Summary
Illegal bushmeat traffic is an important threat to biodiversity conservation of several endangered species and may contribute to the emergence and spread of infectious diseases in humans. The hunting, manipulation and consumption of wildlife‐based products, especially those of primate origin, may be a threat to human health; however, few studies have investigated the role of bushmeat trade and consumption as a potential source of human infections to date. In this study, we report the screening of viral pathogens in African simian game seized by French customs at Toulouse Blagnac Airport. Epifluorescence microscopy revealed the presence of virus‐like particles in the samples, and further metagenomic sequencing of the DNA and RNA viromes confirmed the presence of sequences related to the Siphoviridae, Myoviridae and Podoviridae bacteriophage families; some of them infecting bacterial hosts that could be potentially pathogenic for humans. To increase the sensitivity of detection, twelve pan‐generic PCRs targeting several viral zoonoses were performed, but no positive signal was detected. A large‐scale inventory of bacteria, viruses and parasites is urgently needed to globally assess the risk for human health of the trade, manipulation and consumption of wildlife‐related bushmeat.
Keywords: bushmeat, wild game, virus, zoonoses, epifluorescence microscopy, viral metagenomics, pan‐generic PCR
Introduction
Human zoonoses are directly acquired from animals or indirectly via arthropod bites and are an increasing public health problem. More than two‐thirds of emerging human pathogens are of zoonotic origin, and of them, more than 70% originate from wildlife (Cutler et al., 2010;One Health Initiative). Hunted wild animals are not only an important source of protein for many poor rural populations (Brashares et al., 2011); they are also an important part of some cultures, for animist practices or to reflect a luxury status in industrialized countries, for example. Usually composed of rodent‐borne, antelope‐borne, primate‐borne, and turtle‐borne meat, bushmeat is a threat to both biodiversity conservation of several endangered species such as primates and pangolins (Effiom et al., 2013) and public health. The traffic also contributes to decrease the proteins availability in some poor rural regions (Brashares et al., 2011). All industrialized countries are affected by the illegal traffic of bushmeat, as reported by Falk et al. in Switzerland (Falk et al., 2013) and by Bair‐Brake et al. (2014) in the United States. In 2013 in France, customs reported 647 seizures of endangered species (wildlife and flora), of which Convention on International Trade in Endangered Species (CITES)‐listed animals were estimated at 39% of seized bushmeat carcasses (Chaber et al., 2010, French customs annual report). The international illegal traffic of animals has been estimated to total five tons of wild game, livestock meat and fish, carried by passengers arriving from sub‐Saharan Africa at Paris Charles de Gaulle Airport (France) (Chaber et al., 2010), of which little is seized and directly destroyed by customs and veterinary services each year. Equivalent traffic probably occur from other countries (Asia or South American continents, for example), but no report was released yet regarding the detailed origins of customs seizures of illegal game entering in France.
In some countries, bats are hunted for their meat, resulting in the infection of humans, as previously reported (Kamins et al., 2014; Pernet et al., 2014). In addition, several known examples of primate hunting have led to the emergence of human epidemics, such as Ebola haemorrhagic fever and AIDS in which the hunting and manipulation of infected carcasses were shown to be the origins of epidemics (Wolfe et al., 2004). However, few studies have investigated the role of simian bushmeat trade, manipulation and/or consumption as a potential source of human infections. The carriage of pathogens by simian bushmeat, such as parasites (Pourrut et al., 2011), bacteria (Bachand et al., 2012) or viruses (Aghokeng et al., 2010; Smith et al., 2012), was previously reported. For example, in the United States, Smith et al. have reported the presence of retroviruses and herpesviruses in non‐human primate tissue samples confiscated at different airports (Smith et al., 2012). In France, Chaber et al. have reported data suggesting the presence of foodborne pathogens, such as Listeria monocytogenes, and carcinogenic concentrations of benzo(a)pyrene in smoked bushmeat confiscated at the Paris Charles de Gaulle and Toulouse Blagnac Airports (Chaber and Cunningham, 2016).
With the progress of molecular biology, PCR‐based methods, mainly pan‐generic PCR, have become the main techniques for virus discovery; however, these techniques require prior knowledge of closely related viral genomes. Next‐generation sequencing (NGS) techniques make it possible to sequence all viral genomes in a given sample without any previous knowledge about their nature, using a combination of sequence‐independent amplification and high‐throughput sequencing. These techniques, which are known as viral metagenomics, are being used exponentially more frequently to characterize the viral diversities of complex environments, such as animal samples (Temmam et al., 2014). Although molecular‐biology‐based methods are not able to discriminate infectious or inactivated viral particles, PCR‐ and NGS‐based methods can provide crucial information onto the different viral species that are present in a sample. In this study, we report the first DNA and RNA viral metagenomic analysis and screening of zoonotic viral pathogens in illegally imported African simian bushmeat seized by French customs at Toulouse Blagnac Airport.
Materials and Methods
Specimens
A total of four specimens of non‐human primates originating from the Central African Republic were confiscated by French customs at Toulouse Blagnac Airport in September 2013. The four animals had been cut into two halves, eviscerated and smoked before importation into France (Fig. 1).
Figure 1.

Primate bushmeat specimens seized at the Toulouse Blagnac Airport.
Samples were taken from the axillary and popliteal regions, containing lymph nodes and muscle tissues. The tissue samples (samples STE0011 to STE0014) were then directly stored at −20°C by the customs officials until further analyses.
Sample pre‐treatment, epifluorescence microscopy and virome preparation
Dilacerated tissues (0.5 cm3) were rehydrated overnight at +4°C in 2 ml of sterile EMEM (Life Technologies, Saint Aubin, France) and then crushed with two 3‐mm tungsten beads and a TissueLyser at 25 Hz for 2 min (Qiagen, Courtaboeuf, France). The clarified supernatant was subsequently used as a template for virome preparation.
DNA and RNA viromes of sample no STE0011 were prepared as previously described (Temmam et al., 2015). Briefly, the clarified supernatant was filtered through a 0.45‐μm filter (Millipore, Molsheim, France), and free nucleic acids were digested at 37°C for 1 h with the following cocktail of nucleases: 20 U Exonuclease I (New England Biolabs, Évry, France), 25 U Benzonase® (Merck Millipore, Molsheim, France), 25 U RNase A (Roche Diagnostics, Meylan, France), 20 U Turbo DNase (Life Technologies) and 10 μl of 10× Turbo DNase buffer. The digested supernatant was then deposited onto a discontinuous 66%–30% sucrose gradient and ultracentrifuged in an MLS50 Beckman‐Coulter rotor at 130 000 g for 2 h at +4°C. The viral fraction was harvested at the interphase between the 66% and 30% sucrose layers using a 23‐G needle.
One hundred microlitres of the purified fraction was harvested to assess the virus‐like particle (VLP) concentration by fluorescence microscopy, as previously described by Thurber et al. (2009). All fluorescence images were acquired with a Leica SP5 inverted confocal microscope with four lasers, a 100× objective and a numerical aperture of 1.4.
DNA and RNA were extracted from the purified viral fraction with Trizol LS® reagent (Life Technologies). Two microlitres of DNA was randomly amplified in 2 independent reactions using a Genomiphi V3 kit (GE Healthcare, Vélizy‐Villacoublay, France) according to the manufacturer's recommendations. Total RNA was processed by random reverse transcription as previously described by Froussard (1992) using Superscript III Reverse Transcriptase (Life Technologies). cDNA was subsequently used as a template for the Klenow reaction and randomly amplified with 2.5 U of Long Amp Taq DNA Polymerase (New England Biolabs) in a final volume of 25 μl, as previously described (Temmam S. ‘Host‐associated metagenomics: a guide to generating infectious RNA viromes’, in revision).
DNA and RNA amplification products were purified twice with Agencourt AMPure Beads (Beckman‐Coulter, Villepinte, France) according to the manufacturer's protocol, eluted to a final volume of 15 μl and sequenced using MiSeq Technology with the paired‐end and barcode strategies according to a Nextera XT library kit in a 2 × 300 bp format (Illumina Inc., San Diego, USA).
Bioinformatic analyses of viromes
Raw reads were imported in pairs into CLC Genomics Workbench 6.0.1 program (CLC Bio, Aarhus, Denmark) and trimmed according to their quality score (Illumina pipeline 1.8 and later), the presence of ambiguities (a maximum of 2 ambiguities), length (reads <50 nt long were discarded) and the adaptors and primers used for random PCR. The pre‐processed viral metagenomes are publicly available on Metavir server (http://metavir-meb.univ-bpclermont.fr) with the identifiers ‘STE0011_DNA’ and ‘STE0011_RNA’ under the project ‘Simian_bushmeat’ and on MG‐RAST server (http://metagenomics.anl.gov/) with the identifiers 4604107.3 and 4604109.3 for the DNA and RNA viromes, respectively.
Cleaned paired reads were assembled into contigs with CLC Genomics program using the following parameters: a word size of 20 nt, minimum contig length of 200 nt, mismatch cost of 2, insertion/deletion cost of 3, length fraction of 0.5 and similarity fraction of 0.8. Contigs and non‐assembled reads were then compared to the NCBI nucleotide database using the BLASTN algorithm, with a minimum coverage of 50%, minimum identity of 50% and E‐value <10–5. Sequences having no significant hits according to the criteria described above were classified as ‘unknown’.
To enhance the detection of viral reads, cleaned paired reads were compared to the NCBI RefSeq viral database using BLASTX program with a minimum coverage of 50%, minimum identity of 50% and E‐value <10−5. Taxonomic assignation of the reads was performed at the family level, and bacterial target hosts of bacteriophages were taxonomically determined at the genus level. Reads taxonomically classified by BLASTX were then mapped against reference genomes using CLC Genomics Workbench 6.0.1 program (CLC Bio, Aarhus, Denmark) to verify their correct taxonomic assignation, with the following parameters: a minimal length fraction of 0.25, minimal similarity fraction of 0.7, mismatch cost of 2 and insertion/deletion cost of 3.
Pan‐generic PCRs
Total nucleic acids were extracted from an aliquot of 100 μl of the purified viral fraction with a BioRobot EZ1 and EZ1 Virus Mini Kit (Qiagen) in a final volume of 60 μl, and 20 μl was subsequently reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Life Technologies) and random hexamers, according to the manufacturer's recommendations.
To control the efficiency of the PCR and the presence of inhibitors originating from the smoked tissue sample extractions, PCR targeting 18S rRNA was performed with 2 μl of cDNA, as previously described (Breitbart and Rohwer, 2005).
A total of 12 pan‐generic PCRs, nested PCR or real‐time PCR were conducted (Table S1) to screen for the presence of several zoonotic viruses possibly infecting humans, including paramyxoviruses (Tong et al., 2008), coronaviruses (de Souza Luna et al., 2007), flaviviruses (Moureau et al., 2007), phleboviruses (Sánchez‐Seco et al., 2003), nairoviruses (Rodriguez et al., 1997), poxviruses (Sánchez‐Seco et al., 2006), alphaviruses (Sánchez‐Seco et al., 2001), hantaviruses (Klempa et al., 2006), orthobunyaviruses (Lambert and Lanciotti, 2009), arenaviruses (Bowen et al., 1997), filoviruses (Zhai et al., 2007) and herpesviruses (VanDevanter et al., 1996), according to the cited authors' recommendations for primer concentrations and annealing conditions. Briefly, 5 μl of cDNA was used in a final volume of 25 μl for the first round of PCR, along with HotStar Taq DNA Polymerase (Qiagen) or a QuantiTect SYBR Green qPCR Kit (Qiagen), depending on the pan‐generic PCR. A volume of 1 μl of the first‐round PCR product was used for nested PCR when needed. PCR products were analysed on a 2% agarose gel, and bands of the expected size were extracted from the gel, purified with a QIAquick Gel Extraction Kit (Qiagen) and sequenced with a Big Dye Terminator v1.1 Cycle Sequencing Kit (Life Technologies) and an ABI 3130 Genetic Analyzer (Life technologies).
Results
Specimen collection
Four specimens of non‐human primates originating from the Central African Republic were confiscated by French customs at the Toulouse Blagnac Airport in 2013. Their specific determination was impossible due to their conservation statuses (Fig. 1), but they all belonged to the Cercopithecidae family.
Detection and quantification of virus‐like particles by epifluorescence microscopy
The estimated numbers of VLPs were 8.85x106 VLPs/ml, 8.28x104 VLPs/ml, 3.72x106 VLPs/ml and 1.73x107 VLPs/ml for the samples STE0011, STE0012, STE0013 and STE0014, respectively (Fig. 2).
Figure 2.

Fluorescence microscopy of VLPs in the bushmeat sample no STE0014. All images were acquired with a Leica SP5 inverted confocal microscope with four lasers, a 100× objective and a numerical aperture of 1.4. The scale bar represents 20 μm.
DNA and RNA viromes of simian bushmeat sample
The DNA and RNA viromes of sample STE0011 were seq‐uenced with Illumina MiSeq technology. After trimming, the DNA and RNA viromes contained 647 272 and 519 397 paired reads, respectively.
The results of the BLASTN search against the nucleotide NCBI database are presented in Figure 3a and 3c, respectively, for the DNA and RNA viromes. The unknown fractions of the DNA and RNA virome datasets represented 25.55% and 6.43%, respectively. Among the known DNA sequences, bacterial sequences represented 74.84%, eukaryotes comprised 25.14%, and viruses included 0.03% of the total assigned reads. Within the known RNA virome, sequences related to bacteria represented the majority of the reads (more than 92% of the total assigned reads), eukaryotic reads comprised 7.23%, and viral reads included 0.64% of the total assigned reads.
Figure 3.
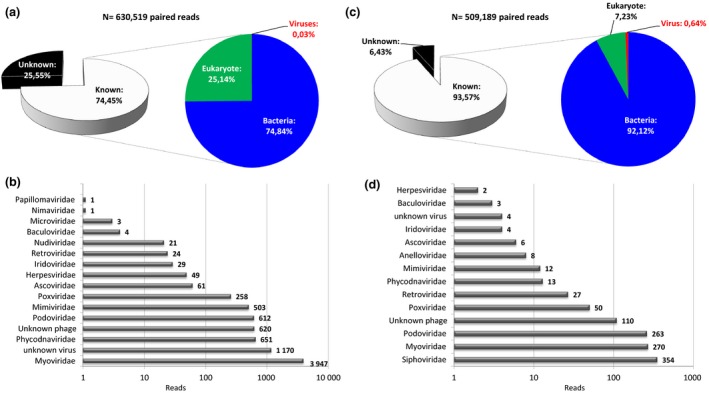
Taxonomic assignation of reads according to a BLASTN search against the NCBI nucleotide database (a and c for the DNA and RNA viromes, respectively) and according to a BLASTX search against the viral RefSeq NCBI database (b and d for the DNA and RNA viromes, respectively).
The eukaryotic reads were related to humans and primates (95.21% and 63.70% of the total eukaryotic reads for the DNA and RNA metagenomes, respectively) in addition to plants (1.48% and 0.43% of the total eukaryotic reads for the DNA and RNA metagenomes, respectively) and arthropods (0.01% and 11.90% of the total eukaryotic reads for the DNA and RNA metagenomes, respectively). Parasite (mainly nematodes and helminths) eukaryotic reads were also detected in the DNA and RNA viromes (0.96% and 21.26%, respectively), which probably remained from undigested free DNA. Among them, Spirometra erinaceieuropaei platyhelminth, a worm commonly infecting domestic animals that causes several diseases in humans, was quite abundant (n = 1503 reads) in the DNA metagenome. Additionally, Haemonchus placei, a nematode mainly infecting cattle in tropical areas, was detected in 558 reads; 125 reads were related to Wuchereria bancrofti, a nematode responsible for lymphatic filariasis in humans, and 5 and 8 reads of the DNA virome were assigned to Leishmania and Plasmodium spp., respectively. Finally, we observed the presence of 2983 sequences representing Schistosoma rodhaini in the RNA metagenome, a trematode responsible for infections in small mammals.
The results of the BLASTX search against the RefSeq viral database are presented in Figure 3B and 3d for the DNA and RNA viromes, respectively. More than two‐thirds of the total viral reads of the DNA virome belonged to bacteriophages, the majority of which were from Siphoviridae (51.38% of the viral reads) and Myoviridae (24.13% of the viral reads). Podoviridae, Microviridae and non‐classified phages were less abundant, comprising only 3.74%, 0.02% and 3.79% of the total viral reads, respectively (Fig. 3b). The majority of bacterial genera infected by Myoviridae belonged to the Bacillus sp. genus (46.31%), followed by Lactobacillus sp. (12.09%), Enterococcus sp. (9.12%), Cronobacter sp. (7.83%) and Enterobacteria sp. (6.89%) (Figure S1). In addition, more than 70% of the bacterial genera infected by Podoviridae were Planktothrix sp. (71.90%) and Cellulophaga sp. (16.01%). Most of the Siphoviridae bacterial hosts belonged to the Enterococcus sp. (44.46%), Staphylococcus sp. (15.51%), Lactococcus sp. (14.87%) and Listeria sp. (7.76%) genera (Figure S1).
Sequences belonging to the Phycodnaviridae (n = 651 reads), Iridoviridae (n = 29 reads), Nudiviridae (n = 21 reads) and Baculoviridae (n = 4 reads) families (Figure 3b) were also detected. Iridoviridae, Nudiviridae and Baculoviridae are insect‐infecting viruses, whereas Phycodnaviridae are large viruses infecting algae. Several reads belonging to other Megavirales, an order of large viruses that infect eukaryotic hosts (Colson et al., 2013), were present in the DNA virome, belonging to the Mimiviridae family (n = 503 reads), the non‐classified Faustovirus (n = 982 reads) and Pandoravirus (n = 89 reads) (Figure 3b). Further verification of the taxonomic assignation of reads in the Poxviridae (n = 258 reads), Ascoviridae (n = 61 reads), Herpesviridae (n = 49 reads) and Papillomaviridae (n = 1 read) families (Figure 3b) by mapping against reference genomes showed that they corresponded to repeated patterns and thus could not be confidently attributed to these families.
The sequences of the RNA virome were related to Siphoviridae, Myoviridae and Podoviridae bacteriophages or Mimiviridae, Phycodnaviridae and Ascoviridae. These sequences probably reflected the DNA remaining in the RNA fraction during Trizol LS® extraction (Figure 3d). Similar to the DNA virome, the presence of Poxviridae was verified by mapping the reads against reference genomes, which did not result in good taxonomic assignation. Reads belonging to the Retroviridae and Anelloviridae families may reflect the presence of residual simian genomic material and blood, respectively. Only two reads of RNA viruses belonging to the Tymoviridae family were detected in the RNA virome. Tymoviridae are non‐enveloped plant‐infecting RNA viruses. The presence of such viruses may be linked with the diets of the primates, that is the plants, fruits or insects that they ingest (Figure 3d).
Pan‐generic PCR screening of zoonotic viruses
Several studies have shown that the NGS sequencing of viromes is a less sensitive technique than PCR (Cheval et al., 2011; Frey et al., 2014). Moreover, due to the limited depth of sequencing and the high abundance of bacteriophages detected by metagenomics, the presence of rare viral species, especially zoonotic viruses, may have been missed. Thus, we used pan‐generic PCR to screen the four bushmeat samples for the presence of most human viral zoonoses, including paramyxoviruses, coronaviruses, flaviviruses, phleboviruses, nairoviruses, alphaviruses, hantaviruses, orthobunyaviruses, arenaviruses, filoviruses, poxviruses and herpesviruses; however, none of the four specimens tested positive using these 12 pan‐generic PCRs.
The presence of inhibitors and the efficiencies of the nucleic acid extractions from the smoked tissue samples were controlled by performing PCR targeting 18S rRNA. The four samples tested positive (Figure S2), indicating the good efficiency of the amplification of DNA originating from smoked tissues.
Discussion
Quantifying the global wildlife trade is hard since it ranges from live to dead animals, from local barter to major international routes, and is almost always conducted illegally or through informal networks (Karesh et al., 2005). However, illegal bushmeat traffic may contribute to the emergence and spread of infectious diseases in humans and need to be addressed. The origin of AIDS epidemics (Gao et al., 1999), case reports of hepatitis E originating from wild boars (Vasickova et al., 2007; Meng et al., 2009), brucellosis (CDC, 2009), Ebola previous epidemics (Pourrut et al., 2005) and trichinellosis (Roy et al., 2003) are some examples of foodborne illnesses acquired after hunting of wild animals (Karesh et al., 2012). Indeed, some human foodborne pathologies may either be caused by the consumption or during the preparation (i.e. butchering, cutting or washing) of meat originating from infected animals. In France, important quantities of meat enter illegally each year but only few are seized by customs (Chaber et al., 2010) because these are out of the priorities of customs officers. Moreover, since 2014 Ebola epidemics and the potential risks of contamination for customs workers, the difficulties to sample bushmeat for research purposes before their immediate destruction have increased. In this context, it is important to obtain the necessary support from relevant authorities to seize and analyse wild game before their destruction. A transparent partnership between customs and health authorities will provide the best opportunity for improving the effectiveness of efforts to control the risk of international bushmeat trade.
In this study, we were able to obtain four non‐human primate samples that were first screened for the presence of VLPs using epifluorescence microscopy. Estimated viral loads were high, ranging from 104–107 VLPs/ml depending on the sample, but these may have been overestimated due to the use of a non‐specific fluorochrome which can also stain membrane‐derived vesicles or cell debris (Forterre et al., 2013). Observations confirmed the absence of bacterial contamination in the treated samples.
We thus decided to go further and sequence the purified viral fraction of one bushmeat sample. Viral metagenomic analyses performed on the simian game showed that a large majority of the sequences were related to bacteriophages belonging to the Siphoviridae, Myoviridae and Podoviridae families. Regardless of infectivity, the bacterial hosts of these viruses mainly belong to the Firmicutes and Proteobacteria phyla, which include environmental bacteria in addition to potential human pathogenic species, such as some of the Staphylococcus sp., Listeria sp. and Enterococcus sp. For example, Cronobacter spp. (formerly Enterobacter sakazakii) includes a group of Gram‐negative bacteria ubiquitously found in the environment. Cronobacter spp. are particularly resistant to osmotic stress and elevated temperatures, explaining their ability to survive in a wide variety of dried foods, such as infant formulas (Edelson‐Mammel et al., 2005; Estuningsih et al., 2006). They are emerging foodborne pathogens (Estuningsih et al., 2006; Healy et al., 2010) that may be responsible for life‐threatening infections in newborns, immunocompromised adults and the elderly (Bowen and Braden, 2006; Gosney et al., 2006; Mullane et al., 2007; Than and Tang, 2007). The presence of Cronobacter and other pathogenic bacteria, and the risk of human infection caused by bushmeat manipulation and consumption have to be addressed. The source of bacterial contamination of the bushmeat, due, for example, to non‐hygienically handlings (during hunting, carcass evisceration, smoking treatments or during unsafe transportation conditions), or due to the presence of several bacteria in the original tissue samples, is actually unknown. But whatever the origin of micro‐organisms, their presence results in the same potential risk of transmission to humans. A recent study by Chaber et al. reported the detection of viable aerobic bacteria above levels considered safe for human consumption, and unsafe levels of carcinogens in fish (Chaber and Cunningham, 2016). An exhaustive inventory of the bacterial communities present in bushmeat samples, for example, by high‐throughput sequencing of the 16S rRNA gene and by isolation, would help to clarify and identify the potential risks of introducing emerging bacterial pathogens by the illegal import of bushmeat.
Recent studies have demonstrated that metagenomic analyses are less sensitive than PCR‐based methods for detecting viruses present in low abundance. Indeed, the detection limit for NGS techniques is estimated to be 103 to 104 genomic copies (Cheval et al., 2011; Frey et al., 2014), while PCR‐based methods, especially those based on a nested format, can detect up to one genomic copy. Moreover, analysis of the rarefaction curve of bushmeat metagenomes revealed a plateau (Figure S3), indicating that full sequence diversity and characterization were not achieved. Thus, we screened for the presence of several zoonotic viruses by pan‐generic PCR. No human viral pathogens were detected using the 12 pan‐generic PCRs targeting the most common viral zoonoses, although most PCRs were in nested format and presented low limits of detection. In addition, the control PCR targeting the 18S rRNA gene demonstrated that the negative results were not due to the presence of inhibitors and/or degraded nucleic acids after smoking treatments. These findings could suggest that either the twelve targeted viral genera were not present in the studied populations at the time of sampling or the viral particles and their genomic content were degraded during the smoking and drying of the bushmeat, resulting in their increased sensitivity to nucleases used to purify viromes. In addition, the small sampling size could explain this negative result, especially if a low prevalence of the targeted viruses occurs.
To our knowledge, no outbreaks directly linked to the consumption of contaminated bushmeat (e.g. of primate, rodent or antelope origins) have been officially reported in France, but outbreaks may have been underestimated as the illegal origin of the products may prevent diseased consumers to declare consumption. Although smoking and thoroughly cooking the meat may reduce the risk of consumption, human contamination may still occur during handling infected meat that escaped sanitary controls. This risk is even higher for people manipulating and consuming fresh carcasses (Wolfe et al., 2005; Subramanian, 2012; Paige et al., 2014) which is of great concerns as international traffic of bushmeat also frequently involves fresh products. Large‐scale studies targeting the bacterial, viral and parasitic levels from wild‐caught game of different geographical and/or animal origins and, whenever possible, using large cohorts are urgently needed to identify risks for human health. These studies should be accompanied with extensive diffusion of information to exposed populations (regarding viral, bacterial and parasitic biohazard) about precautions that have to be taken while manipulating/consuming bushmeat. In addition, the effects of transformation treatments (such as smoking) on the external and internal contamination of meat have also to be addressed.
Supporting information
Figure S1. Bacterial genera infected by bacteriophages detected in the virome dataset.
Figure S2. Agarose gel electrophoresis of the 18S rRNA PCR performed on the four bushmeat samples.
Figure S3. Rarefaction curve of DNA and RNA metagenomes according to a similarity cut‐off of 75% for clustering, generated through metavir software (Roux et al., 2011).
Table S1. Primers used in this study.
Acknowledgements
The authors thank Dr. Sonia Kaeuffer from the DDSV (France) for taking care of the material, David Danède from DREAL Midi‐Pyrénées (France) for his help in obtaining permits of sampling protected animals seized at the international Toulouse Blagnac Airport and Pascal Weber for his help with confocal microscopy. This work was partially supported by ANR‐13‐JSV6‐0004.
References
- Aghokeng, A. F. , Ayouba A., Mpoudi‐Ngole E., Loul S., Liegeois F., Delaporte E., and Peeters M., 2010: Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross‐species transmissions. Infect. Genet. Evol. 10, 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachand, N. , Ravel A., Onanga R., Arsenault J., and Gonzalez J. P., 2012: Public health significance of zoonotic bacterial pathogens from bushmeat sold in urban markets of Gabon, Central Africa. J. Wildl. Dis. 48, 785–789. [DOI] [PubMed] [Google Scholar]
- Bair‐Brake, H. , Bell T., Higgins A., Bailey N., Duda M., Shapiro S., Eves H. E., Marano N., and Galland G., 2014: Is that a rodent in your luggage? A mixed method approach to describe bushmeat importation into the United States. Zoonoses Public Health 61, 97–104. [DOI] [PubMed] [Google Scholar]
- Bowen, A. B. , and Braden C. R., 2006: Invasive Enterobacter sakazakii disease in infants. Emerg. Infect. Dis. 12, 1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, M. D. , Peters C. J., and Nichol S. T., 1997: Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol. Phylogenet. Evol. 8, 301–316. [DOI] [PubMed] [Google Scholar]
- Brashares, J. S. , Golden C. D., Weinbaum K. Z., Barrett C. B., and Okello G. V., 2011: Economic and geographic drivers of wildlife consumption in rural Africa. Proc. Natl Acad. Sci. USA 108, 13931–13936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart, M. , and Rohwer F., 2005: Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39, 729–736. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) , 2009: Brucella suis infection associated with feral swine hunting ‐ three states, 2007‐2008. MMWR Morb. Mortal. Wkly Rep. 58, 618–621. [PubMed] [Google Scholar]
- Chaber, A. L. , and Cunningham A., 2016: Public health risks from illegally imported african bushmeat and smoked fish: public health risks from african bushmeat and smoked fish. EcoHealth, 13, 135–138. [DOI] [PubMed] [Google Scholar]
- Chaber, A. L. , Allebone‐Webb S., Lignereux Y., Cunningham A., and Rowcliffe J. M., 2010: The scale of illegal meat importation from Africa to Europe via Paris. Conserv. Lett. 3, 1–7. [Google Scholar]
- Cheval, J. , Sauvage V., Frageul L., Dacheux L., Guignon G., Dumey N., Pariente K., Rousseaux C., Dorange F., Berthet N., Brisse S., Moszer I., Bourhy H., Manuguerra J. C., Lecuit M., Burguiere A., Caro V., and Eloit M., 2011: Evaluation of high‐throughput sequencing for identifying known and unknown viruses in biological samples. J. Clin. Microbiol. 49, 3268–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, P. , de Lamballerie X., Yutin N., Asgari S., Bigot Y., Bideshi D. K., Cheng X. W., Federici B. A., Van Etten J. L., Koonin E. V., La Scola B., and Raoult D., 2013: “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 158, 2517–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S. J. , Fooks A. R., and van der Poel W. H., 2010: Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerg. Infect. Dis. 16, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson‐Mammel, S. G. , Porteous M. K., and Buchanan R. L., 2005: Survival of Enterobacter sakazakii in a dehydrated powdered infant formula. J. Food Prot. 68, 1900–1902. [DOI] [PubMed] [Google Scholar]
- Effiom, E. O. , Nuñez‐Iturri G., Smith H. G., Ottosson U., and Olsson O., 2013: Bushmeat hunting changes regeneration of African rainforests. Proc. Biol. Sci. 280, 20130246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estuningsih, S. , Kress C., Hassan A. A., Akineden O., Schneider E., and Usleber E., 2006: Enterobacteriaceae in dehydrated powdered infant formula manufactured in Indonesia and Malaysia. J. Food Prot. 69, 3013–3017. [DOI] [PubMed] [Google Scholar]
- Falk, H. , Dürr S., Hauser R., Wood K., Tenger B., Lörtscher M., and Schüpbach‐Regula G., 2013: Illegal import of bushmeat and other meat products into Switzerland on commercial passenger flights. Rev. Sci. Tech. 32, 727–739. [DOI] [PubMed] [Google Scholar]
- Forterre, P. , Soler N., Krupovic M., Marguet E., and Ackermann H. W., 2013: Fake virus particles generated by fluorescence microscopy. Trends Microbiol. 21, 1–5. [DOI] [PubMed] [Google Scholar]
- French customs annual report , Available at http://www.douane.gouv.fr/Portals/0/fichiers/datadouane/publication-douane/bilans-resultats/resultats-2013.pdf(accessed September 29, 2015)
- Frey, K. G. , Herrera‐Galeano J. E., Redden C. L., Luu T. V., Servetas S. L., Mateczun A. J., Mokashi V. P., and Bishop‐Lilly K. A., 2014: Comparison of three next‐generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood. BMC Genom. doi: 10.1186/1471-2164-15-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froussard, P. , 1992: A random‐PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 20, 2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F. , Bailes E., Robertson D. L., Chen Y., Rodenburg C. M., Michael S. F., Cummins L. B., Arthur L. O., Peeters M., Shaw G. M., Sharp P. M., and Hahn B. H., 1999: Origin of HIV‐1 in the chimpanzee Pan troglodytes troglodytes . Nature 397, 436–441. [DOI] [PubMed] [Google Scholar]
- Gosney, M. A. , Martin M. V., Wright A. E., and Gallagher M., 2006: Enterobacter sakazakii in the mouths of stroke patients and its association with aspiration pneumonia. Eur. J. Intern. Med. 17, 185–188. [DOI] [PubMed] [Google Scholar]
- Healy, B. , Cooney S., O'Brien S., Iversen C., Whyte P., Nally J., Callanan J. J., and Fanning S., 2010: Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog. Dis. 7, 339–350. [DOI] [PubMed] [Google Scholar]
- Kamins, A. O. , Rowcliffe J. M., Ntiamoa‐Baidu Y., Cunningham A. A., Wood J. L., and Restif O., 2014: Characteristics and risk perceptions of Ghanaians potentially exposed to bat‐borne zoonoses through bushmeat. EcoHealth 12, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karesh, W. B. , Cook R. A., Bennett E. L., and Newcomb J., 2005: Wildlife trade and global disease emergence. Emerg. Infect. Dis. 11, 1000–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karesh, W. B. , Loh E., and Machalaba C., 2012: Food safety: a view from the wild side In “Improving Food Safety Through A One Health Approach: Workshop Summary”, National Academies Press, US. [PubMed] [Google Scholar]
- Klempa, B. , Fichet‐Calvet E., Lecompte E., Auste B., Aniskin V., Meisel H., Denys C., Koivogui L., ter Meulen J., and Krüger D. H., 2006: Hantavirus in African wood mouse, Guinea. Emerg. Infect. Dis. 12, 838–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, A. J. , and Lanciotti R. S., 2009: Consensus amplification and novel multiplex sequencing method for S segment species identification of 47 viruses of the Orthobunyavirus, Phlebovirus, and Nairovirus genera of the family Bunyaviridae . J. Clin. Microbiol. 47, 2398–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. J. , Lindsay D. S., and Sriranganathan N., 2009: Wild boars as sources for infectious diseases in livestock and humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2697–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moureau, G. , Temmam S., Gonzalez J. P., Charrel R. N., Grard G., and de Lamballerie X., 2007: A real‐time RT‐PCR method for the universal detection and identification of flaviviruses. Vector Borne Zoonotic Dis. 7, 467–477. [DOI] [PubMed] [Google Scholar]
- Mullane, N. R. , Iversen C., Healy B., Walsh C., Whyte P., Wall P. G., Quinn T., and Fanning S., 2007: Enterobacter sakazakii, an emerging bacterial pathogen with implications for infant health. Minerva Pediatr. 59, 137–148. [PubMed] [Google Scholar]
- One Health Initiative . Available at: http://www.onehealthinitiative.com/ (accessed February 5, 2016).
- Paige, S. B. , Frost S. D., Gibson M. A., Jones J. H., Shankar A., Switzer W. M., Ting N., and Goldberg T. L., 2014: Beyond bushmeat: animal contact, injury, and zoonotic disease risk in western Uganda. EcoHealth 11, 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet, O. , Schneider B. S., Beaty S. M., LeBreton M., Yun T. E., Park A., Zachariah T. T., Bowden T. A., Hitchens P., Ramirez C. M., Daszak P., Mazet J., Freiberg A. N., Wolfe N. D., and Lee B., 2014: Evidence for henipavirus spillover into human populations in Africa. Nat. Commun. 5, 5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourrut, X. , Kumulungui B., Wittmann T., Moussavou G., Delicat A., Yaba P., Nkoghe D., Gonzalez J. P., and Leroy E. M., 2005: The natural history of Ebola virus in Africa. Microbes Infect. 7, 1005–1014. [DOI] [PubMed] [Google Scholar]
- Pourrut, X. , Diffo J. L., Somo R. M., Bilong Bilong C. F., Delaporte E., LeBreton M., and Gonzalez J. P., 2011: Prevalence of gastrointestinal parasites in primate bushmeat and pets in Cameroon. Vet. Parasitol. 175, 187–191. [DOI] [PubMed] [Google Scholar]
- Rodriguez, L. L. , Maupin G. O., Ksiazek T. G., Rollin P. E., Khan A. S., Schwarz T. F., Lofts R. S., Smith J. F., Noor A. M., Peters C. J., and Nichol S. T., 1997: Molecular investigation of a multisource outbreak of Crimean‐Congo hemorrhagic fever in the United Arab Emirates. Am. J. Trop. Med. Hyg. 57, 512–518. [DOI] [PubMed] [Google Scholar]
- Roux, S. , Faubladier M., Mahul A., Paulhe N., Bernard A., Debroas D., and Emault F., 2011: Metavir: a web server dedicated to virome analysis. Bioinformatics 27, 3074–3075. [DOI] [PubMed] [Google Scholar]
- Roy, S. L. , Lopez A. S., and Schantz P. M., 2003: Trichinellosis surveillance–United States, 1997‐2001. MMWR Surveill. Summ. 52, 1–8. [PubMed] [Google Scholar]
- Sánchez‐Seco, M. P. , Rosario D., Quiroz E., Guzmán G., and Tenorio A., 2001: A generic nested‐RT‐PCR followed by sequencing for detection and identification of members of the Alphavirus genus. J. Virol. Methods 95, 153–161. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Seco, M. P. , Echevarría J. M., Hernández L., Estévez D., Navarro‐Marí J. M., and Tenorio A., 2003: Detection and identification of Toscana and other phleboviruses by RT‐nested‐PCR assays with degenerated primers. J. Med. Virol. 71, 140–149. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Seco, M. P. , Hernández L., Eiros J. M., Negredo A., Fedele G., and Tenorio A., 2006: Detection and identification of orthopoxviruses using a generic nested PCR followed by sequencing. Br. J. Biomed. Sci. 63, 79–85. [DOI] [PubMed] [Google Scholar]
- Smith, K. M. , Anthony S. J., Switzer W. M., Epstein J. H., Seimon T., Jia H., Sanchez M. D., Huynh T. T., Galland G. G., Shapiro S. E., Sleeman J. M., McAloose D., Stuchin M., Amato G., Kolokotronis S. O., Lipkin W. I., Karesh W. B., Daszak P., and Marano N., 2012: Zoonotic viruses associated with illegally imported wildlife products. PLoS ONE. 7, e29505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Luna, L. K. , Heiser V., Regamey N., Panning M., Drexler J. F., Mulangu S., Poon L., Baumgarte S., Haijema B. J., Kaiser L., and Drosten C., 2007: Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription‐PCR and nonfluorescent low‐density microarray. J. Clin. Microbiol. 45, 1049–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, M. , 2012: Zoonotic disease risk and the bushmeat trade: assessing awareness among hunters and traders in Sierra Leone. EcoHealth 9, 471–482. [DOI] [PubMed] [Google Scholar]
- Temmam, S. , Davoust B., Berenger J. M., Raoult D., and Desnues C., 2014: Viral metagenomics on animals as a tool for the detection of zoonoses prior to human infection? Int. J. Mol. Sci. 15, 10377–10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmam, S. , Monteil‐Bouchard S., Robert C., Pascalis H., Michelle C., Jardot P., Charrel R., Raoult D., and Desnues C., 2015: Host‐associated metagenomics: a guide to generating infectious RNA viromes. PLoS ONE doi: 10.1371/journal.pone.0139810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than, H. A. , and Tang T., 2007: Enterobacter sakazakii bacteraemia with multiple splenic abscesses in a 75‐year‐old woman: a case report. Age Ageing 36, 595–596. [DOI] [PubMed] [Google Scholar]
- Thurber, R. V. , Haynes M., Breitbart M., Wegley L., and Rohwer F., 2009: Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4, 470–483. [DOI] [PubMed] [Google Scholar]
- Tong, S. , Chern S. W., Li Y., Pallansch M. A., and Anderson L. J., 2008: Sensitive and broadly reactive reverse transcription‐PCR assays to detect novel paramyxoviruses. J. Clin. Microbiol. 46, 2652–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDevanter, D. R. , Warrener P., Bennett L., Schultz E. R., Coulter S., Garber R. L., and Rose T. M., 1996: Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34, 1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasickova, P. , Psikal I., Chalupa P., Holub M., Svoboda R., and Pavlik I., 2007: Hepatitis E virus: A review. Vet. Med. 52, 365–384. [Google Scholar]
- Wolfe, N. D. , Prosser T. A., Carr J. K., Tamoufe U., Mpoudi‐Ngole E., Torimiro J. N., LeBreton M., McCutchan F. E., Birx D. L., and Burke D. S., 2004: Exposure to nonhuman primates in rural Cameroon. Emerg. Infect. Dis. 10, 2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, N. D. , Daszak P., Kilpatrick A. M., and Burke D. S., 2005: Bushmeat hunting, deforestation, and prediction of zoonotic disease emergence. Emerg. Infect. Dis. 11, 1822–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J. , Palacios G., Towner J. S., Jabado O., Kapoor V., Venter M., Grolla A., Briese T., Paweska J., Swanepoel R., Feldmann H., Nichol S. T., and Lipkin W. I., 2007: Rapid molecular strategy for filovirus detection and characterization. J. Clin. Microbiol. 45, 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Bacterial genera infected by bacteriophages detected in the virome dataset.
Figure S2. Agarose gel electrophoresis of the 18S rRNA PCR performed on the four bushmeat samples.
Figure S3. Rarefaction curve of DNA and RNA metagenomes according to a similarity cut‐off of 75% for clustering, generated through metavir software (Roux et al., 2011).
Table S1. Primers used in this study.


