Abstract
The characterization of functional yet nonprotein coding (nc) RNAs has expanded the role of RNA in the cell from a passive player in the central dogma of molecular biology to an active regulator of gene expression. The misregulation of ncRNA function has been linked with a variety of diseases and disorders ranging from cancers to neurodegeneration. However, a detailed molecular understanding of how ncRNAs function has been limited; due, in part, to the difficulties associated with obtaining high‐resolution structures of large RNAs. Tertiary structure determination of RNA as a whole is hampered by various technical challenges, all of which are exacerbated as the size of the RNA increases. Namely, RNAs tend to be highly flexible and dynamic molecules, which are difficult to crystallize. Biomolecular nuclear magnetic resonance (NMR) spectroscopy offers a viable alternative to determining the structure of large RNA molecules that do not readily crystallize, but is itself hindered by some technical limitations. Recently, a series of advancements have allowed the biomolecular NMR field to overcome, at least in part, some of these limitations. These advances include improvements in sample preparation strategies as well as methodological improvements. Together, these innovations pave the way for the study of ever larger RNA molecules that have important biological function.
This article is categorized under:
RNA Structure and Dynamics > RNA Structure, Dynamics, and Chemistry
Regulatory RNAs/RNAi/Riboswitches > Regulatory RNAs
RNA Structure and Dynamics > Influence of RNA Structure in Biological Systems
Keywords: chemical probing, cryo‐electron microscopy, nuclear magnetic resonance spectroscopy, RNA regulation, RNA structure, X‐ray crystallography
Overview of important sample preparation and methodological advancements that facilitate the study of large RNA structure and dynamics by nuclear magnetic resonance spectroscopy. These innovations pave the way for the study of previously intractable systems.
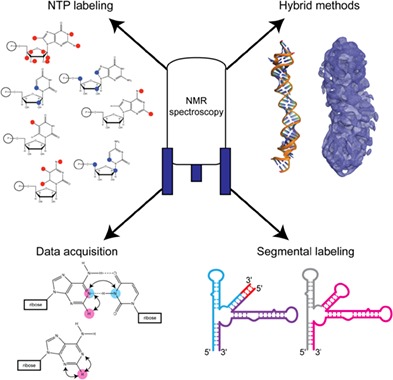
1. INTRODUCTION
Canonically, the central dogma limits the role of RNA in the cell to being either a messenger, transfer, or ribosomal RNA, all of which involve converting the genetic information encoded in the genome into a protein product. However, several classes of nonprotein coding (nc) RNAs play regulatory roles in diverse biological processes. More recently, the discovery of functional long ncRNAs has expanded our understanding of the roles that RNA can play in the cell. Though several classes of ncRNAs are active players in translation—ribosomal RNAs make up the bulk of the translation machinery while transfer RNAs shuttle amino acids into the ribosome for incorporation into the growing polypeptide chain, there is a growing appreciation of the functional importance of other ncRNA classes as well as noncoding regions of mRNAs. These elements have been shown to be key players in the regulation of gene transcription and translation, epigenetic control, and RNA turnover (Bonasio & Shiekhattar, 2014; Guil & Esteller, 2015; J. T. Lee, 2012).
Numerous studies have highlighted how complex structural folds in RNA molecules confer specific functions including riboswitches (Knappenberger, Reiss, & Strobel, 2018; Liberman et al., 2015; Peselis & Serganov, 2018; Vicens et al., 2018; B. Zhao, Guffy, Williams, & Zhang, 2017; also reviewed by Jones & Ferre‐D'Amare, 2017; Roth & Breaker, 2009), ribozymes (Chan et al., 2018; Costa, Walbott, Monachello, Westhof, & Michel, 2016; Meyer et al., 2014; L. A. Nguyen, Wang, & Steitz, 2017; Qu et al., 2016; Suslov et al., 2015; C. Zhao, Rajashankar, Marcia, & Pyle, 2015; also reviewed by Pyle, 2016; Ren, Micura, & Patel, 2017), and viral elements (Akiyama et al., 2016; Au et al., 2015; Imai, Kumar, Hellen, D'Souza, & Wagner, 2016; Keane et al., 2015). Additionally, recent studies have identified highly structured ncRNAs as important regulators in various cellular processes and linked misregulation of ncRNAs to important human diseases (Esteller, 2011). For these reasons, ncRNAs are attracting increased attention in both drug design and intermolecular recognition (Warner, Hajdin, & Weeks, 2018). Structural study of these important RNAs is becoming increasingly necessary to understand how highly structured RNAs function and interact with other RNAs, proteins, and small molecule ligands. Elucidation of three‐dimensional structures and conformational landscapes provides detailed information of their regulatory mechanisms.
In this review, we will give a broad overview of different methods for structure determination and discuss some recent advancements in both sample preparation and data acquisition that have advanced the study of large RNAs by nuclear magnetic resonance (NMR) spectroscopy.
2. COMMON METHODS FOR RNA STRUCTURE DETERMINATION
2.1. Chemical probing
First developed more than 30 years ago, chemical probing of RNA can be a useful tool to rapidly gain nucleotide‐level structural information (Ehresmann et al., 1987; Peattie & Gilbert, 1980; Stern, Moazed, & Noller, 1988). Base‐pairing, hydrogen bonding, nucleotide accessibility and other secondary structure‐related characteristics, can be obtained by treating RNAs with specific chemical reagents (typically dimethyl sulfate [DMS] and/or one or more SHAPE reagents) and probing for the sites of modification by sequencing (Cordero, Kladwang, VanLang, & Das, 2012; Merino, Wilkinson, Coughlan, & Weeks, 2005; Tian & Das, 2016; Weeks, 2010). Reactivity data derived from chemical probing studies can be converted to pseudo free energy terms and then incorporated into a secondary structure prediction algorithms to bolster the quality of the resulting structure predictions (Cordero et al., 2012; Deigan, Li, Mathews, & Weeks, 2009; Reuter & Mathews, 2010). Chemical probing remains the best method for high‐throughput analysis of secondary structure within large (even genome‐wide) RNAs (Bevilacqua, Ritchey, Su, & Assmann, 2016; Watts et al., 2009; Zubradt et al., 2017). However, chemical probing provides limited insight into RNA tertiary structure (Homan et al., 2014) and only approximations of fast time scale dynamics of the molecule (Gherghe, Shajani, Wilkinson, Varani, & Weeks, 2008).
2.2. X‐ray crystallography
Biomolecular structures at atomic resolution provide key insights into the molecular determinants of interactions, the mechanisms of catalysis, and molecular topology, among others. Generally, X‐ray crystallography is the most widely used method of structure determination for biological macromolecules. Indeed, protein structure elucidation has been overwhelmingly conducted using X‐ray crystallography, with approximately 90% of protein structures having been determined using X‐ray crystallography (Berman et al., 2000; http://www.rcsb.org). Crystallography, as the name implies, requires the formation of an ordered crystal capable of diffracting X‐rays. This necessitates conformational homogeneity within the crystal lattice. Therefore, X‐ray crystallography has proven very useful for those RNA molecules that adopt relatively rigid conformation, for instance, riboswitches and catalytic ribozymes (Liberman & Wedekind, 2012; Marcia & Pyle, 2012; Pyle, 2016; Roth & Breaker, 2009; Toor, Keating, Taylor, & Pyle, 2008; Toor, Rajashankar, Keating, & Pyle, 2008). However, many RNA molecules, particularly larger RNAs, are flexible and may sample multiple conformations and thus less likely to crystallize in the absence of stabilizing cofactors like protein binding partners. X‐ray crystallography has therefore played an important, but significantly smaller (approximately 60% of RNA structures were determined using X‐ray crystallography) role in the three‐dimensional structure determination of RNAs relative to proteins (Berman et al., 2000; http://www.rcsb.org).
2.3. Cryo‐electron microscopy
Single particle cryo‐electron microscopy (cryo‐EM) is a powerful technique for structure elucidation of large biomolecules and biomolecular machines, and recent technological advances have catapulted this technique to the forefront of structural biology (Kuhlbrandt, 2014a, 2014b; Nogales & Scheres, 2015; Vinothkumar & Henderson, 2016). Cryo‐EM importantly allows for the visualization of dynamic biological processes by direct visualization of biomolecules with multiple structural states (Dong et al., 2019; Nakanishi, Kishikawa, Tamakoshi, Mitsuoka, & Yokoyama, 2018; Roh et al., 2017). However, to date, cryo‐EM has largely been applied to protein only or protein‐nucleic acid complexes (e.g., the ribosome and spliceosome). Cryo‐EM has been used to study the overall size and shape of very large RNAs, reporting at very low resolution on secondary structure and branching pattern of these RNAs (Garmann et al., 2015; Gopal, Zhou, Knobler, & Gelbart, 2012). There are a few structures of relatively small RNAs derived from cryo‐EM data, (Baird et al., 2010; Zhang et al., 2018) but these low resolution density maps harbor limited structural detail requiring higher resolution models determined via other methods to glean mechanistic insight. A recent software developed by the Das lab facilitates the building of RNA coordinates into low resolution cryo‐EM density (Kappel et al., 2018). While this technique is described for protein‐RNA complex datasets, it is likely applicable to RNA‐only density maps.
2.4. NMR spectroscopy
NMR spectroscopy is another common method for determining RNA structure at atomic resolution. NMR spectroscopy is particularly well‐suited for the study of biomolecules that are not prone to crystallization or those which require specific sample conditions to maintain the structure. Compared to proteins, NMR spectroscopy has played a large role in the structure determination of RNA molecules, roughly 40% of RNA structures have been solved using this method (Berman et al., 2000; http://www.rcsb.org). Importantly, NMR spectroscopy provides far more than just structural information, as it allows the dynamics of the biomolecule to be studied as well.
While NMR is a powerful tool for studying RNA structure and dynamics at the atomic level, there are some very real challenges/limitations when using NMR spectroscopy to determine the structure and dynamics of large RNAs. (a) Labor intensive. Structure elucidation by NMR spectroscopy requires that most resonances are assigned before beginning any structural analysis. This is a time‐consuming process both as it relates to data acquisition and data analysis. (b) Signal overlap. RNAs are built from four nucleotides which have similar chemical structures. This results in a lack of dispersion in the chemical shifts (also known as signal degeneracy). (c) Unfavorable relaxation properties. Large RNA molecules tumble very slowly in solution leading to severe broadening of signals and significant losses in signal intensity. These challenges have generally restricted the size of RNA that can be studied by NMR spectroscopy. Of the RNA structures determined by NMR spectroscopy, the average size is 30 nucleotides with only six structures of RNA larger than 100 nucleotides (nt) (Figure 1) (Berman et al., 2000; http://www.rcsb.org).
Figure 1.
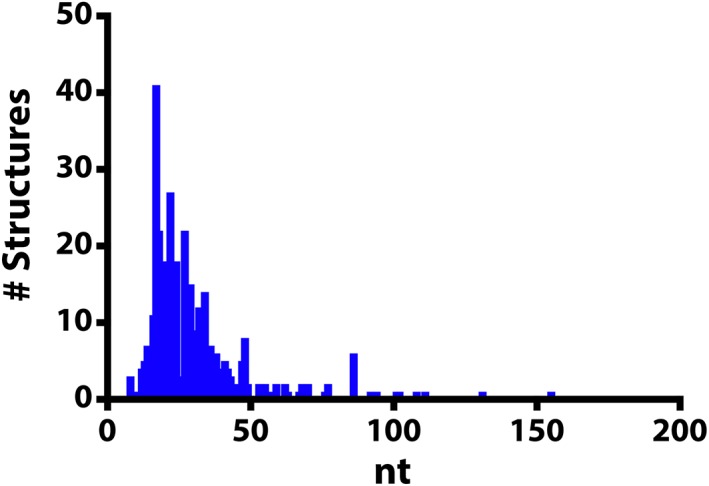
Histogram of RNA structures that were determined using nuclear magnetic resonance spectroscopy. The size of the RNA molecule (nucleotides, nt) is correlated with the number of structures reported in the Protein Data Bank (http://rcsb.org, June 2018)
While solution NMR spectroscopy has a number of technical challenges, it also has some distinct advantages. (a) Tertiary structure information. While secondary structure is importantly represented in the three‐dimensional structure of RNAs, the tertiary structure provides key insights into the function of the RNA. (b) Study the native conformation of an RNA. Because NMR is not predicated on growing crystals of the sample, there are no concerns that the structure might be affected by crystal packing or the conditions under which crystals can be grown. (c) Access to information about dynamics. While a number of biophysical techniques can be applied to study dynamic processes in RNA molecules (Hao et al., 2018; P. Nguyen & Qin, 2012; Panja, Hua, Zegarra, Ha, & Woodson, 2017; Ritchie & Woodside, 2015), NMR is uniquely poised to study the dynamics of biomolecules at atomic resolution and over a wide range of timescales (Al‐Hashimi & Walter, 2008; Marion, 2013).
Structure determination by NMR spectroscopy is first and foremost predicated on having assignments for all (or most) atoms within the molecule. Chemical shift assignments can be made via analysis of a variety of data and are useful in the analysis of nuclear Overhauser effect (NOE) distance restraints, among others. The experimental restraints can then be combined with empirical restraints to generate initial geometry‐based structural models. These initial structural models can then be refined using a simulated annealing strategy to generate an ensemble of 20 lowest energy conformers. In order to simplify, or in some cases make possible, the analysis and collection of data for very large RNAs, a number of both methodological and technical improvements have recently been developed. Some of the key contributions are highlighted below.
3. SAMPLE PREPARATION STRATEGIES TO AID IN STRUCTURAL DETERMINATION OF LARGE RNAs
3.1. Deuterium labeling
Because of the limited chemical shift dispersion that RNAs exhibit, NMR spectra of unlabeled large RNAs are extremely crowded, prohibiting unambiguous resonance assignment (Figure 2a). There are a number of ways to “edit” a spectrum, separating chemical shifts by a second NMR‐active nucleus (typically 15N or 13C). However, the large 13C‐1H dipolar coupling has a significant effect on T2 relaxation mechanisms leading to broad line widths thereby limiting the sensitivity of heteronuclear correlation experiments, particularly for large RNAs that have large rotational correlation times (A. L. Hansen & Al‐Hashimi, 2006; Lu, Miyazaki, & Summers, 2010; Tolbert et al., 2010).
Figure 2.
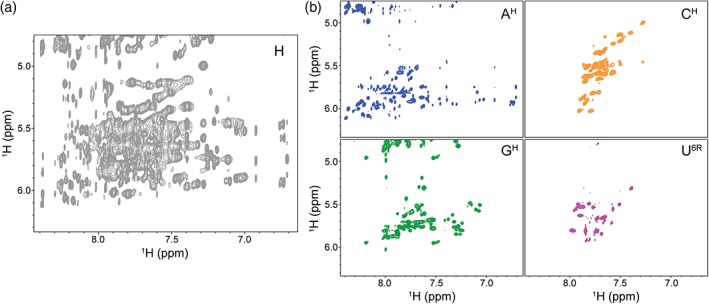
Deuterium labeling greatly improves spectral quality and simplicity. (a) 1H‐1H NOESY spectrum of the full‐protiated (H, unlabeled) 155 nucleotides HIV‐1 core encapsidation signal (Keane et al., 2015) is characterized by severe signal overlap and broad line widths. (b) Selective deuteration of the sample results in spectra of significantly higher quality. There is minimal signal overlap and resonances are now effectively identifiable by the respective labeling scheme. Four different labeling schemes are shown: AH (adenosines protiated, cytosines, guanosines, and uracils deuterated), CH (cytosines protiated, adenosines, guanosines, and uracils deuterated), GH (guanosines protiated, adenosines, cytosines, and uracils deuterated), U6R (uracils protiated on the ribose and at C5, adenosines, cytosines, and guanosines deuterated)
One attractive alternative to the traditional heteronuclear experiments is to “edit” a spectrum by the nucleotide‐specific incorporation of deuterium atoms throughout the RNA. Using this approach, protons are replaced with deuterium and because deuterium resonates at a different frequency than protons, the signals are not observed in a proton detected 1H‐1H NOESY spectrum. The spectral complexity is therefore significantly reduced (Figure 2b). In addition to the reduced spectral overlap, incorporation of deuterium limits pathways for 1H‐1H spin diffusion resulting in narrower line widths. Because this method relies on removing signals rather than separating the signals into a second or third dimension, no one sample contains sufficient structural information (Figure 2b) and many different samples, each with a unique labeling scheme, must be prepared to obtain full sequence coverage of the RNA. This approach is labor intensive and requires considerable resources and spectrometer time. Nevertheless, this method is universally applicable and has been used to study a variety of systems including viral RNAs (D'Souza, Dey, Habib, & Summers, 2004; D'Souza & Summers, 2004; Keane et al., 2015; Lu et al., 2011; Miller, Yildiz, Lo, Wang, & D'Souza, 2014) and tetraloop‐receptor complexes (Davis et al., 2005). Notably, this approach was recently used to probe the structure of the 230 kDa HIV 5′ leader and provide insight into its dimerization mechanism (Keane et al., 2016). Partially or fully deuterated ribonucleotide triphosphates (rNTPs) can be purchased or produced in the laboratory using either chemical or enzymatic methods (Huang, Yu, LeProust, & Gao, 1997; Lu et al., 2010; Scott, Tolbert, & Williamson, 2000). rNTPs may be deuterated at nonexchangeable positions within the base and/or the ribose and can subsequently be used in the enzymatic synthesis of RNA using T7 RNA polymerase in place of fully protiated rNTPs (Batey, Battiste, & Williamson, 1995; Batey, Inada, Kujawinski, Puglisi, & Williamson, 1992; Scott et al., 2000).
3.2. Incorporation of 15N and 13C labels
In much the same way that partially or fully deuterated rNTPs can be incorporated into an RNA using in vitro transcription, so too can other common NMR‐active isotopes. Isotopically enriched 13C and 15N rNTPs have widely been used in NMR experiments to study RNA structure, dynamics, and ligand binding (Batey et al., 1995; Nikonowicz et al., 1992). Traditionally, these studies have been applied to relatively small RNAs. The ability to site‐specifically incorporate 13C labels within an RNA ribose and/or base makes NMR spectroscopy a powerful tool for dynamics studies of large RNAs, expanding the types of experiments that can be conducted and simplifying analysis (see data acquisition and analysis advancements below). Compared to uniform 13C incorporation, site specific incorporation of 13C labels allows for reduced spectral crowding while eliminating the strong 13C‐13C scalar couplings thereby increasing signal‐to‐noise ratios and facilitating direct carbon detection experiments (Alvarado et al., 2014; Longhini et al., 2016; Lukavsky & Puglisi, 2005; Marchant, Bax, & Summers, 2018). Drawing inspiration from earlier synthetic strategies (SantaLucia, Shen, Cai, Lewis, & Tinoco, 1995), the Dayie lab has developed a chemo‐enzymatic method to produce isotopically enriched pyrimidines (Alvarado et al., 2014) and purines (Longhini et al., 2016) with different 13C and/or 15N patterns. The incorporation of site specifically labeled nucleotides into large RNAs strongly reduced spectral crowding and removed 13C‐13C J‐coupling, allowing study of large RNAs by TROSY (Pervushin, Riek, Wider, & Wuthrich, 1997) experiments and enabling new method for RNA NOESY assignment with improved signal intensity of C6 and C8 protons. In addition, dynamics studies of RNA could also benefit from using those selectively labeled nucleotides (LeBlanc, Longhini, Tugarinov, & Dayie, 2018).
3.3. Position‐selective labeling of RNA
As discussed above, the uniform incorporation of specifically labeled nucleotides can greatly facilitate NMR studies of large RNAs. However, the ability to incorporate a labeled nucleotide in a position‐specific rather than nucleotide‐specific manner could also be advantageous. Some complementary biophysical techniques, like fluorescence resonance energy transfer and electron paramagnetic resonance spectroscopy, require labeling at specific positions rather than uniformly at a nucleotide type throughout the whole molecule. Chemical synthesis is commonly used to make RNAs with position‐specific labeling; however, it is generally limited to relatively small RNAs (<60 nt) (Paredes, Evans, & Das, 2011). It is possible to enzymatically ligate several smaller synthetic RNAs together; however, the low efficiency of the ligation reactions limits the overall yield of full‐length product. In order to circumvent these issues, the Wang lab developed a hybrid solid‐solution phase transcription method, position‐selective labeling of RNA (PLOR), that can theoretically synthesize any RNA with labeling at any specific position using commercially available nucleotides (Liu et al., 2015, 2016, 2018). The biggest advantage of this approach over other methods is that it can prepare large selectively labeled RNA samples at a scale appropriate for NMR studies (Liu et al., 2016). The use of PLOR to position‐specifically incorporate a labeled nucleotide into a large RNA has many potential benefits including breaking ambiguity in resonance assignments and facilitating the identification of long‐range interactions.
4. FRAGMENTATION METHODS
A very common approach to tackling resonance assignments of large RNAs is a “divide and conquer” strategy, where the large RNA (Figure 3a) is broken down into individual folded domains (Figure 3b) (Imai et al., 2016; Keane et al., 2015; Ziegeler, Cevec, Richter, & Schwalbe, 2012). These small, individual domains are amenable to rapid data collection and resonance assignment. The assignment data can then be “grafted” to the full‐length data to help deconvolute the more complicated data. While this approach works in many situations, it is dependent on the fragments adopting the same structure in isolation as they do in the full‐length construct. It is possible for the fragments to not fold properly, or to be poor representations of the region in the larger context. However, we generally find this approach exceptionally helpful in the early stages of full‐length resonance assignments.
Figure 3.
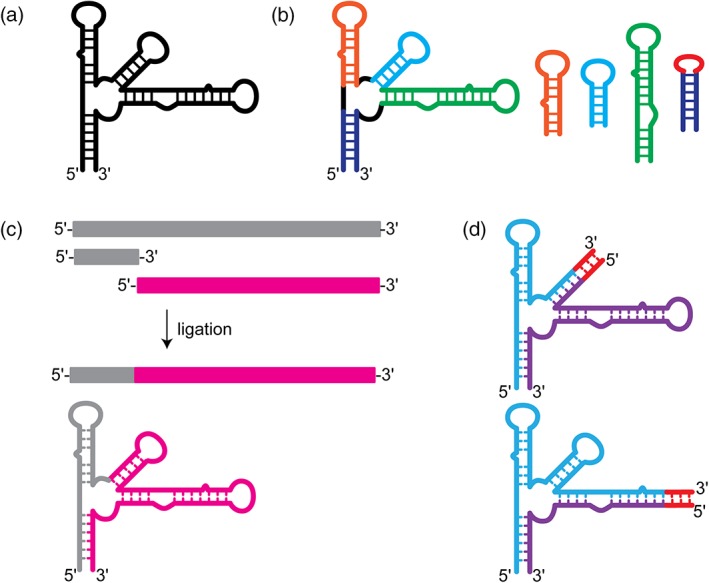
Methods for fragmentation or segmental labeling of large RNAs. (a) Secondary structure of a large RNA with a complex multi‐helix junction. (b) “Divide and conquer” approach. Small oligo RNAs are designed to mimic the structure of particular regions within the large RNA. These oligo RNAs are well‐suited for rapid chemical shift assignment by traditional methods. (c) Enzymatic ligation allows for segmental labeling of a large RNAs. Two RNAs are independently synthesized and can therefore be independently labeled. The two RNAs can then be ligated together to make a full‐length construct with only one region of the RNA containing nuclear magnetic resonance‐active isotope labeling. (d) Fragmentation‐based segmental labeling. The RNA of interest is fragmented at a hairpin loop. The loop sequence is replaced (in some cases) with a run of intermolecular C–G base pairs which serve as an annealing handle. This approach in theory can be applied at any hairpin loop structure within a large RNA molecules
Another approach that is widely used is an enzymatic ligation approach using either T4 DNA or RNA ligase to join either chemically synthesized or in vitro transcribed RNAs with different labeling strategies (Figure 3c) (Duss, Maris, von Schroetter, & Allain, 2010; I. Kim, Lukavsky, & Puglisi, 2002; Lu et al., 2011; Nelissen et al., 2008; Tzakos, Easton, & Lukavsky, 2007; Xu, Lapham, & Crothers, 1996). These methods require extensive optimization and troubleshooting and are notorious for low yield of product, although significant improvements have been made to improve the yield, eliminate specific sequence requirements, and streamline the process (Duss et al., 2010). The Allain lab developed a robust and versatile segmental labeling method that uses ribozymes and RNaseH cleavage to generate fragments of RNA for subsequent ligation. This approach was demonstrated on the 72 nucleotide RsmZ RNA, which was fragmented into four components for reassembly in NMR spectral analysis (Duss et al., 2010). This method offers several distinct advantages over other methods for segmental labeling of RNAs. First, this method is flexible—new ligation sites can be introduced by performing the RNaseH cleavage with a new splint‐directed site. Second, only two pools of full‐length RNA, isotopically labeled and unlabeled, are needed—cloning of subfragments is not necessary. Finally, the cleavage and ligation strategy are efficient even for highly structured RNAs, making this approach virtually universal.
One alternative to traditional enzymatic‐based segmental labeling strategies is a fragmentation‐based approach that relies on annealing rather than ligation to join the fragments together (Figure 3d). This approach was pioneered in the Summers lab and was a requisite for the unambiguous identification of an unexpected long‐range base pairing interaction within the HIV‐1 packaging signal (Keane et al., 2015). In this work, the authors replaced a hairpin loop with a run of intermolecular C–G base pairs, fragmenting the RNA into two discrete pieces that could be transcribed (and therefore isotopically labeled) independently and annealed together post purification (Keane et al., 2015). A similar approach was used to study a 122 nucleotide transcriptional intermediate of the type I‐A 2′‐deoxyguanosine‐sensing transcriptional riboswitch (Helmling et al., 2017). Here, a similar approach was used, a break in the sequence was introduced by removing a loop; however, a G–C handle was not included and may not be necessary for all sequences (Helmling et al., 2017). This approach is versatile as a fragmentation site can in principle be incorporated at the loop of any hairpin element, which are ubiquitous elements in RNA structures. A caveat to this approach is that individual fragments might form a stable structure that inhibits intermolecular base pairing. This approach might not work for all RNAs or at all hairpin loops and therefore must be rigorously tested to ensure proper formation of the native structure.
5. DATA ACQUISITION AND ANALYSIS ADVANCEMENTS
5.1. Detection of hydrogen bonding
One of the most telling characteristics of RNA structure is the hydrogen bonding pattern, the base pairing within the molecule. An NMR spectrum that reports on the base pairing within an RNA can serve as a “fingerprint” of the molecule. Traditionally, hydrogen bonding within small RNAs has been inferred through NOE (through space) correlations of imino protons (Varani, Aboulela, & Allain, 1996; Wüthrich, 1986). Hydrogen bonding can also be identified directly using a scalar coupling HNN‐COSY (through bond) experiment (Dingley & Grzesiek, 1998; Pervushin et al., 1998). Additional improvements in sensitivity can be gained by implementation of longitudinal‐relaxation‐enhancement techniques such as BEST‐HNN‐COSY experiments which selectively excite only desired nuclei which shortens experimental acquisition time and improves signal:noise (Farjon et al., 2009). However, all of these experiments rely on the detection of the imino proton, a proton that is readily exchangeable with solvent and subject to conformational dynamics at elevated temperatures (Gao & Patel, 1987; J. H. Lee, Jucker, & Pardi, 2008; Wagner, Rinnenthal, Narberhaus, & Schwalbe, 2015). The imino protons of transient base pairs can be broadened beyond detection due to exchange with solvent therefore these experiments are typically conducted at temperatures below room temperature (typically around 10 °C). Large RNAs are generally not amenable to study at low temperature due to the slower molecular tumbling. Nevertheless, imino assignments have been made for several large RNAs including the 111 nt U2/U6 snRNA complex (Burke, Sashital, Zuo, Wang, & Butcher, 2012), a 67 nt region of the encephalomyocarditis virus IRES (Imai et al., 2016), and the 112 nt adenine riboswitch (Reining et al., 2013).
The Sattler lab made important improvements to the hydrogen bond correlation experiments, utilizing band‐selective pulses coupled with BEST relaxation enhancement to enable detection of hydrogen bonds in dynamic regions of RNA (Dallmann et al., 2013). These methodological improvements allow for work at high temperature without exchange concerns, because the detection occurs via nonexchangeable protons (Figure 4) (Dallmann et al., 2013).
Figure 4.
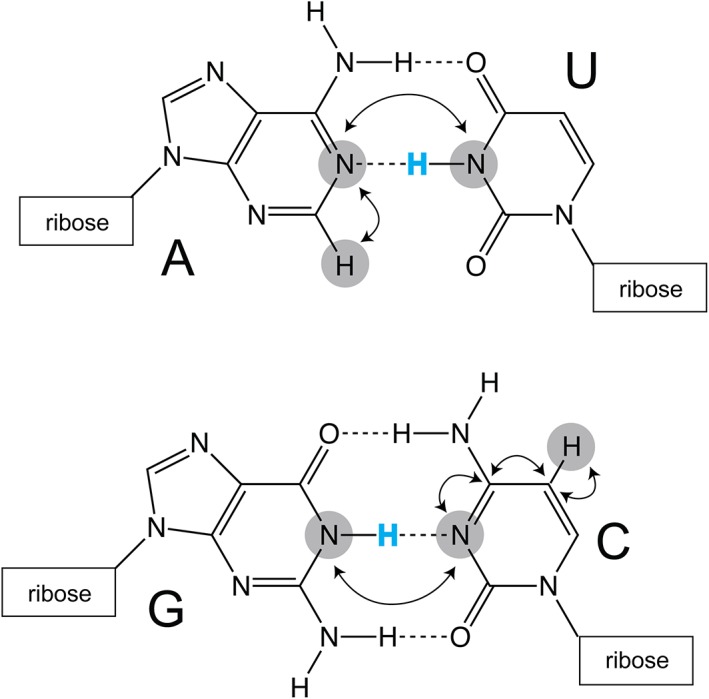
Detection of hydrogen bonds in canonical A–U and G–C base pairs. Traditionally, imino protons (blue) are used to infer base pairing within helical structural elements. Improvements to pulse sequences, implemented by the Sattler lab, allow for the detection of hydrogen‐bonding on nonexchangeable C2 (adenosine) and C5 (cytosine and uracil) protons (Dallmann et al., 2013). Magnetization transfer pathways are indicated with arrows
5.2. Global orientation of helical elements
Distance restraints (which are obtained via the analysis of NOE spectroscopy experiments) provide high‐resolution local structural information, however cannot generally define the relative orientation of secondary structure elements within a large RNA. Residual dipolar couplings (RDCs) provide important orientation restraints and inclusion of these restraints in structure refinement greatly improves the overall accuracy of the structure (Bermejo, Clore, & Schwieters, 2016). RDC measurements are conducted both in the absence and presence of an alignment media (Pf1 phage is common for RNA samples) (Clore, Starich, & Gronenborn, 1998; M. R. Hansen, Hanson, & Pardi, 2000), and are measured on a variety of carbon–hydrogen and/or nitrogen–hydrogen correlations within the base and ribose (M. R. Hansen, Mueller, & Pardi, 1998; Yan, Corpora, Pradhan, & Bushweller, 2002; Zidek, Wu, Feigon, & Sklenar, 2001). RDCs provide important restraints in RNA structure determination; however, slightly fewer than half of NMR‐derived medium to large RNA structures (RNAs larger than 50 nt) used RDC measurements in structure refinement (Au et al., 2015; Barnwal et al., 2016; Burke et al., 2012; Cornilescu et al., 2016; D'Souza et al., 2004; Davis et al., 2005; Houck‐Loomis et al., 2011; Jain, Morgan, Rife, Salemi, & Tolbert, 2016; Kang, Eichhorn, & Feigon, 2014; N. K. Kim, Zhang, & Feigon, 2014; Lukavsky, Kim, Otto, & Puglisi, 2003; Lukavsky & Puglisi, 2005; Miller et al., 2014; Ulyanov et al., 2006). The incorporation of 13C into large RNAs results in severe line broadening, limiting the usefulness of this approach for the study of larger RNAs. Marchant et al. developed a new method for recording RDC measurements in large RNAs that capitalizes on the large chemical shift dispersion and narrow line widths of adenosine C2 protons (Marchant et al., 2018). This method requires the preparation of RNA with 15N‐adenosine and uses a variable flip angle HMQC to obtain RDC measurements and was applied to the study of a 232 nt element corresponding to the HIV‐1 Reve response element (Marchant et al., 2018).
5.3. RNA dynamics
Dynamic conformational exchange is important for the function of regulatory RNAs in molecular processes (Al‐Hashimi & Walter, 2008; Dethoff, Chugh, Mustoe, & Al‐Hashimi, 2012; Xue et al., 2015). One major strength of NMR spectroscopy is the ability to capture dynamics information, reporting on the local conformational fluctuations at single nucleotide resolution (Bothe et al., 2011; Dethoff, Chugh, et al., 2012). Conformational dynamics of macromolecules occurs over a broad range of time scale, from picosecond to second, and formation of new base pairs in RNA conformational exchange lies in the scale of microsecond to second (Bothe et al., 2011; Rinnenthal et al., 2011). Numerous NMR methods have been developed to investigate biomolecular dynamics at different time scales (Bailor et al., 2007; Kloiber, Spitzer, Tollinger, Konrat, & Kreutz, 2011; Vallurupalli, Sekhar, Yuwen, & Kay, 2017; B. Zhao, Hansen, & Zhang, 2014). Here, we will highlight three approaches for the quantification of conformational dynamic exchange in RNA molecules.
The chemical shift saturation transfer (CEST) NMR experiments, which utilize magnetization “interaction” of one state with another state of molecules, has been developed to look into slow conformational exchange of biomolecules (reviewed in Vallurupalli et al., 2017). Recently, the Zhang lab optimized carbon CEST experiments for nucleic acids and applied these methods as well as R1ρ experiments (see below) to the study of a 47 nt fluoride riboswitch (B. Zhao et al., 2014). In an effort to fully characterize riboswitch function, structural insight into the ligand free state is essential. Their work revealed that the unliganded riboswitch formed a pseudoknot‐like structure that is lowly populated, reminiscent of the ligand‐bound structure, and was undetectable via conventional approaches (B. Zhao et al., 2014). CEST experiments do not require specialized isotopic labeling strategies and can be conducted on RNAs that are uniformly 15N/13C labeled.
Relaxation dispersion (RD) NMR experiments are frequently used to characterize excited states (ESs) of biomolecules (reviewed in Al‐Hashimi, 2013; Bothe et al., 2011; Xue et al., 2015). One such RD experiment is the R1ρ experiment (reviewed in Bothe et al., 2011; Palmer & Massi, 2006). R1ρ experiments can provide structural and functional insight for low‐populated ESs that have short lifetimes. This approach was used to characterize the dynamics of a 31 nt portion of ribosomal RNA, the A‐site, which must dynamically sample multiple conformations to decode a messenger RNA (Dethoff, Petzold, Chugh, Casiano‐Negroni, & Al‐Hashimi, 2012; A. L. Hansen, Nikolova, Casiano‐Negroni, & Al‐Hashimi, 2009). R1ρ experiments have also provided insight into the roles of structured RNA elements within the HIV‐1 genome, including the transactivation response (TAR) element apical loop and the primary dimer initiation site (SL1) (Dethoff, Petzold, et al., 2012). The Al‐Hashimi lab characterized the ES of the 14 nt TAR RNA apical loop, in which many of the loop nucleotides are sequestered in base pairing (Dethoff, Petzold, et al., 2012). The sequestration of loop nucleotides in the TAR ES inhibits binding by the viral transactivator protein and human cyclin T1, activating HIV‐1 genome transcription (Dethoff, Petzold, et al., 2012). The 36 nt SL1 exhibited significant conformational exchange, consistent with not one, but two excited state structures and the authors propose that these ESs play important roles in intermolecular dimerization (Dethoff, Petzold, et al., 2012). Similar to CEST experiments, R1ρ experiments can be carried out using uniformly 15N/13C‐labeled or unlabeled (A. L. Hansen et al., 2009) RNAs but have thus far been applied to the study of relatively small RNAs.
Another common RD‐based experiment used to characterize protein and nucleic acid dynamics in the microsecond‐to‐millisecond timescale is the Carr–Purcell–Meiboom–Gill (CPMG) experiment (Yamazaki, Muhandiram, & Kay, 1994). CPMG can help extract parameters like population, life time and chemical shifts of ES in slow and intermediate exchange. Extensive studies have been conducted in protein dynamics by CPMG, but in nucleic acids CPMG study is highly limited due to the intrinsic characteristics of DNA and RNA molecules, such as 13C‐13C J coupling and signal degeneracy (Johnson & Hoogstraten, 2008; Lundstrom, Hansen, & Kay, 2008). Recently, using nucleotides that are selectively labeled with 13C and/or 15N at specific atom positions (as described above), CPMG was used to study the dynamics of relatively large RNAs including the 59 nt frame‐shifting element from a human coronavirus (Longhini et al., 2016). The combination of these newly optimized NMR experiments as well as efficient synthesis of position‐selective labeled nucleotides, conformational exchange dynamics of larger RNAs will become less challenging and novel mechanisms of RNA regulation may be unveiled.
5.4. Validation and automation of chemical shift assignments
One of the main hindrances of NMR analysis of large RNAs is the arduous task of assigning chemical shifts. Chemical shift assignment is a prerequisite for any quantitative analysis of NMR data for either structure determination or dynamics studies. In an effort to establish the sequence and structural dependence on chemical shift, Johnson and colleagues analyzed the nonexchangeable H8, H2, H6, H5, H1′, H2′, and H3′ chemical shifts for RNAs that have been deposited in the publicly available Biological Magnetic Resonance Data Bank and have associated structure coordinates (Barton, Heng, Johnson, & Summers, 2013; Brown, Summers, & Johnson, 2015). Here, they sought to identify structural features of a given base pair triplet and examined how the chemical shift of a given nuclei is affected by changes in the local structural environment. These database prediction tools can facilitate the validation of experimentally derived assignments and aid in the manual assignment of NMR data collected on large RNA molecules.
6. OUTLOOK—HYBRID METHODOLOGIES
While advancements have been made to facilitate the study of large RNA structure and dynamics by NMR spectroscopy, complimentary structural techniques have proven incredibly useful for providing additional global structural restraints. Small‐angle X‐ray scattering (SAXS) has been widely used in combination with NMR spectroscopy to refine NMR‐derived structural ensembles (Burke et al., 2012; Cornilescu et al., 2016; Grishaev, Ying, Canny, Pardi, & Bax, 2008; Imai et al., 2016; Jain et al., 2016; Zuo et al., 2010). In fact, studies show that SAXS can be combined with sparse RDC restraints to accurately refine idealized/modeled RNA structures (Grishaev et al., 2008; Wang et al., 2009). Another approach is to refine NMR structures with a cryo‐electron microscopy density map. This approach has been applied to both relatively large and small RNAs (Gong, Schwieters, & Tang, 2015; Miyazaki et al., 2010; Zhang et al., 2018) and will likely be a necessary complement to structural studies as the size of the RNA being investigated increases.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLES
https://doi.org/10.1002/wrna.1482
ACKNOWLEDGMENTS
We are grateful to Dr Xiao Heng and Dr Michael F. Summers for careful reading of this manuscript. This work was supported by the University of Michigan.
Zhang H, Keane SC. Advances that facilitate the study of large RNA structure and dynamics by nuclear magnetic resonance spectroscopy. WIREs RNA. 2019;10:e1541. 10.1002/wrna.1541
Funding information University of Michigan
REFERENCES
- Akiyama, B. M. , Laurence, H. M. , Massey, A. R. , Costantino, D. A. , Xie, X. , Yang, Y. , … Kieft, J. S. (2016). Zika virus produces noncoding RNAs using a multi‐pseudoknot structure that confounds a cellular exonuclease. Science, 354(6316), 1148–1152. 10.1126/science.aah3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Hashimi, H. M. (2013). NMR studies of nucleic acid dynamics. Journal of Magnetic Resonance, 237, 191–204. 10.1016/j.jmr.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Hashimi, H. M. , & Walter, N. G. (2008). RNA dynamics: It is about time. Current Opinion in Structural Biology, 18(3), 321–329. 10.1016/j.sbi.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado, L. J. , LeBlanc, R. M. , Longhini, A. P. , Keane, S. C. , Jain, N. , Yildiz, Z. F. , … Dayie, T. K. (2014). Regio‐selective chemical‐enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. Chembiochem, 15(11), 1573–1577. 10.1002/cbic.201402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au, H. H. , Cornilescu, G. , Mouzakis, K. D. , Ren, Q. , Burke, J. E. , Lee, S. , … Jan, E. (2015). Global shape mimicry of tRNA within a viral internal ribosome entry site mediates translational reading frame selection. Proceedings of the National Academy of Sciences of the United States of America, 112(47), E6446–E6455. 10.1073/pnas.1512088112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailor, M. H. , Musselman, C. , Hansen, A. L. , Gulati, K. , Patel, D. J. , & Al‐Hashimi, H. M. (2007). Characterizing the relative orientation and dynamics of RNA A‐form helices using NMR residual dipolar couplings. Nature Protocols, 2(6), 1536–1546. 10.1038/nprot.2007.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, N. J. , Ludtke, S. J. , Khant, H. , Chiu, W. , Pan, T. , & Sosnick, T. R. (2010). Discrete structure of an RNA folding intermediate revealed by cryo‐electron microscopy. Journal of the American Chemical Society, 132(46), 16352–16353. 10.1021/ja107492b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwal, R. P. , Loh, E. , Godin, K. S. , Yip, J. , Lavender, H. , Tang, C. M. , & Varani, G. (2016). Structure and mechanism of a molecular rheostat, an RNA thermometer that modulates immune evasion by Neisseria meningitidis . Nucleic Acids Research, 44(19), 9426–9437. 10.1093/nar/gkw584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, S. , Heng, X. , Johnson, B. A. , & Summers, M. F. (2013). Database proton NMR chemical shifts for RNA signal assignment and validation. Journal of Biomolecular NMR, 55(1), 33–46. 10.1007/s10858-012-9683-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R. T. , Battiste, J. L. , & Williamson, J. R. (1995). Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods in Enzymology, 261, 300–322. [DOI] [PubMed] [Google Scholar]
- Batey, R. T. , Inada, M. , Kujawinski, E. , Puglisi, J. D. , & Williamson, J. R. (1992). Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Research, 20(17), 4515–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H. M. , Westbrook, J. , Feng, Z. , Gilliland, G. , Bhat, T. N. , Weissig, H. , … Bourne, P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo, G. A. , Clore, G. M. , & Schwieters, C. D. (2016). Improving NMR structures of RNA. Structure, 24(5), 806–815. 10.1016/j.str.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua, P. C. , Ritchey, L. E. , Su, Z. , & Assmann, S. M. (2016). Genome‐wide analysis of RNA secondary structure. Annual Review of Genetics, 50, 235–266. 10.1146/annurev-genet-120215-035034 [DOI] [PubMed] [Google Scholar]
- Bonasio, R. , & Shiekhattar, R. (2014). Regulation of transcription by long noncoding RNAs. Annual Review of Genetics, 48, 433–455. 10.1146/annurev-genet-120213-092323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe, J. R. , Nikolova, E. N. , Eichhorn, C. D. , Chugh, J. , Hansen, A. L. , & Al‐Hashimi, H. M. (2011). Characterizing RNA dynamics at atomic resolution using solution‐state NMR spectroscopy. Nature Methods, 8(11), 919–931. 10.1038/nmeth.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. D. , Summers, M. F. , & Johnson, B. A. (2015). Prediction of hydrogen and carbon chemical shifts from RNA using database mining and support vector regression. Journal of Biomolecular NMR, 63(1), 39–52. 10.1007/s10858-015-9961-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J. E. , Sashital, D. G. , Zuo, X. , Wang, Y. X. , & Butcher, S. E. (2012). Structure of the yeast U2/U6 snRNA complex. RNA, 18(4), 673–683. 10.1261/rna.031138.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, R. T. , Peters, J. K. , Robart, A. R. , Wiryaman, T. , Rajashankar, K. R. , & Toor, N. (2018). Structural basis for the second step of group II intron splicing. Nature Communications, 9(1), 4676 10.1038/s41467-018-06678-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore, G. M. , Starich, M. R. , & Gronenborn, A. M. (1998). Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod‐shaped viruses. Journal of the American Chemical Society, 120(40), 10571–10572. 10.1021/ja982592f [DOI] [Google Scholar]
- Cordero, P. , Kladwang, W. , VanLang, C. C. , & Das, R. (2012). Quantitative dimethyl sulfate mapping for automated RNA secondary structure inference. Biochemistry, 51(36), 7037–7039. 10.1021/bi3008802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu, G. , Didychuk, A. L. , Rodgers, M. L. , Michael, L. A. , Burke, J. E. , Montemayor, E. J. , … Butcher, S. E. (2016). Structural analysis of multi‐helical RNAs by NMR‐SAXS/WAXS: Application to the U4/U6 di‐snRNA. Journal of Molecular Biology, 428(5 Pt A), 777–789. 10.1016/j.jmb.2015.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M. , Walbott, H. , Monachello, D. , Westhof, E. , & Michel, F. (2016). Crystal structures of a group II intron lariat primed for reverse splicing. Science, 354(6316), aaf9258 10.1126/science.aaf9258 [DOI] [PubMed] [Google Scholar]
- Dallmann, A. , Simon, B. , Duszczyk, M. M. , Kooshapur, H. , Pardi, A. , Bermel, W. , & Sattler, M. (2013). Efficient detection of hydrogen bonds in dynamic regions of RNA by sensitivity‐optimized NMR pulse sequences. Angewandte Chemie (International Ed. in English), 52(40), 10487–10490. 10.1002/anie.201304391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J. H. , Tonelli, M. , Scott, L. G. , Jaeger, L. , Williamson, J. R. , & Butcher, S. E. (2005). RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop‐receptor complex. Journal of Molecular Biology, 351(2), 371–382. 10.1016/j.jmb.2005.05.069 [DOI] [PubMed] [Google Scholar]
- Deigan, K. E. , Li, T. W. , Mathews, D. H. , & Weeks, K. M. (2009). Accurate SHAPE‐directed RNA structure determination. Proceedings of the National Academy of Sciences of the United States of America, 106(1), 97–102. 10.1073/pnas.0806929106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethoff, E. A. , Chugh, J. , Mustoe, A. M. , & Al‐Hashimi, H. M. (2012). Functional complexity and regulation through RNA dynamics. Nature, 482(7385), 322–330. 10.1038/nature10885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethoff, E. A. , Petzold, K. , Chugh, J. , Casiano‐Negroni, A. , & Al‐Hashimi, H. M. (2012). Visualizing transient low‐populated structures of RNA. Nature, 491(7426), 724–728. 10.1038/nature11498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingley, A. J. , & Grzesiek, S. (1998). Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. Journal of the American Chemical Society, 120(33), 8293–8297. 10.1021/ja981513x [DOI] [Google Scholar]
- Dong, Y. , Zhang, S. , Wu, Z. , Li, X. , Wang, W. L. , Zhu, Y. , … Mao, Y. (2019). Cryo‐EM structures and dynamics of substrate‐engaged human 26S proteasome. Nature, 565(7737), 49–55. 10.1038/s41586-018-0736-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, V. , Dey, A. , Habib, D. , & Summers, M. F. (2004). NMR structure of the 101‐nucleotide core encapsidation signal of the Moloney murine leukemia virus. Journal of Molecular Biology, 337(2), 427–442. 10.1016/j.jmb.2004.01.037 [DOI] [PubMed] [Google Scholar]
- D'Souza, V. , & Summers, M. F. (2004). Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature, 431(7008), 586–590. 10.1038/nature02944 [DOI] [PubMed] [Google Scholar]
- Duss, O. , Maris, C. , von Schroetter, C. , & Allain, F. H. (2010). A fast, efficient and sequence‐independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Research, 38(20), e188 10.1093/nar/gkq756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann, C. , Baudin, F. , Mougel, M. , Romby, P. , Ebel, J. P. , & Ehresmann, B. (1987). Probing the structure of RNAs in solution. Nucleic Acids Research, 15(22), 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller, M. (2011). Non‐coding RNAs in human disease. Nature Reviews. Genetics, 12(12), 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Farjon, J. , Boisbouvier, J. , Schanda, P. , Pardi, A. , Simorre, J. P. , & Brutscher, B. (2009). Longitudinal‐relaxation‐enhanced NMR experiments for the study of nucleic acids in solution. Journal of the American Chemical Society, 131(24), 8571–8577. 10.1021/ja901633y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. L. , & Patel, D. J. (1987). NMR studies of A.C mismatches in DNA dodecanucleotides at acidic pH. Wobble A(anti).C(anti) pair formation. The Journal of Biological Chemistry, 262(35), 16973–16984. [PubMed] [Google Scholar]
- Garmann, R. F. , Gopal, A. , Athavale, S. S. , Knobler, C. M. , Gelbart, W. M. , & Harvey, S. C. (2015). Visualizing the global secondary structure of a viral RNA genome with cryo‐electron microscopy. RNA, 21(5), 877–886. 10.1261/rna.047506.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe, C. M. , Shajani, Z. , Wilkinson, K. A. , Varani, G. , & Weeks, K. M. (2008). Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. Journal of the American Chemical Society, 130(37), 12244–12245. 10.1021/ja804541s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Z. , Schwieters, C. D. , & Tang, C. (2015). Conjoined use of EM and NMR in RNA structure refinement. PLoS One, 10(3), e0120445 10.1371/journal.pone.0120445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal, A. , Zhou, Z. H. , Knobler, C. M. , & Gelbart, W. M. (2012). Visualizing large RNA molecules in solution. RNA, 18(2), 284–299. 10.1261/rna.027557.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishaev, A. , Ying, J. , Canny, M. D. , Pardi, A. , & Bax, A. (2008). Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data. Journal of Biomolecular NMR, 42(2), 99–109. 10.1007/s10858-008-9267-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil, S. , & Esteller, M. (2015). RNA‐RNA interactions in gene regulation: The coding and noncoding players. Trends in Biochemical Sciences, 40(5), 248–256. 10.1016/j.tibs.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Hansen, A. L. , & Al‐Hashimi, H. M. (2006). Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: Towards new long‐range orientational constraints. Journal of Magnetic Resonance, 179(2), 299–307. 10.1016/j.jmr.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Hansen, A. L. , Nikolova, E. N. , Casiano‐Negroni, A. , & Al‐Hashimi, H. M. (2009). Extending the range of microsecond‐to‐millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R‐1 rho NMR spectroscopy. Journal of the American Chemical Society, 131(11), 3818–3819. 10.1021/ja8091399 [DOI] [PubMed] [Google Scholar]
- Hansen, M. R. , Hanson, P. , & Pardi, A. (2000). Pf1 filamentous phage as an alignment tool for generating local and global structural information in nucleic acids. Journal of Biomolecular Structure & Dynamics, 17, 365–369. 10.1080/07391102.2000.10506642 [DOI] [PubMed] [Google Scholar]
- Hansen, M. R. , Mueller, L. , & Pardi, A. (1998). Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nature Structural Biology, 5(12), 1065–1074. 10.1038/4176 [DOI] [PubMed] [Google Scholar]
- Hao, Y. , Bohon, J. , Hulscher, R. , Rappe, M. C. , Gupta, S. , Adilakshmi, T. , & Woodson, S. A. (2018). Time‐resolved hydroxyl radical footprinting of RNA with X‐rays. Current Protocols in Nucleic Acid Chemistry, 73(1), e52 10.1002/cpnc.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmling, C. , Wacker, A. , Wolfinger, M. T. , Hofacker, I. L. , Hengesbach, M. , Furtig, B. , & Schwalbe, H. (2017). NMR structural profiling of transcriptional intermediates reveals riboswitch regulation by metastable RNA conformations. Journal of the American Chemical Society, 139(7), 2647–2656. 10.1021/jacs.6b10429 [DOI] [PubMed] [Google Scholar]
- Homan, P. J. , Favorov, O. V. , Lavender, C. A. , Kursun, O. , Ge, X. , Busan, S. , … Weeks, K. M. (2014). Single‐molecule correlated chemical probing of RNA. Proceedings of the National Academy of Sciences of the United States of America, 111(38), 13858–13863. 10.1073/pnas.1407306111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck‐Loomis, B. , Durney, M. A. , Salguero, C. , Shankar, N. , Nagle, J. M. , Goff, S. P. , & D'Souza, V. M. (2011). An equilibrium‐dependent retroviral mRNA switch regulates translational recoding. Nature, 480(7378), 561–564. 10.1038/nature10657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Yu, P. , LeProust, E. , & Gao, X. (1997). An efficient and economic site‐specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Research, 25(23), 4758–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, S. , Kumar, P. , Hellen, C. U. , D'Souza, V. M. , & Wagner, G. (2016). An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nature Structural & Molecular Biology, 23(9), 859–864. 10.1038/nsmb.3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, N. , Morgan, C. E. , Rife, B. D. , Salemi, M. , & Tolbert, B. S. (2016). Solution structure of the HIV‐1 intron splicing silencer and its interactions with the UP1 domain of heterogeneous nuclear ribonucleoprotein (hnRNP) A1. The Journal of Biological Chemistry, 291(5), 2331–2344. 10.1074/jbc.M115.674564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J. E., Jr. , & Hoogstraten, C. G. (2008). Extensive backbone dynamics in the GCAA RNA tetraloop analyzed using 13C NMR spin relaxation and specific isotope labeling. Journal of the American Chemical Society, 130(49), 16757–16769. 10.1021/ja805759z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C. P. , & Ferre‐D'Amare, A. R. (2017). Long‐range interactions in riboswitch control of gene expression. Annual Review of Biophysics, 46, 455–481. 10.1146/annurev-biophys-070816-034042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, M. , Eichhorn, C. D. , & Feigon, J. (2014). Structural determinants for ligand capture by a class II preQ1 riboswitch. Proceedings of the National Academy of Sciences of the United States of America, 111(6), E663–E671. 10.1073/pnas.1400126111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel, K. , Liu, S. , Larsen, K. P. , Skiniotis, G. , Puglisi, E. V. , Puglisi, J. D. , … Das, R. (2018). De novo computational RNA modeling into cryo‐EM maps of large ribonucleoprotein complexes. Nature Methods, 15(11), 947–954. 10.1038/s41592-018-0172-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane, S. C. , Heng, X. , Lu, K. , Kharytonchyk, S. , Ramakrishnan, V. , Carter, G. , … Summers, M. F. (2015). RNA structure. Structure of the HIV‐1 RNA packaging signal. Science, 348(6237), 917–921. 10.1126/science.aaa9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane, S. C. , Van, V. , Frank, H. M. , Sciandra, C. A. , McCowin, S. , Santos, J. , … Summers, M. F. (2016). NMR detection of intermolecular interaction sites in the dimeric 5′‐leader of the HIV‐1 genome. Proceedings of the National Academy of Sciences of the United States of America, 113(46), 13033–13038. 10.1073/pnas.1614785113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I. , Lukavsky, P. J. , & Puglisi, J. D. (2002). NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. Journal of the American Chemical Society, 124(32), 9338–9339. [DOI] [PubMed] [Google Scholar]
- Kim, N. K. , Zhang, Q. , & Feigon, J. (2014). Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function. Nucleic Acids Research, 42(5), 3395–3408. 10.1093/nar/gkt1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloiber, K. , Spitzer, R. , Tollinger, M. , Konrat, R. , & Kreutz, C. (2011). Probing RNA dynamics via longitudinal exchange and CPMG relaxation dispersion NMR spectroscopy using a sensitive 13C‐methyl label. Nucleic Acids Research, 39(10), 4340–4351. 10.1093/nar/gkq1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappenberger, A. J. , Reiss, C. W. , & Strobel, S. A. (2018). Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA. eLife, 7, e36381. 10.7554/eLife.36381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt, W. (2014a). Biochemistry. The resolution revolution. Science, 343(6178), 1443–1444. 10.1126/science.1251652 [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt, W. (2014b). Cryo‐EM enters a new era. eLife, 3, e03678 10.7554/eLife.03678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc, R. M. , Longhini, A. P. , Tugarinov, V. , & Dayie, T. K. (2018). NMR probing of invisible excited states using selectively labeled RNAs. Journal of Biomolecular NMR, 71(3), 165–172. 10.1007/s10858-018-0184-3 [DOI] [PubMed] [Google Scholar]
- Lee, J. H. , Jucker, F. , & Pardi, A. (2008). Imino proton exchange rates imply an induced‐fit binding mechanism for the VEGF165‐targeting aptamer, Macugen. FEBS Letters, 582(13), 1835–1839. 10.1016/j.febslet.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. T. (2012). Epigenetic regulation by long noncoding RNAs. Science, 338(6113), 1435–1439. 10.1126/science.1231776 [DOI] [PubMed] [Google Scholar]
- Liberman, J. A. , Suddala, K. C. , Aytenfisu, A. , Chan, D. , Belashov, I. A. , Salim, M. , … Wedekind, J. E. (2015). Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome‐binding site regulated by fast dynamics. Proceedings of the National Academy of Sciences of the United States of America, 112(27), E3485–E3494. 10.1073/pnas.1503955112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman, J. A. , & Wedekind, J. E. (2012). Riboswitch structure in the ligand‐free state. WIREs RNA, 3(3), 369–384. 10.1002/wrna.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Holmstrom, E. , Yu, P. , Tan, K. , Zuo, X. , Nesbitt, D. J. , … Wang, Y. X. (2018). Incorporation of isotopic, fluorescent, and heavy‐atom‐modified nucleotides into RNAs by position‐selective labeling of RNA. Nature Protocols, 13(5), 987–1005. 10.1038/nprot.2018.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Holmstrom, E. , Zhang, J. , Yu, P. , Wang, J. , Dyba, M. A. , … Wang, Y. X. (2015). Synthesis and applications of RNAs with position‐selective labelling and mosaic composition. Nature, 522(7556), 368–372. 10.1038/nature14352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Yu, P. , Dyba, M. , Sousa, R. , Stagno, J. R. , & Wang, Y. X. (2016). Applications of PLOR in labeling large RNAs at specific sites. Methods, 103, 4–10. 10.1016/j.ymeth.2016.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhini, A. P. , LeBlanc, R. M. , Becette, O. , Salguero, C. , Wunderlich, C. H. , Johnson, B. A. , … Dayie, T. K. (2016). Chemo‐enzymatic synthesis of site‐specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Research, 44(6), e52 10.1093/nar/gkv1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, K. , Heng, X. , Garyu, L. , Monti, S. , Garcia, E. L. , Kharytonchyk, S. , … Summers, M. F. (2011). NMR detection of structures in the HIV‐1 5′‐leader RNA that regulate genome packaging. Science, 334(6053), 242–245. 10.1126/science.1210460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, K. , Miyazaki, Y. , & Summers, M. F. (2010). Isotope labeling strategies for NMR studies of RNA. Journal of Biomolecular NMR, 46(1), 113–125. 10.1007/s10858-009-9375-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky, P. J. , Kim, I. , Otto, G. A. , & Puglisi, J. D. (2003). Structure of HCV IRES domain II determined by NMR. Nature Structural Biology, 10(12), 1033–1038. 10.1038/nsb1004 [DOI] [PubMed] [Google Scholar]
- Lukavsky, P. J. , & Puglisi, J. D. (2005). Structure determination of large biological RNAs. Methods in Enzymology, 394, 399–416. 10.1016/S0076-6879(05)94016-0 [DOI] [PubMed] [Google Scholar]
- Lundstrom, P. , Hansen, D. F. , & Kay, L. E. (2008). Measurement of carbonyl chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy: Comparison between uniformly and selectively 13C labeled samples. Journal of Biomolecular NMR, 42(1), 35–47. 10.1007/s10858-008-9260-4 [DOI] [PubMed] [Google Scholar]
- Marchant, J. , Bax, A. , & Summers, M. F. (2018). Accurate measurement of residual dipolar couplings in large RNAs by variable flip angle NMR. Journal of the American Chemical Society, 140(22), 6978–6983. 10.1021/jacs.8b03298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia, M. , & Pyle, A. M. (2012). Visualizing group II intron catalysis through the stages of splicing. Cell, 151(3), 497–507. 10.1016/j.cell.2012.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion, D. (2013). An introduction to biological NMR spectroscopy. Molecular & Cellular Proteomics, 12(11), 3006–3025. 10.1074/mcp.O113.030239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino, E. J. , Wilkinson, K. A. , Coughlan, J. L. , & Weeks, K. M. (2005). RNA structure analysis at single nucleotide resolution by selective 2′‐hydroxyl acylation and primer extension (SHAPE). Journal of the American Chemical Society, 127(12), 4223–4231. 10.1021/ja043822v [DOI] [PubMed] [Google Scholar]
- Meyer, M. , Nielsen, H. , Olieric, V. , Roblin, P. , Johansen, S. D. , Westhof, E. , & Masquida, B. (2014). Speciation of a group I intron into a lariat capping ribozyme. Proceedings of the National Academy of Sciences of the United States of America, 111(21), 7659–7664. 10.1073/pnas.1322248111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S. B. , Yildiz, F. Z. , Lo, J. A. , Wang, B. , & D'Souza, V. M. (2014). A structure‐based mechanism for tRNA and retroviral RNA remodelling during primer annealing. Nature, 515(7528), 591–595. 10.1038/nature13709 [DOI] [PubMed] [Google Scholar]
- Miyazaki, Y. , Irobalieva, R. N. , Tolbert, B. S. , Smalls‐Mantey, A. , Iyalla, K. , Loeliger, K. , … Summers, M. F. (2010). Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo‐electron tomography. Journal of Molecular Biology, 404(5), 751–772. 10.1016/j.jmb.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi, A. , Kishikawa, J. I. , Tamakoshi, M. , Mitsuoka, K. , & Yokoyama, K. (2018). Cryo EM structure of intact rotary H(+)‐ATPase/synthase from Thermus thermophilus . Nature Communications, 9(1), 89 10.1038/s41467-017-02553-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen, F. H. , van Gammeren, A. J. , Tessari, M. , Girard, F. C. , Heus, H. A. , & Wijmenga, S. S. (2008). Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic Acids Research, 36(14), e89 10.1093/nar/gkn397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L. A. , Wang, J. , & Steitz, T. A. (2017). Crystal structure of Pistol, a class of self‐cleaving ribozyme. Proceedings of the National Academy of Sciences of the United States of America, 114(5), 1021–1026. 10.1073/pnas.1611191114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, P. , & Qin, P. Z. (2012). RNA dynamics: Perspectives from spin labels. WIREs RNA, 3(1), 62–72. 10.1002/wrna.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonowicz, E. P. , Sirr, A. , Legault, P. , Jucker, F. M. , Baer, L. M. , & Pardi, A. (1992). Preparation of 13C and 15N labelled RNAs for heteronuclear multi‐dimensional NMR studies. Nucleic Acids Research, 20(17), 4507–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales, E. , & Scheres, S. H. (2015). Cryo‐EM: A unique tool for the visualization of macromolecular complexity. Molecular Cell, 58(4), 677–689. 10.1016/j.molcel.2015.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, A. G. , & Massi, F. (2006). Characterization of the dynamics of biomacromolecules using rotating‐frame spin relaxation NMR spectroscopy. Chemical Reviews, 106(5), 1700–1719. 10.1021/cr0404287 [DOI] [PubMed] [Google Scholar]
- Panja, S. , Hua, B. , Zegarra, D. , Ha, T. , & Woodson, S. A. (2017). Metals induce transient folding and activation of the twister ribozyme. Nature Chemical Biology, 13(10), 1109–1114. 10.1038/nchembio.2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes, E. , Evans, M. , & Das, S. R. (2011). RNA labeling, conjugation and ligation. Methods, 54(2), 251–259. 10.1016/j.ymeth.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Peattie, D. A. , & Gilbert, W. (1980). Chemical probes for higher‐order structure in RNA. Proceedings of the National Academy of Sciences of the United States of America, 77(8), 4679–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin, K. , Ono, A. , Fernandez, C. , Szyperski, T. , Kainosho, M. , & Wuthrich, K. (1998). NMR scaler couplings across Watson‐Crick base pair hydrogen bonds in DNA observed by transverse relaxation optimized spectroscopy. Proceedings of the National Academy of Sciences of the United States of America, 95(24), 14147–14151. 10.1073/pnas.95.24.14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin, K. , Riek, R. , Wider, G. , & Wuthrich, K. (1997). Attenuated T2 relaxation by mutual cancellation of dipole‐dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proceedings of the National Academy of Sciences of the United States of America, 94(23), 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peselis, A. , & Serganov, A. (2018). ykkC riboswitches employ an add‐on helix to adjust specificity for polyanionic ligands. Nature Chemical Biology, 14(9), 887–894. 10.1038/s41589-018-0114-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle, A. M. (2016). Group II intron self‐splicing. Annual Review of Biophysics, 45, 183–205. 10.1146/annurev-biophys-062215-011149 [DOI] [PubMed] [Google Scholar]
- Qu, G. , Kaushal, P. S. , Wang, J. , Shigematsu, H. , Piazza, C. L. , Agrawal, R. K. , … Wang, H. W. (2016). Structure of a group II intron in complex with its reverse transcriptase. Nature Structural & Molecular Biology, 23(6), 549–557. 10.1038/nsmb.3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reining, A. , Nozinovic, S. , Schlepckow, K. , Buhr, F. , Furtig, B. , & Schwalbe, H. (2013). Three‐state mechanism couples ligand and temperature sensing in riboswitches. Nature, 499(7458), 355–359. 10.1038/nature12378 [DOI] [PubMed] [Google Scholar]
- Ren, A. , Micura, R. , & Patel, D. J. (2017). Structure‐based mechanistic insights into catalysis by small self‐cleaving ribozymes. Current Opinion in Chemical Biology, 41, 71–83. 10.1016/j.cbpa.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, J. S. , & Mathews, D. H. (2010). RNA structure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, 129 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnenthal, J. , Buck, J. , Ferner, J. , Wacker, A. , Furtig, B. , & Schwalbe, H. (2011). Mapping the landscape of RNA dynamics with NMR spectroscopy. Accounts of Chemical Research, 44(12), 1292–1301. 10.1021/ar200137d [DOI] [PubMed] [Google Scholar]
- Ritchie, D. B. , & Woodside, M. T. (2015). Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Current Opinion in Structural Biology, 34, 43–51. 10.1016/j.sbi.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh, S. H. , Hryc, C. F. , Jeong, H. H. , Fei, X. , Jakana, J. , Lorimer, G. H. , & Chiu, W. (2017). Subunit conformational variation within individual GroEL oligomers resolved by Cryo‐EM. Proceedings of the National Academy of Sciences of the United States of America, 114(31), 8259–8264. 10.1073/pnas.1704725114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A. , & Breaker, R. R. (2009). The structural and functional diversity of metabolite‐binding riboswitches. Annual Review of Biochemistry, 78, 305–334. 10.1146/annurev.biochem.78.070507.135656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia, J., Jr. , Shen, L. X. , Cai, Z. , Lewis, H. , & Tinoco, I., Jr. (1995). Synthesis and NMR of RNA with selective isotopic enrichment in the bases. Nucleic Acids Research, 23(23), 4913–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, L. G. , Tolbert, T. J. , & Williamson, J. R. (2000). Preparation of specifically 2H‐ and 13C‐labeled ribonucleotides. Methods in Enzymology, 317, 18–38. [DOI] [PubMed] [Google Scholar]
- Stern, S. , Moazed, D. , & Noller, H. F. (1988). Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods in Enzymology, 164, 481–489. [DOI] [PubMed] [Google Scholar]
- Suslov, N. B. , DasGupta, S. , Huang, H. , Fuller, J. R. , Lilley, D. M. , Rice, P. A. , & Piccirilli, J. A. (2015). Crystal structure of the Varkud satellite ribozyme. Nature Chemical Biology, 11(11), 840–846. 10.1038/nchembio.1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, S. , & Das, R. (2016). RNA structure through multidimensional chemical mapping. Quarterly Reviews of Biophysics, 49, e7 10.1017/S0033583516000020 [DOI] [PubMed] [Google Scholar]
- Tolbert, B. S. , Miyazaki, Y. , Barton, S. , Kinde, B. , Starck, P. , Singh, R. , … Summers, M. F. (2010). Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H‐13C residual dipolar coupling on refinement. Journal of Biomolecular NMR, 47(3), 205–219. 10.1007/s10858-010-9424-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor, N. , Keating, K. S. , Taylor, S. D. , & Pyle, A. M. (2008). Crystal structure of a self‐spliced group II intron. Science, 320(5872), 77–82. 10.1126/science.1153803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor, N. , Rajashankar, K. , Keating, K. S. , & Pyle, A. M. (2008). Structural basis for exon recognition by a group II intron. Nature Structural & Molecular Biology, 15(11), 1221–1222. 10.1038/nsmb.1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzakos, A. G. , Easton, L. E. , & Lukavsky, P. J. (2007). Preparation of large RNA oligonucleotides with complementary isotope‐labeled segments for NMR structural studies. Nature Protocols, 2(9), 2139–2147. 10.1038/nprot.2007.306 [DOI] [PubMed] [Google Scholar]
- Ulyanov, N. B. , Mujeeb, A. , Du, Z. , Tonelli, M. , Parslow, T. G. , & James, T. L. (2006). NMR structure of the full‐length linear dimer of stem‐loop‐1 RNA in the HIV‐1 dimer initiation site. The Journal of Biological Chemistry, 281(23), 16168–16177. 10.1074/jbc.M601711200 [DOI] [PubMed] [Google Scholar]
- Vallurupalli, P. , Sekhar, A. , Yuwen, T. , & Kay, L. E. (2017). Probing conformational dynamics in biomolecules via chemical exchange saturation transfer: A primer. Journal of Biomolecular NMR, 67(4), 243–271. 10.1007/s10858-017-0099-4 [DOI] [PubMed] [Google Scholar]
- Varani, G. , Aboulela, F. , & Allain, F. H. T. (1996). NMR investigation of RNA structure. Progress in Nuclear Magnetic Resonance Spectroscopy, 29, 51–127. 10.1016/0079-6565(96)01028-X [DOI] [Google Scholar]
- Vicens, Q. , Mondragon, E. , Reyes, F. E. , Coish, P. , Aristoff, P. , Berman, J. , … Batey, R. T. (2018). Structure‐activity relationship of flavin analogues that target the flavin mononucleotide riboswitch. ACS Chemical Biology, 13(10), 2908–2919. 10.1021/acschembio.8b00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar, K. R. , & Henderson, R. (2016). Single particle electron cryomicroscopy: Trends, issues and future perspective. Quarterly Reviews of Biophysics, 49, e13 10.1017/S0033583516000068 [DOI] [PubMed] [Google Scholar]
- Wagner, D. , Rinnenthal, J. , Narberhaus, F. , & Schwalbe, H. (2015). Mechanistic insights into temperature‐dependent regulation of the simple cyanobacterial hsp17 RNA thermometer at base‐pair resolution. Nucleic Acids Research, 43(11), 5572–5585. 10.1093/nar/gkv414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Zuo, X. , Yu, P. , Xu, H. , Starich, M. R. , Tiede, D. M. , … Wang, Y. X. (2009). A method for helical RNA global structure determination in solution using small‐angle X‐ray scattering and NMR measurements. Journal of Molecular Biology, 393(3), 717–734. 10.1016/j.jmb.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, K. D. , Hajdin, C. E. , & Weeks, K. M. (2018). Principles for targeting RNA with drug‐like small molecules. Nature Reviews Drug Discovery, 17(8), 547–558. 10.1038/nrd.2018.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, J. M. , Dang, K. K. , Gorelick, R. J. , Leonard, C. W. , Bess, J. W., Jr. , Swanstrom, R. , … Weeks, K. M. (2009). Architecture and secondary structure of an entire HIV‐1 RNA genome. Nature, 460(7256), 711–716. 10.1038/nature08237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, K. M. (2010). Advances in RNA structure analysis by chemical probing. Current Opinion in Structural Biology, 20(3), 295–304. 10.1016/j.sbi.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich, K. (1986). NMR of proteins and nucleic acids. New York, New York: Wiley‐Interscience. [Google Scholar]
- Xu, J. , Lapham, J. , & Crothers, D. M. (1996). Determining RNA solution structure by segmental isotopic labeling and NMR: Application to Caenorhabditis elegans spliced leader RNA 1. Proceedings of the National Academy of Sciences of the United States of America, 93(1), 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Y. , Kellogg, D. , Kimsey, I. J. , Sathyamoorthy, B. , Stein, Z. W. , McBrairty, M. , & Al‐Hashimi, H. M. (2015). Characterizing RNA excited states using NMR relaxation dispersion. Methods in Enzymology, 558, 39–73. 10.1016/bs.mie.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, T. , Muhandiram, R. , & Kay, L. E. (1994). Nmr experiments for the measurement of carbon relaxation properties in highly enriched, uniformly 13C, 15N‐labeled proteins—Application to 13C(alpha) carbons. Journal of the American Chemical Society, 116(18), 8266–8278. 10.1021/ja00097a037 [DOI] [Google Scholar]
- Yan, J. L. , Corpora, T. , Pradhan, P. , & Bushweller, J. H. (2002). MQ‐HCN‐based pulse sequences for the measurement of 13C1'‐1H1', 13C1'‐15N, 1H1'‐15N, 13C1'‐13C2', 1H1'‐13C2', 13C6/8‐1H6/8, 13C6/8‐15N, 1H6/8‐15N, 13C6‐13C5, 1H6‐13C5 dipolar couplings in 13C, 15N‐labeled DNA (and RNA). Journal of Biomolecular NMR, 22(1), 9–20. 10.1023/A:1013876105589 [DOI] [PubMed] [Google Scholar]
- Zhang, K. , Keane, S. C. , Su, Z. , Irobalieva, R. N. , Chen, M. , Van, V. , … Chiu, W. (2018). Structure of the 30 kDa HIV‐1 RNA dimerization signal by a hybrid Cryo‐EM, NMR, and molecular dynamics approach. Structure, 26, 490–498.e3. 10.1016/j.str.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Guffy, S. L. , Williams, B. , & Zhang, Q. (2017). An excited state underlies gene regulation of a transcriptional riboswitch. Nature Chemical Biology, 13(9), 968–974. 10.1038/nchembio.2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Hansen, A. L. , & Zhang, Q. (2014). Characterizing slow chemical exchange in nucleic acids by carbon CEST and low spin‐lock field R(1rho) NMR spectroscopy. Journal of the American Chemical Society, 136(1), 20–23. 10.1021/ja409835y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Rajashankar, K. R. , Marcia, M. , & Pyle, A. M. (2015). Crystal structure of group II intron domain 1 reveals a template for RNA assembly. Nature Chemical Biology, 11(12), 967–972. 10.1038/nchembio.1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidek, L. , Wu, H. H. , Feigon, J. , & Sklenar, V. (2001). Measurement of small scalar and dipolar couplings in purine and pyrimidine bases. Journal of Biomolecular NMR, 21(2), 153–160. 10.1023/A:1012435106858 [DOI] [PubMed] [Google Scholar]
- Ziegeler, M. , Cevec, M. , Richter, C. , & Schwalbe, H. (2012). NMR studies of HAR1 RNA secondary structures reveal conformational dynamics in the human RNA. Chembiochem, 13(14), 2100–2112. 10.1002/cbic.201200401 [DOI] [PubMed] [Google Scholar]
- Zubradt, M. , Gupta, P. , Persad, S. , Lambowitz, A. M. , Weissman, J. S. , & Rouskin, S. (2017). DMS‐MaPseq for genome‐wide or targeted RNA structure probing in vivo. Nature Methods, 14(1), 75–82. 10.1038/nmeth.4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, X. , Wang, J. , Yu, P. , Eyler, D. , Xu, H. , Starich, M. R. , … Wang, Y. X. (2010). Solution structure of the cap‐independent translational enhancer and ribosome‐binding element in the 3' UTR of turnip crinkle virus. Proceedings of the National Academy of Sciences of the United States of America, 107(4), 1385–1390. 10.1073/pnas.0908140107 [DOI] [PMC free article] [PubMed] [Google Scholar]


