Abstract
Viruses are obligatory intracellular parasites that require the host machinery to replicate. During their replication cycle, viral RNA intermediates can be recognized and degraded by different antiviral mechanisms that include RNA decay, RNA interference, and RNase L pathways. As a consequence of viral RNA degradation, infected cells can accumulate virus‐derived small RNAs at high levels compared to cellular molecules. These small RNAs are imprinted with molecular characteristics that reflect their origin. First, small RNAs can be used to reconstruct viral sequences and identify the virus from which they originated. Second, other molecular features of small RNAs such as size, polarity, and base preferences depend on the type of viral substrate and host mechanism of degradation. Thus, the pattern of small RNAs generated in infected cells can be used as a molecular footprint to identify and characterize viruses independent on sequence homology searches against known references. Hence, sequencing of small RNAs obtained from infected cells enables virus discovery and characterization using both sequence‐dependent strategies and novel pattern‐based approaches. Recent studies are helping unlock the full application of small RNA sequencing for virus discovery and characterization. WIREs RNA 2016, 7:824–837. doi: 10.1002/wrna.1361
This article is categorized under:
-
1
RNA Processing > Processing of Small RNAs
-
2
RNA Turnover and Surveillance > Turnover/Surveillance Mechanisms
-
3
RNA Methods > RNA Analyses In Vitro and In Silico

INTRODUCTION
Viruses are intracellular obligatory parasites that repurpose the host cell machinery to replicate. Viruses can create modified intracellular compartments referred to as viral factories where they concentrate resources required for replication and are shielded from host antiviral mechanisms.1, 2 However, because viruses do not encode their own ribosomes, viral messenger RNAs (mRNAs) require access to cellular ribosomes to be translated. When viral mRNAs traffic to cellular ribosomes, they are exposed to host defense mechanisms more than other products generated during virus replication. Viral genomic RNAs are also excellent targets of host surveillance mechanism as they can be significantly different from cellular molecules. Thus, viral nucleic acids are a common target of several host antiviral mechanisms.3, 4, 5, 6 Targeting of viral RNAs often results in the generation of virus‐derived small RNAs (vsRNAs) that can be detected during infection in fungi, plants, arthropods, and mammals.7, 8, 9, 10, 11, 12, 13 Although vsRNAs are commonly observed in different organisms, their relative abundance can vary substantially.8 In insects, for example, viral sequences are 10‐fold enriched in the small RNA fraction compared to what was observed in the long RNA pool from infected cells.10 In contrast, viral sequences in lungs of infected mice were underrepresented by a factor of 100 in the pool of small compared with long RNAs.10 This relative enrichment or depletion is likely caused by differential targeting of viruses by RNA surveillance mechanisms. Nevertheless, targeting of viral transcripts and genomes is a common feature of different antiviral mechanisms and the production of vsRNAs can be highly informative. An analogy can be made about the indirect information about viruses obtained from vsRNAs and the mammalian antibody response. As initially proposed by Rivers and coworkers, virus‐specific antibody responses can be indicative of an infection without requiring direct detection of the virus.14 Similarly, vsRNAs generated by host pathways can also provide information about the infection without the requirement for direct detection of the virus.
HOST PATHWAYS THAT GENERATE vsRNAs
Viruses have evolved efficient mechanisms to escape host defense and ensure their survival. However, viral transcripts and genomes are often significantly different from cellular molecules, which allows them to be targeted by host pathways. Viral RNAs can be recognized by different RNA surveillance mechanisms, which often lead to their degradation and the generation of vsRNAs. RNA interference (RNAi), RNA decay, and RNase L pathways are good examples of such RNA surveillance mechanisms that have been extensively reviewed elsewhere.3, 4, 5, 6, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 Here, we provide a few examples of how RNA degradation pathways may generate small RNA products with distinct molecular characteristics (Figure 1).
Figure 1.
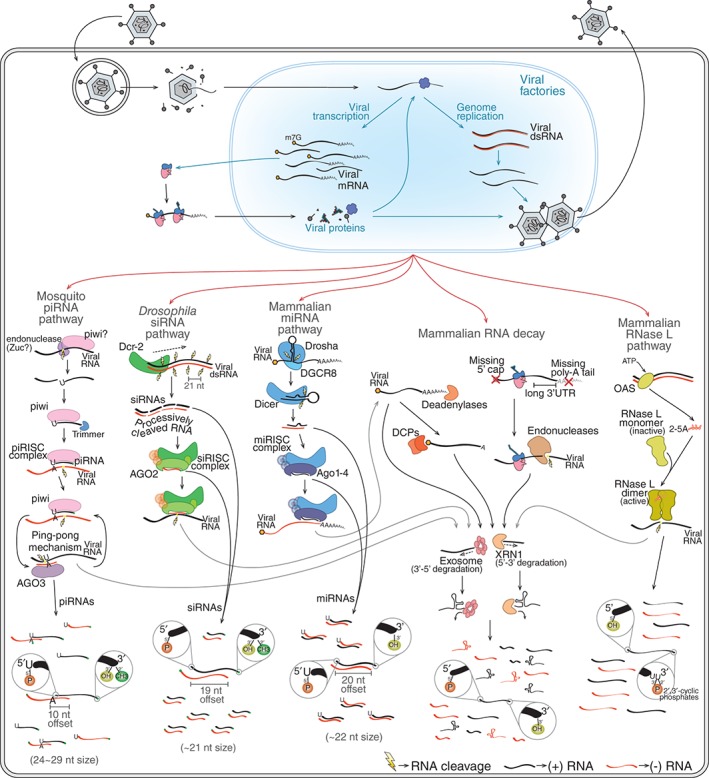
The generation of virus‐derived small RNAs by host pathways. During infection, virus genomes and transcripts are sometimes exposed and can be recognized by different host RNA surveillance mechanisms. RNA decay, RNA interference (RNAi), and RNase L pathways are good examples of such mechanisms. Mammalian RNA decay involves different mechanisms such as deadenylation‐dependent and nonsense‐mediated decay (NMD). The former involves shortening of the poly(A) tail by deadenylases followed by removal of the 5′ cap by mRNA‐decapping enzymes (DCPs). NMD is initiated by recognition of aberrant mRNAs that are cleaved by endonucleases. In both cases, the initial cleavage allows the mRNA to be targeted 5′→3′ by XRN1 and 3′→5′ by the exosome complex. The mammalian RNase L pathway is triggered when viral dsRNA is recognized by OAS enzymes that catalyze the production of 2′‐5′ oligoadenylates (2‐5A). These molecules induce dimerization and activation of RNase L that cleaves single‐stranded RNAs mostly at U‐rich regions. Different RNAi mechanisms can generate vsRNAs during viral infection including the mammalian miRNA pathway, Drosophila small interfering RNA (siRNA) pathway, and mosquito piRNA pathway. The Drosophila siRNA pathway is activated by Dcr‐2‐mediated recognition of viral dsRNA that is processes progressively to generate phased duplex siRNAs. siRNA duplexes are loaded onto AGO2 to generate siRISC that will find and cleave complementary RNAs. The mammalian miRNA pathway is initiated by the recognition of structure regions within long transcripts in the nucleus by the RNase III Drosha. This enzyme, in association with a partner protein known as DGCR8, cleaves the primary transcript to excise a short hairpin (~65 nt) that is then exported to the cytoplasm. There, the hairpin is further processed by Dicer to generate miRNA duplexes of ~22 nt that will be loaded onto different mammalian Argonaute proteins (Ago1–4) to form miRISC. This complex targets complementary regions within the 3′ UTR of mRNAs leading to translation inhibition. The mosquito piRNA pathway is triggered by the recognition of single‐stranded RNA precursors in a manner dependent on a specialized group of Argonautes known as PIWI proteins usually associated with an endonuclease known as Zucchini (Zuc). This initial recognition triggers processing of the precursor into primary piRNAs that remain associated with PIWI proteins to form the piRISC. This complex carries out cleavage of complementary RNA and can also initiate the production of more piRNAs. These secondary piRNAs require an amplification loop, referred to as the ping‐pong mechanism, that involves another Argonaute protein known as AGO3. Different RNA surveillance mechanisms may work together to generate vsRNAs. For example, RNA fragments generated by RNAi and RNase L pathways can be further targeted by RNA decay mechanisms. RNA surveillance mechanisms generate vsRNAs that have unique molecular characteristics such as terminal modifications, size, strand bias, and nucleotide preferences shown in the figure.
RNA Decay
RNA decay mechanisms play an important role in the quality control and turnover of cellular transcripts.16, 18, 21 Cellular transcripts are characterized by bona fide stability determinants such as the 5′ cap and 3′ poly(A) tails that are required to optimize translation.26 Transcripts that lack canonical features are targeted by several RNA decay mechanisms.16, 18 In the deadenylation‐dependent mRNA decay, the poly(A) tail of transcripts is shortened by the action of different deadenylases generating an unprotected 3′ end that can be degraded 3′→5′ by a large complex of exonucleases known as exosome.16, 27 After poly(A) shortening, the transcript can also be degraded by removal of the 5′ cap by mRNA‐decapping enzymes (DCPs), which allows the transcript to be targeted by XRN1, an exoribonuclease that carries out 5′→3′ degradation. In addition, nonsense‐mediated decay (NMD) is a highly conserved deadenylation‐independent mechanism capable of detecting aberrant mRNAs with short open reading frames and premature termination codons.18 NMD triggers endonucleolytic cleavage of target transcripts that is followed by degradation promoted by XRN1 and the exosome complex.18, 28 RNA degradation by XRN1 and the exosome generates not only nucleosides but also small RNA fragments with 3′ OH and/or 5′ monophosphate groups depending on the structure and secondary modifications found in the substrate.28, 29
In addition to quality control of cellular transcripts, RNA decay mechanisms have been shown to play an important role in the recognition and targeting of viral RNAs in mammals30, 31 (Figure 1). Viral RNAs often lack essential features that characterize cellular mRNAs, such as 5′ cap and poly(A) tails, due to their complex genomic structure.17 Besides, viruses may also produce transcripts with short open reading frames and long 3′ UTRs or unusual RNA products due to the activity of prolific and error‐prone RNA polymerases.6, 18 These features help to explain how viral RNAs are often detected as aberrant transcripts by RNA decay mechanism.17 RNA decay mechanisms help monitor proper translation in infected cells, thus playing important roles in the antiviral response. In addition to RNA decay, other specialized antiviral pathways can also target viral RNAs and generate vsRNAs in eukaryotes.
RNA Interference
RNAi is a conserved mechanism of sequence‐specific regulation of gene expression found in most eukaryotes. Initiation of RNAi requires recognition of an RNA substrate by specialized nucleases that are often type III RNases such as Dicers responsible for the biogenesis of small RNAs. During the effector phase of RNAi, these small RNAs associate with Argonaute proteins to form the RNA‐induced silencing complex (RISC). RISC will utilize the small RNA sequence to select target RNAs via Watson–Crick base pairing, which will be degraded by Argonaute‐mediated endonucleolytic cleavage.20 RNA fragments generated by RISC cleavage are short lived likely because they are further degraded by regular RNA decay mechanism. Small RNAs generated by both Dicer and Argonaute have 5′ monophosphates and 3′ OH groups. However, small RNAs generated during the initiation phase that associate with Argonaute proteins may be further stabilized by secondary modifications such as 2′O‐methylation.
There are different RNAi pathways in eukaryotes that differ in their mechanism of activation, small RNA biogenesis, and action.22, 23 In animals, there are at least three separate RNAi mechanisms that can generate vsRNAs during viral infection: the microRNA (miRNA), piwi‐interacting RNA (piRNA), and small interfering RNA (siRNA) pathways (Figure 1). Each different RNAi pathway requires specific types of Argonaute–small RNA complexes.32
The miRNA pathway is triggered by the recognition of short hairpins (~65 nt) formed by secondary structures found in long single‐stranded RNAs.33 These internal hairpins are processed in two steps by two double‐stranded RNA (dsRNA)‐specific ribonucleases, Drosha and Dicer, to generate small RNA duplexes of ~20–23 nt that will be subsequently loaded onto an Argonaute protein to form miRISC.34 This complex will target complementary regions in the 3′ UTR of cellular mRNA leading to translation inhibition and mRNA destabilization.
Activation of the siRNA pathway is triggered by Dicer‐mediated recognition of long dsRNA that is processed progressively to generate phased duplex small RNAs of ~20–23 nt.35 siRNA duplexes are loaded onto specialized Argonaute proteins to generate siRISC that remains associated with one of the strands.36 Mature siRISC is a multiple turnover enzyme that can efficiently catalyze endonucleolytic cleavage of RNA targets.37, 38
Unlike miRNA and siRNA, the piRNA pathway is triggered by the recognition of single‐stranded RNA precursors seemingly independent on the presence of secondary structures.39 This single‐stranded RNA precursor is processed by an endonuclease that generates phased single‐stranded small RNAs that are ~24–30 nt long.40 The generation of these primary piRNAs requires a distinct subgroup of animal Argonautes known as PIWI proteins.40 Primary piRNAs can also trigger the production of secondary piRNAs by a self‐amplifying cycle dependent on PIWI proteins known as the ping‐pong mechanism.41, 42 The complex formed by piRNAs and PIWI proteins known as piRISC can mediate transcriptional silencing as well as target RNA degradation.39 In contrast to siRNAs and miRNAs, piRNAs are commonly enriched in the germline and reproductive tissues of different animals.41, 43, 44, 45
RNAi pathways can have different roles during viral infection. The siRNA pathway is directly involved in the antiviral immunity in most eukaryotes including fungi, animals, and plants; although in mammals, this seems to be restricted to undifferentiated cells.12, 46 In contrast, production of virus‐derived miRNAs and piRNAs during viral infection appears to be more restricted. Activation of the piRNA pathway by virus infection has been reported in insects, although its antiviral role remains unclear.10, 13, 47, 48, 49, 50, 51 Generation of virus‐derived miRNAs seems to be restricted to some animal viruses that exploit this pathway to control the expression of viral and cellular transcripts and help their replication.24, 25
Mammalian RNase L
Mammals have developed an antiviral system based on the degradation of viral RNAs by a specialized ribonuclease known as RNase L19 (Figure 1). RNase L is found in a latent state in healthy cells and can be rapidly activated during virus infection.19 The RNase L pathway is initiated by a group of enzymes known as 2′‐5′‐oligoadenylate synthetases (OAS) that can bind to dsRNA generated during viral infection. Once activated by dsRNA, these enzymes catalyze the polymerization of cellular ATP to generate 2′‐5′‐oligoadenylates (2‐5A) that act as second messengers.52 RNase L directly binds to 2‐5A, which induces its dimerization and activation.53, 54 Active RNase L will degrade single‐stranded RNAs within infected cells to generate small RNAs of varied sizes that contain 5′‐hydroxyl and 2′,3′‐cyclic phosphoryl group characteristic of metal ion‐independent ribonucleases.55, 56 Although both cellular and viral RNAs can be targeted by RNase L, there is evidence that the enzyme has some capacity to selectively target viral molecules.57 In addition to its direct antiviral activity, RNase L products may also work in a positive feedback loop where the RNA helicase RIG‐I is activated by small RNA products of RNase L degradation.55 The RNase L system plays an important role in the antiviral response against several viruses in mammals.58, 59, 60, 61
WHAT WE CAN LEARN FROM vsRNAs
Different RNA surveillance mechanisms contribute to the degradation of viral molecules and generation of vsRNAs in infected cells. Small RNAs have molecular characteristics that reflect the virus and host pathways from which they originated. Thus, the pattern of vsRNAs can be analyzed and provide extensive information about their source.
Reconstruction of Viral Sequences From vsRNAs
Small RNAs derived from viral transcripts and genomes can be used to reconstitute the sequences from which they originate. Thus, the presence of viral sequences in small RNA libraries prepared from an organism provides evidence for the presence of viruses. However, owing to their limited size, vsRNAs require prior assembly into longer contiguous sequences (contigs) before they can be identified by comparisons to known viral sequences present in reference databases (Figure 2). This strategy has been successfully applied to identification and characterization of viruses in plants, fungi, and animals.7, 10, 11, 50, 62 Despite these successful efforts using small RNAs, sequencing of long RNAs has been a preferred strategy in studies aiming at identification of viruses within infected hosts.63, 64, 65, 66
Figure 2.
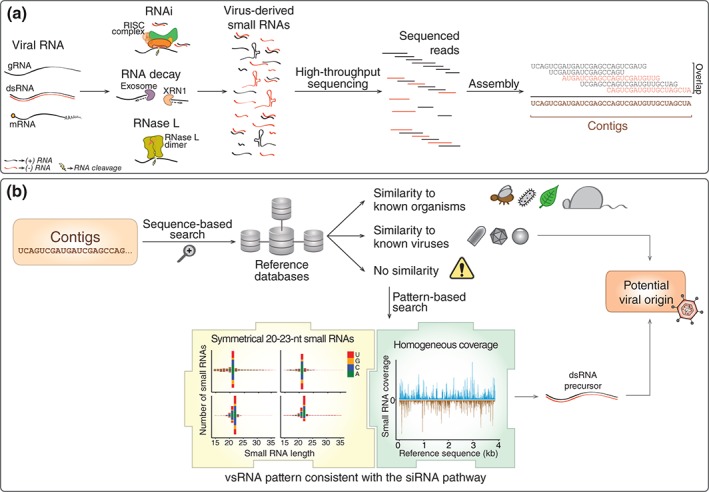
Identification of viral sequences utilizing virus‐derived small RNAs. (a) Virus‐derived small RNAs are generated by different host pathways during infection. These small RNAs can be sequenced and assembled into long contiguous sequences (contigs) reflecting the substrate from which they originate. (b) Classically, the origin of contigs from large‐scale sequencing studies is inferred by sequence similarities by comparisons to known references in databases. Thus, contigs are identified when they show similarity to sequences from known viruses or other organisms. However, a large number of contigs in large‐scale sequencing studies do not show similarity to any known reference sequence. In these cases, the molecular pattern of small RNAs derived from any contig sequence can be used. The presence of a clear small RNA symmetrical size preference between 20 and 23 nt at similar abundance on both strands of the contig suggests they originated from a double‐stranded RNA (dsRNA) precursor that was processed by the small interfering RNA (siRNA) pathway. This can be indicative of the contig origin because this molecular signature is reasonably specific to viruses. Thus, a small RNA profile can be used to identify potential viral contigs without any prior information about the sequence itself.
As large‐scale sequencing of small and long RNAs have both been used to identify viruses within infected hosts, our group has directly compared strategies for virus identification. In Aedes aegypti mosquitoes, we observed that the identification of viruses by small RNA sequencing performed significantly better than long RNAs. Small RNAs provided more sequence coverage of the viral genome in less processing time. Efficient generation of small RNAs by RNAi pathways has been proposed as the main reason for the optimized detection of viruses by small RNA sequencing.50, 62 Indeed, we observed that small RNA products from siRNA and piRNA pathways tended to optimize the assembly of longer viral contigs likely due to extensive overlap between sequences. Viral sequences also showed a 10‐fold natural enrichment in the small RNA fraction compared to long RNAs in the same mosquito sample.10 Interestingly, small RNAs from lungs of coronavirus‐infected mice, where no activation of RNAi pathways was observed, showed a 100‐fold depletion of viral sequences relative to long RNAs.10 Nevertheless, even in this situation, viral contigs represented 30% of contigs assembled from small RNAs compared to 5% of long RNAs. Thus, small RNAs seem to favor assembly of viral contigs even when they represent a small fraction of the total.10 This can help to save computational time used for the analysis of large‐scale sequencing data that can be the bottleneck for many metagenomic studies. Thus, some advantages of virus detection using small RNAs compared to long RNAs are not restricted to situations where the RNAi pathway is activated.10
Construction and sequencing of small RNA libraries requires little sample manipulation and processing steps, which can be simpler compared to long RNAs.67, 68 Long RNA libraries often require extensive depletion of ribosomal RNAs that may otherwise represent ~80% of all RNA species within an organism.68 Long RNA libraries may also utilize polyA‐enriched RNA without the need for prior depletion of ribosomal RNA but this can compromise the detection of viruses whose RNAs are not polyadenylated.69 Notably, some insects, such as Drosophila melanogaster and Lutzomyia longipalpis, encode an abundant small RNA corresponding to the 2S ribosomal RNA that requires depletion before preparation of small RNA libraries.10, 70 Even in this case, this corresponds to a single sequence that can be depleted with a single complementary probe more efficiently than large ribosomal RNA subunits that require multiple probes. It is noteworthy that the preparation of small RNAs libraries directly from total RNA could interfere with the detection of viruses.71, 72 Hence, sequencing of small RNAs allows efficient detection of viral sequences although more studies are required to determine the power and extent of its application.
Virus Discovery Using vsRNA Patterns
Different host pathways generate vsRNAs with unique molecular characteristics such as size, polarity, and base enrichment. The siRNA pathway is likely the best example on how molecular patterns of small RNAs can be explored. Viruses often produce dsRNA molecules during their replication cycle, which is considered a hallmark of viral infection73 (Figure 1). In most eukaryotes, the siRNA pathway is activated when Dicer directly recognizes and processes viral dsRNA into 20–23 nt long duplex small RNAs.4 These siRNAs usually do not have strong nucleotide preferences and cover symmetrically both strands of the dsRNA trigger. Canonical siRNAs also have 3′ 2‐nt overhangs and ~19–21 nt overlap between the two strands. Thus, these molecular characteristics are a signature of activation of the siRNA pathway that has been consistently detected for a variety of viruses in fungi, animals, and plants9, 12, 73, 74, 75, 76 (Figure 3(a)). As the activation of the siRNA pathway is so strongly associated with viral infection, the detection of small RNAs containing molecular patterns consistent with siRNAs could indicate a viral origin (Figure 2(b)). We and others have used this premise to discover novel viruses based on the ability of their sequences to generate siRNAs.10, 11 Importantly, this strategy has enabled the discovery of divergent viral sequences with no similarities to known viruses in reference databases.10 The detection of sequences that generate siRNAs can be an efficient strategy to find viruses although there are potential limitations that can affect its sensitivity and specificity.
Figure 3.
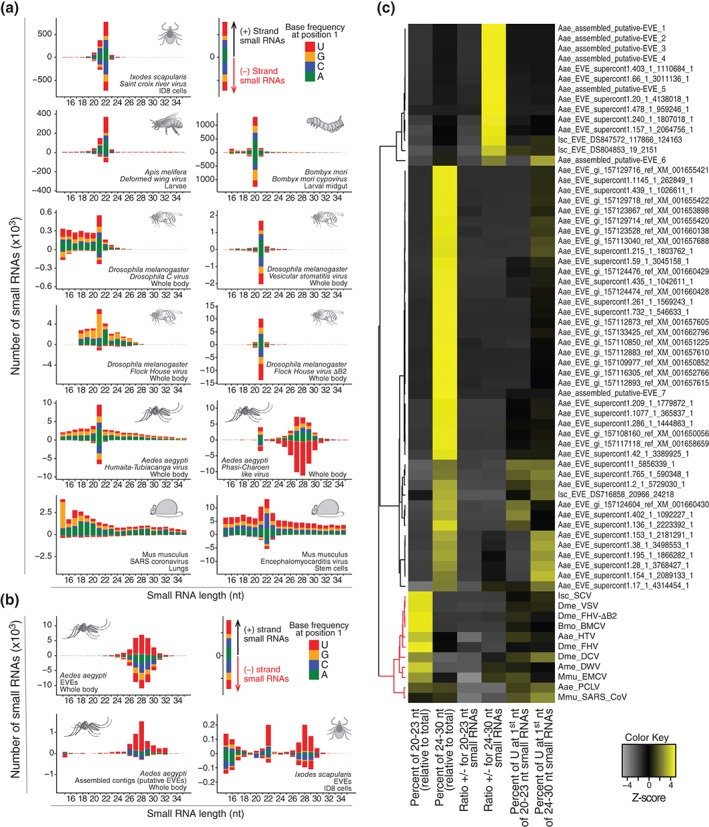
Activation of the small interfering RNA (siRNA) pathway is a common and specific response to virus infection. (a) Virus‐derived small RNAs in different animals often show a profile consistent with the activation of the siRNA pathway. Virus‐derived siRNAs range from ~20 to 23 nt and are symmetrical in polarity and base preferences. (b) Endogenous viral elements (EVEs) represent remnants of virus sequences integrated into animal genomes and also generate small RNAs. EVE‐derived small RNAs are often ~24–30 nt, asymmetrical in polarity, and base preferences that is consistent with a piRNA signature. (c) Size, polarity, and nucleotide preferences were determined for small RNAs derived from EVEs and active viruses. These molecular footprints were then used to perform hierarchical clustering of different EVEs and viruses. The small RNA pattern clearly separates clusters containing viruses (in red) and EVEs (in black). Notably, within the cluster of viruses, we observe two small subclusters. Virus grouped in the larger subcluster show a classical siRNA signature even when the pathway is partially inhibited such as for FHV and DCV in Drosophila. The second subcluster contains the vsRNA profile of coronavirus in mouse lungs and PCLV in mosquitoes, both of which show a more divergent profile from the siRNA signature. The data in this figure were obtained from the analysis of small RNA libraries from published studies (accession numbers: SRR1803378 , SRR1803382 , ERR555100, ERR274423, ERR654010, SRR1803383 , GSM792688, GSM792692, SRR452408, and SRR640612).10, 12, 77, 78, 79, 80, 81, 82
First, there are endogenous sources of dsRNA capable of generating siRNAs.36 Endogenous siRNAs arise from regions of convergent transcription, structured RNAs, or repetitive elements. In each of these cases, it would not be difficult to ascertain the origin of siRNAs because these are easily differentiated from viruses. Convergent transcription and structured RNAs arise from annotated genes and repetitive elements can be identified using repeat filters. However, animal genomes have also integrated endogenous viral elements (EVEs) that represent remnants of viral sequences.80, 83 We analyzed the small RNA profile observed for EVEs in different organisms compared to active viruses. EVE‐derived small RNAs were observed but show clearly different molecular patterns compared to viruses in the same organism (Figure 3). Interestingly, EVEs seemed to favor the generation of small RNAs with molecular characteristics of piRNAs rather than siRNAs.
Second, some viruses have developed viral suppressors of the siRNA pathway (VSRs) that can significantly affect the pattern of vsRNAs. VSRs allow for accumulation of viral RNA that can be degraded by other host mechanisms.10, 46, 77 Patterns of vsRNA distinct from canonical siRNAs have been observed upon suppression of the siRNA pathway by VSRs.10, 77, 84 For example, Drosophila adults infected with Flock house virus (FHV) show accumulation of small RNAs in the positive strand with a broad size distribution and only a very small peak at 21 nt in the negative strand77 (Figure 3(a)). FHV encodes a potent VSR known as B2 that successfully protects its dsRNAs from access by Dicer‐2 and prevents activation of the siRNA pathway. The viral RNA genome accumulates at higher levels than the antigenome and is likely degraded by other nucleases, which explains the bias observed in small RNAs covering the positive strand and the broad size profile. Accordingly, in the absence of B2, the pattern of small RNAs observed in FHV‐infected flies is consistent with the activation of the siRNA pathway77 (Figure 3(a)).
Lastly, technical artifacts from small RNA sequencing strategies could interfere with the detection of siRNA signatures. Biased cloning due to inefficient adaptor ligation or sequestration of antisense small RNAs by the sense target can result in a distortion of vsRNA profile.71, 72 However, it is unclear how much the detection of a canonical siRNA profile can be compromised by technical biases. In arthropods, for example, a canonical siRNA profile is observed more often than not, even using standard strategies for small RNA library construction (Figure 3(a)).
Hence, small RNA sequencing is a promising strategy to identify viruses, despite potential limitations that require further analysis.
Using vsRNA Patterns to Extract Information About the Infection
The pattern of vsRNAs is a molecular footprint that can be explored beyond the activation of the siRNA pathway. Each RNA surveillance mechanisms may show cell‐type‐ or tissue‐specific expression and differentially target viral RNA intermediates. Thus, virus tropism and the abundance of different viral RNA targets will have a direct influence on the pattern of small RNAs produced by host pathways. Consequently, the pattern of vsRNAs generated during infection can provide information about their origin, which reflects both virus and host features (Figure 4).
Figure 4.
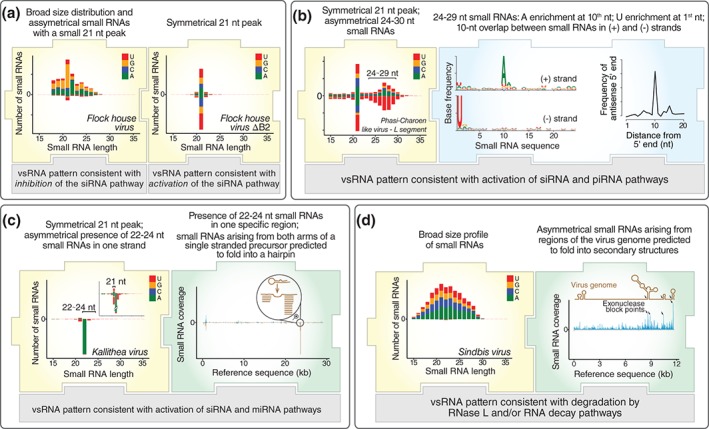
Molecular footprints of virus‐derived small RNAs. In addition to utilizing a classical small interfering RNA (siRNA) signature to identify potential viral sequences, the analysis of molecular characteristics of small RNAs derived from viral contigs can also provide further information about virus biology. The profile of virus‐derived small RNAs sometimes diverges from the canonical siRNA signature but still have unique molecular features that tend to reflect intrinsic properties of the virus such as tissue tropism, presence of inhibitors (VSR), and secondary structures or modifications in viral RNA. (a) The partial or complete absence of an expected siRNA signature often indicates that the virus escapes the siRNA pathway. This is the case of FHV in Drosophila, whose profile is fairly divergent from the canonical siRNA profiles unless a mutant virus incapable of producing the B2 VSR is analyzed. (b) The presence of longer virus‐derived small RNAs of ~24–30 nt that show a 10‐nt overlap between opposite strands is a signature of piRNAs. As these are often enriched in reproductive organs, this signature may be used to infer virus infection of ovaries as observed for PCLV in Aedes mosquitoes. (c) Animal viruses can sometimes show hotspots of small RNA coverage originating exclusively from one strand. These tend to come from genomic regions that are predicted to fold onto hairpins where the 20–24 nt long small RNAs arise from the stems. This small RNA signature suggests that this region is responsible for the generation of virus‐derived miRNAs as observed for Kallithea virus in Drosophila. (d) In the absence of a clear signature of the siRNA pathway, virus‐derived small RNAs may show a broad size distribution with highly heterogeneous coverage of the viral genome. This pattern suggests extensive viral RNA degradation with accumulation of small RNAs in regions corresponding to secondary structures or chemical modifications as observed for Sindbis virus in human cells. The small RNA data shown in this figure were obtained from the analysis of small RNA libraries from published studies (accession numbers GSM792688, GSM792692, SRR1803378 , SRR1914952 , and GSM1185388 ).10, 11, 61, 77
Some viruses have developed VSRs to escape the siRNA pathway as we described for the B2 protein of FHV. Interestingly, another insect virus, Drosophila C virus (DCV), encodes a VSR that binds long dsRNA and prevents Dcr‐2‐mediated processing.85 The profile of vsRNAs observed in DCV‐infected flies shows a peak in 21 nt long coming from both strands but also accumulation of small RNAs with smaller sizes derived from the coding strand.10, 84 The VSR encoded by DCV is not as potent as B2 and presumably still allows detectable activation of the siRNA pathway.86 This situation generates superimposed vsRNAs pattern combining products of different host pathways that could still allow virus identification based on a canonical siRNA pattern. Nevertheless, it is interesting to speculate that once a virus is identified, the absence of virus‐derived siRNAs in infected hosts may be interpreted as active inhibition by VSRs as shown for FHV (Figure 4(a)). In addition, the pattern of vsRNAs may also provide insights into the mechanism of action and potency of the VSRs considering the differences we observed between the small RNA profile generated by DCV and FHV (Figure 3(a)).
RNAi mechanisms other than the siRNA pathway could also be explored. The piRNA pathway, for example, is not broadly activated during viral infections. At the moment, virus‐derived piRNAs have only been observed in insect cell lines including Drosophila ovary, culicoides, and mosquito cell lines.10, 13, 49, 50, 51 In vivo, virus‐derived piRNAs have only been observed in A. albopictus and A. aegypti mosquitoes.10, 13 Molecular characteristics of piRNAs are quite distinct from siRNAs, as the former are ~24–30 nt long and show enrichment for U at the 5′ of antisense RNAs and A in 10th position of sense RNAs. piRNAs in opposite strands may also show an overlap of 10 nt between their 5′ ends, which is signature of the ping‐pong amplification mechanism.10, 51 In terms of their origin, piRNAs may show asymmetrical coverage of the viral genome or more homogenous distribution similar to siRNAs.10, 13 It is unclear why the activation of the piRNA pathway is restricted to certain viruses and how it is initiated during viral infection. Indeed, viruses that generate virus‐derived piRNAs have RNA genomes and replicate in the cytoplasm while the endogenous piRNA pathway is initiated from DNA‐dependent transcripts in the nucleus, at least in Drosophila.15 Our own data suggest that a strong activation of the piRNA pathway requires that the virus infects reproductive tissues where components of this pathway are commonly enriched in insects.10, 41, 87 We observed that the same A. aegypti mosquitoes infected by two different viruses, Phasi Charoen like‐virus (PCLV) and Humaita‐Tubiacanga virus (HTV), only the former showed abundant production of piRNAs which correlated with ovary infection (Figure 3(a)).10 In contrast, A. albopictus mosquitoes infected with Chikungunya virus showed production of virus‐derived piRNAs in the head and thorax of infected insects.13 However, in this case, the production of piRNAs was very inefficient as virus‐derived siRNAs were 120‐fold more abundant than piRNAs. Although more studies are required, a strong production of virus‐derived piRNAs may suggest that the virus has tropism for reproductive tissues in the insect (Figure 4(b)). Thus, the activation of the piRNA pathway could be very informative about tissue tropism but cannot be used as a viral signature as it is not a broad response to viral infection. Indeed, we still lack good evidence that the piRNA pathway has an antiviral role because it has little impact on virus replication in mosquito cell lines.48
Virus‐derived miRNAs can also be observed during infection but are usually produced by the virus to regulate the expression of viral or cellular genes.25 miRNA are distinct from siRNAs and piRNAs because they originate uniquely from specific regions of the virus genome and are restricted to just one of the strands.11 In terms of molecular features, miRNAs are ~20–24 nt long with a strong enrichment for U at 5′ end. The production of virus‐derived miRNAs has mostly been reported for animal DNA viruses.24, 25 Thus, in a somewhat more limited manner, the detection of these molecular patterns arising from regions of the viral genome could be used to infer whether a virus is capable of generating its own miRNAs (Figure 4(c)).
Non‐RNAi pathways, such as RNA decay and RNase L, also leave molecular footprints on vsRNAs that could be potentially explored. Indeed, these nucleases have substrate specificities that generate nonrandom patterns.28, 88 Ribonucleases from RNA decay pathways such as XRN1 and the exosome attack from their substrates from the extremities, which tends to reduce the RNA to a few nucleotides. However, these exonucleases can be impaired by highly structured regions or internal modifications within the target.89, 90 As a result, RNA decay nucleases often produce vsRNAs of different sizes that tend to accumulate near the end of the substrate or close to regions with secondary structure.29, 91 In the case of RNase L, small RNA products also have a very broad distribution as this endonuclease targets U‐rich sequences whose abundance may vary within target RNAs. In addition, RNA decay and RNase L pathways may act together to generate complex patterns of vsRNAs in infected cells. In human embryonic kidney 293 cells and green monkey vero cells, the size profile of small RNAs derived from Sindbis virus was remarkably consistent even in the absence of detectable activation of RNAi pathways.61 RNase L was required for the generation of some vsRNAs although they did not seem to be direct products of this nuclease. Indeed, the generation of Sindbis vsRNAs required the joint action of RNase L and other nucleases such as XRN1. In this case, vsRNAs seemed to accumulate in regions of the Sindbis genome containing posttranscriptional modifications that inhibited further degradation.61 Similarly, in human HeLa cells, the degradation of the poliovirus RNA is further targeted by RNase L together with other nucleases, which resulted in a complex pattern of vsRNAs generated across the virus genome.92 Thus, small RNAs generated by RNA decay and RNase L seem to reflect structured or modified regions within viral RNAs (Figure 4(d)). Indeed, the consistency of vsRNA profiles observed in different mammalian cells even without activation of RNAi suggests that the pattern does reflect stable virus characteristics.
Hence, complex vsRNA patterns can be very informative even when they result from the combined action of different nucleases. We are only beginning to understand how to decipher this information.
CONCLUSIONS
In the arms race between host and viruses, different pathways have evolved to target viral RNAs. These pathways contribute to the antiviral response by degrading viral RNAs, which results in the generation of vsRNAs. This common targeting of viral RNAs helps to explain why viral sequences are often abundant within the pool of small RNAs in infected cells. In addition, these tiny molecules are imprinted with molecular characteristics that reflect the host pathway and viral RNA from which they originate. Indeed, information can be extracted from these molecular footprints to trace back their viral origin. Directly, small RNA from infected hosts can be assembled into longer contiguous sequences and used to detect viruses by homology searches against known viral references. Indirectly, molecular patterns of small RNAs that are consistent with the activation of the siRNA pathway can be used as a signature to suggest a viral origin to novel assembled sequences. This latter strategy can help overcome a great limitation of virus discovery by metagenomic strategies, because it does not require sequence similarity searches against know references. Pattern analysis of vsRNAs allows the discovery of more divergent viral sequences with no similarities to known viruses present in reference databases.10, 11 We also showed here that the pattern of small RNAs is able to differentiate between sequences derived from viruses and integrated EVEs, which can be a powerful tool to indicate whether viral sequences originate from an active infection. Furthermore, the pattern of vsRNAs generated by host mechanisms other than the siRNA pathway can also provide information about viruses such as their tissue tropism, coding of inhibitors to the siRNA pathway, and structured regions of the viral genome. Although it might not be universally applicable, these advantages make small RNA sequencing a great strategy to identify and characterize viruses.
The potential use of vsRNA patterns is just beginning to be unlocked. We have recently showed that it is possible to use patterns of small RNAs different from the canonical siRNA signature to detect novel viral sequences.10 As a general idea, if the profile of small RNAs is known for any segment of a viral genome, this can be used as a reference to find other potential sequences from the same virus that show a similar pattern.10 This broadens the diversity of vsRNA patterns that can be utilized beyond the dependence on activation of the siRNA pathway. The use of additional tools could significantly improve our capacity to explore small RNAs patterns. Small RNAs generated by RNA decay, RNAi, and RNase L pathways have clear molecular features that can be utilized to differentiate specific products. Indeed, unique molecular features found in small RNAs generated by RNAi pathways such as 5′ monophosphate or 2′O‐methylation have been extensively explored.93, 94, 95 More recently, characteristics generated by RNase L have also been explored to identify specific products of this nuclease in infected cells.92, 96 These strategies allow more specific analysis of the products from each pathway and can be combined to provide a broader view of small RNAs within infected cells. This expansion in known vsRNA patterns would certainly broaden our references for pattern searches and improve our ability to identify and characterize viruses.
ACKNOWLEDGMENTS
We would like to thank Álvaro Gil Araujo Ferreira, Isaque João da Silva de Faria, and other members of our laboratory for invaluable suggestions and criticism. This work was supported by CAPES, CNPq, and FAPEMIG.
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
RELATED WIREs ARTICLES
https://doi.org/10.1002/wrna.40
REFERENCES
- 1. den Boon JA, Diaz A, Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe 2010, 8:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell 2005, 97:147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marques JT, Carthew RW. A call to arms: coevolution of animal viruses and host innate immune responses. Trends Genet 2007, 23:359–364. [DOI] [PubMed] [Google Scholar]
- 4. Ding S‐W. RNA‐based antiviral immunity. Nat Rev Immunol 2010, 10:632–644. [DOI] [PubMed] [Google Scholar]
- 5. Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res 2011, 31:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rigby RE, Rehwinkel J. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol 2015, 36:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vainio EJ, Jurvansuu J, Streng J, Rajamaki ML, Hantula J, Valkonen JP. Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J Gen Virol 2015, 96:714–725. [DOI] [PubMed] [Google Scholar]
- 8. Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, et al. Six RNA viruses and forty‐one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog 2010, 6:e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, Yu JL, Ding SW. RNAi‐mediated viral immunity requires amplification of virus‐derived siRNAs in Arabidopsis thaliana . Proc Natl Acad Sci USA 2010, 107:484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aguiar ER, Olmo RP, Paro S, Ferreira FV, de Faria IJ, Todjro YM, Lobo FP, Kroon EG, Meignin C, Gatherer D, et al. Sequence‐independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res 2015, 43:6191–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Webster CL, Waldron FM, Robertson S, Crowson D, Ferrari G, Quintana JF, Brouqui JM, Bayne EH, Longdon B, Buck AH, et al. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster . PLoS Biol 2015, 13:e1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science 2013, 342:235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. Production of virus‐derived ping‐pong‐dependent piRNA‐like small RNAs in the mosquito soma. PLoS Pathog 2012, 8:e1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwentker FF, Rivers TM. The antibody response of rabbits to injections of emulsions and extracts of homologous brain. J Exp Med 1934, 60:559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Czech B, Hannon GJ. One loop to rule them all: the ping‐pong cycle and piRNA‐guided silencing. Trends Biochem Sci 2016, 41:324–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen CY, Shyu AB. Mechanisms of deadenylation‐dependent decay. Wiley Interdiscip Rev RNA 2011, 2:167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaglia MM, Glaunsinger BA. Viruses and the cellular RNA decay machinery. Wiley Interdiscip Rev RNA 2010, 1:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang L, Wilkinson MF. Regulation of nonsense‐mediated mRNA decay. Wiley Interdiscip Rev RNA 2012, 3:807–828. [DOI] [PubMed] [Google Scholar]
- 19. Sadler AJ, Williams BR. Interferon‐inducible antiviral effectors. Nat Rev Immunol 2008, 8:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moon SL, Wilusz J. Cytoplasmic viruses: rage against the (cellular RNA decay) machine. PLoS Pathog 2013, 9:e1003762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet 2009, 10:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Batista TM, Marques JT. RNAi pathways in parasitic protists and worms. J Proteomics 2011, 74:1504–1514. [DOI] [PubMed] [Google Scholar]
- 24. Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 2009, 23:1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454:780–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 1991, 5:2108–2116. [DOI] [PubMed] [Google Scholar]
- 27. Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 2005, 12:1054–1063. [DOI] [PubMed] [Google Scholar]
- 28. Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′‐‐>5′ exoribonucleases. Cell 1997, 91:457–466. [DOI] [PubMed] [Google Scholar]
- 29. Chapman EG, Moon SL, Wilusz J, Kieft JS. RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. Elife 2014, 3:e01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garcia D, Garcia S, Voinnet O. Nonsense‐mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 2014, 16:391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balistreri G, Horvath P, Schweingruber C, Zund D, McInerney G, Merits A, Muhlemann O, Azzalin C, Helenius A. The host nonsense‐mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe 2014, 16:403–411. [DOI] [PubMed] [Google Scholar]
- 32. Ipsaro JJ, Joshua‐Tor L. From guide to target: molecular insights into eukaryotic RNA‐interference machinery. Nat Struct Mol Biol 2015, 22:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005, 6:376–385. [DOI] [PubMed] [Google Scholar]
- 34. Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer‐1. Cell 2007, 130:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, Zamore PD. Phosphate and R2D2 restrict the substrate specificity of Dicer‐2, an ATP‐driven ribonuclease. Mol Cell 2011, 42:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol 2010, 17:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 2005, 11:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti‐guide strand of siRNA during RISC activation. Cell 2005, 123:621–629. [DOI] [PubMed] [Google Scholar]
- 39. Le Thomas A, Toth KF, Aravin AA. To be or not to be a piRNA: genomic origin and processing of piRNAs. Genome Biol 2014, 15:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han BW, Wang W, Li C, Weng Z, Zamore PD. Noncoding RNA. piRNA‐guided transposon cleavage initiates Zucchini‐dependent, phased piRNA production. Science 2015, 348:817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137:522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer‐mediated mechanism for repeat‐associated siRNA 5' end formation in Drosophila. Science 2007, 315:1587–1590. [DOI] [PubMed] [Google Scholar]
- 43. Castellano L, Rizzi E, Krell J, Di Cristina M, Galizi R, Mori A, Tam J, De Bellis G, Stebbing J, Crisanti A, et al. The germline of the malaria mosquito produces abundant miRNAs, endo‐siRNAs, piRNAs and 29‐nt small RNAs. BMC Genomics 2015, 16:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007, 129:69–82. [DOI] [PubMed] [Google Scholar]
- 45. Roovers EF, Rosenkranz D, Mahdipour M, Han CT, He N, Chuva de Sousa Lopes SM, van der Westerlaken LA, Zischler H, Butter F, Roelen BA, et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep 2015, 10:2069–2082. [DOI] [PubMed] [Google Scholar]
- 46. Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science 2013, 342:231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miesen P, Girardi E, van Rij RP. Distinct sets of PIWI proteins produce arbovirus and transposon‐derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res 2015, 43:6545–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schnettler E, Donald CL, Human S, Watson M, Siu RW, McFarlane M, Fazakerley JK, Kohl A, Fragkoudis R. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J Gen Virol 2013, 94:1680–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schnettler E, Ratinier M, Watson M, Shaw AE, McFarlane M, Varela M, Elliott RM, Palmarini M, Kohl A. RNA interference targets arbovirus replication in Culicoides cells. J Virol 2013, 87:2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu Q, Luo Y, Lu R, Lau N, Lai EC, Li W‐X, Ding S‐W. Virus discovery by deep sequencing and assembly of virus‐derived small silencing RNAs. Proc Natl Acad Sci USA 2010, 107:1606–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vodovar N, Bronkhorst AW, van Cleef KW, Miesen P, Blanc H, van Rij RP, Saleh MC. Arbovirus‐derived piRNAs exhibit a ping‐pong signature in mosquito cells. PLoS One 2012, 7:e30861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2'‐specific and double‐stranded RNA‐activated interferon‐induced antiviral protein 2'‐5'‐oligoadenylate synthetase. Mol Cell 2003, 12:1173–1185. [DOI] [PubMed] [Google Scholar]
- 53. Zhou A, Hassel BA, Silverman RH. Expression cloning of 2‐5A‐dependent RNAase: a uniquely regulated mediator of interferon action. Cell 1993, 72:753–765. [DOI] [PubMed] [Google Scholar]
- 54. Dong B, Silverman RH. 2‐5A‐dependent RNase molecules dimerize during activation by 2‐5A. J Biol Chem 1995, 270:4133–4137. [DOI] [PubMed] [Google Scholar]
- 55. Malathi K, Dong B, Gale M Jr, Silverman RH. Small self‐RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007, 448:816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Floyd‐Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2'‐5')oligoadenylate‐‐dependent endonuclease. Science 1981, 212:1030–1032. [DOI] [PubMed] [Google Scholar]
- 57. Li XL, Blackford JA, Hassel BA. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol 1998, 72:2752–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Malathi K, Saito T, Crochet N, Barton DJ, Gale M Jr, Silverman RH. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA 2010, 16:2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sawicki DL, Silverman RH, Williams BR, Sawicki SG. Alphavirus minus‐strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R. J Virol 2003, 77:1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. RNase L plays a role in the antiviral response to West Nile virus. J Virol 2006, 80:2987–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Girardi E, Chane‐Woon‐Ming B, Messmer M, Kaukinen P, Pfeffer S. Identification of RNase L‐dependent, 3'‐end‐modified, viral small RNAs in Sindbis virus‐infected mammalian cells. MBio 2013, 4:e00698‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kreuze JF, Perez A, Untiveros M, Quispe D, Fuentes S, Barker I, Simon R. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: a generic method for diagnosis, discovery and sequencing of viruses. Virology 2009, 388:1–7. [DOI] [PubMed] [Google Scholar]
- 63. Chen LX, Hu M, Huang LN, Hua ZS, Kuang JL, Li SJ, Shu WS. Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J 2015, 9:1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cook S, Chung BY, Bass D, Moureau G, Tang S, McAlister E, Culverwell CL, Glucksman E, Wang H, Brown TD, et al. Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts. PLoS One 2013, 8:e80720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Coffey LL, Page BL, Greninger AL, Herring BL, Russell RC, Doggett SL, Haniotis J, Wang C, Deng X, Delwart EL. Enhanced arbovirus surveillance with deep sequencing: Identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology 2014, 448:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, Qin XC, Xu J, Holmes EC, Zhang YZ. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative‐sense RNA viruses. Elife 2015, 4:eLife.05979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 2008, 44:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nagalakshmi U, Waern K, Snyder M. RNA‐Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol 2010, Chapter 4:Unit 4.11.1‐13. [DOI] [PubMed] [Google Scholar]
- 69. Radford AD, Chapman D, Dixon L, Chantrey J, Darby AC, Hall N. Application of next‐generation sequencing technologies in virology. J Gen Virol 2012, 93:1853–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5' precision of both microRNAs and their miRNA* strands in flies. Curr Biol 2008, 18:147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Harris CJ, Molnar A, Muller SY, Baulcombe DC. FDF‐PAGE: a powerful technique revealing previously undetected small RNAs sequestered by complementary transcripts. Nucleic Acids Res 2015, 43:7590–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Raabe CA, Tang TH, Brosius J, Rozhdestvensky TS. Biases in small RNA deep sequencing data. Nucleic Acids Res 2014, 42:1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double‐stranded RNA is produced by positive‐strand RNA viruses and DNA viruses but not in detectable amounts by negative‐strand RNA viruses. J Virol 2006, 80:5059–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Marques JT, Wang JP, Wang X, de Oliveira KP, Gao C, Aguiar ER, Jafari N, Carthew RW. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog 2013, 9:e1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, et al. Broad RNA interference‐mediated antiviral immunity and virus‐specific inducible responses in Drosophila. J Immunol 2013, 190:650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. Characterization of hypovirus‐derived small RNAs generated in the chestnut blight fungus by an inducible DCL‐2‐dependent pathway. J Virol 2008, 82:2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Han Y‐H, Luo Y‐J, Wu Q, Jovel J, Wang X‐H, Aliyari R, Han C, Li W‐X, Ding S‐W. RNA‐based immunity terminates viral infection in adult Drosophila in the absence of viral suppression of RNA interference: characterization of viral small interfering RNA populations in wild‐type and mutant flies. J Virol 2011, 85:13153–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schnettler E, Tykalova H, Watson M, Sharma M, Sterken MG, Obbard DJ, Lewis SH, McFarlane M, Bell‐Sakyi L, Barry G, et al. Induction and suppression of tick cell antiviral RNAi responses by tick‐borne flaviviruses. Nucleic Acids Res 2014, 42:9436–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Peng X, Gralinski L, Ferris MT, Frieman MB, Thomas MJ, Proll S, Korth MJ, Tisoncik JR, Heise M, Luo S, et al. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. MBio 2011, 2:e00198‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fort P, Albertini A, Van‐Hua A, Berthomieu A, Roche S, Delsuc F, Pasteur N, Capy P, Gaudin Y, Weill M. Fossil rhabdoviral sequences integrated into arthropod genomes: ontogeny, evolution, and potential functionality. Mol Biol Evol 2012, 29:381–390. [DOI] [PubMed] [Google Scholar]
- 81. Kolliopoulou A, Van Nieuwerburgh F, Stravopodis DJ, Deforce D, Swevers L, Smagghe G. Transcriptome analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PLoS One 2015, 10:e0121447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor‐mediated, or in vitro, transmission. PLoS Pathog 2014, 10:e1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet 2010, 6:e1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sabin LR, Zheng Q, Thekkat P, Yang J, Hannon GJ, Gregory BD, Tudor M, Cherry S. Dicer‐2 processes diverse viral RNA species. PLoS One 2013, 8:e55458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster . Genes Dev 2006, 20:2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Berry B, Deddouche S, Kirschner D, Imler JL, Antoniewski C. Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS One 2009, 4:e5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Akbari OS, Antoshechkin I, Amrhein H, Williams B, Diloreto R, Sandler J, Hay BA. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 2013, 3:1493–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jinek M, Coyle SM, Doudna JA. Coupled 5' nucleotide recognition and processivity in Xrn1‐mediated mRNA decay. Mol Cell 2011, 41:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121:713–724. [DOI] [PubMed] [Google Scholar]
- 90. Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 2006, 127:1223–1237. [DOI] [PubMed] [Google Scholar]
- 91. Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, et al. A highly structured, nuclease‐resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4:579–591. [DOI] [PubMed] [Google Scholar]
- 92. Cooper DA, Jha BK, Silverman RH, Hesselberth JR, Barton DJ. Ribonuclease L and metal‐ion‐independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic Acids Res 2014, 42:5202–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Endres MW, Cook RT, Gregory BD. A high‐throughput sequencing‐based methodology to identify all uncapped and cleaved RNA molecules in eukaryotic genomes. Methods Mol Biol 2011, 732:209–223. [DOI] [PubMed] [Google Scholar]
- 94. Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol 2004, 8:570–579. [DOI] [PubMed] [Google Scholar]
- 95. Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell 2010, 143:703–709. [DOI] [PubMed] [Google Scholar]
- 96. Cooper DA, Banerjee S, Chakrabarti A, Garcia‐Sastre A, Hesselberth JR, Silverman RH, Barton DJ. RNase L targets distinct sites in influenza A virus RNAs. J Virol 2015, 89:2764–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]


