Abstract
Epitranscriptomics, the study of posttranscriptional chemical moieties placed on RNA, has blossomed in recent years. This is due in part to the emergence of high‐throughput detection methods as well as the burst of discoveries showing biological function of select chemical marks. RNA modifications have been shown to affect RNA structure, localization, and functions such as alternative splicing, stabilizing transcripts, nuclear export, cap‐dependent and cap‐independent translation, microRNA biogenesis and binding, RNA degradation, and immune regulation. As such, the deposition of chemical marks on RNA has the unique capability to spatially and temporally regulate gene expression. The goal of this article is to present the exciting convergence of the epitranscriptomic and virology fields, specifically the deposition and biological impact of N7‐methylguanosine, ribose 2′‐O‐methylation, pseudouridine, inosine, N6‐methyladenosine, and 5‐methylcytosine epitranscriptomic marks on gene expression of RNA viruses.
This article is categorized under:
RNA in Disease and Development > RNA in Disease
RNA Interactions with Proteins and Other Molecules > Protein–RNA Interactions: Functional Implications
Keywords: 5‐methylcytosine, epitranscriptomics, innate immunity, inosine, N6‐methyladenosine, N7‐methylguanosine, pseudouridine, ribose 2′‐O‐methylation, RNA virus
RNA modifications and RNA viruses.
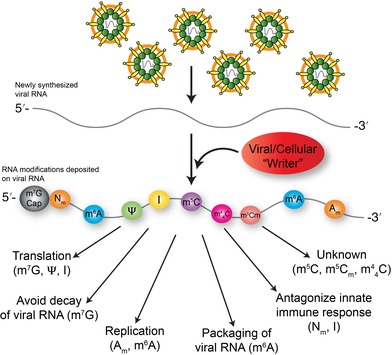
1. OVERVIEW OF POSTTRANSCRIPTIONAL MODIFICATIONS ON RNA
RNA has a breathtaking variety of biological functions, which far exceeds its classical role as a carrier of genetic information. This range of functions is particularly exemplified in RNA viruses where the genome directs translation of viral proteins, functions as a template for the synthesis of new viral genomes and/or messenger RNAs (mRNAs), and is packaged in viral particles for subsequent infections. These functions are facilitated by specific structures or configurations in the viral RNA (Nicholson & White, 2015), and interactions with viral and cellular RNA binding proteins. Thus, to ensure a successful infection cycle, viruses intimately interact with, subvert and/or avoid cellular RNA transcription, splicing, translation, storage, and decay pathways (Cross, Michalski, Miller, & Wilusz, 2019). More recently, the deposition of chemical moieties on viral RNA has been shown to be a pivotal regulator of spatiotemporal viral gene expression (Gonzales‐van Horn & Sarnow, 2017).
RNA may be posttranscriptionally modified with different chemical moieties or marks. To date, more than 150 RNA modifications with very distinct chemical and structural properties have been discovered on all four canonical ribonucleotides as well as on noncanonical nucleotides such as pseudouridine and inosine (Boccaletto et al., 2018; Cantara et al., 2011; Limbach, Crain, & McCloskey, 1994; Machnicka et al., 2012). RNA modifications were first described in the 1960s and 1970s and initial studies focused on highly abundant RNAs such as ribosomal RNA (rRNA) and transfer RNA (tRNA). Because of the dearth of effective analytical tools, incomplete knowledge of the enzymes involved in the biogenesis of these marks, and the lack of a robust framework for understanding their regulatory and metabolic significance, the biological functions of the vast majority of RNA modifications are still largely unknown. The emergence however, of sensitive high‐throughput techniques combined with select available reagents (Schwartz & Motorin, 2017), and the identification of some of the biogenic enzymes that deposit (writers), remove (erasers) or facilitate function (readers) have enabled studies investigating the role of RNA modifications or the epitranscriptome in cellular RNAs. RNA modifications have been shown to impact numerous posttranscriptional RNA processes. Here we focus on how RNA viruses are influenced by RNA modifications. In particular we present an overview of the viral and cellular enzymes involved in depositing chemical moieties on viral RNA, as well as the consequence of specific RNA modifications on distinct steps in the infectious cycle and the innate immune response.
2. VIRUS‐DERIVED RNA MODIFICATIONS
In eukaryotic cells, cellular mRNAs contain an essential modified nucleotide at the 5′ end, namely the N7‐methylguanosine (m7G) cap, followed by one or two 2′‐O‐methylated nucleotides (Figure 1) (Wei, Gershowitz, & Moss, 1975). The m7G cap on cellular mRNAs is predominantly added cotranscriptionally in the nucleus (Topisirovic, Svitkin, Sonenberg, & Shatkin, 2011), although RNAs that have lost the m7G cap can also be recapped in the cytoplasm (Otsuka, Kedersha, & Schoenberg, 2009; Trotman, Giltmier, Mukherjee, & Schoenberg, 2017). The addition of the cap structure occurs via three consecutive reactions (Figure 1b) (Topisirovic et al., 2011). First, the γ‐phosphate on a newly synthesized RNA (pppNp‐RNA, where N is the first nucleotide of the RNA, and p denotes the phosphate groups) is cleaved by a 5′ RNA triphosphatase to a diphosphate RNA (ppNp‐RNA) intermediate (Figure 1b). Next, a guanylyltransferase covalently interacts with GTP via the α‐phosphate to produce a GTPase‐Gp intermediate and then links the GMP molecule to the 5′ diphosphate RNA via a 5′→5′ linkage to produce GpppNp‐RNA or the Cap 0 structure (Figure 1b). Last, using S‐adenosyl‐l‐methionine (SAM) as a methyl donor guanine‐N7‐methyltransferase (N7 MTase) deposits a methyl group on the guanosine at the N7 position to produce m7GpppNp (Figure 1b). The capping reaction is completed by a nucleoside‐2′‐O‐methyltransferase (2′‐O MTase) that methylates the 2′‐O position on the ribose of the first, and sometimes second, nucleotide to produce the Cap 1 or Cap 2 structure (Figure 1b). In the nucleus, the addition of the cap protects mRNAs from degradation by 5′ exoribonucleases, directs pre‐mRNA splicing and facilitates export out of the nucleus (Topisirovic et al., 2011). In the cytoplasm, the 5′‐cap ensures efficient translation, and stabilizes and protects mRNAs from 5′ exoribonucleases (Topisirovic et al., 2011). Moreover the 2′‐O methyl modification on the ribose of the penultimate nucleotide functions as a marker of “self,” such that RNAs lacking this modification trigger the innate immune response (Hyde & Diamond, 2015).
Figure 1.
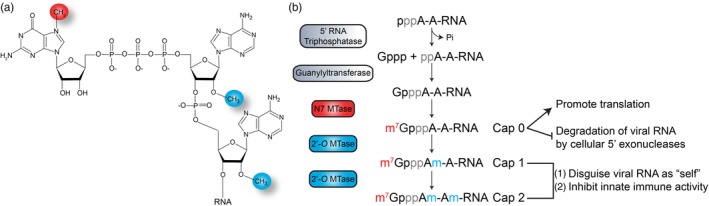
5′‐Cap on messenger RNA (mRNA). (a) Schematic of N7‐methylguanosine cap attached to nucleotides 1 and 2 of RNA that are 2′‐O‐methylated. (b) Overview of the canonical pathway for installation of Cap 0, Cap 1, and Cap 2 structures at the 5′‐end of an mRNA. The functional significance of the Cap 0, Cap 1, and Cap 2 are shown
2.1. Viral methyltransferases direct addition of m7G caps
Given the critical role of the cap structure on cellular mRNAs, it is hardly surprising that many viruses similarly install a cap on the viral RNA. Interestingly, the m7G modified nucleotide blocking the 5′‐end was first identified in viral RNAs prior to the identification on cellular mRNAs (Furuichi, Morgan, Muthukrishnan, & Shatkin, 1975; Furuichi, Shatkin, Stavnezer, & Bishop, 1975; Rose, 1975; Wei & Moss, 1975). To cap viral RNAs, viruses use different strategies such as usurping the nuclear capping machinery, “stealing” caps from cellular mRNAs, and encoding viral capping enzymes (Decroly & Canard, 2017; Decroly, Ferron, Lescar, & Canard, 2012; Dong, Zhang, & Shi, 2008; Ramanathan, Robb, & Chan, 2016). For a comprehensive overview of the mechanisms and functions of viral RNA caps we refer the reader to the excellent reviews by Decroly et al. (2012), Decroly and Canard (2017) and Ramanathan et al. (2016). A number of viruses with cytoplasmic infectious cycles virally encode a methyltransferase protein that adds a guanosine cap, although the mode by which the guanosine is added and methylated differs with each virus. Curiously, the RNA from some alphaviruses, a flavivirus, and human immunodeficiency virus type 1 (HIV‐1) have also been shown to harbor not only the canonical m7G cap but also di‐ and tri‐methylated guanosine caps (Dong, Ren, Li, & Shi, 2008; Hsuchen & Dubin, 1976; van Duijn, Kasperaitis, Ameling, & Voorma, 1986; Yedavalli & Jeang, 2010). Regardless of the capping mechanism, the monomethylated m7G cap facilitates efficient translation of viral proteins and protects the viral RNAs from degradation by cellular exonucleases (Figure 1b, Table 1) (Decroly et al., 2012; Ramanathan et al., 2016).
Table 1.
List of RNA modifications and functions associated with RNA viruses
| RNA virus | Function | References |
|---|---|---|
| N7‐methylguanosine (m7G) | ||
| Viruses within Corona‐, Arteri‐, Rhabdo‐, Filo‐, Paramyxo‐, Pox‐, and Reo‐, Retro‐, Togaviridae families and flavivirus genus | • m7G in 5′‐cap promotes translation and RNA stability | Decroly and Canard (2017), Decroly et al. (2012) |
| Zika virus, Dengue virus, HCV, Poliovirus, HIV‐1 and Murine leukemia virus | • To be determined for m7G modifications within genome | Courtney et al. (2019), McIntyre et al. (2018) |
| 2′‐O‐methylated nucleotides (Nm) | ||
| Viruses within Corona‐, Arteri‐, Rhabdo‐, Filo‐, Paramyxo‐, Pox‐, and Reoviridae families and flavivirus genus | • Nm at 5′‐end: Escape innate immune sensing | Hyde and Diamond (2015) |
| HIV‐1 | • Internal Nm: Escape innate immune sensing | McIntyre et al. (2018), Ringeard, Marchand, Decroly, Motorin, and Bennasser (2019) |
| Dengue virus | • Internal Am restricts elongation of viral RNA‐dependent RNA polymerase | Dong et al. (2012) |
| Pseudouridine (Ψ) | ||
| Turnip yellow mosaic virus and Brome mosaic virus | • Possibly promotes RNA structure | Baumstark and Ahlquist (2001), Becker (1998) |
| Tobacco mosaic virus | • Ψ within tRNATyr anticodon loop terminates translation of viral polymerase | Zerfass and Beier (1992) |
| Zika virus, Dengue virus, HCV, Poliovirus, and HIV‐1 | • To be determined | McIntyre et al. (2018) |
| Inosine (I) | ||
| Measles virus and Respiratory syncytial virus |
• Introduces a mutation and affects mRNA coding capacity • Causes frameshift which terminates synthesis of viral proteins • Suppresses innate immune response |
Cattaneo et al. (1988), Martínez, Dopazo, and Melero (1997), Pfaller, Donohue, Nersisyan, Brodsky, and Cattaneo (2018), Rueda, García‐Barreno, and Melero (1994) |
| Hepatitis δ virus | • Maintains balance between replication and virus production | Casey (2006) |
| HIV‐1 | • Introduces a mutation and affects codon and splicing of viral RNA | Doria, Neri, Gallo, Farace, and Michienzi (2009), McIntyre et al. (2018), Phuphuakrat et al. (2008) |
| HCV | • Affects viral gene expression | Taylor, Puig, Darnell, Mihalik, and Feinstone (2005) |
| Zika virus, Dengue virus, and Poliovirus | • To be determined | Khrustalev, Khrustaleva, Sharma, and Giri (2017), McIntyre et al. (2018), Piontkivska, Frederick, Miyamoto, and Wayne (2017) |
| N6‐methyladenosine (m6A) | ||
| HIV‐1 |
• Modulates HIV‐1 gene expression early and late in infection • Affects stability of viral RNA • Influences export of HIV‐1 RNA out of nucleus |
Kennedy et al. (2016), Lichinchi, Gao, et al. (2016), Lu et al. (2018), Tirumuru and Wu (2019), Tirumuru et al. (2016) |
| Avian sarcoma virus and Rous sarcoma virus | • Affects splicing of viral RNA | Beemon and Keith (1977), Dimock and Stoltzfus (1977), Kane and Beemon (1985), Stoltzfus and Dane (1982) |
| Murine leukemia virus | • Affects viral gene expression | Courtney et al. (2019) |
| Influenza virus |
• Promotes infection kinetics • Decreases virus pathogenicity |
Courtney et al. (2017), Krug, Morgan, and Shatkin (1976), Narayan, Ayers, Rottman, Maroney, and Nilsen (1987) |
| HCV | • Negatively regulates the production of new virus particles | Gokhale et al. (2016) |
| Zika virus, Dengue virus, West Nile virus, Yellow fever virus, Poliovirus, and Enterovirus 71 | • Affects viral gene expression | Gokhale et al. (2016), Hao et al. (2019), Lichinchi, Zhao, et al. (2016), McIntyre et al. (2018) |
| 5‐Methylcytosine (m5C) | ||
| Sindbis virus and Drosophila C virus | • m5C methyltransferase (Mt or Dnmt) modulates innate immune responses | Bhattacharya, Newton, and Hardy (2017), Dubin and Stollar (1975), Dubin, Stollar, Hsuchen, Timko, and Guild (1977), Durdevic et al. (2013) |
| Murine leukemia virus | • Affects viral gene expression and infectivity | Courtney et al. (2019) |
| Zika virus, Dengue virus, HCV, Poliovirus, and HIV‐1 | • To be determined | McIntyre et al. (2018) |
Abbreviations: HCV, hepatitis C virus; HIV‐1, human immunodeficiency virus 1.
2.2. Role of ribose 2′‐O‐methylation of viral RNA
2.2.1. 2′‐O‐methylation at the 5′‐end of viral RNA
Methylation of the ribose 2′‐hydroxyl group (2′‐OH) has been known since the 1960s (Baskin & Dekker, 1967). Ribose 2′‐O‐methylation in different cellular and viral RNAs occurs on all four ribonucleotides and on noncanonical nucleotides such as pseudouridine and inosine (Ayadi, Galvanin, Pichot, Marchand, & Motorin, 2019). While the biological function of internal 2′‐O‐methylated nucleotides is incomplete, the installation of this modification on the first and second nucleotides of cellular mRNAs to produce a Cap 1 or Cap 2 structure (Figure 1) serves as a molecular signature of “self” (Table 1) (Hyde & Diamond, 2015). As such many viruses mimic the Cap 1 structure by either reappropriating caps from cellular mRNAs onto viral RNAs, or by encoding a viral 2′‐O methyltransferase (MTase). In particular, the methyltransferase encoded by Corona‐, Arteri‐, Rhabdo‐, Filo‐, Paramyxo‐, Pox‐, and Reoviridae families and some viruses within the Flaviviridae family exhibits two enzymatic functions that N7‐methylate the guanosine cap and 2′‐O‐methylate the ribose 2′‐OH of the first nucleotide of the viral RNA (Decroly et al., 2012). Daffis et al. (2010) elucidated the function of the 2′‐O‐methylation on the first nucleotide of the viral RNA, and showed that the addition of the 2′‐O‐methyl modification at this particular position protected the viral RNA from the innate immune response pathway by camouflaging the viral RNA as a cellular mRNA (Daffis et al., 2010). The innate immune response is a critical cellular pathway that functions to limit pathogens. Particular molecular signatures within pathogens (for example double‐stranded RNA, 5′‐triphosphate RNA, or non‐2′‐O‐methylated RNA) when recognized by cellular sensors initiate signaling cascades resulting in expression of type I interferon and cytokines (McFadden, Gokhale, & Horner, 2017). Secretion of interferon‐α (IFN‐α) and IFN‐β in turn stimulate signaling in neighboring cells by binding the type I interferon receptor (IFNAR). Subsequent propagation of the intracellular signals through Jak kinase and STAT transcription factors results in the expression of interferon stimulated genes (ISGs) with specific antiviral activities. For example, ISGs encoding protein kinase R (PKR), ISG56 (IFIT1), and ISG54 (IFIT2) function to limit translation and consequently influence viral gene expression.
That 2′‐O‐methylation is a marker of “self” was first shown in West Nile virus (WNV). WNV belongs the Flaviviridae family of viruses and has a positive‐sense RNA genome containing a Cap 1 structure. This Cap 1 structure (m7GpppAm) is added by the methyltransferase domain of the viral NS5 protein (Y. Zhou et al., 2007), where specific amino acids within NS5 direct methylation of the guanosine cap and ribose 2′‐OH moiety. Daffis et al. (2010) showed that WNV deficient in 2′‐O MTase activity was attenuated in wild‐type mice but virulent in mice lacking Ifnar, the gene encoding the interferon (IFN) receptor (Daffis et al., 2010). Furthermore, when embryonic fibroblasts isolated from Ifnar‐deficient mice were infected with wild‐type or 2′‐O MTase‐deficient viruses, both viruses induced the same amount of interferon mRNA. These data indicated that the intracellular sensors recognizing the viral RNA and promoting downstream signaling remained intact. Pretreatment of cells with IFN, inhibited mutant WNV more than the wild‐type virus, and this presented the possibility that an IFN‐induced gene might act on non‐2′‐O‐methylated WNV RNA (Daffis et al., 2010). Indeed, Daffis et al. (2010) demonstrated that interferon‐induced protein with tetratricopeptides (IFIT) proteins recognized non‐2′‐O‐methylated RNA genomes from three different viruses namely WNV, poxvirus, and coronavirus, and restricted the viral infection. Recent X‐ray crystallography studies revealed the mechanism by which the IFIT proteins discriminate between “self” and “non‐self” RNA. In particular, IFIT is comprised of tetratricopeptide subdomains that assemble as a clamp around an internal RNA binding tunnel (Abbas et al., 2017). This internal RNA binding tunnel was found to only accommodate RNA with a m7G cap and non‐2′‐O‐methylated nucleotides. Modeling studies revealed that a space constraint within the RNA tunnel excluded a m7G‐capped RNA harboring ribose 2′‐O‐methylation modifications. The cytoplasmic RNA senor, retinoic acid inducible gene I (RIG‐I) recognizes uncapped RNA containing a triphosphate at the 5′‐end of RNA to activate innate immune signaling (Luo, Kohlway, Vela, & Pyle, 2012). RIG‐I, however like IFIT, is also able to differentiate between a capped RNA without and with the penultimate 2′‐O‐methylated nucleotide (Devarkar et al., 2016; Ramanathan et al., 2016; Schuberth‐Wagner et al., 2015). As such by binding non‐2′‐O‐methylated nucleotides at the 5′‐end of RNA, IFIT and RIG‐I proteins discriminate between cellular mRNA and pathogen RNAs (Abbas et al., 2017; Devarkar et al., 2016; Ramanathan et al., 2016; Schuberth‐Wagner et al., 2015). Additionally, by binding a “non‐self” RNA without a 2′‐O‐methyl modification, IFIT proteins also inhibit translation of the viral RNA by competing with cellular cap‐binding enzymes for the m7G‐cap (Habjan et al., 2013; Kimura et al., 2013; Kumar et al., 2014). Notably, the IFIT proteins restrict translation of viral mRNA by interacting with the eukaryotic initiation factor eIF3 that is pivotal in the assembly of translation initiation complexes (Hinnebusch, 2006). IFIT interaction with eIF3 blocks the assembly of the ternary complex containing eIF2, GTP and the methionine initiator tRNA (Met‐tRNAi) and/or the preinitiation complex comprised of the ternary complex, mRNA, and the 40S ribosome (Hui, Bhasker, Merrick, & Sen, 2003; Hui, Terenzi, Merrick, & Sen, 2005; Terenzi, Hui, Merrick, & Sen, 2006). Thus, the 2′‐O‐methyl modification facilitates evasion of antiviral factors induced by the production of type I interferon in response to viral infection (Figure 2).
Figure 2.
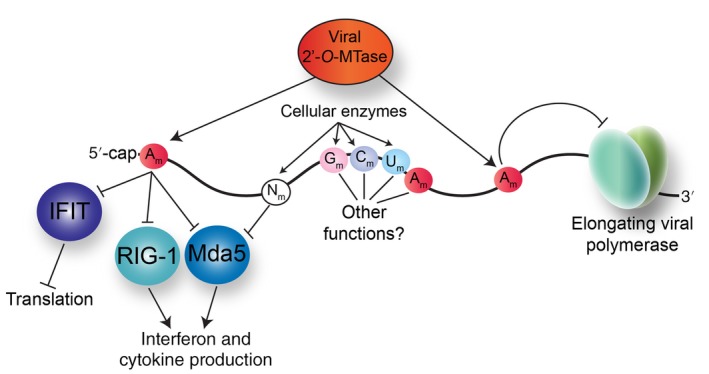
Consequences of ribose 2′‐O‐methyl modifications on RNA virus gene expression. Methylation of the 2′‐O‐position on the ribose can occur on all nucleotides and may be deposited by both viral and cellular 2′‐O‐methyltransferases (2′‐O‐MTase). The 2′‐O‐methylation of the penultimate nucleotide of the 5′‐end of the viral RNA limits innate immune sensing, first by masking the recognition of the viral RNA by the cytosolic RNA sensors RIG‐I and Mda5 to prevent transcription of innate immune response genes, and second by preventing the interaction with IFIT proteins which downregulate translation. 2′‐O‐methylated nucleotides (Nm) within viral RNA also function to mask the viral RNA from cytosolic RNA sensors. Deposition of internal 2′‐O‐methyl groups on adenosines (Am) by the viral 2′‐O‐MTase inhibit the elongation of the viral RNA‐dependent RNA polymerase during replication
The ribose 2′‐O‐methyl modification also masks viral RNA from the cytoplasmic innate immune response RNA sensor melanoma differentiation‐associated protein 5 (Mda5) (Figure 2) (Züst et al., 2011). Similar to WNV, the positive‐sense RNA genome of coronaviruses, a virus in the Coronaviridae family, contains a Cap 1 structure that is introduced by viral m7G and ribose 2′‐O MTases (Bouvet et al., 2010; Chen et al., 2009; Decroly et al., 2008). Targeted mutation of the coronavirus Nsp16 protein ablates viral 2′‐O MTase activity (Decroly et al., 2008). Infection studies comparing wild‐type virus and Nsp16 mutant coronaviruses showed that the mutant viruses had decreased replication kinetics but induced higher levels of IFN‐β (Züst et al., 2011). Infection studies in Ifnar‐deficient macrophages similarly showed high IFN‐β levels. These data together indicated that increased interferon production was in response to a cytosolic RNA sensor, and not a result of the secondary signal arising from interferon production (Züst et al., 2011). Interferon production in macrophages in response to coronavirus infection is dependent on the RNA sensor Mda5 (Roth‐Cross, Bender, & Weiss, 2008; Zalinger, Elliott, Rose, & Weiss, 2015). Therefore, as expected, interferon levels were undetectable in Mda5‐deficient macrophages infected with wild‐type and 2′‐O MTase‐deficient coronaviruses. These data were corroborated by demonstrating that 2′‐O MTase deficient coronaviruses replicated in mice deficient in the RNA sensors Mda5 and Toll‐like receptor 7 (TLR7) to the same extent as in Ifnar‐deficient mice (Züst et al., 2011). Therefore, the ribose 2′‐O‐methyl group on the first nucleotide marks viral RNA as “self” which prevents the activation of the RNA sensors Mda5, TLR7, and RIG‐I, as well as avoids recognition of RNA by the interferon‐induced translation inhibitors, IFIT1 and IFIT2 (Figure 2).
2.2.2. Internal 2′‐O‐methylation of nucleotides in viral RNA
Studies investigating the 2′‐O MTase activities of WNV and the closely related Dengue virus (DENV) NS5 protein found that the NS5 protein 2′‐O‐methylated not just the penultimate nucleotide of the genome (adenosine) but also adenosines within the genome (Dong et al., 2012). The first clue to such activity came from an assay that measured methylation of uncapped RNA, which demonstrated that NS5, or the methyltransferase (MTase) domain of NS5 alone, could specifically methylate polyA, but not polyG, polyC, or polyU oligonucleotides. The NS5 MTase methylated a 12‐nucleotide oligomer containing just adenosine or N6,N6‐dimethyladenosine, but not an oligomer that contained 2′‐O‐methyladenosines (Dong et al., 2012). The internal methylation activity modified homogenous oligonucleotides as well as uncapped truncated viral RNA, and 18S and 28S rRNA, indicating that this activity was not sequence specific. The authors also showed that viral genomes isolated from infected Aedes albopictus cell culture supernatants contained an estimated 3.4 ± 0.5 2′‐O‐methylated adenosines per genome (Dong et al., 2012). Interestingly the internal 2′‐O‐methyladenosines activity required the same K‐D‐K‐E motif required for 2′‐O methylation of the 5′ cap. Transfection and expression of in vitro transcribed viral RNA containing internal 2′‐O‐methyladenosines showed that the RNA modifications modestly decreased viral translation. Notably, however, the installation of 2′‐O‐methyladenosines in the viral RNA significantly impaired replication, and in particular elongation of the viral RNA‐dependent RNA polymerase (Figure 2) (Dong et al., 2012). Incorporation of 2′‐O‐methylated uridine, guanosine, and adenosine into short RNA strongly inhibits cytokine and interferon production resulting from activation of the endosomal immune sensor TLR7 (Petes, Odoardi, & Gee, 2017; Robbins et al., 2007). Therefore, in addition to modulating virus gene expression and interactions with the host, it is also possible that the internal 2′‐O‐methylated adenosines limit innate sensing during virus entry, which for WNV and DENV occurs via the endosome, although such an effect remains to be verified (Cruz‐Oliveira et al., 2015). Interestingly, the ability of the methyltransferase to internally 2′‐O‐methylate adenosines is not unique to WNV and DENV methyltransferases. Indeed, the methyltransferase domain in the polymerase or L (large) protein of Ebola virus, a negative‐sense RNA virus, was also shown to 2′‐O‐methylate internal adenosines, although the function of these internal modifications is unknown (Martin et al., 2018). This internal 2′‐O‐methylation addition by the Ebola virus methyltransferase however appears to be virus‐specific, as another negative‐strand RNA virus, but in a different virus family to Ebola virus, namely human metapneumovirus (hMPV), lacked this activity (Martin et al., 2018).
The RNA from two different viruses within the Retroviridae family namely, HIV‐1 and murine leukemia virus (MLV) was recently shown to contain internal 2′‐O‐methylated nucleotides (Courtney et al., 2019; McIntyre et al., 2018; Ringeard et al., 2019). Notably, these 2′‐O‐methylated nucleotides in the HIV‐1 genomes were shown to be installed by FTSJ3, a cellular 2′‐O MTase (Ringeard et al., 2019). Efficient transcription of HIV‐1 RNA is facilitated by the viral Tat protein that interacts with the transactivation response (TAR) element, an RNA structure at the 5′‐end of the viral genome, which then recruits cellular RNA polymerase II (Ott, Geyer, & Zhou, 2011). The cellular TAR RNA binding protein (TRBP), which is known to interact with HIV‐1 RNA, also forms a complex with FTSJ3 and MAT2A, a SAM metabolism protein (Ringeard et al., 2019). Using in vitro assays, Ringeard et al. (2019) showed that FTSJ3 isolated from cells, or recombinantly expressed, could 2′‐O‐methylate radiolabeled m7GpppG to form m7GpppGm within the context of m7G‐capped HIV‐1 RNA, and an oligonucleotide containing 27‐adenosines or uridines or guanosines, but not an oligonucleotide containing 2′‐O‐methylated adenosines. RiboMethSeq and validation by primer extension analyses showed 2′‐O‐methylated nucleotides were deposited by FTSJ3 at 17 positions within HIV‐1 RNA isolated from viral particles (Ringeard et al., 2019). The function of these internal 2′‐O‐methylated nucleotides was dissected by first isolating viral RNA from virions produced in FTSJ3 knocked‐out cells, and then transfecting this RNA into a pro‐monocytic cell line. HIV‐1 RNA lacking (or having a reduced number of 2′‐O‐methylated nucleotides) dramatically upregulated interferon‐α and interferon‐β mRNA levels by specifically activating the Mda5 cytosolic RNA sensor (Ringeard et al., 2019). Thus, similar to the ribose 2′‐O‐methyl modification of the first nucleotide, internal 2′‐O‐methylations may also enable viruses to escape immune sensing (Figure 2). It is also possible that these 2′‐O‐methylated nucleotides within the HIV‐1 genome might limit reverse transcriptase activity, similar to the internal 2′‐O‐methyladenosines restricting the DENV NS5 RNA‐dependent RNA polymerase (Dong et al., 2012). Such a mechanism however remains to be demonstrated.
Mass spectrometry analyses of DENV, Zika virus (ZIKV), hepatitis C virus (HCV), poliovirus, and HIV‐1 RNA genomes similarly revealed 2′‐O‐methyladenosines, as well as 2′‐O‐methylguanosines, 2′‐O‐methylcytosines, and 2′‐O‐methyluridines on the isolated viral RNAs (Lichinchi, Zhao, et al., 2016; McIntyre et al., 2018). While a percentage of 2′‐O‐methyladenosines detected on the DENV and ZIKV genomes is likely the result of viral methyltransferase activity, the addition of the other 2′‐O‐methylated nucleotides on DENV and ZIKV RNA might be deposited by cellular 2′‐O MTases. The presence of 2′‐O‐methylated nucleotides on HCV and poliovirus RNA isolated from infected cells and virions was surprising, particularly as both viruses replicate in the cytoplasm, have an uncapped RNA genome, and do not encode a methyltransferase. Thus, the deposition of 2′‐O‐methyl marks on the viral RNA is likely the result of cellular 2′‐O MTase activities. Recently fibrillarin, the cellular methyltransferase known to 2′‐O‐methylate rRNA in the nucleolus (Reichow, Hamma, Ferré‐D'Amaré, & Varani, 2007), was shown to be required by Hendra and Nipah viruses, as well as by other viruses within the same Paramyxoviridae family such as measles and mumps virus (Deffrasnes et al., 2016). Specifically, RNAi depletion of fibrillarin decreased viral titers, which could be rescued by overexpressing fibrillarin resistant to siRNA targeting. Using a single‐cycle infection assay and a minigenome assay, Deffrasnes et al. (2016) report that fibrillarin impacted viral RNA synthesis. Future assays will likely determine whether the effect on viral replication is the result of fibrillarin 2′‐O‐methylating Hendra and Nipah virus mRNAs or negative‐sense genomes to affect translation, transcription, and/or replication by the viral polymerase protein. Since fibrillarin 2′‐O‐methylates rRNA and is important for pre‐rRNA processing, it was also possible that depletion of fibrillarin altered the abundance, 2′‐O‐methylation status or composition of functional ribosomes to affect translation of viral proteins and ultimately viral RNA synthesis (Erales et al., 2017; Mauro & Edelman, 2002; Z. Shi et al., 2017; Tollervey, Lehtonen, Carmo‐Fonseca, & Hurt, 1991). Deffrasnes et al. (2016) however, showed that depletion of fibrillarin had no effect on influenza virus infection, suggesting the effect on Hendra virus was not a result of ribosome biogenesis or translation deficiencies.
Beyond the deposition of the 2′‐O‐methyl modification at the 5′‐end of RNA which functions as a marker of “self” (Table 1) (Hyde & Diamond, 2015), the role of internal 2′‐O‐methylation in cellular and viral RNAs is difficult to predict, particularly as methylation of the 2′‐O position on the ribose does not affect base pairing (Prusiner, Yathindra, & Sundaralingam, 1974). 2′‐O‐methylation of nucleotides protects RNA from hydrolytic cleavage and stabilizes nucleotide conformation to limit the shape and flexibility of the RNA strand (Lane & Tamaoki, 1967; Trim & Parker, 1970). In addition to affecting interactions with cellular components, processivity of the viral polymerase and evading immune sensing (Figure 2), 2′‐O‐modification in RNA viruses might also impact actions required for efficient replication such as impair unwinding of viral RNA, or restrict critical changes in RNA configurations that modulate specific steps in the infectious cycle (Iglesias & Gamarnik, 2011; Nicholson & White, 2015). Different high‐throughput sequencing strategies have recently been developed to identify 2′‐O‐methylated nucleotides (Dai et al., 2017; Incarnato et al., 2017; Marchand, Blanloeil‐Oillo, Helm, & Motorin, 2016; Zhu, Pirnie, & Carmichael, 2017), which together with known structures of viral RNA and protein interactions, might provide new insight into the role of this modified nucleotide on viral gene expression.
3. EDITING NUCLEOTIDES WITHIN VIRAL RNAS
RNA editing is a posttranscriptional process that modifies RNA by inserting, deleting, or substituting nucleotides within the nascent RNA. Here we concentrate on the consequences of two RNA editing events on viral gene expression namely the isomerization of uridine‐to‐5‐ribosyl uracil (or pseudouridine; Figure 3) and the deamination of adenosine‐to‐inosine (Figure 4) (Table 1). The writer enzymes responsible for these RNA editing events are known (George, John, & Samuel, 2014; X. Li, Ma, & Yi, 2016; Slotkin & Nishikura, 2013; Spenkuch, Motorin, & Helm, 2014). Taking into account the enzymatic activities required to revert pseudouridine‐to‐uridine and inosine‐to‐adenosine, and that the eraser enzymes for pseudourine and inosine are to date unknown, suggest that these RNA editing events are irreversible. Therefore, compared to reversible RNA modifications, putative irreversible editing on viral RNAs has distinct consequences on viral infection (Table 1).
Figure 3.
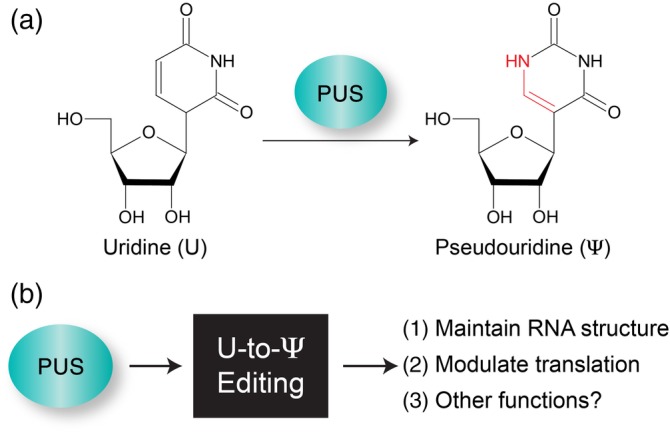
Editing of Uridine‐to‐Pseudouridine. (a) Pseudouridine synthase (PUS) catalyzes the isomerization of uridine (U) to form 5‐ribosyl uracil or pseudouridine (Ψ). (b) Functional outcomes of U‐to‐Ψ editing
Figure 4.
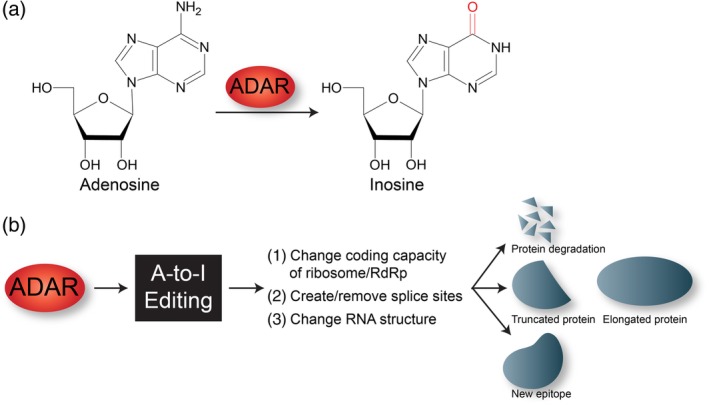
Adenosine‐to‐Inosine editing. (a) ADAR deaminates the C6 position in adenosine (A) to produce inosine (I). (b) A‐to‐I editing in RNA alters RNA metabolism to impact different cellular processes. RdRp, viral RNA‐dependent RNA polymerase
3.1. Uridine‐to‐pseudouridine (Ψ) editing
Pseudouridine is the most abundant RNA modification and is often referred to as the fifth nucleotide (Cohn & Volkin, 1951). Pseudouridine synthases (PUS) form pseudouridine by catalyzing the isomerization of uridine to form 5‐ribosyl uracil (Figure 3) (Cohn, 1959). The isomerization of uridine does not change the Watson–Crick base‐pairing with adenosine, however pseudouridine is also able to base pair with any of the other nucleotides. While pseudouridine was first discovered in 1951 in rRNA and tRNA, the application of next generation sequencing coupled with CMCT (cyclohexyl‐N′‐(2‐morpholinoethyl) carbodiimide metho‐p‐toluene‐sulfonate) labeling has enabled transcriptome‐wide analysis of pseudouridine and the identification of this modification on noncoding RNAs and mRNAs (Carlile et al., 2014; X. Li et al., 2015; Lovejoy, Riordan, & Brown, 2014; Schwartz et al., 2014). Despite the abundance and availability of transcriptome‐wide approaches to map pseudouridine, there are only a few reports identifying pseudouridine in viral RNAs, and the function of pseudouridine in regulating viral gene expression is largely unexplored.
In the plant turnip yellow mosaic virus (TYMV), a virus within the Tymoviridae family, the last 82 nucleotides of the genomic RNA (gRNA) form a “tRNA‐like” domain. Within this “tRNA‐like” domain, one of the stem‐loop structures mimics the T‐arm of tRNA which contains pseudouridine (Becker, 1998). Incubation of the TYMV‐RNA fragment with yeast extract or purified pseudouridine synthases PUS1 and PUS4 resulted in pseudouridylation of the RNA fragment (Becker, 1998). Interestingly, the intergenic region of the RNA3 segment of brome mosaic virus (BMV) within the Bromoviridae family contains a similar T‐pseudouridine stem loop structure of RNA (Baumstark & Ahlquist, 2001). Baumstark and Ahlquist showed that in BMV‐infected yeast and plants the intergenic region is pseudouridylated at a position that would correspond to the modified nucleotide in tRNA (Baumstark & Ahlquist, 2001). Notably, mass spectrometry profiling of RNA modification landscapes on the genomes of positive‐sense RNA viruses isolated from infected cells and virions showed that pseudouridine is an abundant modification on viral RNAs (McIntyre et al., 2018). Interestingly, PUS proteins were identified in a CRISPR screen for host factors targeting viruses in the Flaviviridae family such as HCV and DENV (Marceau et al., 2016). Although the PUS proteins were not the leading antiviral factors identified in the screen, these data lend support for future studies examining the impact of pseudouridine during viral infection.
Although epitranscriptomic marks installed on viral RNAs are the focus of this overview it is worth noting that RNA modifications within cellular RNAs such as tRNAs may also influence viral gene expression. For instance, reverse transcription of HIV RNA is initiated by the interaction of the cellular tRNALys3 with the primer binding site within the HIV genome (Mak & Kleiman, 1997), where the interaction between the HIV RNA and the tRNALys3 was found to be stabilized by the pseudouridine within the cellular tRNA (Bilbille et al., 2009). Additionally, cellular tRNAs may suppress or promote translation of viral proteins. For example, a virus within the Virgaviridae family namely Tobacco mosaic virus has a weak UAG stop codon between two open reading frames where read‐through is required to produce the putative RNA‐dependent polymerase (Zerfass & Beier, 1992). Interestingly, tRNATyr isolated from tobacco and yeast contains pseudouridine in the anticodon loop (GΨA). Rather than promoting read‐through, the interaction of this tRNATyr with the UAG stop codon counterintuitively terminates translation and thus inhibits the production of the larger protein. Zerfass and Beier (1992) showed that suppression of the translation of the viral polymerase was specifically the result of the pseudouridine within the anticodon of the tRNATyr.
In addition to pseudouridine being an abundant RNA modification, stress has also been shown to shown to affect the levels of pseudouridines (Carlile et al., 2014; X. Li et al., 2015), suggesting this RNA modification is inducible. Indeed, virus infections induce a stress response (Lloyd, 2013), and epitranscriptome‐wide analysis of mock‐ and RNA virus‐infected cells revealed undulations in pseudouridine abundance during infection (McIntyre et al., 2018). In mRNAs, pseudouridines were identified within the 5′ and 3′ untranslated regions (UTRs) as well as within the coding region (Carlile et al., 2014). Considering this broad distribution of pseudouridine within an RNA, how might this RNA modification impact viral gene expression? Similar to tRNAs (Davis, 1995; Newby & Greenbaum, 2002), pseudouridines within viral RNAs might regulate or stabilize for RNA structure or affect interactions with tRNAs to modulate the viral proteome by frameshifting, misreading, or suppressing stop or nonsense codons. Incorporation of pseudouridines into synthetic mRNA facilitates immune evasion (Karikó et al., 2008), stabilizes mRNAs (Anderson et al., 2011; Karikó et al., 2008), impacts translation, and limits activation of the double‐stranded RNA protein kinase (Anderson et al., 2010; Aspden & Jackson, 2010), the stress‐ and interferon‐induced kinase that regulates cellular translation during stress by phosphorylating the translation initiation factor eIF2α (Dalet, Gatti, & Pierre, 2015). Even these limited functions would have broad impacts on viral gene expression (Table 1). That the enzymes responsible for installing pseudouridine are known and high throughput approaches are available to identify the positions of pseudouridines within viral RNA presents unique and exciting opportunities to investigate the function this abundant nucleotide within different viral systems.
3.2. Adenosine‐to‐inosine editing
Adenosine‐to‐inosine (A‐to‐I) editing is another abundant RNA modification (Danecek et al., 2012; J. B. Li et al., 2009; Pfaller et al., 2018). The enzyme adenosine deaminase acting on RNA (ADAR) catalyzes this A‐to‐I editing event in regions of double stranded RNA (dsRNA) by deaminating the C6 position of adenine (A) to produce inosine (I) (Figure 4) (George et al., 2014; Samuel, 2011; Slotkin & Nishikura, 2013). Because inosine acts in a similar manner to guanosine (G), A‐to‐I conversions may have significant biological implications such as changing the coding capacity of the ribosome or viral RNA dependent RNA polymerases, creating or removing splice sites, and altering RNA structure (Figure 4). Consequently, viral infection may be promoted or limited by A‐to‐I editing of the viral RNA (Table 1). While there are three ADAR enzymes (ADAR‐1, ‐2, and ‐3) in mammalian cells, only ADAR‐1 and ‐2 have been shown to have A‐to‐I editing capabilities (Samuel, 2011). There are additionally two ADAR‐1 isoforms; ADAR‐1 p110 is constitutively expressed and localizes in the nucleus and ADAR‐1 p150, which is induced by interferon signaling, localizes in both the cytoplasm and nucleus. While not the focus of this overview, it is important to note that ADAR‐1 also functions with other proteins synthesized in response to interferon signaling to modulate viral infection (Samuel, 2011).
3.2.1. A‐to‐I editing of paramyxovirus RNA
Infection with measles virus, a negative‐strand RNA virus in the Paramyxoviridae family, is typically acute. However, a rare and late outcome of acute measles infection is subacute sclerosing panencephalitis (SSPE), where the virus establishes a persistent infection in the central nervous system (Griffin, Lin, & Nelson, 2018). Notably, SSPE patients have high titers against all measles virus proteins except against the matrix (M) protein, the nonglycosylated viral membrane protein (Hall, Lamb, & Choppin, 1979). Immunofluorescence studies of SSPE brain material similarly showed low expression of the M protein (Liebert, Baczko, Budka, & Ter Meulen, 1986). While M is not required for measles virus replication, the low abundance or absence of M expression inhibits the viral nucleocapsids from colocalizing with the viral glycoproteins at the cell membrane. Instead, the nucleocapsids accumulate in the cytoplasm and nucleus, thus promoting virus persistence by restricting virus spread (Patterson et al., 2001). Sequence analysis of M genes cloned from brain autopsies of SSPE patients revealed that 50% of uridine (U) residues in the M gene were changed to cytosine (C) (Cattaneo et al., 1988). The consequence therefore of these biased hypermutations included frameshift mutations and the introduction of a stop codon, which decreased M protein expression (Figure 4) (Cattaneo et al., 1988). ADAR was shown to be responsible for introducing the clusters of A‐to‐I (G) (or U‐to‐C, depending on RNA strand polarity) transitions during transcription and replication of measles virus (Bass, Weintraub, Cattaneo, & Billeter, 1989; Pfaller et al., 2015; Pfaller, Radeke, Cattaneo, & Samuel, 2014). ADAR has been reported to edit genomes of other viruses in the Paramyxoviridae family such as human respiratory syncytial virus (hRSV), parainfluenza virus 5 and hMPV (Martínez & Melero, 2002; Rima et al., 2014; Samuel, 2011; van den Hoogen et al., 2014). Similar to measles virus, biased hypermutations were identified by sequence analysis in the glycoprotein (G) of hRSV changing the amino acid sequences and an antibody epitope within the G protein (Figure 4) (Martínez et al., 1997; Rueda et al., 1994). That ADAR‐1 is able to edit paramyxovirus genomes is intriguing particularly because the viral RNA is tightly associated with the nucleocapsid proteins (Gutsche et al., 2015). Thus, the double‐stranded RNA structures recognized by ADAR‐1 are unlikely to form and to be edited from A‐to‐I. It is however possible that if nucleocapsid‐RNA genome interactions were disrupted, the viral genomes may be edited (Pfaller et al., 2014, 2015, 2018). Interestingly, the installation of inosine masks RNA from detection by the cytoplasmic sensor protein of dsRNA Mda5, thus preventing the synthesis of interferon and other inflammatory proteins (Liddicoat et al., 2015; Mannion et al., 2014). Thus, A‐to‐I editing in viral RNAs can introduce mutations that affect biological outcomes as well as suppress the innate immune response (Table 1) (Pfaller et al., 2018).
3.2.2. A‐to‐I editing of the hepatitis δ virus RNA
Perhaps the viral lifecycle most intricately tied to A‐to‐I editing is that of hepatitis δ virus (HDV), the only virus in the Deltaviridae family. Coinfection of HDV with hepatitis B virus (HBV) increases the risk of sever liver damage in HBV patients (Lempp, Ni, & Urban, 2016). HDV is a subviral pathogen in that the RNA genome encodes only one protein namely hepatitis delta antigen (HDAg), yet HDV is disseminated by co‐opting the HBV envelope proteins which encapsidates the HDV RNA (Rizzetto et al., 1980). The HDAg is an essential protein that directs two distinct steps in the virus life cycle, namely replication and assembly. These steps are regulated by distinct forms of HDAg. HDAg‐S, the small form of the protein, modulates replication, while the large form (HDAg‐L) inhibits replication and promotes virus assembly (Kuo, Chao, & Taylor, 1989; Ryu, Bayer, & Taylor, 1992). The circular negative‐sense RNA HDV genome is replicated through an RNA intermediate, the antigenome, which also serves as a template for transcription of viral mRNA. Notably, a significant number of the antigenomes were found to have a very specific uridine‐to‐cytidine substitution (Casey, Bergmann, Brown, & Gerin, 1992). An in vitro assay, using a double‐stranded RNA deaminase isolated from Xenopus levis eggs, radiolabeled in vitro transcribed antigenome RNA and thin layer chromatography, demonstrated inosine within the HDV RNA (Polson, Bass, & Casey, 1996). Using an editing assay and site‐directed mutagenesis studies, adenosine 1012 on the antigenome was shown to be specifically changed to inosine (Casey & Gerin, 1995; Polson, Ley, Bass, & Casey, 1998). In converting adenosine to inosine, which like guanosine preferentially base‐pairs with cytosine, the UAG amber stop codon of HDAg‐S is changed to UIG (or UGG), a tryptophan codon, thus permitting the translation of the elongated protein HDAg‐L (Figure 4). An important contribution to this specific editing comes from the structure of the HDV RNA. Notably, the HDV genome contains significant complementarity within the circular RNA to form a predicted rod‐like dsRNA structure, where stabilizing or destabilizing the secondary structure modulates the extent of A‐to‐I editing (Jayan & Casey, 2005). Additionally, the extent of editing is influenced by the nucleotides surrounding the edited adenosine, as well as by the nucleotides 3′ of the editing site (Casey et al., 1992; Jayan & Casey, 2005). Editing was shown to occur during the replication step or synthesis of the antigenome (Sato, Cornillez‐Ty, & Lazinski, 2004). Thus, the secondary RNA structure and extent of editing influence HDAg‐L expression and synthesis of virus production. Because HDAg‐L inhibits replication, HDV maintains an exquisite balance between replication, A‐to‐I editing, and virus production (Table 1) (Sato et al., 2004). Assays using a nonreplicating HDV antigenome reporter and overexpression of ADAR‐1 and ADAR‐2 revealed that both deaminases are able to edit the HDV antigenome (Sato, Wong, & Lazinski, 2001). Subsequent studies however, using RNAi to differentially deplete cells of ADAR‐2 or ADAR‐1 p150, showed that ADAR‐2 minimally, if at all, edited HDV RNA. In contrast, introduction of inosine into the HDV RNA was inhibited following depletion of both ADAR‐1 p110 and p150. Depletion of ADAR‐1 and overexpression of either ADAR‐1 p110 or p150 revealed that the nuclear form of ADAR‐1 p110 more efficiently promoted editing of the antigenome than ADAR‐1 p150 (Wong & Lazinski, 2002). ADAR‐1 p150 is only expressed following interferon induction, and the protein abundance is a fraction of the ADAR‐1 p110 form, yet ADAR‐1 p150 also edits the HDV RNA to promote HDAg‐L expression (and downregulation of HDV replication). As such, pegylated interferon‐α has been used as an antiviral treatment of HDV, although newer drugs targeting other steps in the infectious cycles, and putatively fewer adverse effects, are being developed (Koh, Da, & Glenn, 2019; Mentha, Clément, Negro, & Alfaiate, 2019).
3.2.3. A‐to‐I editing of HIV‐1 RNA
The positive‐sense RNA genome of HIV‐1, a lentivirus within the Retroviridae family, is reverse transcribed into a double‐stranded DNA provirus that is integrated into the host DNA (Hu & Hughes, 2012). HIV‐1 RNAs, like cellular mRNAs, are transcribed by the cellular RNA polymerase II and may be edited by ADAR (Doria et al., 2009; McIntyre et al., 2018; Phuphuakrat et al., 2008). Indeed inosines were first identified in the stem structure of the TAR element following injection of the in vitro transcribed TAR RNA into the nucleus of Xenopus oocytes and thin layer chromatography analysis (Sharmeen, Bass, Sonenberg, Weintraub, & Groudine, 1991). Deletion and mutagenesis studies identified the position of the edited adenosine, and editing was found to be dependent on Tat, the HIV‐1 activator of gene expression (Sharmeen et al., 1991). Overexpression of ADAR‐1 and ADAR‐2 has been shown to increase the abundance of HIV‐1 proteins (Doria et al., 2009; Phuphuakrat et al., 2008). This effect on HIV‐1 protein levels was shown to be dependent on the editing activity of ADAR. Conversely, depletion of ADAR with target‐specific siRNAs depleted HIV‐1 protein levels. The effect of ADAR on the viral protein levels was shown to be independent of the editing function and more likely a consequence of ADAR modulating translation by inhibiting the RNA‐activated protein kinase, PKR (Doria et al., 2009). Notably, this effect of ADAR on HIV‐1 protein levels was more robust than the impact of ADAR with editing capabilities. Modulation of ADAR‐1 abundance affected the levels of unspliced HIV‐1 RNA in the cytoplasm. Sequence analysis showed that A‐to‐G (I) changes in the 5′ UTRs of all HIV‐1 transcripts including within the TAR and Rev response element (RRE), and Tat coding sequence (Doria et al., 2009; Phuphuakrat et al., 2008). Introduction of specific A‐to‐G substitutions in the RRE resulted in a codon change, increased the levels of the capsid (p24) protein and unspliced RNA, as well as the production of HIV‐1 particles. Immunoprecipitation studies revealed that ADAR associates with both unspliced and spliced HIV‐1 transcripts (Doria et al., 2009). Surprisingly however, these particles had low infectivity likely because of effects on reverse transcription. A‐to‐I editing has also been reported in another retrovirus namely Rous‐associated virus type 1 (Felder et al., 1994). In particular the U3 sequence, within the long untranslated repeat, contains a high number of A‐to‐G (I) substitutions. Although, the conversions did not affect transcription or replication of the viral genome, the polyadenylation signal was changed resulting in read‐through transcription to include cellular gene sequences (Felder et al., 1994). A‐to‐I editing impacts retrovirus gene expression, however the extent to which this occurs within the virus family and whether the molecular consequences are virus‐specific remain to be determined.
3.2.4. A‐to‐I editing in single‐stranded positive‐sense viral RNA genomes
Inosine has also been identified in genomes of single‐stranded positive‐sense RNA viruses within the Flaviviridae family such as HCV, ZIKV, and DENV, and Picornaviridae family namely poliovirus (McIntyre et al., 2018; Taylor et al., 2005). Notably, A‐to‐I editing in HCV has been shown to exhibit antiviral activity (Taylor et al., 2005). In particular, interferon treatment of the hepatoma cell line stably expressing an HCV replicon decreased HCV RNA and protein levels. Reduced HCV protein levels were likely due to the effect of interferon‐mediated decrease in cap‐independent translation that is, translation from the HCV internal ribosomal entry site (IRES). Taylor et al. (2005) showed that overexpression of PKR (a regulator of translation induced by interferon) decreased replicon expression and overexpression of a dominant‐negative inactive PKR mutant modestly rescued IRES‐mediated activity in the replicon cell line. Interestingly however, the expression of an RNA that inhibits ADAR activity robustly rescued IRES‐mediated activity and HCV replicon RNA (Taylor et al., 2005). Sequence analysis of the HCV replicon RNA isolated from interferon‐treated cells showed mutative A‐to‐I editing, and thin‐layer chromatography demonstrated an increase in inosine in total RNA isolated from the replicon cells (Taylor et al., 2005). Together the data indicate that HCV RNA might be modified by ADAR, and that inhibiting ADAR activity promotes HCV gene expression. Bioinformatic analyses of ZIKV genomes from the African versus Asian lineages revealed more guanosine nucleotides, which may have arisen from A‐to‐I editing (Khrustalev et al., 2017; Piontkivska et al., 2017). These putative ADAR editing sites were found in sites of predicted secondary structure, and on both the positive and negative (a replication intermediate) RNA strands (Khrustalev et al., 2017). Knockout of ADAR in mouse embryonic fibroblasts induces an aberrant immune response and increased apoptosis (Liddicoat et al., 2015; Mannion et al., 2014). As such, ADAR editing of the ZIKV genomes has been proposed to be an evolutionary driver as well as a possibly contributor to the neuropathologies linked to the outbreak of ZIKV in the Americas (Lazear & Diamond, 2016).
A‐to‐I editing is frequently identified by bioinformatic analyses of A‐to‐I or A‐to‐G substitutions within sequencing data. Using this approach, it is clear that the genomes of many RNA viruses contain inosine (Samuel, 2011). Although, the consequence of A‐to‐I editing has only been investigated for a handful of RNA viruses, orthogonal validation of editing sites by chemical modification of inosine and high‐throughput sequence analyses (Cattenoz, Taft, Westhof, & Mattick, 2013; Knutson, Ayele, & Heemstra, 2018; Sakurai, Yano, Kawabata, Ueda, & Suzuki, 2010), and mechanistic studies will uncover the function of this abundant posttranscription modification in other RNA viruses.
4. MODIFICATION OF VIRAL RNA BY CELLULAR ENZYMES
4.1. N6‐methyladenosine
N6‐methyladenosine (m6A) is an abundant RNA modification found within cellular and viral RNAs (Gokhale & Horner, 2017; Gonzales‐van Horn & Sarnow, 2017; Kennedy, Courtney, Tsai, & Cullen, 2017; Roundtree, Evans, Pan, & He, 2017; H. Shi, Wei, & He, 2019). This dynamic RNA modification is deposited on RNA by the writer or methyltransferase enzymes METTL3 and METTL14 together with Wilms tumor 1‐associated protein and KIAA1429 (Figure 5) (Meyer & Jaffrey, 2017; H. Shi et al., 2019). Likewise, the methyl group may be erased by the demethylases FTO (fat mass and obesity associated protein) and ALKBH5 (α‐ketoglutarate‐dependent dioxygenase AlkB homology 5) (Figure 5) (Jia et al., 2011; Zheng et al., 2013). The nuclear and cytoplasmic RNA binding and reader proteins, YTH‐domain family of proteins (YTHDC1, YTHDC2, YTHDF1, YTHDF3, and YTHDF3) direct the posttranscriptional functions of m6A (Figure 5) (H. Shi et al., 2019). Compared to other RNA modifications, the m6A field is well established. This is in large part because of the availability of a specific anti‐m6A antibody and the application of immunoprecipitation and RNA‐seq approaches together with the identification of the writer, eraser, and reader enzymes (Figure 5). As a result there are now a number of studies describing a role for m6A in virus gene expression (Gokhale & Horner, 2017; Gonzales‐van Horn & Sarnow, 2017; Kennedy et al., 2017). The focus of this overview is on the impact of RNA modifications on RNA viruses. We however note a number of excellent studies describing a role for m6A during infection with DNA viruses such as simian virus 40 (SV40), adenovirus, HBV, herpes simplex virus, Karposi's sarcoma‐associated herpesvirus and Epstein–Barr virus (Canaani, Kahana, Lavi, & Groner, 1979; Finkel & Groner, 1983; Hesser, Karijolich, Dominissini, He, & Glaunsinger, 2018; Imam et al., 2018; Lang et al., 2019; Moss, Gershowitz, Stringer, Holland, & Wagner, 1977; Moss & Koczot, 1976; Sommer et al., 1976; Tan et al., 2017; Tsai, Courtney, & Cullen, 2018; Ye, Chen, & Nilsen, 2017).
Figure 5.
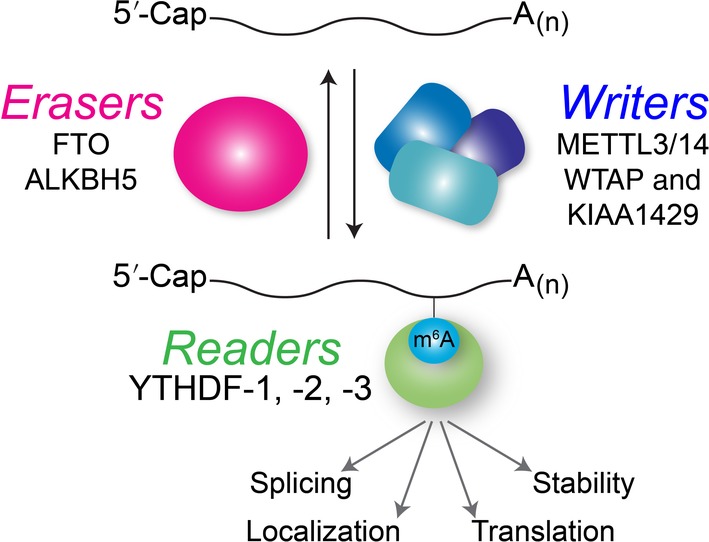
N6‐methyladenosine modification on messenger RNA and functions that are modulated by writer, eraser and reader proteins
4.1.1. Role of m6A in RNA viruses that replicate in the nucleus
The genome of influenza virus, a virus within the Orthomyxoviridae family, is comprised of eight segments of single‐stranded negative‐sense RNA (Bouvier & Palese, 2008). Early studies demonstrated that 55–60% of the influenza virus complementary RNA or mRNA contained methyladenosine, and a third of these modifications were found to be internal m6A (Krug et al., 1976). Moreover, analysis of the different influenza virus mRNAs revealed an uneven distribution of these m6A modifications (Narayan et al., 1987). The role of m6A on influenza virus RNA was recently investigated by Courtney et al. (2017), where knockout of the METTL3 writer decreased influenza virus protein and mRNA levels and virus titers, while overexpression of the YTHDF reader proteins promoted virus gene expression. RNA‐seq analyses of m6A and YTHDF‐binding sites showed putative modification sites in both the viral mRNAs and negative‐sense genomic RNA. Notably, the m6A sites were enriched towards the 3′‐end of the viral RNA consistent with m6A topology in cellular mRNAs (Courtney et al., 2017). Collective mutation of m6A sites decreased the infection kinetics and decreased virus pathogenicity, which together indicated that m6A on the viral RNA positively regulates influenza virus gene expression (Figure 6, Table 1) (Courtney et al., 2017).
Figure 6.
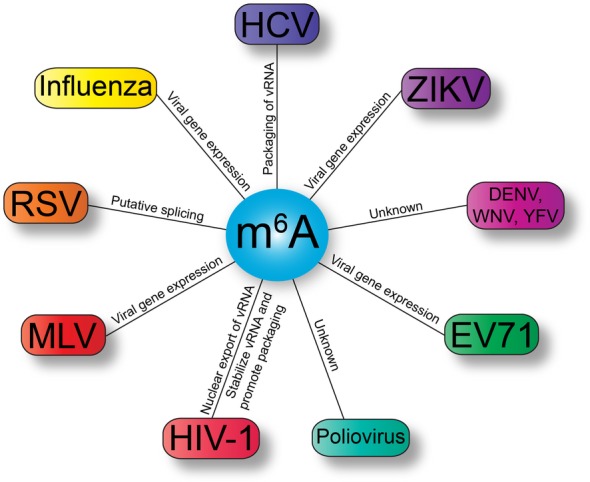
Viruses with N6‐methyladenosine (m6A) modifications in the viral RNA, and putative functions. Virus families shown include: Flaviviridae (purple) namely hepatitis C virus (HCV), Zika virus (ZIKV), Dengue virus (DENV), West Nile virus (WNV), and yellow fever virus (YFV); Picornaviridae (green) with enterovirus 71 (EV71) and poliovirus; Retroviridae (red) namely human immunodeficiency virus type 1 (HIV‐1), murine leukemia virus (MLV) and Rous sarcoma virus (RSV); and Influenza virus (yellow) within Orthomyxoviridae. vRNA, viral RNA
m6A writer enzymes recognize and modify adenosines within a consensus motif. Interestingly, this motif was first described from studies examining internal methylation on a virus in the Retroviridae family, avian sarcoma virus (ASV) (Dimock & Stoltzfus, 1977). Indeed, radiolabeling methyl‐moieties on B77 ASV RNA with [methyl‐3H]methionine showed that in addition to the Cap 1 structure, 14–16 internal methyl modification were m6A (Stoltzfus & Dimock, 1976). RNase digestion of radiolabeled RNA and chromatography demonstrated that adenosine within the motif (G>A)‐m6A‐C was methylated (Dimock & Stoltzfus, 1977). RNA‐seq studies have more precisely defined the motif as DRm6ACH (where D may be G, A or U, R represents the purines G and A, and H may be U, C or A) (Dominissini et al., 2012; Liu et al., 2014). Similar studies undertaken with another retrovirus Rous sarcoma virus (RSV) also identified 10–15 m6A modifications. These modifications were not randomly distributed, instead the majority of sites occurred within 500–4,000 nucleotides of the 3′‐end of the RNA genome (Beemon & Keith, 1977). Indeed RNA‐seq topology studies of cellular mRNAs have demonstrated that m6A sites are enriched in the 3′ UTR near termination sites, 5′ UTR and long exons (Dominissini et al., 2012; Meyer et al., 2012; J. Zhou et al., 2015). A subsequent biochemical study described seven m6A sites in the RSV RNA, and while the modification was dynamic, a number of the modified sites were upstream of consensus splice acceptor sequences. These data together with the accumulation of unspliced viral RNA following incubation of cells with cycloleucine, an inhibitor of internal methylation (Dimock & Stoltzfus, 1978; Stoltzfus & Dane, 1982) raised the possibility that m6A might modulate splicing of RSV RNA (Figure 6) (Kane & Beemon, 1985).
While m6A was identified on the RNAs of both RNA and DNA viruses in the 1970s (Beemon & Keith, 1977; Canaani et al., 1979; Hashimoto & Green, 1976; Krug et al., 1976), a renaissance in viral epitranscriptomics occurred following recent studies examining m6A and HIV‐1 gene expression (Kennedy et al., 2016; Lichinchi, Gao, et al., 2016; Lu et al., 2018; Tirumuru et al., 2016). Immunoprecipitations and RNA‐seq studies identified numerous m6A sites in the HIV‐1 genome. Interestingly, m6A was found in the regulatory 5′ and 3′ long terminal repeats (Kennedy et al., 2016; Tirumuru et al., 2016), Env and Rev genes and the RRE (Kennedy et al., 2016; Lichinchi, Gao, et al., 2016; Tirumuru et al., 2016). Knockdown or overexpression studies of the m6A writer, eraser, and reader proteins were undertaken to dissect the role of m6A in HIV‐1 infection. Mechanistically, installation of m6A on the HIV‐1 genome was found to modulate HIV‐1 gene expression at early and late steps during infection (Lichinchi, Gao, et al., 2016; Lu et al., 2018; Tirumuru et al., 2016). By changing the abundance of the reader proteins and measuring early and late reverse transcription (RT) products, Tirumuru et al. (2016) demonstrated that m6A influences steps prior to the import of cDNA into the nucleus and integration into the cellular DNA. By overexpressing the reader proteins, more HIV‐1 genomic RNA was immunoprecipitated with the reader proteins yet the overall abundance of viral RNA decreased at early times following infection, suggesting that m6A affects the stability of the viral RNA (Figure 6) (Tirumuru et al., 2016). Additional investigations will establish whether the presence of m6A alone or as a result of reader proteins binding the m6A moiety hinders reverse transcription (RT) activity such that aborted RT products are degraded or whether the recognition of the viral RNA by the reader proteins targets HIV‐1 RNA for turnover. Indeed, the spatiotemporal mechanism of this function will be interesting to uncover particularly as the RT activity putatively occurs within the confines of the p24 capsid shell (Campbell & Hope, 2015) and the reader proteins are not packaged into viral particles (Tirumuru et al., 2016). Lichinchi, Gao, et al. (2016) identified two conserved m6A sites within a stem loop of the RRE. This stem loop is critical for binding the Rev protein and directing export of the viral RNA out of the nucleus. Both sites were verified by primer extension assay using either AMV or Tth reverse transcriptase (RT), where the presence of m6A in RNA stalls the Tth RT reaction. Furthermore, by modulating the writer and eraser levels, and thus the level of m6A modification on the genome, the extent of Rev binding to the RRE was altered. Notably, adenosine‐to‐guanosine mutation to disrupt the m6A consensus motifs, demonstrated that ablation of the second m6A site decreased the genomic RNA levels and nuclear export (Figure 6) (Lichinchi, Gao, et al., 2016). Two putative m6A binding sites the HIV‐1 5′ UTR were identified at or within proximity of the primer‐binding site (PBS) and dimer initiation sequence (DIS), respectively. PBS binding of cellular tRNALys is critical for new rounds of reverse transcription, while the DIS facilitates encapsidation of two HIV‐1 genomes. Disruption of either or both m6A sites modestly affected newly synthesized HIV‐1 proteins, however markedly impacted subsequent HIV‐1 infections. These data, together with the HIV‐1 RNA‐dependent interaction of m6A reader proteins with the unprocessed Gag protein suggest that m6A might also influence virus assembly (Figure 6) (Lu et al., 2018). Twenty putative m6A sites were recently identified in the RNA of another retrovirus, MLV (Courtney et al., 2019). Mutation of three m6A sites in the Env gene decreased intracellular and virion proteins, but not viral RNA levels when expressed from the proviral expression plasmid, and viral gene expression was reduced in subsequent infections. Overexpression of the YTHDF2 reader also increased MLV proteins, demonstrating a role for m6A in MLV gene expression (Figure 6) (Courtney et al., 2019).
4.1.2. Role of m6A in RNA viruses that replicate in the cytoplasm
Because m6A is predominantly installed on RNA posttranscriptionally in the nucleus, viruses that replicate exclusively in the cytoplasm were thought to lack m6A modifications. However, recent studies show that the RNA genomes of a number of viruses in the Flaviviridae and Picornaviridae families are m6A‐modified (Figure 6, Table 1) (Gokhale et al., 2016; Hao et al., 2019; Lichinchi, Zhao, et al., 2016; McIntyre et al., 2018). During HCV infection, manipulation of writer and eraser abundance altered intracellular HCV protein levels, virus titers, and the amount of HCV RNA in the extracellular media thus affecting the production of new virus particles (Gokhale et al., 2016). Notably, writer, eraser, and reader protein abundance did not influence virus replication. Interestingly, the reader proteins were found to colocalize with the core protein at lipid droplets, which are well characterized sites of virus assembly suggesting that the m6A regulatory proteins might modulate virus assembly (Gokhale et al., 2016; Vieyres & Pietschmann, 2019). Fourteen putative m6A sites were identified along the viral RNA, with no apparent gene or distribution bias. Interestingly, mutation of four putative m6A sites in the envelope E1 gene dramatically increased virus titers without affecting HCV protein levels or translation or replication of the HCV genome. Disruption of m6A sites in the E1 gene decreased the amount of HCV RNA immunoprecipitated by the YTHDF2 reader but promoted an increased interaction with the core protein that surrounds the viral RNA (Gokhale et al., 2016). Therefore, this elegant study by Gokhale et al. (2016) demonstrated that the interaction of reader proteins with m6A‐modified HCV RNA regulated the packaging of viral RNA into new virus particles (Figure 6). m6A immunoprecipitation and RNA‐seq, and mass spectrometry, showed that the genomes of other viruses within the Flaviviridae family such as ZIKV, DENV, WNV, and yellow fever virus are also m6A‐modified (Gokhale et al., 2016; Lichinchi, Zhao, et al., 2016; McIntyre et al., 2018). Additional studies altering the abundance of the writer, eraser, and reader proteins during ZIKV and enterovirus 71 infection have shown m6A modulates viral gene expression (Figure 6) (Hao et al., 2019; Lichinchi, Zhao, et al., 2016).
4.2. 5‐Methylcytosine
Methylated cytosine is a common epitranscriptomic mark found in DNA and different species of RNA. This RNA modification in tRNA and rRNA has been shown to impact structure and function of both types of RNA molecules (Trixl & Lusser, 2019). Classical analytical approaches such as chromatography and mass spectrometry, and more recent high‐throughput sequencing technologies have been used to identify 5‐methylcytosine (m5C) within RNA (Trixl & Lusser, 2019). Although m5C studies using bisulfite RNA‐seq analysis reveal no clear RNA motif within mRNAs, m5C‐modified nucleotides were found to be enriched in regions that were CG‐rich, immediately downstream of the AUG start codon, and in 3′ UTRs (Yang et al., 2017). Notably for studies elucidating biological function, the enzymes critical for m5C deposition and function are known. In particular, RNA m5C methyltransferases belong to two specific subgroups of methyltransferases namely DNMT2 or NOL/NOP2/sun (NSUN) that use SAM as the methyl donor group (Bujnicki, Feder, Ayres, & Redman, 2004; Motorin, Lyko, & Helm, 2009). In mammals, seven NSUN proteins have been identified of which NSUN2 was found to specifically deposit m5C on mRNAs (Squires et al., 2012). While the m5C eraser enzyme is unknown, the RNA transport adaptor protein, Aly/REF export factor (ALYREF or THOC4) was identified as a putative reader enzyme that binds m5C sites within mRNA (Yang et al., 2017).
Early studies investigating the methylation status of Sindbis virus (SINV), a single‐stranded positive‐sense RNA virus in the Togaviridae family and alphavirus genus, used electrophoresis to separate 14C‐uridine‐labeled RNA and 3H‐methylated chemical groups. Dubin & Stollar, 1975 and Dubin et al. (1977) demonstrated that the SINV RNA was extensively methylated. In particular, one fourth to one half of all the internal methyl marks on intracellular viral RNA were attributed to m5C (Dubin et al., 1977), and m5C was found to exclusively decorate the subgenome that encodes the structural proteins required for assembly of new virus particles (Dubin & Stollar, 1975). Notably however, viral RNA packaged into virus particles lacked m5C marks. The position and function of m5C in SINV RNA are at present unknown, although a recent report demonstrated that Mt2, a gene encoding a m5C methyltransferase, was upregulated following infection of Drosophila with SINV (Bhattacharya et al., 2017). The goal of the study by Bhattacharya et al. (2017) was to elucidate the cellular mechanism by which the intracellular endosymbiont Wolbachia pipientis controlled virus infection in arthropods. Using Drosophila flies and a cell line as a model for SINV infection in mosquitoes, the authors established that Wolbachia‐infection downregulated SINV protein and RNA levels and decreased virus infectivity. An analysis of Drosophila immune response genes in mock‐infected flies, and in the absence or presence of Wolbachia demonstrated a significant upregulation of Mt2. Indeed modulation of Mt2 levels, by either knocked‐out or shRNA‐depletion approaches, increased SINV infectivity (Bhattacharya et al., 2017). Considering that intracellular SINV RNA, but not packaged viral RNA, is decorated with m5C it is possible that this epitranscriptomic mark is a critical spatiotemporal or innate immune response regulator of virus gene expression. Notably, an earlier report by Durdevic et al. (2013) revealed that Mt2 (or Dnmt) played a role in Drosophila innate immune responses. In particular, the authors showed that viral RNA and protein significantly increased following infection of Dnmt2 mutant flies with Drosophila C virus (DCV), a positive‐sense RNA virus within the Dicistroviridae family (Durdevic et al., 2013). Dnmt was also found to interact with DCV RNA raising the possibility that the viral genome is m5C‐modified and this modification plays a role in the Drosophila innate immune response (Durdevic et al., 2013).
Infection with DENV, ZIKV, HCV, poliovirus, and HIV‐1 alters the m5C RNA landscape in cells. Moreover m5C is present on the viral RNA isolated from cells and from released virus particles (McIntyre et al., 2018). Using mass spectrometry to identify epitranscriptomic marks as well as m5C‐seq analyses Courtney et al. (2019) recently showed that MLV genomic RNA, isolated from virions and cells, harbored a number of different epitranscriptomic marks. To elucidate the function of these putative m5C modification sites, mutations were introduced at four positions within the polymerase gene to putatively inhibit deposition of m5C (Courtney et al., 2019). RNA‐seq validation of mutant MLV genomic RNA showed that m5C marks at the four mutated sites were indeed absent. Interestingly, the abundance of m5C modifications when analyzed by RNA‐seq showed an overall decrease of m5C across the mutant viral genome. Transfection of the wild‐type and m5C mutant proviral expression vectors demonstrated that mutation of the DNA sequence had a small effect on the production of MLV Gag p65 and p30 proteins and no effect on the genomic RNA levels. An analysis of the proteins and RNA levels in MLV particles released into the media from transfection of the provirus DNA plasmid, demonstrated a two‐fold decrease on the levels of Gag proteins without affecting genomic RNA abundance. Courtney et al. (2019) determined that disruption of the four m5C sites in the polymerase gene did not affect virus particle assembly; however, the absence (and overall reduction of m5C marks) decreased subsequent virus infectivity. Consistent with these data, RNAi depletion of the writer enzyme NSUN2 also modestly decreased MLV Gag protein levels, while overexpression of Flag‐tagged NSUN2 had no effect (Courtney et al., 2019). The modest effects on MLV gene expression might in part be a result of the discrepancy between the mass spectrometry and m5C‐seq analyses, where on the MLV genomic RNA 13 and 40 m5C marks were identified by each respective detection approach. It is feasible that some of the m5C sites identified by m5C‐seq analyses might represent 5,2′‐O‐dimethylcytosine (m5Cm) requiring regulation by additional methyltransferases. Regardless, the data support a role for m5C in MLV infection. Interestingly, a subsequent round of infection of the newly produced mutant virions revealed a decrease in infectivity. This observation raises the possibility that similar to ribose 2′‐O‐methylation and inosine m5C masks the viral RNA from innate immune sensing. Alternatively, m5C within MLV genomes influences RNA structure or another critical step such as reverse transcription. Notably detection strategies to map m5C sites are available, and the m5C writer and reader enzymes are known, which together open new prospects to unravel the role of m5C during virus infection.
4.3. The viral epitranscriptome
The availability of antibodies and reactive chemicals to some RNA modifications together with RNA‐seq has enabled studies on the role of a few RNA modifications during virus infection. However with more than 150 known RNA modifications (Boccaletto et al., 2018; Cantara et al., 2011), the viral epitranscriptome is likely far more complex than we currently understand. To uncover the viral epitranscriptome, a systems‐wide approach with highly sensitivity and an agnostic identification of all possible RNA modifications is required. To this end, McIntyre et al. (2018) employed the unbiased platform of mass spectrometry to investigate RNA modification landscapes on the entire transcriptome harvested from mock‐ and virus‐infected cells. Using RNA affinity captures and mass spectrometry, the RNA genomes isolated from ZIKV, DENV, HCV, poliovirus, and HIV‐1 were shown to be decorated with a stunning array of RNA modifications (McIntyre et al., 2018). More than 30 different RNA modifications were identified on the viral RNA genomes. Notably, extrapolation of m6A abundance relative to the number of adenosines within the viral RNAs demonstrated comparable levels of m6A when identified by mass spectrometry versus RNA‐seq approaches. For example, on HIV‐l RNA encapsidated in virus particles, McIntyre et al. (2018) estimated 11 m6A modifications per genome, and RNA‐seq studies by Lichinchi, Gao, et al. (2016) reported 12 m6As per genome. Other notable modifications on the viral genomes included 2′‐O‐methylated nucleotides, 1‐methyladenosine, m5C, N4‐acetylcytosine an abundant modification that was recently shown to impact cellular mRNA translation (Arango et al., 2018), as well as m7G (McIntyre et al., 2018). Indeed, the identification of m7G in HCV and poliovirus RNA genomes is surprising particularly as both viruses lack 5′‐cap structures. However, a new chemical‐assisted sequencing approach demonstrated internal m7G in mRNAs and a role for m7G in translation (Zhang et al., 2019). Using mass spectrometry, Courtney et al. (2019) also showed a number of different RNA modifications on the MLV genome. There are challenges towards decrypting the RNA code on viral genomes. For example, RNA‐seq studies are limited by the availability of reagents to specific RNA modifications, and nucleotide resolution of RNA modifications sites has yet to be established for mass spectrometric analysis of large RNAs, such as an RNA virus genome. Moreover, manipulation of the levels of RNA modifications may be difficult as many of the enzymes involved in the biogenesis and function of these modifications are largely unknown. Despite these challenges, the astounding number of modifications on RNA virus genomes highlights that viral epitranscriptomics is an exciting and largely unchartered field.
5. CONCLUSIONS
The influence of RNA modifications on viral infection and host response is breathtaking. Indeed, the range of functions of a single RNA modification such as m6A on virus infection demonstrates that epitranscriptomic marks are pivotal regulators of viral gene expression. As sensitive high‐throughput techniques and reagent availability emerge, together with the identification of different writers, erasers and readers, the interplay between the host and viral epitranscriptomes will be uncovered. Moreover, the effects on viral gene expression following overexpression or depletion of writer, eraser, and reader enzymes need to be carefully evaluated to determine whether the outcome is a direct or indirect consequence of changes in the viral or host epitranscriptome. Such knowledge will be critical to deciphering the regulatory functions of RNA modifications or the epitranscriptomic code. This code will broaden our understanding of virus–cell interactions, identify new antiviral targets, and facilitate the development of novel therapeutics. Given that viruses have proven to be exquisite explorers of cellular and molecular biology, elucidating the viral epitranscriptome will likely also shed new light on the functions of the epitranscriptome in cellular RNA processes and increase our understanding of the myriad of health conditions linked to RNA malfunction.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Rachel Netzband: Conceptualization; data curation; writing‐original draft; writing‐review and editing. Cara T. Pager: Conceptualization; data curation; funding acquisition; project administration; writing‐original draft; writing‐review and editing.
RELATED WIREs ARTICLES
https://dx.doi.org/10.1002/wrna.1536
https://dx.doi.org/10.1002/wrna.1510
ACKNOWLEDGMENTS
We thank Dr Gabriele Fuchs for her thoughtful feedback and suggestions on the manuscript. This work was supported by grants to C.T.P. from the National Institutes for Health (NIH) (R21 AI133617 and R01 GM123050). R.N. received a Predoctoral Fellowship from the University at Albany and The RNA Institute.
Netzband R, Pager CT. Epitranscriptomic marks: Emerging modulators of RNA virus gene expression. WIREs RNA. 2020;11:e1576 10.1002/wrna.1576
Funding information National Institutes for Health (NIH), Grant/Award Numbers: R01 GM123050, R21 AI133617
REFERENCES
- Abbas, Y. M. , Laudenbach, B. T. , Martínez‐Montero, S. , Cencic, R. , Habjan, M. , Pichlmair, A. , … Nagar, B. (2017). Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′‐O methylations. Proceedings of the National Academy of Sciences of the United States of America, 114(11), E2106–E2115. 10.1073/pnas.1612444114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, B. R. , Muramatsu, H. , Jha, B. K. , Silverman, R. H. , Weissman, D. , & Karikó, K. (2011). Nucleoside modifications in RNA limit activation of 2′‐5′‐oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Research, 39(21), 9329–9338. 10.1093/nar/gkr586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, B. R. , Muramatsu, H. , Nallagatla, S. R. , Bevilacqua, P. C. , Sansing, L. H. , Weissman, D. , & Karikó, K. (2010). Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Research, 38(17), 5884–5892. 10.1093/nar/gkq347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango, D. , Sturgill, D. , Alhusaini, N. , Dillman, A. A. , Sweet, T. J. , Hanson, G. , … Oberdoerffer, S. (2018). Acetylation of cytidine in mRNA promotes translation efficiency. Cell, 175(7), 1872–1886.e24. 10.1016/j.cell.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspden, J. L. , & Jackson, R. J. (2010). Differential effects of nucleotide analogs on scanning‐dependent initiation and elongation of mammalian mRNA translation in vitro. RNA, 16(6), 1130–1137. 10.1261/rna.1978610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi, L. , Galvanin, A. , Pichot, F. , Marchand, V. , & Motorin, Y. (2019). RNA ribose methylation (2′‐O‐methylation): Occurrence, biosynthesis and biological functions. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms, 1862(3), 253–269. 10.1016/j.bbagrm.2018.11.009 [DOI] [PubMed] [Google Scholar]
- Baskin, F. , & Dekker, C. A. (1967). A rapid and specific assay for sugar methylation in ribonucleic acid. The Journal of Biological Chemistry, 242(22), 5447–5449. [PubMed] [Google Scholar]
- Bass, B. L. , Weintraub, H. , Cattaneo, R. , & Billeter, M. A. (1989). Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double‐stranded RNA. Cell, 56(3), 331 10.1016/0092-8674(89)90234-1 [DOI] [PubMed] [Google Scholar]
- Baumstark, T. , & Ahlquist, P. (2001). The brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC‐stem loop and is modified in vivo. RNA, 7(11), 1652–1670. [PMC free article] [PubMed] [Google Scholar]
- Becker, H. (1998). Pseudouridine and ribothymidine formation in the tRNA‐like domain of turnip yellow mosaic virus RNA. Nucleic Acids Research, 26(17), 3991–3997. 10.1093/nar/26.17.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemon, K. , & Keith, J. (1977). Localization of N6‐methyladenosine in the Rous sarcoma virus genome. Journal of Molecular Biology, 113(1), 165–179. 10.1016/0022-2836(77)90047-X [DOI] [PubMed] [Google Scholar]
- Bhattacharya, T. , Newton, I. L. G. , & Hardy, R. W. (2017). Wolbachia elevates host methyltransferase expression to block an RNA virus early during infection. PLoS Pathogens, 13(6), e1006427 10.1371/journal.ppat.1006427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbille, Y. , Vendeix, F. A. P. , Guenther, R. , Malkiewicz, A. , Ariza, X. , Vilarrasa, J. , & Agris, P. F. (2009). The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs. Nucleic Acids Research, 37(10), 3342–3353. 10.1093/nar/gkp187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto, P. , MacHnicka, M. A. , Purta, E. , Pitkowski, P. , Baginski, B. , Wirecki, T. K. , … Bujnicki, J. M. (2018). MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Research, 46(D1), D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet, M. , Debarnot, C. , Imbert, I. , Selisko, B. , Snijder, E. J. , Canard, B. , & Decroly, E. (2010). In vitro reconstitution of SARS‐coronavirus mRNA cap methylation. PLoS Pathogens, 6(4), e1000863 10.1371/journal.ppat.1000863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, N. M. , & Palese, P. (2008). The biology of influenza viruses. Vaccine, 26(Suppl. 4), D49–D53. 10.1016/j.vaccine.2008.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki, J. M. , Feder, M. , Ayres, C. L. , & Redman, K. L. (2004). Sequence–structure–function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Research, 32(8), 2453–2463. 10.1093/nar/gkh564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, E. M. , & Hope, T. J. (2015). HIV‐1 capsid: The multifaceted key player in HIV‐1 infection. Nature Reviews Microbiology, 13(8), 471–483. 10.1038/nrmicro3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani, D. , Kahana, C. , Lavi, S. , & Groner, Y. (1979). Identification and mapping of N6‐methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Research, 6(8), 2879–2899. 10.1093/nar/6.8.2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantara, W. A. , Crain, P. F. , Rozenski, J. , McCloskey, J. A. , Harris, K. A. , Zhang, X. , … Agris, P. F. (2011). The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Research, 39(Suppl. 1), D195–D201. 10.1093/nar/gkq1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile, T. M. , Rojas‐Duran, M. F. , Zinshteyn, B. , Shin, H. , Bartoli, K. M. , & Gilbert, W. V. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature, 515(7525), 143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, J. L. (2006). RNA editing in hepatitis delta virus. Current Topics in Microbiology and Immunology, 307, 67–89. [DOI] [PubMed] [Google Scholar]
- Casey, J. L. , Bergmann, K. F. , Brown, T. L. , & Gerin, J. L. (1992). Structural requirements for RNA editing in hepatitis delta virus: Evidence for a uridine‐to‐cytidine editing mechanism. Proceedings of the National Academy of Sciences of the United States of America, 89(15), 7149–7153. 10.1073/pnas.89.15.7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, J. L. , & Gerin, J. L. (1995). Hepatitis D virus RNA editing: Specific modification of adenosine in the antigenomic RNA. Journal of Virology, 69(12), 7593–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo, R. , Schmid, A. , Eschle, D. , Baczko, K. , ter Meulen, V. , & Billeter, M. A. (1988). Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell, 55(2), 255–265. 10.1016/0092-8674(88)90048-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattenoz, P. B. , Taft, R. J. , Westhof, E. , & Mattick, J. S. (2013). Transcriptome‐wide identification of A>I RNA editing sites by inosine specific cleavage. RNA, 19(2), 257–270. 10.1261/rna.036202.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Cai, H. , Pan, J. , Xiang, N. , Tien, P. , Ahola, T. , & Guo, D. (2009). Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proceedings of the National Academy of Sciences of the United States of America, 106(9), 3484–3489. 10.1073/pnas.0808790106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, W. E. (1959). 5‐Ribosyl uracil, a carbon–carbon ribofuranosyl nucleoside in ribonucleic acids. Biochimica et Biophysica Acta, 32, 569–571. 10.1016/0006-3002(59)90644-4 [DOI] [PubMed] [Google Scholar]
- Cohn, W. E. , & Volkin, E. (1951). Nucleoside‐5′‐phosphates from ribonucleic acid. Nature, 167(4247), 483–484. 10.1038/167483a0 [DOI] [Google Scholar]
- Courtney, D. G. , Chalem, A. , Bogerd, H. P. , Law, B. A. , Kennedy, E. M. , Holley, C. L. , & Cullen, B. R. (2019). Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. MBio, 10(3), e01209–e01219 10.1128/mBio.01209-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney, D. G. , Kennedy, E. M. , Dumm, R. E. , Bogerd, H. P. , Tsai, K. , Heaton, N. S. , & Cullen, B. R. (2017). Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host and Microbe, 22(3), 377–386.e5. 10.1016/j.chom.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, S. T. , Michalski, D. , Miller, M. R. , & Wilusz, J. (2019). RNA regulatory processes in RNA virus biology. WIREs: RNA, 10, e1536 10.1002/wrna.1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz‐Oliveira, C. , Freire, J. M. , Conceição, T. M. , Higa, L. M. , Castanho, M. A. R. B. , & Da Poian, A. T. (2015). Receptors and routes of dengue virus entry into the host cells. FEMS Microbiology Reviews, 39(2), 155–170. 10.1093/femsre/fuu004 [DOI] [PubMed] [Google Scholar]
- Daffis, S. , Szretter, K. J. , Schriewer, J. , Li, J. , Youn, S. , Errett, J. , … Diamond, M. S. (2010). 2′‐O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature, 468(7322), 452–456. 10.1038/nature09489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Q. , Moshitch‐Moshkovitz, S. , Han, D. , Kol, N. , Amariglio, N. , Rechavi, G. , … He, C. (2017). Nm‐seq maps 2′‐O‐methylation sites in human mRNA with base precision. Nature Methods, 14(7), 695–698. 10.1038/nmeth.4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalet, A. , Gatti, E. , & Pierre, P. (2015). Integration of PKR‐dependent translation inhibition with innate immunity is required for a coordinated anti‐viral response. FEBS Letters, 589(14), 1539–1545. 10.1016/j.febslet.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Danecek, P. , Nellåker, C. , McIntyre, R. E. , Buendia‐Buendia, J. E. , Bumpstead, S. , Ponting, C. P. , … Adams, D. J. (2012). High levels of RNA‐editing site conservation amongst 15 laboratory mouse strains. Genome Biology, 13(4), R26 10.1186/gb-2012-13-4-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D. R. (1995). Stabilization of RNA stacking by pseudouridine. Nucleic Acids Research, 23(24), 5020–5026. 10.1093/nar/23.24.5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly, E. , & Canard, B. (2017). Biochemical principles and inhibitors to interfere with viral capping pathways. Current Opinion in Virology, 24, 87–96. 10.1016/j.coviro.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly, E. , Ferron, F. , Lescar, J. , & Canard, B. (2012). Conventional and unconventional mechanisms for capping viral mRNA. Nature Reviews Microbiology, 10(1), 51–65. 10.1038/nrmicro2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly, E. , Imbert, I. , Coutard, B. , Bouvet, M. , Selisko, B. , Alvarez, K. , … Canard, B. (2008). Coronavirus nonstructural protein 16 is a Cap‐0 binding enzyme possessing (nucleoside‐2′O)‐methyltransferase activity. Journal of Virology, 82(16), 8071–8084. 10.1128/jvi.00407-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffrasnes, C. , Marsh, G. A. , Foo, C. H. , Rootes, C. L. , Gould, C. M. , Grusovin, J. , … Wang, L. F. (2016). Genome‐wide siRNA screening at biosafety level 4 reveals a crucial role for fibrillarin in Henipavirus infection. PLoS Pathogens, 12(3), e1005478 10.1371/journal.ppat.1005478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarkar, S. C. , Wang, C. , Miller, M. T. , Ramanathan, A. , Jiang, F. , Khan, A. G. , … Marcotrigiano, J. (2016). Structural basis for m7G recognition and 2′‐O‐methyl discrimination in capped RNAs by the innate immune receptor RIG‐I. Proceedings of the National Academy of Sciences of the United States of America, 113(3), 596–601. 10.1073/pnas.1515152113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimock, K. , & Stoltzfus, C. M. (1977). Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry, 16(3), 471–478. 10.1021/bi00622a021 [DOI] [PubMed] [Google Scholar]
- Dimock, K. , & Stoltzfus, C. M. (1978). Cycloleucine blocks 5′‐terminal and internal methylations of avian sarcoma virus genome RNA. Biochemistry, 17(17), 3627–3632. 10.1021/bi00610a032 [DOI] [PubMed] [Google Scholar]
- Dominissini, D. , Moshitch‐Moshkovitz, S. , Schwartz, S. , Salmon‐Divon, M. , Ungar, L. , Osenberg, S. , … Rechavi, G. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature, 485(7397), 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Dong, H. , Chang, D. C. , Hua, M. H. C. , Lim, S. P. , Chionh, Y. H. , Hia, F. , … Shi, P. Y. (2012). 2′‐O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathogens, 8(4), e1002642 10.1371/journal.ppat.1002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. , Ren, S. , Li, H. , & Shi, P. Y. (2008). Separate molecules of West Nile virus methyltransferase can independently catalyze the N7 and 2′‐O methylations of viral RNA cap. Virology, 377(1), 1–6. 10.1016/j.virol.2008.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. , Zhang, B. , & Shi, P. Y. (2008). Flavivirus methyltransferase: A novel antiviral target. Antiviral Research, 80(1), 1–10. 10.1016/j.antiviral.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria, M. , Neri, F. , Gallo, A. , Farace, M. G. , & Michienzi, A. (2009). Editing of HIV‐1 RNA by the double‐stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Research, 37(17), 5848–5858. 10.1093/nar/gkp604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin, D. T. , & Stollar, V. (1975). Methylation of Sindbis virus “26S” messenger RNA. Biochemical and Biophysical Research Communications, 66(4), 1373–1379. 10.1016/0006-291X(75)90511-2 [DOI] [PubMed] [Google Scholar]
- Dubin, D. T. , Stollar, V. , Hsuchen, C. C. , Timko, K. , & Guild, G. M. (1977). Sindbis virus messenger RNA: The 5′‐termini and methylated residues of 26 and 42 S RNA. Virology, 77(2), 457–470. 10.1016/0042-6822(77)90471-8 [DOI] [PubMed] [Google Scholar]
- Durdevic, Z. , Hanna, K. , Gold, B. , Pollex, T. , Cherry, S. , Lyko, F. , & Schaefer, M. (2013). Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Reports, 14(3), 269–275. 10.1038/embor.2013.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erales, J. , Marchand, V. , Panthu, B. , Gillot, S. , Belin, S. , Ghayad, S. E. , … Diaz, J.‐J. (2017). Evidence for rRNA 2′‐O‐methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proceedings of the National Academy of Sciences of the United States of America, 114(49), 12934–12939. 10.1073/pnas.1707674114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder, M.‐P. , Laugier, D. , Yatsula, B. , Dezélée, P. , Calothy, G. , & Marx, M. (1994). Functional and biological properties of an avian variant long terminal repeat containing multiple A to G conversions in the U3 sequence. Journal of Virology, 68(8), 4759–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, D. , & Groner, Y. (1983). Methylations of adenosine residues (m6A) in pre‐mRNA are important for formation of late simian virus 40 mRNAs. Virology, 131(2), 409–425. 10.1016/0042-6822(83)90508-1 [DOI] [PubMed] [Google Scholar]
- Furuichi, Y. , Morgan, M. , Muthukrishnan, S. , & Shatkin, A. J. (1975). Reovirus messenger RNA contains a methylated, blocked 5′‐terminal structure: m‐7G(5′)ppp(5′)G‐MpCp. Proceedings of the National Academy of Sciences, 72(1), 362–366. 10.1073/pnas.72.1.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi, Y. , Shatkin, A. J. , Stavnezer, E. , & Bishop, J. M. (1975). Blocked, methylated 5′‐terminal sequence in avian sarcoma virus RNA. Nature, 257(5527), 618–620. 10.1038/257618a0 [DOI] [PubMed] [Google Scholar]
- George, C. X. , John, L. , & Samuel, C. E. (2014). An RNA editor, adenosine deaminase acting on double‐stranded RNA (ADAR1). Journal of Interferon & Cytokine Research, 34(6), 437–446. 10.1089/jir.2014.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale, N. S. , & Horner, S. M. (2017). RNA modifications go viral. PLoS Pathogens, 13(3), e1006188 10.1371/journal.ppat.1006188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale, N. S. , McIntyre, A. B. R. , McFadden, M. J. , Roder, A. E. , Kennedy, E. M. , Gandara, J. A. , … Horner, S. M. (2016). N6‐methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host and Microbe, 20(5), 654–665. 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales‐van Horn, S. R. , & Sarnow, P. (2017). Making the mark: The role of adenosine modifications in the life cycle of RNA viruses. Cell Host and Microbe, 21(6), 661–669. 10.1016/j.chom.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, D. E. , Lin, W.‐H. W. , & Nelson, A. N. (2018). Understanding the causes and consequences of measles virus persistence. F1000Research, 7, 237 10.12688/f1000research.12094.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche, I. , Desfosses, A. , Effantin, G. , Ling, W. L. , Haupt, M. , Ruigrok, R. W. H. , … Schoehn, G. (2015). Near‐atomic cryo‐EM structure of the helical measles virus nucleocapsid. Science, 348(6235), 704–707. 10.1126/science.aaa5137 [DOI] [PubMed] [Google Scholar]
- Habjan, M. , Hubel, P. , Lacerda, L. , Benda, C. , Holze, C. , Eberl, C. H. , … Pichlmair, A. (2013). Sequestration by IFIT1 impairs translation of 2′O‐unmethylated capped RNA. PLoS Pathogens, 9(10), e1003663 10.1371/journal.ppat.1003663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, W. W. , Lamb, R. A. , & Choppin, P. W. (1979). Measles and subacute sclerosing panencephalitis virus proteins: Lack of antibodies to the M protein in patients with subacute sclerosing panencephalitis. Proceedings of the National Academy of Sciences of the United States of America, 76(4), 2047–2051. 10.1073/pnas.76.4.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, H. , Hao, S. , Chen, H. , Chen, Z. , Zhang, Y. , Wang, J. , … Guan, W. (2019). N6‐methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Research, 47(1), 362–374. 10.1093/nar/gky1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, S. I. , & Green, M. (1976). Multiple methylated cap sequences in adenovirus type 2 early mRNA. Journal of Virology, 20(2), 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesser, C. R. , Karijolich, J. , Dominissini, D. , He, C. , & Glaunsinger, B. A. (2018). N6‐methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma‐associated herpesvirus infection. PLoS Pathogens, 14(4), e1006995 10.1371/journal.ppat.1006995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, A. G. (2006). eIF3: A versatile scaffold for translation initiation complexes. Trends in Biochemical Sciences, 31(10), 553–562. 10.1016/j.tibs.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Hsuchen, C. C. , & Dubin, D. T. (1976). Di‐ and trimethylated congeners of 7‐methylguanine in Sindbis virus mRNA. Nature, 264(5582), 190–191. 10.1038/264190a0 [DOI] [PubMed] [Google Scholar]
- Hu, W.‐S. , & Hughes, S. H. (2012). HIV‐1 reverse transcription. Cold Spring Harbor Perspectives in Medicine, 2(10), a006882–a006882. 10.1101/cshperspect.a006882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, D. J. , Bhasker, C. R. , Merrick, W. C. , & Sen, G. C. (2003). Viral stress‐inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2·GTP·Met‐tRNAi. Journal of Biological Chemistry, 278(41), 39477–39482. 10.1074/jbc.M305038200 [DOI] [PubMed] [Google Scholar]
- Hui, D. J. , Terenzi, F. , Merrick, W. C. , & Sen, G. C. (2005). Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. Journal of Biological Chemistry, 280(5), 3433–3440. 10.1074/jbc.M406700200 [DOI] [PubMed] [Google Scholar]
- Hyde, J. L. , & Diamond, M. S. (2015). Innate immune restriction and antagonism of viral RNA lacking 2′‐O methylation. Virology, 479–480, 66–74. 10.1016/j.virol.2015.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias, N. G. , & Gamarnik, A. V. (2011). Dynamic RNA structures in the dengue virus genome. RNA Biology, 8(2), 249–257. 10.4161/hv.8.2.14992 [DOI] [PubMed] [Google Scholar]
- Imam, H. , Khan, M. , Gokhale, N. S. , McIntyre, A. B. R. , Kim, G. W. , Jang, J. Y. , … Siddiqui, A. (2018). N6‐methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proceedings of the National Academy of Sciences of the United States of America, 115(35), 8829–8834. 10.1073/pnas.1808319115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incarnato, D. , Anselmi, F. , Morandi, E. , Neri, F. , Maldotti, M. , Rapelli, S. , … Oliviero, S. (2017). High‐throughput single‐base resolution mapping of RNA 2‐O‐methylated residues. Nucleic Acids Research, 45(3), 1433–1441. 10.1093/nar/gkw810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayan, G. C. , & Casey, J. L. (2005). Effects of conserved RNA secondary structures on hepatitis delta virus genotype I RNA editing, replication, and virus production. Journal of Virology, 79(17), 11187–11193. 10.1128/JVI.79.17.11187-11193.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, G. , Fu, Y. , Zhao, X. , Dai, Q. , Zheng, G. , Yang, Y. , … He, C. (2011). N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nature Chemical Biology, 7(12), 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, S. E. , & Beemon, K. (1985). Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Molecular and Cellular Biology, 5(9), 2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó, K. , Muramatsu, H. , Welsh, F. A. , Ludwig, J. , Kato, H. , Akira, S. , & Weissman, D. (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular Therapy, 16(11), 1833–1840. 10.1038/mt.2008.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, E. M. , Bogerd, H. P. , Kornepati, A. V. R. , Kang, D. , Ghoshal, D. , Marshall, J. B. , … Cullen, B. R. (2016). Posttranscriptional m6A editing of HIV‐1 mRNAs enhances viral gene expression. Cell Host and Microbe, 19(5), 675–685. 10.1016/j.chom.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, E. M. , Courtney, D. G. , Tsai, K. , & Cullen, B. R. (2017). Viral Epitranscriptomics. Journal of Virology, 91(9), e02263–e02216. 10.1128/JVI.02263-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrustalev, V. V. , Khrustaleva, T. A. , Sharma, N. , & Giri, R. (2017). Mutational pressure in Zika virus: Local ADAR‐editing areas associated with pauses in translation and replication. Frontiers in Cellular and Infection Microbiology, 7, 44 10.3389/fcimb.2017.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, T. , Katoh, H. , Kayama, H. , Saiga, H. , Okuyama, M. , Okamoto, T. , … Takeda, K. (2013). Ifit1 inhibits Japanese encephalitis virus replication through binding to 5′ capped 2′‐O unmethylated RNA. Journal of Virology, 87(18), 9997–10003. 10.1128/JVI.00883-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson, S. D. , Ayele, T. M. , & Heemstra, J. M. (2018). Chemical labeling and affinity capture of inosine‐containing RNAs using acrylamidofluorescein. Bioconjugate Chemistry, 29(9), 2899–2903. 10.1021/acs.bioconjchem.8b00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, C. , Da, B. L. , & Glenn, J. S. (2019). HBV/HDV coinfection: A challenge for therapeutics. Clinics in Liver Disease, 23(3), 557–572. 10.1016/j.cld.2019.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug, R. M. , Morgan, M. A. , & Shatkin, A. J. (1976). Influenza viral mRNA contains internal N6‐methyladenosine and 5′‐terminal 7‐methylguanosine in cap structures. Journal of Virology, 20(1), 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P. , Sweeney, T. R. , Skabkin, M. A. , Skabkina, O. V. , Hellen, C. U. T. , & Pestova, T. V. (2014). Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′‐terminal regions of cap0‐, cap1‐ and 5′ppp‐mRNAs. Nucleic Acids Research, 42(5), 3228–3245. 10.1093/nar/gkt1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, M. Y. , Chao, M. , & Taylor, J. (1989). Initiation of replication of the human hepatitis delta virus genome from cloned DNA: Role of delta antigen. Journal of Virology, 63(5), 1945–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, B. G. , & Tamaoki, T. (1967). Studies of the chain termini and alkali‐stable dinucleotide sequences in 16 s and 28 s ribosomal RNA from L cells. Journal of Molecular Biology, 27(2), 335–348. 10.1016/0022-2836(67)90024-1 [DOI] [PubMed] [Google Scholar]
- Lang, F. , Singh, R. K. , Pei, Y. , Zhang, S. , Sun, K. , & Robertson, E. S. (2019). EBV epitranscriptome reprogramming by METTL14 is critical for viral‐associated tumorigenesis. PLoS Pathogens, 15(6), e1007796 10.1371/journal.ppat.1007796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear, H. M. , & Diamond, M. S. (2016). Zika virus: New clinical syndromes and its emergence in the Western hemisphere. Journal of Virology, 90(10), 4864–4875. 10.1128/jvi.00252-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempp, F. A. , Ni, Y. , & Urban, S. (2016). Hepatitis delta virus: Insights into a peculiar pathogen and novel treatment options. Nature Reviews Gastroenterology & Hepatology, 13(10), 580–589. 10.1038/nrgastro.2016.126 [DOI] [PubMed] [Google Scholar]
- Li, J. B. , Levanon, E. Y. , Yoon, J. K. , Aach, J. , Xie, B. , LeProust, E. , … Church, G. M. (2009). Genome‐wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science, 324(5931), 1210–1213. 10.1126/science.1170995 [DOI] [PubMed] [Google Scholar]
- Li, X. , Ma, S. , & Yi, C. (2016). Pseudouridine: The fifth RNA nucleotide with renewed interests. Current Opinion in Chemical Biology, 33, 108–116. 10.1016/J.CBPA.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhu, P. , Ma, S. , Song, J. , Bai, J. , Sun, F. , & Yi, C. (2015). Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nature Chemical Biology, 11(8), 592–597. 10.1038/nchembio.1836 [DOI] [PubMed] [Google Scholar]
- Lichinchi, G. , Gao, S. , Saletore, Y. , Gonzalez, G. M. , Bansal, V. , Wang, Y. , … Rana, T. M. (2016). Dynamics of the human and viral m(6)A RNA methylomes during HIV‐1 infection of T cells. Nature Microbiology, 1(4), 16011 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi, G. , Zhao, B. S. , Wu, Y. , Lu, Z. , Qin, Y. , He, C. , & Rana, T. M. (2016). Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host and Microbe, 20(5), 666–673. 10.1016/j.chom.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat, B. J. , Piskol, R. , Chalk, A. M. , Ramaswami, G. , Higuchi, M. , Hartner, J. C. , … Walkley, C. R. (2015). RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science, 349(6252), 1115–1120. 10.1126/science.aac7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebert, U. G. , Baczko, K. , Budka, H. , & Ter Meulen, V. (1986). Restricted expression of measles virus proteins in brains from cases of subacute sclerosing panencephalitis. Journal of General Virology, 67(11), 2435–2444. 10.1099/0022-1317-67-11-2435 [DOI] [PubMed] [Google Scholar]
- Limbach, P. A. , Crain, P. F. , & Mccloskey, J. A. (1994). Summary: The modified nucleosides of RNA. Nucleic Acids Research, 22(12), 2183–2196. 10.1093/nar/22.12.2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Yue, Y. , Han, D. , Wang, X. , Fu, Y. , Zhang, L. , … He, C. (2014). A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nature Chemical Biology, 10(2), 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R. E. (2013). Regulation of stress granules and P‐bodies during RNA virus infection. WIREs: RNA, 4(3), 317–331. 10.1002/wrna.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy, A. F. , Riordan, D. P. , & Brown, P. O. (2014). Transcriptome‐wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae . PLoS ONE, 9(10), e110799 10.1371/journal.pone.0110799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W. , Tirumuru, N. , St. Gelais, C. , Koneru, P. C. , Liu, C. , Kvaratskhelia, M. , … Wu, L. (2018). N 6‐Methyladenosine‐binding proteins suppress HIV‐1 infectivity and viral production. Journal of Biological Chemistry, 293(34), 12992–13005. 10.1074/jbc.RA118.004215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, D. , Kohlway, A. , Vela, A. , & Pyle, A. M. (2012). Visualizing the determinants of viral RNA recognition by innate immune sensor RIG‐I. Structure, 20(11), 1983–1988. 10.1016/j.str.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka, M. A. , Milanowska, K. , Osman Oglou, O. , Purta, E. , Kurkowska, M. , Olchowik, A. , … Grosjean, H. (2012). MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Research, 41(D1), D262–D267. 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak, J. , & Kleiman, L. (1997). Primer tRNAs for reverse transcription. Journal of Virology, 71(11), 8087–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion, N. M. , Greenwood, S. M. , Young, R. , Cox, S. , Brindle, J. , Read, D. , … O'Connell, M. A. (2014). The RNA‐editing enzyme ADAR1 controls innate immune responses to RNA. Cell Reports, 9(4), 1482–1494. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau, C. D. , Puschnik, A. S. , Majzoub, K. , Ooi, Y. S. , Brewer, S. M. , Fuchs, G. , … Carette, J. E. (2016). Genetic dissection of Flaviviridae host factors through genome‐scale CRISPR screens. Nature, 535(7610), 159–163. 10.1038/nature18631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand, V. , Blanloeil‐Oillo, F. , Helm, M. , & Motorin, Y. (2016). Illumina‐based RiboMethSeq approach for mapping of 2′‐O‐Me residues in RNA. Nucleic Acids Research, 44(16), e135–e135. 10.1093/nar/gkw547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, B. , Coutard, B. , Guez, T. , Paesen, G. C. , Canard, B. , Debart, F. , … Decroly, E. (2018). The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Research, 46(15), 7902–7912. 10.1093/nar/gky637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, I. , Dopazo, J. , & Melero, J. A. (1997). Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. Journal of General Virology, 78(10), 2419–2429. 10.1099/0022-1317-78-10-2419 [DOI] [PubMed] [Google Scholar]
- Martínez, I. , & Melero, J. A. (2002). A model for the generation of multiple A to G transitions in the human respiratory syncytial virus genome: Predicted RNA secondary structures as substrates for adenosine deaminases that act on RNA. Journal of General Virology, 83(6), 1445–1455. 10.1099/0022-1317-83-6-1445 [DOI] [PubMed] [Google Scholar]
- Mauro, V. P. , & Edelman, G. M. (2002). The ribosome filter hypothesis. Proceedings of the National Academy of Sciences of the United States of America, 99(19), 12031–12036. 10.1073/pnas.192442499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden, M. J. , Gokhale, N. S. , & Horner, S. M. (2017). Protect this house: Cytosolic sensing of viruses. Current Opinion in Virology, 22, 36–43. 10.1016/j.coviro.2016.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre, W. , Netzband, R. , Bonenfant, G. , Biegel, J. M. , Miller, C. , Fuchs, G. , … Pager, C. T. (2018). Positive‐sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Research, 46(11), 5776–5791. 10.1093/nar/gky029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentha, N. , Clément, S. , Negro, F. , & Alfaiate, D. (2019). A review on hepatitis D: From virology to new therapies. Journal of Advanced Research, 17, 3–15. 10.1016/j.jare.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K. D. , & Jaffrey, S. R. (2017). Rethinking m6A readers, writers, and erasers. Annual Review of Cell and Developmental Biology, 33, 319–342. 10.1146/annurev-cellbio-100616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K. D. , Saletore, Y. , Zumbo, P. , Elemento, O. , Mason, C. E. , & Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell, 149(7), 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, B. , Gershowitz, A. , Stringer, J. R. , Holland, L. E. , & Wagner, E. K. (1977). 5′‐Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. Journal of Virology, 23(2), 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, B. , & Koczot, F. (1976). Sequence of methylated nucleotides at the 5′ terminus of adenovirus specific RNA. Journal of Virology, 17(2), 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin, Y. , Lyko, F. , & Helm, M. (2009). 5‐Methylcytosine in RNA: Detection, enzymatic formation and biological functions. Nucleic Acids Research, 38(5), 1415–1430. 10.1093/nar/gkp1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan, P. , Ayers, D. F. , Rottman, F. M. , Maroney, P. A. , & Nilsen, T. W. (1987). Unequal distribution of N6‐methyladenosine in influenza virus mRNAs. Molecular and Cellular Biology, 7(4), 1572–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby, M. I. , & Greenbaum, N. L. (2002). Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proceedings of the National Academy of Sciences of the United States of America, 99(20), 12697–12702. 10.1073/pnas.202477199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, B. L. , & White, K. A. (2015). Exploring the architecture of viral RNA genomes. Current Opinion in Virology, 12, 66–74. 10.1016/j.coviro.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Otsuka, Y. , Kedersha, N. L. , & Schoenberg, D. R. (2009). Identification of a cytoplasmic complex that adds a Cap onto 5′‐monophosphate RNA. Molecular and Cellular Biology, 29(8), 2155–2167. 10.1128/MCB.01325-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott, M. , Geyer, M. , & Zhou, Q. (2011). The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host and Microbe, 10(5), 426–435. 10.1016/j.chom.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, J. B. , Cornu, T. I. , Redwine, J. , Dales, S. , Lewicki, H. , Holz, A. , … Oldstone, M. B. A. (2001). Evidence that the hypermutated M protein of a subacute sclerosing panencephalitis measles virus actively contributes to the chronic progressive CNS disease. Virology, 291(2), 215–225. 10.1006/viro.2001.1182 [DOI] [PubMed] [Google Scholar]
- Petes, C. , Odoardi, N. , & Gee, K. (2017). The Toll for trafficking: Toll‐like receptor 7 delivery to the endosome. Frontiers in Immunology, 8, 1075 10.3389/fimmu.2017.01075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller, C. K. , Donohue, R. C. , Nersisyan, S. , Brodsky, L. , & Cattaneo, R. (2018). Extensive editing of cellular and viral double‐stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1 p150. PLoS Biology, 16(11), e2006577 10.1371/journal.pbio.2006577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller, C. K. , Mastorakos, G. M. , Matchett, W. E. , Ma, X. , Samuel, C. E. , & Cattaneo, R. (2015). Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA‐1‐like hypermutations. Journal of Virology, 89(15), 7735–7747. 10.1128/jvi.01017-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller, C. K. , Radeke, M. J. , Cattaneo, R. , & Samuel, C. E. (2014). Measles virus C protein impairs production of defective copyback double‐stranded viral RNA and activation of protein kinase R. Journal of Virology, 88(1), 456–468. 10.1128/jvi.02572-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuphuakrat, A. , Kraiwong, R. , Boonarkart, C. , Lauhakirti, D. , Lee, T.‐H. , & Auewarakul, P. (2008). Double‐stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. Journal of Virology, 82(21), 10864–10872. 10.1128/JVI.00238-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontkivska, H. , Frederick, M. , Miyamoto, M. M. , & Wayne, M. L. (2017). RNA editing by the host ADAR system affects the molecular evolution of the Zika virus. Ecology and Evolution, 7(12), 4475–4485. 10.1002/ece3.3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, A. G. , Bass, B. L. , & Casey, J. L. (1996). RNA editing of hepatitis delta virus antigenome by dsRNA‐adenosine deaminase. Nature, 380(6573), 454–456. 10.1038/380454a0 [DOI] [PubMed] [Google Scholar]
- Polson, A. G. , Ley, H. L. , Bass, B. L. , & Casey, J. L. (1998). Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Molecular and Cellular Biology, 18(4), 1919–1926. 10.1128/mcb.18.4.1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner, P. , Yathindra, N. , & Sundaralingam, M. (1974). Effect of ribose O(2′)‐methylation on the conformation of nucleosides and nucleotides. BBA Section Nucleic Acids and Protein Synthesis, 366(2), 115–123. 10.1016/0005-2787(74)90325-6 [DOI] [PubMed] [Google Scholar]
- Ramanathan, A. , Robb, G. B. , & Chan, S. H. (2016). mRNA capping: Biological functions and applications. Nucleic Acids Research, 44(16), 7511–7526. 10.1093/nar/gkw551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichow, S. L. , Hamma, T. , Ferré‐D'Amaré, A. R. , & Varani, G. (2007). The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Research, 35(5), 1452–1464. 10.1093/nar/gkl1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rima, B. K. , Gatherer, D. , Young, D. F. , Norsted, H. , Randall, R. E. , & Davison, A. J. (2014). Stability of the parainfluenza virus 5 genome revealed by deep sequencing of strains isolated from different hosts and following passage in cell culture. Journal of Virology, 88(7), 3826–3836. 10.1128/JVI.03351-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringeard, M. , Marchand, V. , Decroly, E. , Motorin, Y. , & Bennasser, Y. (2019). FTSJ3 is an RNA 2′‐O‐methyltransferase recruited by HIV to avoid innate immune sensing. Nature, 565(7740), 500–504. 10.1038/s41586-018-0841-4 [DOI] [PubMed] [Google Scholar]
- Rizzetto, M. , Hoyer, B. , Canese, M. G. , Shih, J. W. , Purcell, R. H. , & Gerin, J. L. (1980). Delta agent: Association of delta antigen with hepatitis B surface antigen and RNA in serum of delta‐infected chimpanzees. Proceedings of the National Academy of Sciences of the United States of America, 77(10), 6124–6128. 10.1073/pnas.77.10.6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, M. , Judge, A. , Liang, L. , McClintock, K. , Yaworski, E. , & MacLachlan, I. (2007). 2′‐O‐methyl‐modified RNAs act as TLR7 antagonists. Molecular Therapy, 15(9), 1663–1669. 10.1038/sj.mt.6300240 [DOI] [PubMed] [Google Scholar]
- Rose, J. K. (1975). Heterogneeous 5′‐terminal structures occur on vesicular stomatitis virus mRNAs. The Journal of Biological Chemistry, 250(20), 8098–8104. [PubMed] [Google Scholar]
- Roth‐Cross, J. K. , Bender, S. J. , & Weiss, S. R. (2008). Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. Journal of Virology, 82(20), 9829–9838. 10.1128/jvi.01199-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree, I. A. , Evans, M. E. , Pan, T. , & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169(7), 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda, P. , García‐Barreno, B. , & Melero, J. A. (1994). Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A‐G substitutions (hypermutations). Virology, 198(2), 653–662. [DOI] [PubMed] [Google Scholar]
- Ryu, W. S. , Bayer, M. , & Taylor, J. (1992). Assembly of hepatitis delta virus particles. Journal of Virology, 66(4), 2310–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai, M. , Yano, T. , Kawabata, H. , Ueda, H. , & Suzuki, T. (2010). Inosine cyanoethylation identifies A‐to‐I RNA editing sites in the human transcriptome. Nature Chemical Biology, 6(10), 733–740. 10.1038/nchembio.434 [DOI] [PubMed] [Google Scholar]
- Samuel, C. E. (2011). Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology, 411(2), 180–193. 10.1016/j.virol.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S. , Cornillez‐Ty, C. , & Lazinski, D. W. (2004). By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. Journal of Virology, 78(15), 8120–8134. 10.1128/JVI.78.15.8120-8134.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S. , Wong, S. K. , & Lazinski, D. W. (2001). Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. Journal of Virology, 75(18), 8547–8555. 10.1128/jvi.75.18.8547-8555.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth‐Wagner, C. , Ludwig, J. , Bruder, A. K. , Herzner, A. M. , Zillinger, T. , Goldeck, M. , … Schlee, M. (2015). A conserved histidine in the RNA sensor RIG‐I controls immune tolerance to N1‐2′O‐methylated self RNA. Immunity, 43(1), 41–51. 10.1016/j.immuni.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S. , Bernstein, D. A. , Mumbach, M. R. , Jovanovic, M. , Herbst, R. H. , León‐Ricardo, B. X. , … Regev, A. (2014). Transcriptome‐wide mapping reveals widespread dynamic‐regulated pseudouridylation of ncRNA and mRNA. Cell, 159(1), 148–162. 10.1016/J.CELL.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S. , & Motorin, Y. (2017). Next‐generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biology, 14(9), 1124–1137. 10.1080/15476286.2016.1251543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmeen, L. , Bass, B. , Sonenberg, N. , Weintraub, H. , & Groudine, M. (1991). Tat‐dependent adenosine‐to‐inosine modification of wild‐type transactivation response RNA. Proceedings of the National Academy of Sciences of the United States of America, 88(18), 8096–8100. 10.1073/pnas.88.18.8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Wei, J. , & He, C. (2019). Where, when, and how: Context‐dependent functions of RNA methylation writers, readers, and erasers. Molecular Cell, 74(4), 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Z. , Fujii, K. , Kovary, K. M. , Genuth, N. R. , Röst, H. L. , Teruel, M. N. , & Barna, M. (2017). Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome‐wide. Molecular Cell, 67(1), 71–83.e7. 10.1016/j.molcel.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin, W. , & Nishikura, K. (2013). Adenosine‐to‐inosine RNA editing and human disease. Genome Medicine, 5(11), 105 10.1186/gm508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, S. , Salditt‐Georgieff, M. , Bachenheimer, S. , Darnell, J. E. , Furuichi, Y. , Morgan, M. , & Shatkin, A. J. (1976). The methylation of adenovirus‐specific nuclear and cytoplasmic RNA. Nucleic Acids Research, 3(3), 749–766. 10.1093/nar/3.3.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spenkuch, F. , Motorin, Y. , & Helm, M. (2014). Pseudouridine: Still mysterious, but never a fake (uridine)! RNA Biology, 11(12), 1540–1554. 10.4161/15476286.2014.992278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires, J. E. , Patel, H. R. , Nousch, M. , Sibbritt, T. , Humphreys, D. T. , Parker, B. J. , … Preiss, T. (2012). Widespread occurrence of 5‐methylcytosine in human coding and non‐coding RNA. Nucleic Acids Research, 40(11), 5023–5033. 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus, C. M. , & Dane, R. W. (1982). Accumulation of spliced avian retrovirus mRNA is inhibited in S‐adenosylmethionine‐depleted chicken embryo fibroblasts. Journal of Virology, 42(3), 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus, C. M. , & Dimock, K. (1976). Evidence of methylation of B77 avian sarcoma virus genome RNA subunits. Journal of Virology, 18(2), 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, B. , Liu, H. , Zhang, S. , Da Silva, S. R. , Zhang, L. , Meng, J. , … Gao, S. J. (2017). Viral and cellular N6‐methyladenosine and N6,2′‐O‐dimethyladenosine epitranscriptomes in the KSHV life cycle. Nature Microbiology, 3(1), 108–120. 10.1038/s41564-017-0056-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D. R. , Puig, M. , Darnell, M. E. R. , Mihalik, K. , & Feinstone, S. M. (2005). New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. Journal of Virology, 79(10), 6291–6298. 10.1128/JVI.79.10.6291-6298.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi, F. , Hui, D. J. , Merrick, W. C. , & Sen, G. C. (2006). Distinct induction patterns and functions of two closely related interferon‐inducible human genes, ISG54 and ISG56. Journal of Biological Chemistry, 281(45), 34064–34071. 10.1074/jbc.M605771200 [DOI] [PubMed] [Google Scholar]
- Tirumuru, N. , & Wu, L. (2019). HIV‐1 envelope proteins up‐regulate N6‐methyladenosine levels of cellular RNA independently of viral replication. Journal of Biological Chemistry, 294(9), 3249–3260. 10.1074/jbc.RA118.005608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru, N. , Zhao, B. S. , Lu, W. , Lu, Z. , He, C. , & Wu, L. (2016). N6‐methyladenosine of HIV‐1 RNA regulates viral infection and HIV‐1 Gag protein expression. eLife, 5 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D. , Lehtonen, H. , Carmo‐Fonseca, M. , & Hurt, E. C. (1991). The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre‐rRNA processing in yeast. The EMBO Journal, 10(3), 573–583. Retrieved from. http://www.ncbi.nlm.nih.gov/pubmed/1825809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic, I. , Svitkin, Y. V. , Sonenberg, N. , & Shatkin, A. J. (2011). Cap and cap‐binding proteins in the control of gene expression. WIREs: RNA, 2(2), 277–298. 10.1002/wrna.52 [DOI] [PubMed] [Google Scholar]
- Trim, A. R. , & Parker, J. E. (1970). Preparation, purification and analyses of thirteen alkali‐stable dinucleotides from ribonucleic acid. Biochemical Journal, 116(4), 589–598. 10.1042/bj1160589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trixl, L. , & Lusser, A. (2019). The dynamic RNA modification 5‐methylcytosine and its emerging role as an epitranscriptomic mark. WIREs: RNA, 10(1), e1510 10.1002/wrna.1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman, J. B. , Giltmier, A. J. , Mukherjee, C. , & Schoenberg, D. R. (2017). RNA guanine‐7 methyltransferase catalyzes the methylation of cytoplasmically recapped RNAs. Nucleic Acids Research, 45(18), 10726–10739. 10.1093/nar/gkx801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, K. , Courtney, D. G. , & Cullen, B. R. (2018). Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathogens, 14(2), e1006919 10.1371/journal.ppat.1006919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen, B. G. , van Boheemen, S. , de Rijck, J. , van Nieuwkoop, S. , Smith, D. J. , Laksono, B. , … Fouchier, R. A. M. (2014). Excessive production and extreme editing of human metapneumovirus defective interfering RNA is associated with type I IFN induction. Journal of General Virology, 95(PART 8), 1625–1633. 10.1099/vir.0.066100-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn, L. P. , Kasperaitis, M. , Ameling, C. , & Voorma, H. O. (1986). Additional methylation at the N(2)‐position of the cap of 26S Semliki Forest virus late mRNA and initiation of translation. Virus Research, 5(1), 61–66. [DOI] [PubMed] [Google Scholar]
- Vieyres, G. , & Pietschmann, T. (2019). HCV pit stop at the lipid droplet: Refuel lipids and put on a lipoprotein coat before exit. Cell, 8(3), 233 10.3390/cells8030233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C.‐M. , Gershowitz, A. , & Moss, B. (1975). Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell, 4(4), 379–386. 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- Wei, C. M. , & Moss, B. (1975). Methylated nucleotides block 5′‐terminus of vaccinia virus messenger RNA. Proceedings of the National Academy of Sciences of the United States of America, 72(1), 318–322. 10.1073/pnas.72.1.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, S. K. , & Lazinski, D. W. (2002). Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proceedings of the National Academy of Sciences of the United States of America, 99(23), 15118–15123. 10.1073/pnas.232416799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Yang, Y. , Sun, B.‐F. , Chen, Y.‐S. , Xu, J.‐W. , Lai, W.‐Y. , … Yang, Y.‐G. (2017). 5‐Methylcytosine promotes mRNA export – NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Research, 27(5), 606–625. 10.1038/cr.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, F. , Chen, E. R. , & Nilsen, T. W. (2017). Kaposi's sarcoma‐associated herpesvirus utilizes and manipulates RNA N6‐adenosine methylation to promote lytic replication. Journal of Virology, 91(16), e00466–e00417. 10.1128/JVI.00466-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli, V. S. R. K. , & Jeang, K.‐T. (2010). Trimethylguanosine capping selectively promotes expression of Rev‐dependent HIV‐1 RNAs. Proceedings of the National Academy of Sciences of the United States of America, 107(33), 14787–14792. 10.1073/pnas.1009490107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalinger, Z. B. , Elliott, R. , Rose, K. M. , & Weiss, S. R. (2015). MDA5 is critical to host defense during infection with murine coronavirus. Journal of Virology, 89(24), 12330–12340. 10.1128/jvi.01470-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerfass, K. , & Beier, H. (1992). Pseudouridine in the anticodon GψA of plant cytoplasmic tRNA Tyr is required for UAG and UAA suppression in the TMV‐specific context. Nucleic Acids Research, 20(22), 5911–5918. 10.1093/nar/20.22.5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L.‐S. , Liu, C. , Ma, H. , Dai, Q. , Sun, H.‐L. , Luo, G. , … He, C. (2019). Transcriptome‐wide mapping of internal N7‐methylguanosine methylome in mammalian mRNA. Molecular Cell, 74(6), 1304–1316.e8. 10.1016/j.molcel.2019.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, G. , Dahl, J. A. , Niu, Y. , Fedorcsak, P. , Huang, C. M. , Li, C. J. , … He, C. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell, 49(1), 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Wan, J. , Gao, X. , Zhang, X. , Jaffrey, S. R. , & Qian, S. B. (2015). Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature, 526(7574), 591–594. 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Ray, D. , Zhao, Y. , Dong, H. , Ren, S. , Li, Z. , … Li, H. (2007). Structure and function of flavivirus NS5 methyltransferase. Journal of Virology, 81(8), 3891–3903. 10.1128/JVI.02704-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Pirnie, S. P. , & Carmichael, G. G. (2017). High‐throughput and site‐specific identification of 2′‐O‐methylation sites using ribose oxidation sequencing (RibOxi‐seq). RNA, 23(8), 1303–1314. 10.1261/rna.061549.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst, R. , Cervantes‐Barragan, L. , Habjan, M. , Maier, R. , Neuman, B. W. , Ziebuhr, J. , … Thiel, V. (2011). Ribose 2′‐O‐methylation provides a molecular signature for the distinction of self and non‐self mRNA dependent on the RNA sensor Mda5. Nature Immunology, 12(2), 137–143. 10.1038/ni.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]


