, S. , Daft, B. , Ellis, J. A. , Haines, D. M. , … Adair, B. M. (1998). Isolation of porcine circovirus‐like viruses from pigs with a wasting disease in the USA and Europe. Journal of Veterinary Diagnostic Investigation, 10(1), 3–10. [DOI] [PubMed] [Google Scholar]
- Bexton, S. , Wiersma, L. C. , Getu, S. , van Run, P. R. , Verjans, G. M. , Schipper, D. , … Smits, S. L. (2015). Detection of circovirus in foxes with meningoencephalitis, United Kingdom, 2009–2013. Emerging Infectious Diseases, 21(7), 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J. (2014). Porcine circovirus a historical perspective. Veterinary Pathology, 51(2), 315–327. [DOI] [PubMed] [Google Scholar]
- Li, L. , Kapoor, A. , Slikas, B. , Bamidele, O. S. , Wang, C. , Shaukat, S. , … Delwart, E. (2010). Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. Journal of Virology, 84(4), 1674–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. J. , Li, W. T. , Yan, W. D. , Chen, H. C. , & He, Q. G. (2009). Genotype analysis of porcine circovirus 2 in some areas of China. Chinese Journal of Animal and Veterinary Science, 09, 1358–1362. (in Chinese). [Google Scholar]
- Li, L. , McGraw, S. , Zhu, K. , Leutenegger, C. M. , Marks, S. L. , Kubiski, S. , … Pesavento, P. A. (2013). Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerging Infectious Diseases, 19(4), 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole, K. S. , Bollinger, R. C. , Paranjape, R. S. , Gadkari, D. , Kulkarni, S. S. , Novak, N. G. , … Ray, S. C. (1999). Full‐length human immunodeficiency virus type 1 genomes from subtype C‐infected seroconverters in India, with evidence of intersubtype recombination. Journal of Virology 73(1), 152‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankertz, A. , Çaliskan, R. , Hattermann, K. , Hillenbrand, B. , P., Mueller, B. , … Finsterbusch, T. (2004). Molecular biology of Porcine circovirus: Analyses of gene expression and viral replication. Veterinary Microbiology, 98(2), 81–88. [DOI] [PubMed] [Google Scholar]
- Nawagitgul, P. , Morozov, I. , Bolin, S. R. , Harms, P. A. , Sorden, S. D. , & Paul, P. S. (2000). Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. Journal of General Virology, 81(9), 2281–2287. [DOI] [PubMed] [Google Scholar]
- Palinski, R. , Piñeyro, P. , Shang, P. , Yuan, F. , Guo, R. , Fang, Y. , … Hause, B. M. (2016). A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. Journal of Virology, 91(1), e01879–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T. G. , Giannitti, F. , Rossow, S. , Marthaler, D. , Knutson, T. , Li, L. , … Delwart, E. (2016). Detection of a novel circovirus PCV3 in pigs with cardiac and multi‐systemic inflammation. Virology Journal, 13(1), 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner, G. , Hofmeister, R. , & Willems, H. (2015). Genetic variability of porcine circovirus 2 (PCV2) field isolates from vaccinated and non‐vaccinated pig herds in Germany. Veterinary Microbiology, 180(1), 41–48. [DOI] [PubMed] [Google Scholar]
- Rose, N. , Opriessnig, T. , Grasland, B. , & Jestin, A. (2012). Epidemiology and transmission of porcine circovirus type 2 PCV2. Virus Research, 164(1), 78–89. [DOI] [PubMed] [Google Scholar]
- Seo, H. W. , Park, C. , Kang, I. , Choi, K. , Jeong, J. , Park, S. J. , & Chae, C. (2014). Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea. Archives of Virology, 159(11), 3107–3111. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. , & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer, I. , Mields, W. , Wolff, D. , Vagt, M. , & Griem, W. (1986). Studies on epidemiology and pathogenicity of porcine circovirus. Archives of Virology, 91(3–4), 271–276. [DOI] [PubMed] [Google Scholar]
- Tischer, I. , Rasch, R. , & Tochtermann, G. (1974). Characterization of papovavirus and picornavirus‐like particles in permanent pig kidney cell lines. Zentralbl Bakteriol, 226(2), 153–167. [PubMed] [Google Scholar]
- Xiao, C. T. , Halbur, P. G. , & Opriessnig, T. (2012). Complete genome sequence of a novel porcine circovirus type 2b variant present in cases of vaccine failures in the United States. Journal of Virology, 86(22), 12469–12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T. , Chen, F. , Ku, X. , Fan, J. , Zhu, Y. , Ma, H. , … He, Q. (2016). Growth characteristics and complete genomic sequence analysis of a novel pseudorabies virus in China. Virus Genes, 52(4), 474–483. [DOI] [PubMed] [Google Scholar]
- Zhang, K. , & He, Q. (2010). Establishment and clinical application of a multiplex reverse transcription PCR for detection of porcine epidemic diarrhea virus, porcine transmissible gastroenteritis virus and porcine group A rotavirus. Chinese Journal of Animal and Veterinary Science, 41, 1001–1005. [Google Scholar]
- Zhang, W. , Li, L. , Deng, X. , Kapusinszky, B. , & Delwart, E. (2014). What is for dinner? Viral metagenomics of US store bought beef, pork, and chicken. Virology, 468, 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]


