Abstract
Background
Lung transplant patients experience a high risk of airway infections and microbial colonization of the lung due to constant exposure to the environment through inhaled microorganisms, denervation, reduced ciliary transport, and decreased cough.
Methods
In this nationwide prospective study on Swedish lung transplant patients, we evaluated the microbiological panorama of bacteria, fungi, and virus found in bronchoalveolar lavage fluid (BALF) obtained the first year after lung transplantation (LTx). Differences in microbiological findings depending of concomitant signs of infection and background factors were assessed.
Results
A total of 470 bronchoscopies from 126 patients were evaluated. Sixty‐two percent (n = 293) of BALF samples had positive microbiological finding(s). Forty‐six percent (n = 217) had bacterial growth, 29% (n = 137) fungal growth, and 9% (n = 43) were positive in viral PCR. In 38% of BALF samples (n = 181), a single microbe was found, whereas a combination of bacteria, fungi or virus was found in 24% (n = 112) of bronchoscopies. The most common microbiological findings were Candida albicans, Pseudomonas aeruginosa and coagulase negative Staphylococcus (in 42 (33%), 36 (29%), and 25 (20%) patients, respectively). Microbiological findings were similar in BALF from patients with and without signs of lung infection and the frequency of multidrug resistant (MDR) bacteria was low. No significant association was found between background factors and time to first lung infection.
Conclusion
This study gives important epidemiologic insights and reinforces that microbiological findings have to be evaluated in the light of clinical symptoms and endobronchial appearance in the assessment of lung infections in lung transplant patients.
Keywords: bronchoalveolar lavage fluid, lung infections, lung transplantation, microbiology
1. INTRODUCTION
Lung transplantation (LTx) increases as a treatment option for end stage lung disease. Although mortality post transplantation has decreased with modern surgical techniques and pharmaceutical regimes, 5‐year mortality has been reported to be around 50%, of which infections are the predominant cause of death during the first year post LTx.1 The constant exposure to the environment through inhaled microorganisms together with denervation, reduced ciliary transport and decreased cough, increase the risk of lung infections. Moreover, heavy immunosuppressive therapy leads to decreased immune defense. Bronchoalveolar lavage (BAL), including microbiological cultures and PCR, are performed in lung transplant patients for surveillance and in response to clinical symptoms to guide antibiotic therapy. As the microbiological panorama differs by geographical region, especially with regards to multidrug resistant bacteria (MDR), it is important to map the local epidemiology. Previous studies of microbiology in lung transplant patients have focused on infections rather than the total panorama of microbiological findings.2, 3, 4, 5, 6 Moreover, most studies of lung infections in lung transplant patients are single center reports with a small number of patients. In Sweden, LTx is performed at two centers only, Skåne University Hospital, Lund and Sahlgrenska University Hospital, Gothenburg. The primary aim of this nationwide prospective study was to examine the microbiological panorama in BALF of lung transplant patients in Sweden during the first‐year post transplantation and to compare findings in patients with and without signs of lung infection.
2. MATERIAL AND METHODS
2.1. Study setting and patient population
This prospective cohort study included all adult patients accepted for LTx during May 2012 to December 2014 in Gothenburg and October 2012 to December 2014 in Lund. Patients under 18 years of age and patients with post‐operative follow up at other sites were excluded. All patients were followed up for 1 year after transplantation. Written informed consent was obtained from all study participants. The study was approved by the Regional Ethics Committee (Reg nr 433‐08) and performed in accordance with the ethical standards of the 1964 declaration of Helsinki and its later amendments.
2.2. Immunosuppressive and prophylactic strategies
Standard protocol for immunosuppression included induction therapy with ATG (anti‐thymocyte globulin), followed by tacrolimus or ciclosporin, mycophenolate mofetil, and steroids. All lung transplant recipients received Pneumocystis prophylaxis with co‐trimoxazole. Cytomegalovirus (CMV) prophylaxis with valganciclovir was given to all participants, with the exception of patients in Gothenburg when both donor and recipient were CMV negative. In Lund, patients received fungal prophylaxis with fluconazole for 3‐6 months, whereas patients in Gothenburg received oral nystatin for at least 3 weeks. Standard perioperative antibiotic treatment was cefotaxime in Gothenburg and imipenem in Lund. Post‐operative modifications or termination of antibiotic treatment was done according to perioperative donor and recipient cultures.
2.3. Sample collection and microbiology analyses
During the LTx procedure, bacterial and fungal cultures were obtained from the donor and recipient lungs. The recipients were then followed during the first year after LTx with routine bronchoscopies at 3, 6, and 12 months post transplantation, and additional bronchoscopies were performed in response to clinical symptoms. BAL procedure followed standardized protocols. Microbiology analyses were performed at the Microbiological Department, Skåne University Hospital and Sahlgrenska University Hospital according to standard protocols.7, 8 All BALF samples were analyzed with bacterial and fungal cultures, including microscopy for fungal elements and PCR for Mycoplasma pneumoniae, Chlamydia pneumoniae and Legionella pneumophila. MDR Pseudomonas aeruginosa was defined as resistance against meropenem and at least one agent in three or more categories of anti‐pseudomonal antibiotics. The definition is based on criteria published by Magiorakos et al9, with the modification that resistance against meropenem had to be present. A panel of airway viruses was analyzed with multiplex PCR, including; Parainfluenza 1, 2 and 3, Respiratory Syncytial Virus (RSV), Influenza A and B, Adenovirus, Metapneumovirus, Coronavirus (OC 43, 229E, NK‐63, HKU1), Enterovirus, Rhinovirus, and Bocavirus.10 In Lund, additional PCR for herpes simplex virus (HSV) 1 and 2 was performed with an in‐house method. Presence of Pneumocystis jiroveci was analyzed with PCR11 or immunofluorescence of BALF. CMV in blood or plasma was determined with quantitative PCR. All BALF samples from Lund were routinely stained and cultured for mycobacteria. BALF samples from Gothenburgh were analyzed for mycobacteria when there was a pretransplant history of mycobacterial infection.
Transbronchial biopsies (TBB) were analyzed at the Department of Pathology at Skåne University Hospital and Sahlgrenska University Hospital. Rejection was defined and graded A0‐A3 according to the International Society for Heart & Lung Transplantation.12
2.4. Clinical data and definitions
Bronchoscopies with positive microbiological finding(s) were classified as pneumonia in the presence of at least one of the following clinical criteria; new or increased cough, dyspnea, increased sputa, fever >38°C, worsening gas exchange, or white blood cell count >15 × 109/L, together with new (within 7 days before or after bronchoscopy) infiltrates in chest radiology (x‐ray or CT scan). Bronchoscopies with positive microbiological finding(s) were classified as tracheobronchitis in the presence of clinical signs as above with no infiltrates on chest radiology but one or more of the following macroscopic endobronchial abnormalities: inflamed endobronchial mucosa, endobronchial lesion (white/yellow) with/without necrotic changes, and purulent secretion. Pneumonia and tracheobronchitis are referred to as bronchoscopies with “signs of infection” in the text. CMV pneumonitis was defined as airway symptoms as described above, >1000 CMV copies/mL in blood, and findings of inclusion bodies in TBB. A minimum of 30 days between lung infections were allowed to be considered a new episode. Bronchoscopies with positive microbiological finding(s) were classified as “no infection” in the absence of clinical and/or radiologic or macroscopic pathology. Growth of pharyngeal flora was considered as contamination and classified as negative microbiology.
Clinical data were recorded at time of bronchoscopy in a study protocol, and retrieved retrospectively from patient records. Time after transplantation was categorized: <1 month, 1‐3 months, 3‐6 months, 6‐9, and 9‐12 months after LTx. Microbiological findings were grouped into Gram‐negative bacteria (G−), Gram‐positive bacteria (G+), yeast, mold, and virus. P. aeruginosa was analyzed separately.
2.5. Statistical analysis
Numerical data are presented as median and range. Chi‐squared, rank sum, Kruskall‐Wallis tests, logistic regression and analysis of variance were used to assess the distribution of background factors among different bacterial groups, lung infections and time periods. Since participants underwent varying numbers of bronchoscopies in the study, and several patients had recurrent findings of the same pathogen in BALF, the frequency of individual microbes is reported as the total number of patients with the actual finding. Grouped microbiological findings during the first year are presented as percent of patients with a bronchoscopy within the specified time period. In contrast to overall microbiological findings, lung infections are presented as number of episodes and not as a proportion of patients. The association between background data (gender, age at LTx, type of LTx and underlying disease, type of immunosuppression, positive or negative donor cultures) and time to first lung infection was estimated with Cox regression. All statistical tests were two‐sided, and 95% CIs that did not overlap 1.0 and P‐values less than 0.05 were considered statistically significant. Analyses were performed using the STATA/SE (version 13.1 for Windows; StataCorp LP, College Station, TX, USA), Graphpad Prism 6 (GraphPad Software; La Jolla, CA, USA) and SPSS (version 20.0; SPSS, Armonk, NY, USA).
3. RESULTS
3.1. Patient cohort and bronchoscopies
In total, 135 of 146 (92%) patients eligible for inclusion were consecutively included in the study. Nine patients were unable to give informed consent, one patient declined participation and one patient was not included for unknown reasons. Nine patients died before any bronchoscopy was performed, resulting in 126 patients (97 from Gothenburg and 29 from Lund) prospectively followed up for 1 year after LTx.
The participants had a median age of 57 years, 52% were women, 85% underwent double lung transplantation, and chronic obstructive pulmonary disease (COPD) was the most common underlying disease. For detailed patient characteristics, see Table 1. Eighty‐eight patients (70%) received post‐transplant immunosuppression with ciclosporin and 38 (30%) with tacrolimus. Patients with cystic fibrosis were more likely to receive tacrolimus than ciclosporin (72% vs 23%, P < 0.01). No other differences in choice of immunosuppression depending on background diagnosis were found.
Table 1.
Patient characteristics
| Total patients n | 126 |
| Gender male/female n | 61/65 |
| Age at tx. years median (range) | 57 (18‐70) |
| Transplantation | |
| Single n (%) | 18 (14) |
| Double n (%) | 108 (85) |
| Underlying diagnosis n patients (%) | |
| Chronic obstructive pulmonary disease | 33 (26) |
| Pulmonary fibrosis | 31 (25) |
| Cystic fibrosis | 18 (14) |
| Alpha‐1 antitrypsin deficiency | 17 (13) |
| Pulmonary arterial hypertension | 7 (6) |
| Re‐transplantation | 6 (5) |
| Sarcoidosis | 2 (2) |
| Bronchiectasis | 2 (2) |
| Bronchiolitis obliterans syndrome | 2 (2) |
| Emphysema | 2 (2) |
| Other | 6 (5) |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
During the study period, 470 bronchoscopies were performed with a median of three per patient (range 1‐10). Two hundred and eleven bronchoscopies (45%) were routine procedures, whereas 259 (55%) were performed on clinical indication. The bronchoscopies were distributed over the first post‐operative year as shown in Figure 1. All samples were analyzed for bacterial and fungal growth, including microscopy for fungal elements, and with PCR for Mycoplasma, Chlamydia, and Legionella. Multiplex PCR for airway viruses were analyzed in 342 samples (73%). Pneumocystis was analyzed in 416 samples (89%). Nineteen patients (15% of the total study population) had negative BALF cultures/PCRs throughout follow up. Of the remaining 107 patients with positive microbiology, 50 patients had no episode with signs of lung infection. Fifty‐seven patients had at least one episode with signs of lung infection, of which 38 patients had at least one pneumonia and 19 patients had at least one episode of tracheobronchitis but no pneumonia.
Figure 1.
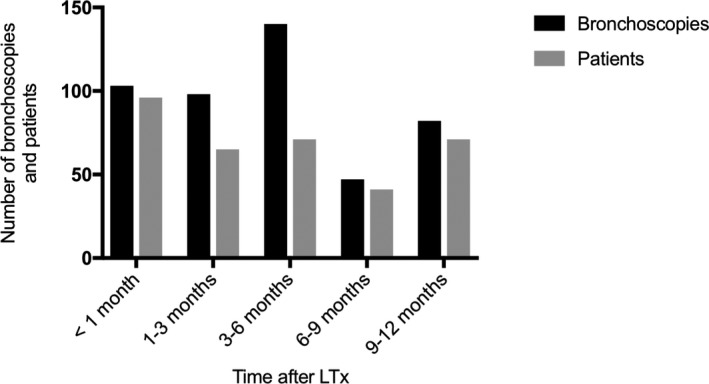
Total number of bronchoscopies and number of patients with a bronchoscopy performed during the specified time interval. In total, 103 (22%) bronchoscopies were performed less than 1 mo after LTx, 98 (21%) 1‐3 mo, 140 (30%) 3‐6 mo, 47 (10%) 6‐9 and 82 (17%) 9‐12 mo after transplantation
3.2. Microbiology results
In total, 177 (38%) BALF samples had negative microbiological results, including 75 samples with growth of pharyngeal flora (Figure 2). These were excluded from further analyses. The remaining 293 (62%) samples had positive microbiological findings, of which 217 (46%) BALF had bacterial growth, 137 (29%) had fungal growth, and 43 (9%) were positive in viral PCR. A single microbe was found in 181 (38%) BALF samples, whereas 112 (24%) BALF had a combination of bacteria, fungi, or virus. Bronchoscopies performed within 3 months after transplantation had more poly‐microbial findings compared to later bronchoscopies (44% vs 33%, P = 0.04).
Figure 2.
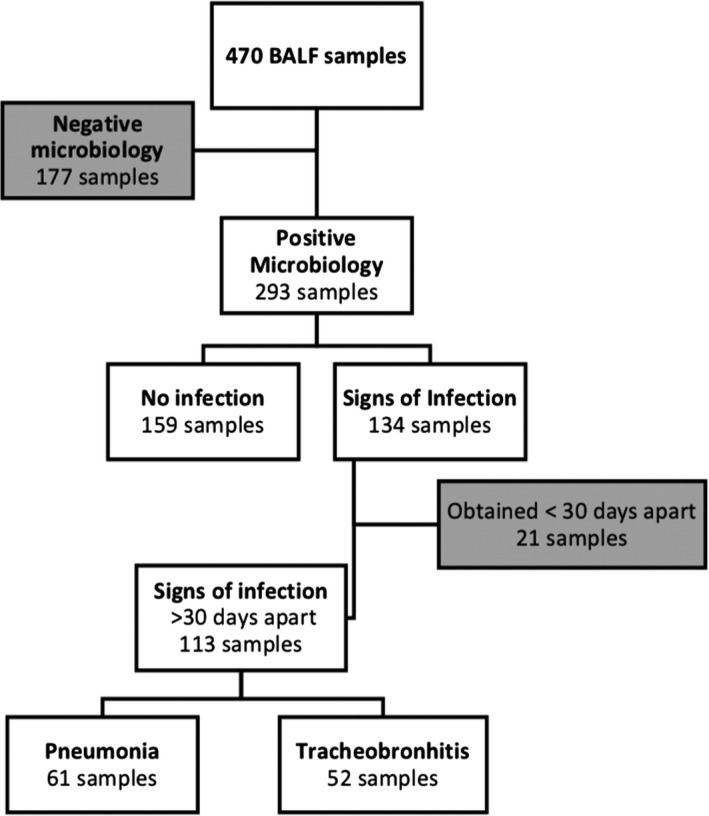
Flowchart of samples in the study cohort and subsequent classification according to microbiological results in BALF and clinical signs of infection at the time of sampling
The most common microbiological findings were Candida albicans, P. aeruginosa and coagulase negative Staphylococcus (in 42 (33%), 36 (29%), and 25 (20%) patients, respectively); see Table 2 for all microbiological findings. Three patients (2%) had growth of extended spectrum beta‐lactamase (ESBL)‐producing bacteria in BALF (two Escherichia coli and one Enterobacter cloacae), one patient (1%) of a multiresistant Burkholderia and nine patients (7%) of multiresistant P. aeruginosa. No methicillin‐resistant Staphylococcus aureus (MRSA), ESBL‐and carbapenemase‐producing Enterobacteriaceae or vancomycin‐resistant Enterococcus (VRE) was found.
Table 2.
(ABC) Microbial findings in BALF during first year expressed as total number of patients (percent of patients, n total=126). Owing to bronchoscopies with several microbiological findings the figures do not sum up to 100%
| n patients (% patients) | |
|---|---|
| A. Bacteria | |
| Gram‐negative | |
| Pseudomonas aeruginosa | 36 (29) |
| Stenotrophomonas | 16 (13) |
| Escherichia coli | 10 (8) |
| Achromobacter | 4 (3) |
| Moraxella catarrhalis | 3 (2) |
| Chryseobacterium | 2 (2) |
| Serratia | 2 (1) |
| Other Gram‐negative | 5 (3) |
| Gram‐positive | |
| Staphylococcus aureus | 22 (17) |
| Coagulase negative staphylococcus | 25 (20) |
| Enterococcus | 8 (6) |
| Enterobacter | 6 (3) |
| Staphylococcus lugdunensis | 2 (2) |
| Pneumococcus | 2 (2) |
| Mycobacterium tuberculosis | 1 (1) |
| Mycobacterium abscessus | 1 (1) |
| B. Fungi | |
| Yeast | |
| Candida albicans | 42 (33) |
| Candida non‐albicans | 17 (13) |
| Mold | |
| Aspergillus fumigatus | 21 (17) |
| Aspergillus non‐fumigatus | 15 (12) |
| Fusarium | 3 (2) |
| Penicillium | 11 (9) |
| Other mold | 3 (2) |
| Pneumocystis a | 5 (3) |
| C. Virus | |
| Coronavirus | 10 (8) |
| Rhinovirus | 9 (7) |
| Parainfluenza virus | 4 (3) |
| Metapneumovirus | 3 (2) |
| Herpes simplex virus 1 | 2 (2) |
| Respiratory syncytial virus | 2 (2) |
| Adenovirus | 2 (2) |
| Other virus | 3 (2) |
Positive in PCR (no sample positive in IF).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The proportion of patients with bacterial findings in BALF increased over the first 6 months and then declined (Figure 3A). Yeast was most common within the first month after LTx, whereas mold peaked at 3‐6 months (Figure 3B). The proportion of viral findings increased over time with a maximum of 20% of patients at 6‐9 months (Figure 3C).
Figure 3.
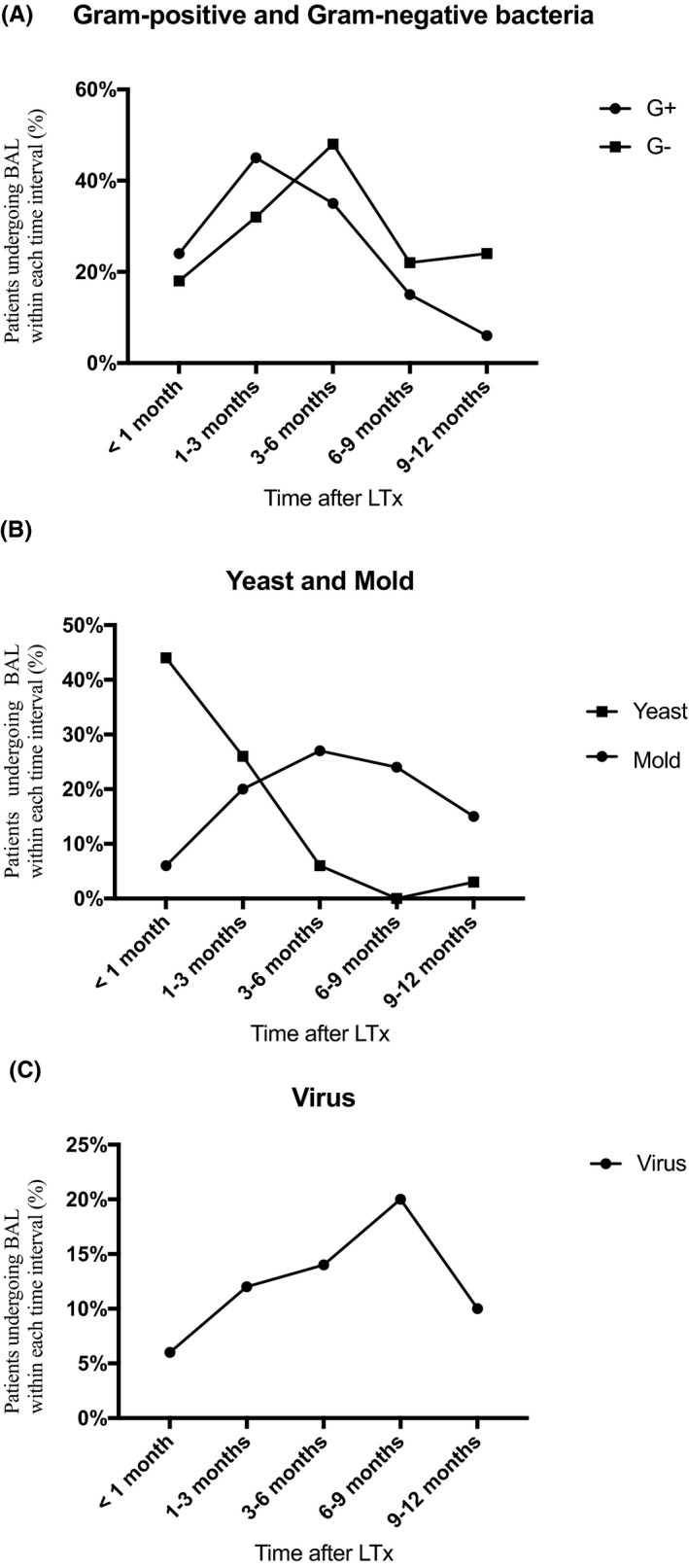
(ABC) Microbiological findings in BALF the first year after LTx expressed as percent of patients with a bronchoscopy performed within each time interval. A, Gram‐negative bacteria (G−) and Gram‐positive (G+) bacteria; B, Yeast and mold; C, Virus
Gram‐positive bacteria were significantly more common in samples from patients receiving immunosuppression with ciclosporin compared to patients receiving tacrolimus (27% and 18%, P = 0.03). In contrast, more Gram‐negative bacteria were found in samples from patients receiving tacrolimus, 30% and 19%, respectively, yielding an OR of 2.0 (95% CI 1.3‐3.3). However, patients with cystic fibrosis were more frequently treated with tacrolimus, and no significant association between Gram‐negative bacteria and tacrolimus was found when adjusting for cystic fibrosis, OR 1.4 (95% CI 0.8‐2.4). No significant differences were noted for mold, yeast, or virus (P = 0.06, P = 0.84 and P = 0.55, respectively).
In the analysis by underlying pulmonary conditions, growth of P. aeruginosa, as well as other Gram‐negative bacteria, were more common findings in patients with cystic fibrosis compared to any other underlying diseases (41% vs 17%, P < 0.01 and 42% vs 19% P = 0.01, respectively). Patients with alpha‐1‐antitrypsin deficiency had significantly more Gram‐positive bacteria (34% vs 22%, P = 0.04), and less Gram‐negative bacteria (11% vs 25%, P < 0.01) and mold (6% vs 23%, P = 0.01) compared to other underlying diagnoses. Patients with underlying pulmonary fibrosis had more Gram‐positive bacteria (30% vs 21%, P = 0.03) and less growth of P. aeruginosa in BALF (8% vs 27%, P < 0.01) compared to other underlying diagnoses.
Peri‐operative recipient cultures were obtained from 121 patients (96%). In all, 34 patients (28%) had positive recipient cultures. Sixteen patients (13%) had growth of P. aeruginosa, and the remaining 18 patients had findings of S. aureus, E. coli, Candida and other Gram‐negatives. All 16 patients with Pseudomonas in recipient cultures had subsequent positive cultures for Pseudomonas at some point during the follow up period. No significant difference in time to first positive Pseudomonas culture was found between patients with (n = 16) and without (n = 20) growth of pseudomonas in recipient cultures (mean time to positive Pseudomonas culture 135 and 140 days, respectively (P = 0.86)).
3.3. Signs of lung infection
In 134 of 293 bronchoscopies with positive microbiological findings, the patients had clinical signs of lung infection (pneumonia or tracheobronchitis) at the time of bronchoscopy. Twenty‐one BALF samples from 17 patients with signs of infection were obtained less than 30 days apart, and thus considered as the same lung infection episode. This left 113 samples, of which 61 samples were from patients with concomitant symptoms consistent with pneumonia and 52 with tracheobronchitis. Six samples had also signs of acute rejection in TBB, which may add to the clinical presentation. Five were grade A1 and one grade A2. The remaining 159 BALF samples with positive microbiological findings were regarded as no infection (Figure 2).
In BALF from patients with concomitant signs of lung infections the most common Gram‐negative, Gram‐positive, fungal, and viral organisms were P. aeruginosa, Stenotrophomonas, coagulase negative Staphylococcus, S. aureus, C. albicans, Aspergillus fumigatus, Rhinovirus and Coronavirus (in 32 (28%), 9 (8%), 13 (11%) 12 (11%), 24 (21%), 9 (8%), 8 (7%), and 6 (5%) samples, respectively). One patient with signs of lung infection had simultaneously >1000 CMV copies/mL in blood and inclusion bodies in TBB and thus considered as a possible CMV pneumonitis. Median time to first episode with signs of lung infection was 37 days (range 5‐383), and the incidence decreased over time during the first year after LTx.
In 32 (28%) of BALF samples from patients with signs of infection, no previous bronchoscopy had been performed within the study. In the remaining 81 (72%) samples, the same microbe was identified in previous BALF in 43 samples (53%), of which 20 (46%) revealed growth of P. aeruginosa.
When comparing microbiological findings with and without signs of infection, the frequency of Gram‐negative bacteria, Gram‐positive bacteria, mold and virus were not significantly different (Table 3). Yeast was significantly more frequent in signs of infection (32% and 19%, P = 0.01). However, Candida was found as a single microbe in only 17 samples (15%) with signs of lung infection, all of which had concomitant macroscopic findings consistent with tracheobronchitis. Fourteen of 17 samples (82%) were obtained within 1 month after LTx and 10 samples (59%) were C. albicans.
Table 3.
Microbiological findings in BALF from patients with and without symptoms of lung infection
| Grouped microbiological findings | No infection n (%) | Signs of infection n (%) | P‐value |
|---|---|---|---|
| Gram‐positive bacteria | 70 (44) | 41 (36) | 0.20 |
| Gram‐negative bacteria | 64 (40) | 48 (43) | 0.71 |
| Yeast | 30 (19) | 36 (32) | 0.01 |
| Mold | 43 (27) | 28 (25) | 0.41 |
| Virus | 22 (14) | 18 (16) | 0.48 |
Samples from patients with positive microbiology and no infection (n total = 159) vs samples from patients with signs of lung infection (n total = 113) within defined microbiological groups. Owing to bronchoscopies with several microbiological findings in BALF the figures do not sum up to 100%.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The total number of bronchoscopies with several microbiological findings did not significantly differ between samples from patients with signs of infection and samples from patients with no infection (34% and 40%, respectively. P = 0.32). Neither did type of immunosuppression significantly differ between samples from patients with and without signs of infection (P = 0.42).
3.4. Donor cultures, background data and lung infections
During the lung transplantation procedure, bacterial and fungal cultures were obtained from donor lungs in 119 patients (94%). Donor cultures were positive in 79 patients (63%), of which 70 (55%) had growth of bacteria and 18 (14%) had fungal growth. The most frequent bacterial and fungal microbiological findings in donor lungs were S. aureus and Candida species (47 (37%) and 17 (13%), respectively). Sixty‐five patients with positive donor cultures underwent bronchoscopy within 1 month post Tx. Cultures from 14 BALF samples (22%) yielded the same microbial species as found in the donor cultures, of which 11 were Candida. In the first post‐operative month, Candida was significantly more common in BALF from patients with Candida in donor cultures compared to BALF from patients with donor lungs negative for Candida (79% and 38%, respectively; P = 0.04).
No significant association was found between background factors and time to first lung infection as estimated by Cox regression; hazard ratio (HR) for positive donor cultures was: 1.5; 0.9‐2.7; HR for gender was 1.5; 0.9‐2.6; HR for age at LTx was 1.0; 0.8‐1.3 per 10‐year increase; HR for single vs double lungs LTx was: 0.9; 0.4‐1.9; HR for type of immunosuppression was: 1.4; 0.8‐2.4; and HR for cystic fibrosis as compared to other underlying diseases was: 1.3; 0.6‐2.7.
In a secondary analysis, patients with no microbial growth in BALF throughout follow up were compared to patients with positive microbial findings. No differences in underlying factors were found, data not reported.
4. DISCUSSION
Lung transplanted patients undergo bronchoalveolar lavage several times during their first year after transplantation. Most studies on microbiology in lung transplant patients evaluate infection episodes, but very few report all microbiological findings in BALF. In this prospective study, we analyzed for the first time the total microbiological panorama found in BALF in Swedish lung transplant recipients. We found that 85% of patients had samples with positive microbiologic finding(s), and only 15% of patients remained negative throughout their bronchoscopies. In line with our findings, Charlson et al13 found that lung transplant recipients have a higher bacterial burden in BALF than healthy control subjects, regardless of underlying indication for transplantation. The single most common bacterium was P. aeruginosa, both overall and in samples with signs of lung infection. P. aeruginosa is previously well described as an important pathogen in lung transplanted patients.2, 5, 14 Not surprisingly, P. aeruginosa, and other Gram‐negative bacteria, were significantly more frequent in patients with cystic fibrosis. The fact that the study participants underwent varying numbers of bronchoscopies may result in overrepresentation of individual microbes. However, by presenting the data as the proportion of patients with the microbiological finding, we have tried to avoid bias.
Gram‐negative bacteria were more common among patients receiving tacrolimus. However, this association was confounded by a high proportion of patients with cystic fibrosis receiving tacrolimus. The total number of infection episodes did not significantly differ between patients receiving tacrolimus compared to ciclosporin. In line with our data, Hachem et al and Zuchelman et al found no difference in frequency of infections depending on type of immunosuppression.15, 16
The low prevalence of MDR bacteria in our study reflects that Sweden still has a favorable situation regarding antimicrobial resistance compared to most countries. In comparison, Kovatz et al described a MRSA prevalence of 25% in lung transplant patients in the first post‐operative year,17 and Tebano et al18 found MDR strains in respiratory samples in 90 of 179 lung transplanted patients (51%). In a recent publication, Rodrigo‐Troyano et al19 describes the serious threat posed by Gram‐negative MDR bacteria in respiratory infections. In fact, management of MDR bacteria in solid organ transplanted patients is a major issue of concern in most countries.20 In our samples, most Gram‐negative bacteria were susceptible to carbapenem treatment and no MDR Gram‐positive bacteria were found. Thus, our results indicate that the peri‐operative antibiotic prophylaxis and later empiric antibiotic therapy at present not necessarily has to include specific targeting of MDR bacteria in Swedish lung transplant patients.
Candida dominated in BALF‐cultures the first post‐operative month. In line with our data, Kovats et al17 report a high Candida burden the first months after LTx. Candida was present in 32% of all samples with concomitant signs of lung infection, but only 15% had Candida as a single pathogen. All had signs of tracheobronchitis, which corresponds to an incidence of Candida tracheobronchitis of 12%, assuming that Candida was the causative pathogen. This number is high compared to a study on 384 heart and/or lung transplant recipients, where an overall incidence of invasive Candida infections of 8.3% was described, of which tracheobronchitis represented 38% and pneumonia 3.1%.21 However, a broad antimicrobial post‐operative treatment the first month after LTx may cause negative bacterial BALF cultures and promote Candida dominance in lung infections in our study.
Forty‐five percent of patients experienced at least one episode with symptoms of lung infection during the first year after transplantation. A limitation of the study is that we did not take additional cultures/PCR from blood, sputum or nasopharyngeal into account. Another limitation is that we did not evaluate patients with culture‐negative samples for signs of infection, which is why the true frequency of lung infections may be underestimated. Although, our results are comparable to other studies that report a frequency of lung infections between 30% and 70%.2, 4, 6, 14, 22, 23 The incidence of lung infections in our study decreased over time. Similar results have been reported in other studies, where the first three post‐operative months have been defined as the critical period for infections in lung transplanted patients, especially for bacterial etiology.5, 24 Consistent with previous studies, we found no significant association between positive donor cultures and later microbiological findings in the absence or presence of lung infections.25, 26
The microbiologic panorama found in BALF from patients with clinical signs of lung infection was similar to that found in samples from patients without symptoms in our study. However, it is important to note that we did not evaluate the probable causative pathogen. Since several microbiologic agents were found in 40% of BALF from patients with signs of lung infection, the causative microbe was difficult to determine. In addition, we lack information about concomitant antimicrobial treatment that may interfere with microbiological results, as well as how treatment of specific pathogens influences the clinical outcome. About half of the patients with signs of lung infection had a previous bronchoscopy with the same microbe found in BALF, suggesting that the microbiology results of a previous bronchoscopy could guide antibiotic treatment in about half of the cases. In particular, previous cultures with P. aeruginosa should be targeted in empiric treatment.
In conclusion, a majority of bronchoscopies from lung transplant patients during the first year after LTx had positive microbiologic findings, often with several pathogens. Previous BAL cultures, in particular findings of P. aeruginosa, could guide empiric treatment of airway infections, which suggests a role for surveillance bronchoscopies. The frequency of MDR bacteria was low in our study, indicating that empiric therapy does not need to include MDR bacteria at present in Sweden. Microbiological findings were similar in BALF from patients with and without signs of lung infection. The results reinforce that microbiological findings have to be evaluated in the light of clinical symptoms and endobronchial appearance in the assessment of lung infections in lung transplant patients.
DISCLOSURES
The authors have no conflicts of interest to declare in relation to the material presented.
AUTHOR CONTRIBUTIONS
ASA planned the study, compiled data, performed statistical calculations, and wrote the manuscript. In accordance with editorial policies and ethical considerations, all authors revised the material, discussed the results, and contributed to the final manuscript.
ACKNOWLEDGEMENTS
We wish to thank the staff at the transplantation and bronchoscopy units in Lund and Gothenburg for assistance with patient inclusion, and Christina Pehrson and Jill Ryd for assistance in collecting clinical data. This work was funded by the Alfred Österlund, Magnus Bergvall, and Mats Kleberg Foundations, Lions research fund in Skåne, The Swedish Heart and Lung Association, the Swedish Government Funds for Clinical Research (ALF), the Royal Physiographic Society of Lund and the Skane County Council's Research and Development foundation.
Stjärne Aspelund A, Hammarström H, Inghammar M, et al. Microbiological findings in bronchoalveolar lavage fluid from lung transplant patients in Sweden. Transpl Infect Dis. 2018;20:e12973 10.1111/tid.12973
REFERENCES
- 1. Yusen RD, Edwards LB, Dipchand AI, et al. The registry of the International Society for heart and lung transplantation: thirty‐third adult lung and heart‐lung transplant report‐2016; focus theme: primary diagnostic indications for transplant. J Heart Lung Transplant. 2016;35:1170‐1184. [DOI] [PubMed] [Google Scholar]
- 2. Campos S, Caramori M, Teixeira R, et al. Bacterial and fungal pneumonias after lung transplantation. Transplant Proc. 2008;40:822‐824. [DOI] [PubMed] [Google Scholar]
- 3. Dudau D, Camous J, Marchand S, et al. Incidence of nosocomial pneumonia and risk of recurrence after antimicrobial therapy in critically ill lung and heart‐lung transplant patients. Clin Transplant. 2014;28:27‐36. [DOI] [PubMed] [Google Scholar]
- 4. Yun JH, Lee SO, Jo KW, et al. Infections after lung transplantation: time of occurrence, sites, and microbiologic etiologies. Korean J Intern Med. 2015;30:506‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parada MT, Alba A, Sepulveda C. Early and late infections in lung transplantation patients. Transplant Proc. 2010;42:333‐335. [DOI] [PubMed] [Google Scholar]
- 6. Gupta MR, Valentine VG, Walker JE Jr, et al. Clinical spectrum of gram‐positive infections in lung transplantation. Transpl Infect Dis. 2009;11:424‐431. [DOI] [PubMed] [Google Scholar]
- 7. The European Committee on Antimicrobial Susceptibility Testing . Breakpoint tables for interpretation of MICs and zonediameters. Version 8.0. 2018. http://www.eucast.org. Accessed March 1, 2018.
- 8. Murray PR. Manual of Clinical Microbiology. Washington, DC: ASM Press; 2007. [Google Scholar]
- 9. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268‐281. [DOI] [PubMed] [Google Scholar]
- 10. Brittain‐Long R, Westin J, Olofsson S, et al. Prospective evaluation of a novel multiplex real‐time PCR assay for detection of fifteen respiratory pathogens‐duration of symptoms significantly affects detection rate. J Clin Virol. 2010;47:263‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dini L, du Plessis M, Frean J, et al. High prevalence of dihydropteroate synthase mutations in Pneumocystis jirovecii isolated from patients with Pneumocystis pneumonia in South Africa. J Clin Microbiol. 2010;48:2016‐2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stewart S, Fishbein MC, Snell GI, et al. Revision of the 1996 working formulation for the standardization of nomenclature in the diagnosis of lung rejection. J Heart Lung Transplant. 2007;26:1229‐1242. [DOI] [PubMed] [Google Scholar]
- 13. Charlson ES, Diamond JM, Bittinger K, et al. Lung‐enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536‐545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aguilar‐Guisado M, Givalda J, Ussetti P, et al. Pneumonia after lung transplantation in the RESITRA Cohort: a multicenter prospective study. Am J Transplant. 2007;7:1989‐1996. [DOI] [PubMed] [Google Scholar]
- 15. Hachem RR, Yusen RD, Chakinala MM, et al. A randomized controlled trial of tacrolimus versus cyclosporine after lung transplantation. J Heart Lung Transplant. 2007;26:1012‐1018. [DOI] [PubMed] [Google Scholar]
- 16. Zuckermann A, Reichenspurner H, Birsan T, et al. Cyclosporine A versus tacrolimus in combination with mycophenolate mofetil and steroids as primary immunosuppression after lung transplantation: one‐year results of a 2‐center prospective randomized trial. J Thorac Cardiovasc Surg. 2003;125:891‐900. [DOI] [PubMed] [Google Scholar]
- 17. Kovats Z, Sütto Z, Muraközy G, et al. Airway pathogens during the first year after lung transplantation: a single‐center experience. Transplant Proc. 2011;43:1290‐1291. [DOI] [PubMed] [Google Scholar]
- 18. Tebano G, Geneve C, Tanaka S, et al. Epidemiology and risk factors of multidrug‐resistant bacteria in respiratory samples after lung transplantation. Transpl Infect Dis. 2016;18:22‐30. [DOI] [PubMed] [Google Scholar]
- 19. Rodrigo‐Troyano A, Sibila O. The respiratory threat posed by multidrug resistant Gram‐negative bacteria. Respirology. 2017;22:1288‐1299. [DOI] [PubMed] [Google Scholar]
- 20. Aguado JM, Silva JT, Fernandez‐Ruiz M, et al. Management of multidrug resistant Gram‐negative bacilli infections in solid organ transplant recipients: SET/GESITRA‐SEIMC/REIPI recommendations. Transplant Rev (Orlando). 2018;32:36‐57. [DOI] [PubMed] [Google Scholar]
- 21. Schaenman JM, Rosso F, Austin JM, et al. Trends in invasive disease due to Candida species following heart and lung transplantation. Transpl Infect Dis. 2009;11:112‐121. [DOI] [PubMed] [Google Scholar]
- 22. Flume PA, Egan TM, Paradowski LJ, et al. Infectious complications of lung transplantation. Impact of cystic fibrosis. Am J Respir Crit Care Med. 1994;149:1601‐1607. [DOI] [PubMed] [Google Scholar]
- 23. Kramer MR, Marshall SE, Starnes VA, et al. Infectious complications in heart‐lung transplantation. Analysis of 200 episodes. Arch Intern Med. 1993;153:2010‐2016. [PubMed] [Google Scholar]
- 24. Bando K, Paradis IL, Komatsu K, et al. Analysis of time‐dependent risks for infection, rejection, and death after pulmonary transplantation. J Thorac Cardiovasc Surg. 1995;109:49‐57; discussion 57‐49. [DOI] [PubMed] [Google Scholar]
- 25. Bonde PN, Patel ND, Borja MC, et al. Impact of donor lung organisms on post‐lung transplant pneumonia. J Heart Lung Transplant. 2006;25:99‐105. [DOI] [PubMed] [Google Scholar]
- 26. Howell CK, Paciullo CA, Lyon GM, et al. Effect of positive perioperative donor and recipient respiratory bacterial cultures on early post‐transplant outcomes in lung transplant recipients. Transpl Infect Dis. 2017;19:e12760. [DOI] [PubMed] [Google Scholar]


