Abstract
Respiratory viruses are a constant concern for all demographics. Examples include established viruses such as respiratory syncytial virus (RSV), the leading cause of respiratory infection in infants and young children, and emerging viruses such as severe acute respiratory syndrome (SARS), which reached near pandemic levels in 2003, or H1N1 (swine) influenza. Despite this prevalence, traditional methods of virus detection are typically labor intensive and require several days to successfully confirm infection. Recently, however, nanoparticle‐based detection strategies have been employed in an effort to develop detection assays that are both sensitive and expedient. Each of these platforms capitalizes on the unique properties of nanoparticles for the detection of respiratory viruses. In this article, several nanoparticle‐based scaffolds are discussed.Gold nanoparticles (AuNPs) have been functionalized with virus specific antibodies or oligonucleotides. In each of these constructs, AuNPs act as both an easily conjugated scaffolding system for biological molecules and a powerful fluorescence quencher. AuNPs have also been immobilized and used as electrochemical transducers. They efficiently serve as a conducting interface of electrocatalyic activity making them a powerful tool in this application. Quantum dots (QDs) posses unique fluorescence properties that have also been explored for their application to virus detection when combined with direct antibody conjugation or streptavidin‐biotin binding systems. QDs have an advantage over many traditional fluorophores because their fluorescence properties can be finely tuned and they are resistant to photobleaching. The development of these nanoparticle‐based detection strategies holds the potential to be a powerful method to quickly and easily confirm respiratory virus infection. WIREs Nanomed Nanobiotechnol 2010 2 277–290
This article is categorized under:
-
1
Diagnostic Tools > In Vitro Nanoparticle-Based Sensing
-
2
Therapeutic Approaches and Drug Discovery > Nanomedicine for Respiratory Disease
Respiratory infections represent a significant threat to a broad range of both human and animal populations. Each year, incidents of human influenza are estimated to cost the United States over 10 billion dollars and result in more than 200,000 hospitalizations and 36,000 deaths.1 Pandemic influenza often results in the deaths of tens of millions of people worldwide. These pandemics are estimated to have direct and indirect costs over 100 billion dollars.1 Influenza, however, is not the only virus to pose a serious threat. Additionally, Respiratory Syncytial Virus (RSV) is the number one cause of serious lower respiratory tract infections in infants and young children worldwide. Infection rates have been estimated as high as 50% within the first year of life with 90,000 annual US hospitalizations in children under 5 years. It has also become an increasing threat to elderly and immunocomprimised populations.2 In 2003, Severe Acute Respiratory Syndrome (SARS) emerged in a global outbreak that resulted in 8000 confirmed cases with a fatality rate of 11%.3 Respiratory infection is not unique to humans. For example, Porcine Reproductive and Respiratory Syndrome (PRRS), caused by the Porcine Reproductive and Respiratory Syndrome Virus (PRRSV), is one of the most economically devastating diseases affecting the swine industry worldwide. In the United States alone, the estimated cost to pork producers is around 600 million dollars annually.4 New strains of respiratory viruses continue to emerge, such as the 2009 H1N1 pandemic, posing new and continual threats.
Efforts to control respiratory infection have focused on prevention through vaccine development.1 However, in the case of some viruses, such as RSV, no vaccine is presently available making detection and treatment of critical importance. Treatment of respiratory viruses can include the administration of one or more antiviral compounds. During the 2003 SARS outbreak, the common antiviral ribavirin was widely distributed and in some cases used in conjunction with the protease inhibitor Kaletra. Although preliminary results suggested that the addition of Kaletra to the traditional drug treatments reduced intubation and mortality rates, it was most successful when administered early. The treatment of RSV is another example where the efficacy of the antiviral medicines available is directly related to the early detection of an infection. Because administration of medication in the early stages of infection is intimately related to the success of treatment for many respiratory viruses, the development of rapid detection methods could prove indispensible in limiting the spread and morbidity of these infectious organisms.
Classical methods for the detection of respiratory viruses are typically costly, labor intensive, and require specialized equipment and expertise. An ideal detection method is sensitive, specific, rapid, and requires the direct detection of virus, viral antigens, or viral RNA. Most of the classical methods for detection contain only one or two of these characteristics. RSV isolation in cell culture has been considered the gold standard for detection in patients. This method is labor intensive and slow, often requiring several days before a positive result can be determined.5, 6, 7 Immunofluorescence and Enzyme‐Linked Immunosorbent Assays (ELISA) are performed more rapidly, but lack the sensitivity required for early detection.5, 6, 7 Serological studies and Polymerase Chain Reaction (PCR) are also commonly utilized methods. Serological studies require seroconversion and consequently cannot be applied to acute infection. PCR, on the other hand, is extremely sensitive, but also sensitive to template contamination and often yield false positive results.6 Although these methods have been used historically to identify respiratory infection, their limitations emphasize the important need for a diagnostic. For this, researchers have turned to new tools from the developing field of nanotechnology, particularly nanoparticles, to devise unique platforms for the detection of respiratory infection.
The field of nanotechnology capitalizes on the unique properties of nanoscale materials, ranging in size from 1 to 100 nm.8 The underlying basis for such nanoscale effects is that every property of a material has a characteristic and critical length associated with it. The fundamental physics and chemistry of a material will change when the dimensions of a solid become comparable to one or more of these characteristic lengths, many of which exist at the nanometer length scale. At this scale, objects take on properties and functions that differ from those seen on the bulk scale.8 Further, nanoscale materials, like nanoparticles, can tune their chemical and spectral properties by changing their size and/or shape. This ability to tune the properties of nanoscale material has been ultilized in electronic and chemical applications such as electrocatalysis, electrochemical sensor devices, and redox recognition.8,9 Recently, the focus has expanded to encompass a new range of biological applications. Nanoparticles, of various compositions, can be effective vehicles for drug delivery. Nanoparticles have also been used to probe the internal cellular structure and dynamics of various biological systems including the vesicular secretion of neurotransmitters and distinguishing healthy colon cells from adenocarcinoma cells.10 The use of magnetic nanoparticles as Magnetic Resonance Imaging (MRI) contrast agents have also led to their use in identifying atherosclerotic lesions in cardiovascular tissue.11 The development of nanoparticles to study these and other biological systems has demonstrated their versatility and functionality. In this article, we review several recent efforts aimed at the detection of respiratory infections. Each of the reviewed articles employs the unique properties of nanoparticles to develop novel methods or variations of existing techniques to enable them to specifically and sensitively detect respiratory viruses.
METAL NANOPARTICLES
Metal nanoparticles have become a critical component in the development of nanotechnology‐based detection for respiratory viruses. Gold Nanoparticles (AuNPs) have garnered much of the attention in this field because of their unique physical and chemical properties. The surface of AuNPs acts as a soft metal ion that easily binds to soft ligands such as thiols. This reactivity provides an efficient strategy to readily functionalize particles with relevant biological molecules.12 Another property of AuNPs is the deep‐red color seen in solutions. This is the result of the Surface Plasmon Band (SPB), a broad absorption band in the visible region between 510 and 580 nm.13 Compared to the bulk gold counterpart, the SPB of AuNPs can be finely tuned because it experiences wavelength shifts based on AuNP size, shape, temperature, and the nature of functionalized ligands. This tunable property has made it the subject in extensive studies of the optical spectroscopic properties of AuNPs. The ligand shell of AuNPs alters the refractive index causing either a red or blue shift, allowing SPB shifts to be used to assess the extent of ligand functionalization or target binding.13 AuNPs also possess the inherent ability to act as a fluorescence quencher because of nonradiative energy transfer from the fluorescence dye to the metal surface.14,15 In the case of chromophore–AuNP composites, the quenching efficiency of AuNPs outperforms that of most organic acceptors. The dominating design factor affecting the efficiency of quenching by the AuNP is the distance between surface of the AuNP and the chromophore.14 In cases where the chromophore is separated from the AuNP by a bulky spacer, such as antibiodies or carbon‐based linker molecules, there is a decrease in quenching that corresponds to the increasing distance between the AuNP surface and the chromophore.14 Each of these properties makes AuNPs interesting platforms for the development of respiratory infection diagnostics.
Antibody Functionalized AuNPs for the FRET‐Based Detection of PRRSV
The ability of the surface of AuNPs to be functionalized with biological molecules for molecular recognition has lead to the development of new tools for the detection of respiratory infection. This has been especially true for systems which allow the AuNP to act as an acceptor molecule in Förster Resonance Energy Transfer (FRET) pairs. FRET pairs have been used to study a variety of biological systems including protein–protein associations, DNA hybridization, and protein conformational change.16,17 However, when paired with traditional organic fluorophores, the incorporation of nanophotonic molecules such as AuNPs yields advantages over traditional organic acceptor molecules. The AuNP is a highly effective fluorescence quencher molecule as the result of a high Stern–Volmer quenching coefficient, leading to subpicomolar analyte detection.18 The AuNP can also dramatically reduce the fluorescence lifetime of fluorescence dyes. Previous research has shown that AuNPs with diameters of 1, 15, or 30 nm can decrease the fluorescence lifetime of a fluorophore by up to 90%. Thus, a FRET immunosensor has been developed by Grant et al. to detect the presence of PRRSV.18,19 In this sensor, the donor fluorophore, Alexa Fluor 546 (AF546), is conjugated to a PRRSV antibody (SDOW17) and protein A is conjugated to 20 nm AuNPs. The two constructs are then incubated to form the intact fluorescence bioprobe. In the presence of the PRRSV, the antibody binds the virus and undergoes a conformational change, resulting in a decreased distance between the fluorophore and the AuNP. The fluorophore is then in close enough proximity to the surface of the AuNP that it is quenched (Figure 1a). In the presence of PRRSV, the AuNP conjugated FRET pairs were substantially more sensitive than similar constructs formed using traditional organic acceptor molecules. Concentration–response curves, obtained by plotting the average biosensor response (fluorescence intensity) against PRRSV concentrations ranging from 0 to 60 particles/µl, revealed a limit of detection of 1741 particles/ml. The specificity of this FRET immunosensor was also explored by exposing the AuNP construct to varying concentrations of both PRRSV and Bovine Serum Albumin (BSA). In the presence of PRRSV, the AuNP sensor had a notable response that was not seen in the presence of BSA (Figure 1b). These studies highlight the sensor's robustness against commonly encountered contaminants.18,19 By utilizing the advantages offered by AuNP, a rapid, specific, and sensitive FRET platform was developed for the detection of PRRSV.18,19
Figure 1.
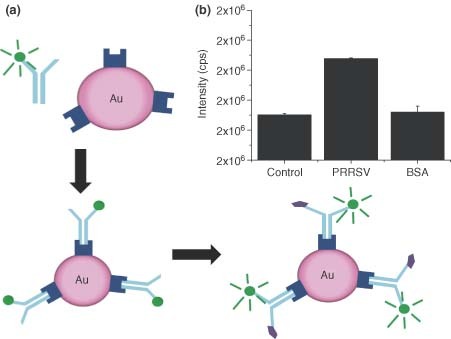
(a) Schematic depicting gold nanoparticles (AuNP)‐based biosensor. Fluorescence labeling is followed by assembly of the biosensor complex via binding of protein A to capture antibody. Antigen binding causes conformational change in the antibody, leading to a detectable Förster resonance energy transfer response. (b) AuNP biosensor specificity is demonstrated by comparing response of porcine reproductive and respiratory syndrome virus sample to a control well and the nonspecific antigen bovine serum albumin (BSA). Negligible response to the BSA by AuNP biosensor indicates minimal nonspecific binding.18.
Molecular Beacon Functionalized Au and Ag Nanowires for the Detection of SARS
A number of oligonucleotide platforms have been explored for virus detection. Molecular Beacons (MBs) are looped strands of DNA which form a hairpin structure with a fluorophore located on one terminus and a fluorescence quencher at the other.20 This platform is similar to the fluorophore/quencher scheme seen in FRET assays. The MB has two major regions, the stem region which holds the MB in its signature hairpin structure, and the loop region which is complement to a specific target strand of DNA or RNA. When in the closed ‘dark’ state, the MB's hairpin structure holds the two termini in close proximity inducing quenching of the fluorophore. However, in the presence of the target strand, the hairpin is opened to the extended conformation sufficiently separating the two termini to overcome quenching. By inducing this open ‘bright’ state, an increase in fluorescence corresponds to the presence of the target sequence (Figure 2a).21 This construct has been employed successfully in the real‐time recognition of DNA in solution, including the detection of pathogens and single nucleotide polymorphisms.21,22 The application of MB to multiplexed detection has been limited by the number of spectrally distinct fluorophores available. Recent attempts to overcome this limitation have focused on using encoded nanoscale substrates to immobilize MBs. The efforts of Sha et al. employ a new approach for the multiplex of MBs based on the use of striped, encoded gold and silver nanowires as quenchers.23 By attaching the MB hairpin to the metal surface with one terminus, this construct capitalizes on the previously discussed quenching properties of the gold and silver surfaces. The hairpin structure forces the fluorophore near the metal surface, subjecting it to metal‐induced quenching and alleviating the need for an organic quencher dye (Figure 2a). To achieve multiplexing capabilities, these functionalized gold and silver nanowires are combined into a nanostructure resembling a barcode.23
Figure 2.
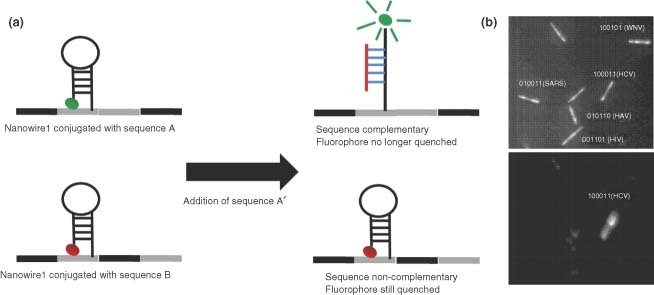
(a) Schematic of Au/Ag nanowire detection system. (b) Representative results from a 5‐plexed virus detection experiment. Each specific nanowire can be identified based on its reflectivity under illumination with 400 nm light (top). The fluorescence of a wire whose complement antigen is present will be high compared to the other wires (bottom).23.
Nanobarcodes are submicron sized metallic nanowires that contain regions of silver and gold fabricated by electrolplating of the inert metals sequentially into alumina templates. When the template is dissolved, striped nanowires are left behind. It is possible to distinguish the gold and silver sections of the nanowires by evaluating the reflectivity of the regions when illuminated with blue light (400 nm).23 When viewed with a microscope, this gives the nanowires a striped appearance. By changing the order of the stripes on nanowires, a library of encoded wires is generated to which biomolecules, including nucleic acids and proteins, can be conjugated. Multiplexed fluorescence‐based assays can be performed by using a fluorescence microscope to simultaneously monitor the reflectivity measurements at 400 nm (to identify the library member) and the optimum fluorescence emissions wavelength (to define the quantitative assay result).23 By applying a binary system to the idenfication of the nanowires where a silver section is defined as a 1 and a gold section as a 0, it was possible to label specific nanowires as indicators for each virus. For example, the nanowire with the label 101010, indicating alternating silver and gold sections, was functionalized with MBs that contained a DNA loop region complement to an SARS genome sequence for a nonstructural polyprotein region. A positive fluorescence signal of this particular nanowire would indicate the presence of the SARS virus. Consequently, several different nanowires can be functionalized for different viruses for the simultaneous detection of multiple analytes such as SARS, Hepatitis a Virus (HAV), Hepatitis C Virus (HCV), West Nile Virus (WNV), and Human Immune Deficiency Virus (HIV). In such a case, oligonucleotides were designed that were complement to the target sequence and contain a self‐complementary stem of 10 bases. These probes were also functionalized with a universal fluorescence TAMRA dye at the 3′ end and a thiol at the 5′ end. The oligonucleotides were conjugated to the metal nanowire through the 5′ thiol, with each nanowire library member being functionalized by an oligonucleotide sequence targeting a specific virus. In the presence of one or more analytes, the complement MB will open giving off a fluorescence signal. In this manner, one nanowire will fire for each analyte present. Decoding of the nanoware striping pattern then indicates which DNA sequence(s) is present (Figure 2b).23
The detection of individual analytes by this 5‐plexed system was demonstrated using synthetic target strands for each virus in several formats.23 Experiments were performed in which each target was added individually and detected, or all five targets were pooled and added simultaneously. The results were controlled with experiments examining noncomplementary target and ‘no target’ samples. In all scenarios, the correct target was easily detected in the five nanowire experiment regardless of the order in which the target was added. By contrast, the noncomplement target and no target control samples yielded negligible signal (Figure 3). These experiments successfully demonstrated a proof of concept for a novel, multiplexed, pathogen detection assay by putting a new twist on the traditional MB construct by combining it with the use of metal‐encoded nanoparticles.23
Figure 3.
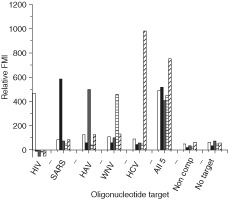
A 5‐plex assay using oligonucleotide targets. The experiment was performed in triplicate and the signals from the noncomplementary target negative control was used to perform a background subtraction. FMI refers to fluorescence mean intensity (arbitrary units). HIV, human immunodeficiency virus (white bars); SARS, severe acute respiratory syndrome (black bars); HAV, hepatitis A virus (gray bars); WNV, West Nile virus (horizontal lines); HCV, hepatitis C virus (diagonal lines).23.
Immobilized AuNP Genosensor for the Detection of the SARS Virus
Although the functionalization properties of AuNPs offer much utility, they possess other properties which have been utilized for the development of sensors for respiratory infections. The metallic nature of AuNPs makes them ideal for electrochemical detection applications. AuNPs have efficient conducting interfaces with electrocatalytic ability and high surface‐to‐volume ratios that make them useful electrode materials.24 Costa‐Garcia et al. combined AuNP‐based amplification scheme with the electrochemical detection of DNA using enzymatic labels for the detection specific to the sequence of DNA from the SARS virus (Figure 4a).25 AuNPs, immobilized on a screen‐printed carbon electrode, served as electrochemical transducers. A probe sequence, complement to the target SARS sequence, is directly conjugated to the AuNPs through a thiol linkage. The target SARS sequence, which will bind to the probe sequence, is biotinylated. This allows the enzymatic label, alkaline phosphatase, to be conjugated to the target sequence through streptavidin–biotin affinity interactions. After excess antigen and streptavidin–alkaline phosphatase are removed by washing, the alkaline phosphatase substrate is added. Alkaline phosphatase catalyzes the dephosphorylation of 3‐indoxyl phosphate to an indoxyl intermediate, an electrochemically active analyte.26 After dephosphorylation by alkaline phosphatase, the indoxyl intermediate is able to reduce silver ions in solution resulting in indigo blue and metallic silver. The deposition of metallic silver is measured by anodic stripping voltammetry.25
Figure 4.
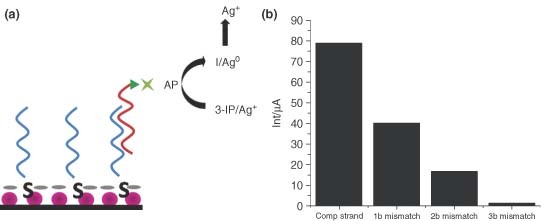
(a) Schematic representation of the genosensor device. (b) Analytical signals obtained for 50 pM of biotinylated target with 25% formamide (to apply stringency conditions) using the genosensor to discriminate between target DNA, 1‐base mismatch, 2‐base mismatch, and 3‐base mismatch. The decrease in signal as a function of degree of mismatch indicates high specificity for the genosensor device.25.
A 30‐mer oligonucleotide sequence was designed for the detection of the SARS virus which corresponds to the gene for a short lysine‐rich region of the nucleocapsid protein that appears to be unique to SARS.25 The detection limit for the SARS target strand was 4.6 pM at a 3σ threshold of the standard deviation of the intercept. The advantages of the AuNP surface were evident when compared to a similar platform prepared with a bulk gold surface. The bulk gold electrode, constructed of gold paste cured at low temperatures, had a limit of detection of 60 pM.25 The 10‐fold increased sensitivity of the nanoparticle electrode over the bulk gold surface was the result of the high surface‐to‐volume ratio found with the nanostructured surface of the particles. The selectivity of this device was also explored by comparing the signal in the presence of the target sequence with signal in the presence of 1‐, 2‐, and 3‐base mismatch sequences.25 Under high stringency conditions, each of the four different strands showed distinct signals that correlated to the degree of mismatch in the target strand (Figure 4b). The unique AuNP‐based design yielded an instrument for the detection of an SARS virus specific DNA sequence that was 10 times more sensitive than non‐nanostructured corollaries and was selective enough to distinguish the target sequences from DNA strands with a single mismatched base.25
QUANTUM DOTS
Quantum Dots (QDs) are nanometer sized crystalline clusters (1–10 nm) made from a variety of semiconductor materials. QDs are characterized by large absorption spectra, but narrow and very symmetric emission bands with full widths at half maximum of 25–35 nm.27 The emission bands can span the spectrum from the ultraviolet to the infrared (365–1350 nm) depending upon the size of the dot. These materials typically have a large absorption cross section and long fluorescence lifetimes (>10 ns).28 Furthermore, they possess a photostability orders of magnitude greater than conventional organic fluorophores. Given their advantages relative to organic fluorophores, QDs have emerged as a new class of fluorescence probes for biological applications.27 The broadband adsorption and narrow, symmetrical, and size‐tunable emission bands of QDs facilitate their use for multiplexed detection of signals. The important consequence of this broad absorption is that one wavelength of light can excite multiple QDs, each with different emission maxima. Highly robust commercial preparations of CdSe/ZnS QDs, emitting in the visible spectrum, match the detection range of many typical imaging devices.29 Further, with the elimination of many chromatic aberration and alignment issues encountered with standard fluorescence microscopy, colocalization studies are possible. Consequently, QDs have been used in a number of biological applications including studies of protein trafficking, DNA detection, and dynamic studies of cell motility. Only recently, however, QDs have been used for the detection or imaging of respiratory viruses.
Streptavidin and Antibody Functionalized QDs for the Detection of RSV
Improved water solubility of QDs has lead to the development of bioconjugates that allow for specific targeting and/or signal enhancement in biological applications.30 Two of the most promising of these bioconjugates have been used for the detection of RSV, streptavidin conjugation, and antibody conjugation.31 Streptavidin–biotin affinity interactions are commonly used in the labeling of a specific biological target with a detectable marker. For the detection of RSV, this interaction was used to label the F and G viral proteins with QDs. The fusion (F) and attachment (G) proteins of RSV are ideal antigenic markers as they are incorporated into the surface of the infected host cell, and the magnitude of these markers is amplified as a function of time because of viral replication.31 By monitoring the appearance and amplification of these markers, QDs were used to detect and monitor RSV infection. Bentzen et al. labeled infected HEp‐2 cells with antibodies specific to the F and G proteins. For the specific identification of the F protein, an antibody mixture of two purified F protein specific mouse monoclonal antibodies was used to label the F protein. This primary antibody was then labeled by a biotinylated secondary antibody (polyclonal goat anti‐mouse). This primary/secondary antibody combination leaves an available biotin molecule. Incubation with streptavidin functionalized QDs resulted in specific labeling of the target (Figure 5).31 A similar protocol was used to label the G protein. With the use of two different colored QDs, it was possible to label both the F and G proteins simultaneously (Figure 6a).31 Both proteins were found to be predominantly located on the surface of the infected cells with significant colocalization of both proteins. The use of QDs allowed the detection of F protein as early as 1 h after infection and uniformly evident 24 h after infection (Figure 6b). At 24 h, it was determined that the limit of detection was 35–50 pfu. These results were compared to antibodies conjugated to Fluorescein Isothiocyanate (FITC), an organic fluorophore.31 The FITC conjugated antibodies could not detect F protein at time points earlier than 24 h and while it could detect the protein at later time points, it required exposures 46 times that of the QD labeled cells for clear imaging and were almost completely photodegraded within 15 min. This clearly demonstrated the improved sensitivity and durability of the use of QDs in this assay as compared to conventional organic dyes.31
Figure 5.
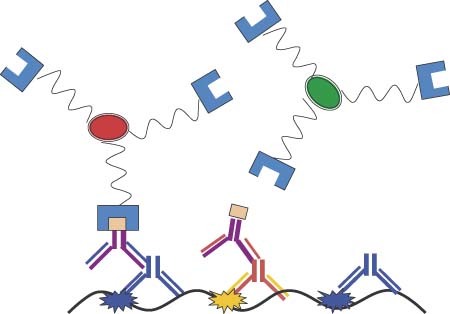
Streptavidin‐based antibody labeling scheme for respiratory syncytial virus F protein and G protein.31.
Figure 6.
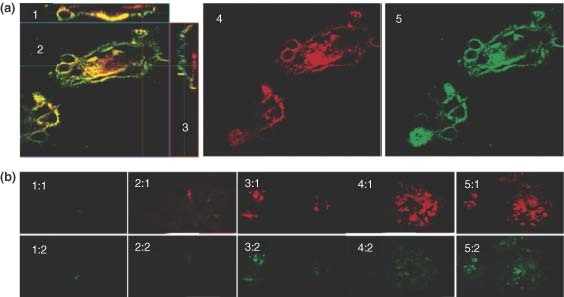
(a) Confocal images showing the sucessful labeling of the F protein32 and G protein18 in respiratory syncytial virus (RSV)‐infected Hep‐2 cells using quantum dots (QDs). The dual labeled sample showed areas of colocalization33 between the two proteins, and analysis of the orthogonal slices XZ2 and YZ23 suggest the proteins are predominately located at the surface of the cells. (b) Fluorescence images of the F protein labeled with QDs at 1 h (1:1), 24 h (2:1), 48 h (3:1), 72 h (4:1), and 96 h (5:1) after infection. The G protein fluorescence can also be seen after subsequent labeling of the same infected monolayer at each time point (1:2–5:2).31.
Although the use of the biotin–streptavidin affinity interactions has proven useful because of the commercial availability of streptavidin functionalized QDs, efforts have been made to remove the secondary labeling step by conjugating target specific antibodies directly to QDs.34 By reducing the labeling process to a single‐step format, this new method has the potential to improve virus detection time, save costs, and reduce background staining that is associated with a multistep process. Tripp et al. have applied this single‐step method to the detection of RSV through the conjugation of an F protein specific monoclonal antibody to a Cadmium Telluride (CdTe) QD.34 To facilitate the conjugation of the antibody to the CdTe QD, the QDs were synthesized with a thioglycolate coating to allow direct conjugation via an EDC/Sulfo‐NHS conjugation reaction. The anti‐F conjugated QDs were then used to explore their specificity and sensitivity both in vitro and in vivo using Vero cells and female BALB mice, respectively. In vitro, there was no significant background signal seen in uninfected control samples.34 By contrast, the RSV‐infected cells showed distinct fluorescence signal because of F protein labeling. This single‐step process was analyzed as potentially reducing the time necessary to quantitate virus titers by plaque assays. It was found that the anti‐F conjugated QDs could detect RSV plaques several days earlier than traditional immunostaining methods (Figure 7).34 To evaluate the efficacy of the anti‐F conjugated QDs to detect RSV infection in vivo, BALB/c mice were infected with 106 pfu RSV and at 4 days postinfection, intravenously administered the anti‐F QDs. Five days postinfection, the lungs of the mice were removed, frozen and imaged by fluorescence microscopy. Uninfected control mice were treated with anti‐F QDs and similarly harvested and imaged. The lungs of both RSV‐infected and uninfected mice which had not been treated with the anti‐F QDs were removed and labeled by traditional Immunohistochemistry (IHC) methods for comparison. The mice which had been treated with anti‐F QDs provided clear and rapid detection of the RSV‐infected lung tissue with very limited background staining when compared to the uninfected controls (Figure 8). Importantly, the staining with the anti‐F QDs was similar in magnitude and location along the epithelial cells of the alveoli to traditional IHC methods (Figure 8).34 This preliminary data demonstrate the potential for antibody conjugated QDs to be useful in the determination of the progression of lung infections such as RSV within the limits of the animal model. Further, the development of a single‐step detection methods based on the direct conjugation of antibodies to QDs can effectively detect RSV infection earlier than the conventional methods used for respiratory infection detection.34
Figure 7.
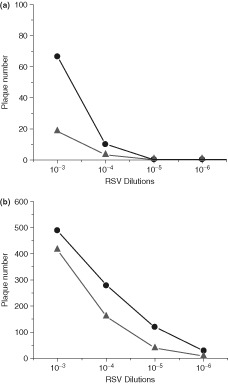
The viral titer of RSV‐infected Vero cells was determined at 3 days postinfection (a) and 5 days postinfection (b) using both conventional immunostaining techniques and the anti‐F QDs. The anti‐F QD labels samples were able to detect infection at 3 days while traditional staining was not. By 5 days however, both methods were able to detect similar degrees of infection. Circles represent the respiratory syncytial virus‐nanoparticle (RSV‐NP) label and triangles represent the conventional label.34.
Figure 8.
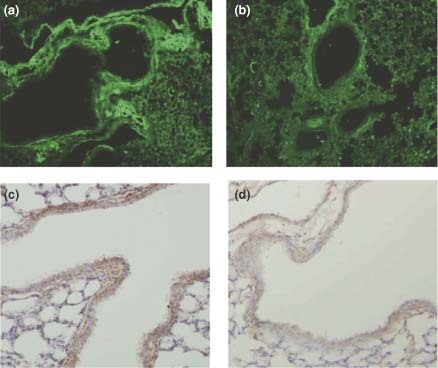
Anti‐F quantum dots (QDs) detection of infected lung tissue from respiratory syncytial virus (RSV)‐infected BALB/c mice (a) compared to uninfected control mice (b). When compared to conventional immunohistochemistry (IHC) staining, the RSV‐infected lungs gave similar regions of positive signal (c) and the background was less than what was seen of the IHC of uninfected control samples (d).34.
Antibody Functionalized QDs for FRET‐Based Detection of PRRSV
QDs have been used in FRET‐based technology for the detection of respiratory viruses. These FRET‐based assays employ QDs as the acceptor molecule and rely on both donor fluorescence quenching and the resulting appearance of QD fluorescence.18 This is similar to traditional two‐fluorophore construct where the secondary fluorescence signal is the result of the excitation of the acceptor molecule. The FRET‐based QD system contains a PRRSV specific antibody conjugated directly to the QD. This PRRSV antibody has been functionalized with a traditional organic fluorophore (AF546) prior to conjugation to the QD; this organic fluorophore will act as the donor molecule and the QD will act as the acceptor molecule.18 When the PRRSV antibody binds to its receptor protein, the excited AF546 will emit a photon which will excite the QD resulting in a decrease in AF546 fluorescence and a corresponding increase in QD fluorescence (Figure 9).18 This construct is similar to the antibody functionalized AuNPs discussed earlier. The primary difference between the two systems is the type of fluorescence change monitored. In the antibody functionalized AuNPs, fluorescence quenching represents a positive signal; in the antibody functionalized QD, a shift in the emission maximum of the fluorescence signal is considered a positive readout. The use of different fluorescence reporting output (QDs vs. AuNP induced quenching) explains the two systems' different sensitivities. Stringer et al. found that with this method, the limit of detection for the PRRSV antibody was 1409 particles/ml, slightly more sensitive than the parallel method developed with AuNP. When the selectivity of the QD biosensor was explored, it was found that the QD system not only gave a strong signal in the presence of PRRSV, but also in the presence of the blocking agent BSA. This nonspecific response can be, in part, attributed to the instability of the QD hydrophilic coating in water.18 Because QDs are intrinsically insoluble in water and must be functionalized for aqueous solubility, it has been assumed that the introduction and nonspecific absorption of a protein to the QD surface can alter the solubilization layer changing the apparent quantum yield. Currently, this process is poorly understood. Although the sensitivity of the QD biosensor probes was higher, it had poor selectivity compared to the AuNP biosensor.18
Figure 9.
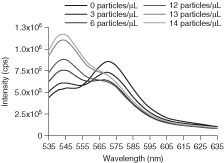
Fluorescence spectra for the quantum dot (QD) biosensor, where Förster resonance energy transfer (FRET) response is indicated by increasing acceptor fluorescence, with porcine reproductive and respiratory syndrome virus (PRRSV) antigen concentrations ranging from 0 to 60 particles/µl.18.
COMBINATION METHODS USING NANOPARTICLES
The use of nanoparticles for the detection of respiratory viruses has not been exclusively devoted to the development of new platforms. Nanoparticles have also been used to expand the capabilities of traditional virus detection techniques. The use of PCR to detect the SARS virus has been employed with relative success; however, the technique has distinct disadvantages. False positive or false negative results can occur when contamination of related organisms with genetic sequences similar to that of the target organism appear. Conventional PCR product detection typically uses electrophoresis on agarose gels, which are subject to toxic hazards, lack of specificity, poor sensitivity, and subjective reading results.35, 36, 37 The development of new techniques, apparatuses, and the addition of new materials to the PCR system attempt to circumvent these drawbacks.38 Nanoscale materials have been introduced to the PCR procedure to increase selectivity.24 The high surface‐to‐volume ratio allows them to be functionalized to enrich the target DNA within samples and decrease false signal from contamination.24 Highly selective nanoparticle PCR strategies utilizing AuNPs have been documented and reviewed elsewhere.39 Superparamagnetic nanoparticles have also been used to specifically detect HCV by PCR‐ELISA.40 Recently, this PCR enhancement technique has been developed for the detection of respiratory viruses, specifically SARS.
Nanoparticle‐Enhanced PCR for the Detection of SARS
The combination of functionalized nanoparticles and a PCR‐based assay has been developed for the sensitive and specific detection of the SARS virus. Gong et al. have developed an assay that employs the use of two types of nanoparticles, silica coated Superparamagnetic Nanoparticles (SMNPs) and Silica Coated Fluorescence Nanoparticles (SFNPs).38 Each of these plays a specific role which when combined increases the specificity and sensitivity of PCR. SMNPs are used to increase the specificity of PCR.38 The addition of the SMNPs is used to extract and enrich the target DNA from the sample. This helps to reduce the false positives reaction that is often a result of inhomogenous DNA in a sample. SMNPs are coupled to a probe complement to the target cDNA, allowing them to bind the target. The particles enriched in target cDNA are removed from the bulk sample solution when a magnetic force is applied. Figure 10a shows the enriched target which is then amplified through PCR amplification scheme and again isolated from the PCR products by binding with the SMNPs. Prior to PCR amplification, gel electrophoresis of both the original sample and post‐SMNP sample was used to assess the ability of the SMNPs to remove target cDNA. The original sample contained two bands, corresponding to the target cDNA and the negative control cDNA. The sample extracted with SMNPs only showed one band corresponding to the target cDNA (Figure 10b).38
Figure 10.
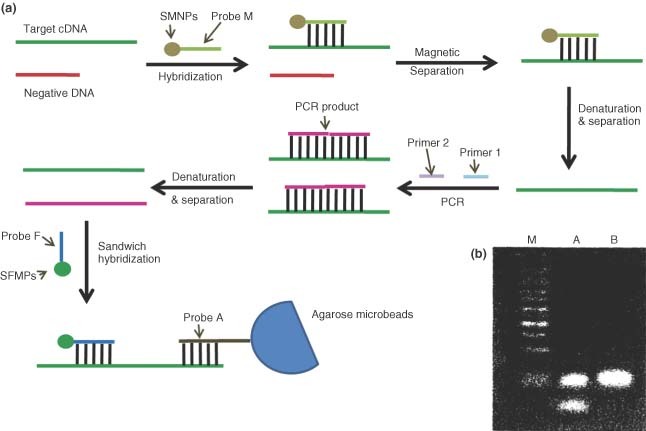
(a) Schematic illustration for severe acute respiratory syndrome (SARS) gene detection by the combination of functionalized nanoparticles and polymerase chain reaction (PCR)‐based assay. (b) The agarose gel electrophoresis for PCR products. Lane M, marker; Lane A, PCR products before treatment with superparamagnetic nanoparticles (SMNPs); Lane B, PCR products after treatment with SMNPs.38.
After amplification using the SMNP enriched sample, the target cDNA was detected using SFNPs as signaling probes. The target cDNA product was hybridized with a short complementary sequence to the end of the target cDNA which was attached to an agarose microbead. This immobilized the target cDNA on the agarose microbeads. The SFNPs were functionalized with a probe oligonucleotide which allows it to hybridize with the immobilized cDNA (Figure 10a). The fluorescence intensity of the SFNP was then used to signal the presence of the PCR target. When the SFNP method was used to detect PCR products, the limit of detection was 2.0 × 103 copies/ml. Compared to gel‐based detection methods, SFNP‐based detection provided an increase in sensitivity of an order of magnitude. This method also eliminated the need for hazardous stains such as ethinium bromide. Together, SMNPs were shown to enhance the specificity of the PCR amplification step while the excellent optical properties of the SFNPs increased the sensitivity of detection.
CONCLUSION
As a result of the threat that emerging respiratory viruses hold, efforts to develop new and improved detection methods have become a subject of intense focus. A recently published study highlighted the challenge by demonstrating that many rapid tests are ineffective at detecting the swine flu (novel H1N1 virus).41 Indeed, the results were so startling that the Centers for Disease Control and Prevention (CDC) updated guidance urging doctors to be cautious in relying on current rapid tests. Nanoparticles have been employed as a tool to overcome many of the limitations of traditional methods. The unique properties of nanoparticles have given them advantages over the typically costly and laborious conventional methods. The ability to be easily functionalized and the fluorescence quenching properties of gold and silver nanoparticles have led to the development of antibody conjugated AuNPs for the detection of PRRSV as well as gold and silver nanobarcodes for multiplexed virus detection. The nanobarcode technology has an advantage over the other detection methods developed here because of the inherent multiplexing capabilities of the system. In particular, the fact that the multiplexing capabilities of this sytem does not depend on the presence of a variety of different fluorescence dyes allows it to be resistant to many of the difficulties of multiplexing found in multicolor systems. However, as with any system that uses traditional organic fluorophores, the risk of photobleaching could lead to a decrease in sensitivity and false negative results.
The fluorescence characteristics of QDs, with broad excitation bands and discrete emissions bands, have resulted in the development of improved FRET‐based detection schemes as well as enhancing antibody‐based immunofluorescence methods. QDs offer an advantage over traditional organic fluorescence dyes because of their resistance to photobleaching, their broad excitation ranges, and their sharp emission spectra. The use of QDs would be particularly advantageous in the use of more than one fluorescence label at a time because the sharp emission spectra minimize signal overlap. Although these unique fluorescence properties of QDs make them a good alternative to traditional organic fluorescence dyes, they have their limitations. The most important limitation is their size. Because of the large size of QDs compared to organic fluorescence dyes, their use in detecting intracellular markers is limited unless extensive fixation and permeabilization techniques are used in conjunction with antibody labeling. However, as respiratory viruses present surface proteins that can be used as antigenic markers, this limitation can be overcome in viral detection with the chose of the right target protein. Thus, the use of QDs in detection technology holds a greater potential than those that continue to employ traditional organic fluorescence dyes.
Although a significant focus on the use of nanoparticles has been to develop novel detection methods, NPs have also been used to improve and expand existing detection methods. The addition of superparamagnetic nanoparticles to a PCR assay was able to increase its specificity during the amplification process and improve PCR product detection through the use of fluorescence nanoparticles. While these methods improve one of the major drawbacks of PCR, which is the specificity of the techniques, they do not address many of the other disadvantages of the PCR system. In particular, the costly and labor intensive aspects of performing PCR cannot be circumvented by the addition of NPs to the PCR system. As a result, despite the NP‐based improvements, the entire PCR system is still limited in its large‐scale applicability. Every approach reviewed in this article has demonstrated the power of nanoparticle technology to enhance the sensitivity and specificity of viral diagnostics. However, as with any new technology, each of these diagnotistic tools is still marked by limitations which open the door for continual efforts to develop the ideal viral detection method.
Acknowledgements
Financial support for this work was provided by NIH (NIBIB) grant R01 EB004537. The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). We thank M.F.Richards for critical comments related to this manuscript.
FURTHER READING
- Driskell J, Kwarta K, Lipert R, Porter M, Neill J, Ridpath J. Low‐Level Detection of Viral Pathogens by a Surface‐Enhanced Raman Scattering Based Immunoassay. Anal Chem 2005, 77:6147–6154. [DOI] [PubMed] [Google Scholar]
- Fatemi K, Ghourchian H, Ziaee A‐A, Samiei S, Hanaee H. Paramagnetic nanoparticle‐based detection of hepatitis B virus using cathodic stripping voltammetry. Biotechnol Appl Biochem 2009, 052:221–225. [DOI] [PubMed] [Google Scholar]
- Fuentes M, Mateo C, Rodriguez A, Casqueiro M, Tercero J, Riese H, Fernandez‐Lafuente R, Guisan JM. Detecting minimal traces of DNA using DNA covalently attached to superparamagnetic nanoparticles and direct PCR‐ELISA. Biosens Bioelectron 2006, 21:1574‐1580. [DOI] [PubMed] [Google Scholar]
- Gao J, Gu H, Xu B. Multifunctional Magnetic Nanoparticles: Design, Synthesis, and Biomedical Applications. Acc Chem Res 2009, 42:1097–1107. [DOI] [PubMed] [Google Scholar]
- Kim E‐Y, Stanton J, Korber B, Krebs K, Bagdan D, Kunstman K, Wu S, Phair J, Mirkin C, Wolinsky S. Detection of HIV‐1 p24A Gag in plasma by a nanoparticle‐based bio‐barcode‐amplification method. Nanomedicine 2008, 3:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Gong JL, Huang Y, Zheng Y, JiangJH, Shen GL, Yu RQ. Biocompatible core‐shell nanoparticle‐based surface‐enhanced Raman scattering probes for detection of DNA related to HIV gene using silica‐coated magnetic nanoparticles as separation tools. Talanta 2007, 72:443‐449. [DOI] [PubMed] [Google Scholar]
- Portney N, Ozkan M. Nano‐oncology: drug delivery, imaging, and sensing. Anal Bioanal Chem 2006, 384:620–630. [DOI] [PubMed] [Google Scholar]
- Algar WR, Massey M, Krull U. The application of quantum dots, gold nanoparticles and molecular switches to optical nucleic‐acid diagnostics. Trends Anal Chem 2009, 28:292–306. [Google Scholar]
- Schuler T, Asmus T, Fritzsche W, Moller R. Screen printing as cost efficient fabrication method for DNA‐chips with electrical readout for detection of viral DNA. Biosens Bioelectron 2009, 24:2077–2084. [DOI] [PubMed] [Google Scholar]
- Su‐Hua H et al. Gold nanoparticle‐based RT‐PCR and real‐time quantitative RT‐PCR assays for detection of Japanese encephalitis virus. Nanotechnology 2008, 19:405101. [DOI] [PubMed] [Google Scholar]
- Valanne A, Huopalahti S, Soukka T, Vainionpaa R, Lovgren T, Harma H. A sensitive adenovirus immunoassay as a model for using nanoparticle label technology in virus diagnostics. J Clin Virol 2005, 33:217–223. [DOI] [PubMed] [Google Scholar]
- Vaseashta A, Dimova‐Malinovska D. Nanostructured and nanoscale devices, sensors and detectors. Sci Technol Adv Mater 2005, 6:312–318. [Google Scholar]
- Zhong W. Nanomaterials in fluorescence‐based biosensing. Anal Bioanal Chem 2009, 394:47–59. [DOI] [PubMed] [Google Scholar]
- Zrenner A et al. Coherent optoelectronics with single quantum dots. J Phys Condens Matter 2008, 20:454210. [Google Scholar]
- Birdi KS. Colloidal systems on the nanometer length scale. Handbook 1 of Surface and Colloid Chemistry. 3rd ed Boca Raton, FL: CRC Press; 2009, 131‐154. [Google Scholar]
RELATED WIREs ARTICLES
REFERENCES
- 1.World Health Organization. Influenze WHO Fact Sheet no. 211, 2003.
- 2. Collins PL, Crowe JE Jr. Respiratory syncytial virus and metapneumovirus In: David PH, Knipe M, eds. Fields Virology, vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2007, 1601–1646. [Google Scholar]
- 3. Samson S, Wong Y, Yuen K‐Y. The managment of coronavirus infections with particular reference to SARS. J Antimicrob Chemother 2008, 62: 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prieto C, Castro JM. Porcine reproductive and respiratory syndrome virus infection in the boar: a review. Theriogenology 2005, 63: 1–16. [DOI] [PubMed] [Google Scholar]
- 5. Hu A, Colella M, Tam JS, Rappaport R, Cheng S‐M. Simultaneous detection, subgrouping, and quantitation of respiratory syncytial virus a and b by real‐time PCR. J Clin Microbiol 2003, 41(1): 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freymuth F, Eugene G, Vabret A, Petitjean J, Gennetay E, et al. Detection of respiratory syncytial virus by reverse transcription‐PCR and hybridization with a DNA enzyme immunoassay. J Clin Microbiol 1995, 33(12): 3352–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gueudin M, Vabret A, Petitjean J, Gouarin S, Brouard J, et al. Quantitation of respiratory syncytial virus RNA in nasal aspirates of children by real‐time RT‐PCR assay. J Virol Meth 2003, 109: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J. Nanomaterial‐based electrochemical biosensors. The Analyst 2005, 130: 421–426. [DOI] [PubMed] [Google Scholar]
- 9. Hu X, Dong S. Metal nanomaterials and carbon nanotubes—synthesis, functionalization, and potential applications towards electrochemistry. J Mater Chem 2008, 18: 1279–1295. [Google Scholar]
- 10. Zhang Q, Li Y, Tsien RW. The dynamic control of kiss‐and_run and vesicular reuse probed with single nanoparticles. Science 2009, 323: 1448–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frias JC, Lipinski MJ, Teresa M, Ibanez AB, Soriano C, et al. Nanoparticles as constrast agents for MRI of atherosclerotic lesions. Clin Med: Cardiol 2008, 2: 173–179. [Google Scholar]
- 12. Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. Mill Valley, CA: University Science Books; 1994. [Google Scholar]
- 13. Daniel M‐C, Astruc D. Gold nanoparticles: assembly, supramolecular chemistry, quantum‐size‐related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev 2004, 104(1): 293–346. [DOI] [PubMed] [Google Scholar]
- 14. Dulkeith E, Ringler M, Klar TA, Feldmann J. Gold nanoparticles quench fluorescence by phase induced radiative rate suppression. Nano Lett 2005, 5(4): 585–589. [DOI] [PubMed] [Google Scholar]
- 15. Dubertret B, Calame M, Libchaber AJ. Single‐mismatch detection using gold‐quenched fluorescent oligonucleotides. Nat Biotechnol 2001, 19(April): 365–370. [DOI] [PubMed] [Google Scholar]
- 16. Clegg RM. Fluorescence Imaging Spectroscopy and Microscopy. New York: John Wiley & Sons; 1996. [Google Scholar]
- 17. Lakowics JR. Principles of Fluorescence Spectroscopy, 2 ed. New York: Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- 18. Stringer RC, Schommer S, Hoehn D, Grant SA. Development of an optical biosensor using gold nanoparticles and quantum dots for the detection of porcine reproductive and respiratory syndrome virus. Sens Actuators B: Chem 2008, 134: 427–431. [Google Scholar]
- 19. Grant SA, Heits B, Kleiboeker S. Development of an optical biosensor utilizing gold nanoparticles to detect porcine productive and prespiratory syndrome virus SensLett 2006, 4(3): 246–252. [Google Scholar]
- 20. Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 1996, 14(3): 303–308. [DOI] [PubMed] [Google Scholar]
- 21. Marras S, Kramer FR, Tyagi S. Multiplex detection of single‐nucleotide variations using molecular beacons. Genet Anal: Biomol Eng 1999, 14(5–6): 151–156. [DOI] [PubMed] [Google Scholar]
- 22. Vet J, Majithia A, Marras S, Tyagi S, Dube S, et al. Multiplex detection of four pathogenic retroviruses using molecular beacons. ProcNatl Acad Sci U S A 1999, 96(11): 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sha MY, Yamanaka M, Walton ID, Norton SM, Stoermer RL, et al. Encoded metal nanoparticle‐based molecular beacons for multiplexe detection of DNA. NanoBiotechnology 2005, 1: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emory SR, Haskins WE, Nie S. Direct observation of size‐dependent optical enhancement in single metal nanoparticles. J Am Chem Soc 1998, 120: 8009–8010. [Google Scholar]
- 25. Martinez‐Paredes G, Gonzalez‐Garcia MB, Costa‐Garcia A. Genosensor for SARS virus detection based on gold nanostructured screen‐printed carbon electrodes. Electroanalysis 2009, 21(3–5): 379–385. [Google Scholar]
- 26. Fanjul‐Bolado P, Hernandez‐Santos D, Gonzalex‐Garcia MB, Costa‐Garcia A. Alkaline phophatase‐catalyzed silver deposition for electrochemical detection. Anal Chem 2007, 79(14): 5272–5277. [DOI] [PubMed] [Google Scholar]
- 27. Bruchez M Jr., Moronne M, Gin P, Weiss S, Paul Alivisatos A. Semiconductor nanocrystals as fluorescent biological labels. Science 1998, 281: 2013. [DOI] [PubMed] [Google Scholar]
- 28. Kim J, Wong C, Scholes G. Exciton fine structure and spin relaxation in semiconductor colloidal quantum dots. Acc Chem Res 0(0). [DOI] [PubMed] [Google Scholar]
- 29. Kim D, Okahara S, Shimura K, Nakayama M. Layer‐by‐layer assembly of colloidal CdS and AzS‐CdS quantum dots and improvement of their photoluminescence properties. J Phys Chem C 2009, 113: 7015–7018. [Google Scholar]
- 30. Larson DR, Zipfel W, Williams RM, Clark SW, Bruchez MP, et al. Water‐soluble quantum dots for multiphoton fluorescence imaging in vivo . Science 2003, 300: 1434–1436. [DOI] [PubMed] [Google Scholar]
- 31. Bentzen EL, House F, Utley TJ, Crowe JE Jr., Wright DW. Progression of respiratory syncytial virus infection monitored by fluorescent quantum dot probes. Nano Lett 2005, 5(4): 591–595. [DOI] [PubMed] [Google Scholar]
- 32. Rossi NL, Giljohann DA, Shad Thaxton C, Lytton‐Jean AKR, Han MS, et al. Oligonucleotide‐Modified gold nanoparticles for intracellular gene regulation. Science 2006, 312: 1027–1030. [DOI] [PubMed] [Google Scholar]
- 33. Pingarron JM, Yanez‐Sedeno P, Gonzalez‐Cortes A. Gold nanoparticle‐based electrochemical biosensors. Electrochem Acta 2008, 53: 5848–5866. [Google Scholar]
- 34. Tripp RA, Alvarez R, Anderson B, Jones L, Weeks C, et al. Bioconjugated nanoparticle detection of respiratory syncytial virus infection. Int J Nanomed 2007, 2(1): 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alvarez J, Sota M, Vivanco AB, Perales I, Cisterna R, et al. Development of a multiplex PCR technique for detection and epidemiological typing of salmonella in human clinical samples. J Clin Microbiol 2004, 42(4): 1734–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu HS, Chiu SC, Tseng TC, Lin SF, Lin JH, et al. Seologic and molecular biologic methods for SARS‐associated coronovirus infection. Emerg Infect Dis 2004, 10: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knoll S, Vogel RF, Niessen L. Identification of Fusarium graminearum in cereal samples by DNA detection test stripsTM . Lett Appl Microbiol 2002, 34(2): 144–148. [DOI] [PubMed] [Google Scholar]
- 38. Gong P, He X, Wang K, Tan W, Xie W, et al. Combination of functionalized nanoparticles and polymerase chain reaction‐based method for SARS‐CoV gene detection. J Nanosci Nanotechnol 2008, 8(1): 293–300. [PubMed] [Google Scholar]
- 39. Li H, Huang J, Lv J, An H, Zhang X, et al. Nanoparticle PCR: nanogold‐assisted PCR with enhanced specificity 13. Angew Chem Int Ed 2005, 44(32): 5100–5103. [DOI] [PubMed] [Google Scholar]
- 40. Fuentes M, Mateo C, Rodriguez A, Casqueir M, Tercero J, et al. Detecting minimal traces of DNA using DNA covalently attached to superparamagnetic nanoparticles and direct PCR‐ELISA. Biosens Bioelectron 2006, 21(8): 1574–1580. [DOI] [PubMed] [Google Scholar]
- 41. Ginocchio CC, Zhang F, Manji R, Arora S, Bornfreund M, et al. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J Clin Vir 2009, 45(3): 191–195. [DOI] [PMC free article] [PubMed] [Google Scholar]


