Abstract
Long noncoding RNAs (lncRNAs) are extensively expressed in mammalian cells and play a crucial role as RNA regulators in various cellular processes. Increasing data reveal that they function in innate antiviral immunity through complex mechanisms. Thousands of lncRNAs are regulated by RNA virus or DNA virus infection. The significant differential expression of lncRNAs is induced by virus or host antiviral signaling mediated by interferons (IFNs) and tumor necrosis factor‐α. In turn, these lncRNAs modulate the host immune response including the pathogen recognition receptor (PRR)‐related signaling, the translocation and activation of transcription factors, the production of IFNs and cytokines, the IFN‐activated JAK‐STAT signaling and the transcription of antiviral IFN‐stimulated genes (ISGs). Using gain‐ or loss‐of‐function analysis, the effect of lncRNAs on viral replication has been investigated to elucidate the essential role of lncRNA in the host–virus interaction. lncRNAs have shown specifically elevated or decreased levels in patients with viral diseases, suggesting the possibility of clinical application as biomarkers. Here we review the current advances of viral infection‐associated host lncRNAs, their functional significance in different aspects of antiviral immune response, the specific mechanisms and unsolved issues. We also summarize the regulation of lncRNAs by viruses, PRR agonists and cytokines. In addition, virus‐encoded lncRNAs and their functional involvement in host–virus interaction are addressed. WIREs RNA 2016, 7:129–143. doi: 10.1002/wrna.1321
This article is categorized under:
-
1
RNA Interactions with Proteins and Other Molecules > Protein–RNA Interactions: Functional Implications
-
2
RNA in Disease and Development > RNA in Disease
INTRODUCTION
Viral infection is an important etiological factor of various infectious diseases including acute illnesses such as influenza pandemics, Middle East respiratory syndrome (MERS), and Ebola virus disease, as well as chronic conditions such as acquired immune deficiency syndrome (AIDS), viral hepatitis and many virus‐associated cancers. Host has evolved deliberated antiviral immune system to eliminate the invading virus. In mammalian cells, thousands of proteins participate in the innate and adaptive antiviral immunity. Although the protein‐mediated antiviral mechanisms have been established for many years, the exquisite immune regulation network to provide appropriate protection with minimal inflammatory damage is still not fully understood. Compared with microRNAs, which are well‐known noncoding RNA regulators of immune genes, long noncoding RNAs (lncRNAs) are newly recognized key components of the complicated antiviral immune system.1, 2, 3, 4, 5, 6, 7, 8, 9
lncRNAs are designated as the transcripts longer than 200 nt without protein‐coding capacity.1, 2, 3 Similar to mRNA, lncRNAs are spliced products of RNA polymerase II or III, 5′‐capped, and with or without 3′‐polyadenylation. Based on their genomic locations relative to adjacent protein‐coding genes, lncRNAs are classified as sense, antisense, bidirectional, intronic, and intergenic lncRNAs. Previously considered as ‘junk’ in the genome, but lncRNAs are recently recognized as critical regulators in various cellular processes.3, 5, 10, 11, 12 The lncRNA genes occupy the majority of pervasively transcribed genomes in human or mouse cells.13 The current GENCODE releases have annotated 15,931 human lncRNA genes (Version 23) and 8359 mouse lncRNA genes (Version M6).14 However, as of July 2015 only a small portion of human and mouse lncRNAs (181 and 114, respectively) have been experimentally characterized as functional RNAs as provided by lncRNAdb.15 Therefore, the functioning of the most lncRNAs remains largely unknown. Recently, increasing evidences have confirmed the crucial roles of lncRNAs in host antiviral response.7, 8, 9, 16, 17 The broad spectrum of activities and versatile regulatory mechanisms of lncRNAs suggest that lncRNAs are key regulators of host immunity during viral infection. For example, NeST RNA is found to epigenetically regulate the transcription of its neighboring IFN‐γ‐encoding gene.8 This finding provides novel insights into complicated mechanisms underlying regulation of IFN‐γ. However, although the functional lncRNAs involved in antiviral innate immunity are explosively accumulated, limited information is available about lncRNAs associated with adaptive immunity. In this review, we will highlight the lncRNAs associated with viral infection, especially focusing on the expression of lncRNAs regulated by virus or host antiviral signaling, together with their functions and underlying mechanisms in the antiviral immunity.
lncRNAs ARE KEY CELLULAR REGULATORS
lncRNAs are an important new class of regulators in a wide range of cellular activities. For example, they are involved in metastasis (HOTAIR),18 X‐chromosome imprint (XIST),19 development (Airn),20 stem cell pluripotency (HOTTIP),21 and immune response (lincRNA‐Cox2).22 Small regulatory noncoding microRNA (about 19–25 nt in length) also participate in many cellular activities. The silencing mechanism of microRNA is to modulate the mRNA degradation or translation via base‐pairing of the target mRNA with specific sequences.23 In comparison with these small ncRNAs, lncRNAs have multiple and flexible mechanisms mediated by their specific sequences or structural motifs that bind with DNA, RNA or protein.2, 12, 24, 25, 26 They act as signals, decoys, guides, and scaffolds to regulate different processes, ranging from chromatin remodeling, transcription, splicing, mRNA stabilization, protein translation to protein translocation (described in details by several reviews).1, 2, 12, 25, 26 It is well‐known that lncRNA HOTAIR epigenetically represses the gene transcription by acting as a scaffold that is involved in recruiting the Polycomb repression complex 2 (PRC2) responsible for the H3K27 methylation, and the LSD1 complex responsible for the H3K4me2 demethylation.18, 24 A recent study reveals that lncRNA NeST critically regulates IFN‐γ transcription by recruiting the WDR5 and MLL/Ash2L/RbBP5 complex to enhance the H3K4me3 modification.8 Moreover, it has been shown that lncRNA THRIL and lincRNA‐Cox2 regulate the transcription of TNF‐α and CCL5, respectively through binding with hnRNP L and hnRNP A/B and A2/B1.22, 27 Interestingly, some lncRNAs, such as NRON and lnc‐DC, regulate transcription by modulating the activities of transcription factors. NRON–protein complex binds transcription factor NFAT in cytoplasm and thus releases activated NFAT for nuclear transportation after stimulation.28 lnc‐DC acts to maintain the phosphorylation state of transcription factor STAT3 and increase its nuclear entry.29 In addition, some lncRNAs regulate gene expression at post‐transcription level. For example, lncRNA BACE1‐AS stabilizes BACE1 mRNA by forming a dsRNA complex through base‐pairing.30 With the help of translation inhibitor Rck, lincRNA‐p21 suppresses translation through partially base‐pairing with target mRNAs.31 lncRNA‐BGL3 functions as a competitive endogenous RNA (ceRNA) for binding microRNAs to regulate PTEN expression.32 On the other hand, it is also worth to note that recent studies provided evidences that putative lncRNAs might harbour small ORFs and can be translated into functional micropeptides.33, 34 Therefore, it is important to determine the coding‐capacity when a novel lncRNA is identified. lncRNAs are the key component of regulatory network maintaining cellular homeostasis, and the dysfunction of lncRNAs is associated with many diseases, such as cancers, degenerative disorders, autoimmune, and viral diseases.5, 16, 18, 19, 20, 21, 27, 32, 35, 36
INNATE IMMUNE RESPONSE TO VIRAL INFECTION
Viral infection of host cells causes viral diseases and also triggers host antiviral innate immune response. Enveloped virus enters the cell membrane through receptor‐mediated endocytosis, followed by viral genome release into the cytoplasm. Then viral genome utilizes cellular machinery and viral enzymes to replicate and assemble new progeny virions. After virus invasion, host cells can quickly sense viral components, known as pathogen‐associated molecular patterns (PAMPs), through pathogen recognition receptors (PRRs) on the cell surface [Toll‐like receptor (TLR) 2 and 4], on the membrane of endosome (TLR 3,7/8, and 9), or within the cytoplasm (RIG‐I, MDA5, DAI, and NLRs) (see Figure 1).37, 38 PRR‐dependent signaling pathways activate transcription factors (TFs) such as IRF3, IRF7, and NF‐κB. After the nuclear import of these TFs, robust interferons (IFNs), chemokines, and other cytokines are expressed. In turn, IFNs bind with their corresponding receptors, leading to the activation of JAK‐STAT signaling. After the nuclear translocation of phosphorylated STATs, they mediate formation of the transcription complex ISGF3 (STAT1, STAT2, and IRF9) that initiates the expression of hundreds of IFN‐stimulated genes (ISGs).37 In addition to the activation of signaling and TFs, the open state of chromatin and the mRNA stabilization are simultaneously required for the transcription of some critical antiviral genes, such as MxA, IFIT1 and IFITM3.39 It has been revealed that pathogenic influenza A virus (IAV) can exploit cellular machinery to control the transcription of a subset of ISGs by altering their histone modifications.40 Viral infection and replication are strongly blocked directly by some critical ISGs, such as MxA, IFITM3, and IFIT3 through various mechanisms.37, 41 However, many cellular proteins are responsible for the negative regulation of IFN production and signaling, such as SOCS1, PIAS, and IFI35, which not only repress the JAK‐STAT pathway but also inhibit PRR.42, 43, 44 Type I IFNs signaling also mediates the development or activation of innate immune cells such as dendritic cells (DCs), NK cells, and T cells, which eliminates the virus‐infected cells directly or indirectly.45 While the critical antiviral roles of proteins have been well‐characterized, little is known about lncRNAs, until recently when increasing evidences have presented and demonstrated that lncRNAs are important players in the battle between virus and host.
Figure 1.
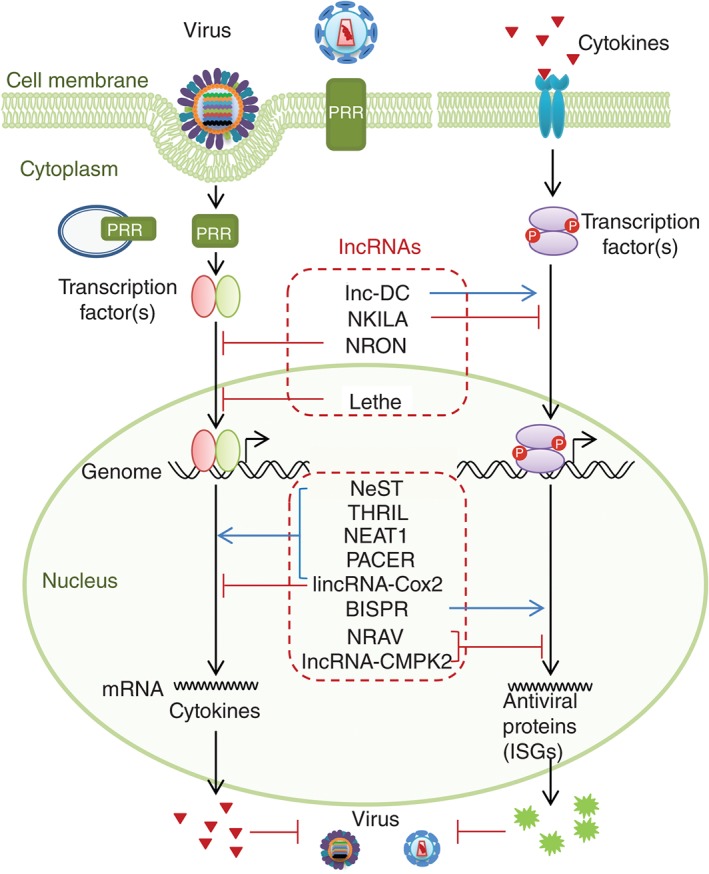
Host lncRNAs regulate multiple steps of the antiviral innate immune response. Upon viral invasion, host cell starts the PRR signaling cascade, which further activates the transcription factors and initiates the expression of cytokines. After binding with corresponding receptors, cytokines trigger the receptor‐associated signaling and the production of antiviral proteins, such as ISG proteins. Some cytoplasmic or nuclear lncRNAs can increase or inhibit the activities of transcription factors, which are listed in the upper dashed red rectangle. Other nuclear lncRNAs can modulate the transcription of cytokines or antiviral genes (ISGs), which are listed in the lower dashed red rectangle.
DIFFERENTIAL EXPRESSION OF lncRNAs DURING VIRAL INFECTION
Over the past 6 years, several whole‐transcriptome investigations have identified over thousands of lncRNAs differentially expressed in severe acute respiratory syndrome coronavirus (SARS‐CoV) infected mice, IAV‐infected human lung cells and enterovirus 71(EV71)‐infected rhabdomyosarcoma (RD) cells.9, 46, 47, 48 Such large amount of lncRNAs regulated by the high‐risk viruses indicates their importance in host response to the viral infection. Using the next‐generation sequencing (NGS) and cDNA library analysis, the first discovery of lncRNA expression in response to positive RNA virus SARS‐CoV infection has identified 504 annotated and 997 nonannotated mouse lncRNAs.46 In vivo and in vitro studies of a subset of these lncRNAs reveal that about half of these lncRNAs are also regulated similarly by negative RNA virus IAV (PR8/H1N1) infection, indicating that lncRNAs might take part in the common host response to viral infection. Solid evidences from IAV and SARS‐CoV infected eight collaborative cross mouse founder strains further confirm that 5329 lncRNAs are differentially expressed and strongly correlated with viral replication and morbidity.48 The module‐based annotation reveals that most lncRNAs are co‐expressed with coding genes associated with lung homeostasis or immune response. The database MONOCLdb (http://www.monocldb.org) provides 5295 mouse lncRNAs and their functional annotations. Similarly, hundreds of lncRNAs are found differentially expressed in IAV (WSN/H1N1) infected human A549 cells.9, 49
Infection with positive‐strand RNA virus EV71, the pathogen of hand, foot, and mouth disease, results in upregulation of 2990 lncRNAs and downregulation of 1876 lncRNAs in human RD cells.47 Many of these lncRNAs have a co‐regulated nearby protein‐coding gene. DAVID functional analysis indicates that these mRNA and lncRNA pairs may play roles in alternative splicing, protein phosphorylation, and acetylation, suggesting their involvement in transcription regulation and signal transduction.
Further investigations are required to define the functional involvement of these lncRNAs in the pathogenesis of these viruses. It is worth noting that the unique expression patterns or dysregulation of lncRNAs might be useful tools in clinical diagnosis or surveillance. In Table 1, shown are some critical lncRNAs that are induced or suppressed by viruses such as SARS‐CoV, IAV, human immunodeficiency virus (HIV), hepatitis B virus (HBV), and hepatitis C virus (HCV). For example, among the 83 differentially expressed lncRNAs in HIV‐1‐infected T cell lines Jurkat and MT4, lncRNA BIC, LIT, MALAT1, and NEAT1 are upregulated, whereas PANDA and SRA are downregulated.56 The high expression of lncRNA HEIH is found associated with HBV‐induced HCC recurrence.61 Both lncRNA‐uc003wbc and lncRNA‐AF085935 are upregulated in HBV patients and HCC patients but to different levels, suggesting their clinical applications as diagnosis biomarkers.62
Table 1.
Host lncRNAs Related With Antiviral Innate Immune Response
| lncRNAs | Stimuli | Differential Expression | Neighbor Coding Genes | Functions/Mechanisms | Subcellular Localization | Refs |
|---|---|---|---|---|---|---|
| NRON (mouse, human) | HIV | Down | MVB12B | Inhibit NFAT through binding with a scaffold complex, which sequesters NFAT in cytoplasm. | Cytoplasm/ NAb whether in nucleus | 28, 50, 51 |
| NKILA (human) | IL‐1β, TNF‐α, breast cancer patienta | UP | PMEPA1 | Inhibit NF‐κB by binding with IKB and preventing the degradation of IKB. | Cytoplasm | 52 |
| lnc‐DC (mouse, human) | Differentiation stimuli from monocyte to DC. | UP | HEATR6, LOC105371849 | Bind to STAT3 in the cytoplasm and block the dephosphorylation of STAT3. | Cytoplasm | 29 |
| NeST/Tmevpg1 (mouse, human) | Theiler's murine encephalitis virus (Th1 cells) | Up | Ifng | Epigenetically activate IFN‐γ expression by binding with WDR5 and altering H3K4me3 at ifng locus, increase Theiler's virus persistence. | Nucleus | 8, 53 |
| lincRNA‐Cox2 (mouse) | Pam3CSK4, R848, LPS, Listeria Monocytogenes (not Poly I:C) | Up | Ptgs2 (Cox2) | Activate and repress distinct immune genes. Repress transcription through interactions with hnRNP A/B and A2/B1. | Nucleus and cytoplasm | 22, 54 |
| THRIL/linc1992 (human) | Pam3CSK4, Kawasaki Diseasea (acute phase) | Down | BRI3BP | Activate TNF‐α transcription by binding with hnRNP L. | Nucleus/NA whether in cytoplasm | 27 |
| Lethe (mouse, human) | TNF‐α, IL‐1β, dexamethasone, LIF, HCV (not vitamin D, estradiol, methyltrienolone, TLR agonists) | Up | Gmeb1, Ythdf2 | Inhibit NF‐κB activity by binding to subunit RelA as a decoy, suppress IL‐6, IL‐8, SOD2, and NFKBia transcription, and promote HCV replication. | Nucleus (80%), half of which on the chromatin | 17, 55 |
| NEAT1/VINC/ AK028745 (mouse, human) | HIV, JEV, RV, poly I:C | UP | FRMD8, MIR612 | Sequester SFPQ in paraspeckles, repress the transcription of IL‐8 and ADARB2, and inhibit HIV production. | Nucleus | 7, 56, 57 |
| PACER (human) | PMA, LPS | UP | COX2 | Enhance the transcription of adjacent COX2 through binding with p50. | Nucleus | 58 |
| NRAV (human) | IAV, SeV, MDRV, HSV | Down | Dynll1 | Epigenetically inhibit the transcription of ISGs, increase IAV replication. | Nucleus and cytoplasm | 9 |
| lncRNA‐CMPK2 (human) | IFN‐α, IFN‐γ, HCVa | Up | CMPK2 | Benefit for HCV replication as a negative regulator of protein‐coding ISGs. | Nucleus | 16 |
| lincRNA VIN (human) | IAV and VSV (not by IBV, RNA mimics, IFN‐β) | Up | LOC100499194, ACTR3 | Support the IAV replication and viral protein synthesis. | Nucleus | 49 |
| lncBST2/BISPR (human) | IFN‐α2, IFN‐λ, IAV (PR8ΔNS1), VSV (M51R), HCVa, | Up | BST2 | Control the expression of BST2 mRNA positively. | Nucleus | 59 |
| lncISG15 (human) | IFN‐α2, IFN‐λ, IAV (PR8ΔNS1), VSV (M51R), HCVa | Up | ISG15 | NAb | Nucleus | 59 |
| ISR2 (human) | IFN‐α2, IFN‐β, HCVa, IAV (PR8ΔNS1), HIVa | Up | GBP6 | NA | Nucleus | 60 |
| ISR8 (human) | IFN‐α2, IFN‐β, HCVa, IAV (PR8ΔNS1) | Up | IRF1 | NA | Nucleus | 60 |
| ISR12 (human) | IFN‐α2, IFN‐β | Up | IL6 | NA | Nucleus | 60 |
Detect differential expression in samples of pathogen‐infected patient.
NA, not available.
In addition to the host lncRNAs regulated by invaded viruses, endogenous retroviruses (ERVs) encoded long noncoding genetic elements are identified to stimulate intracellular sensors.63 It is suggested that the cytosolic DNA sensor cGAS (cGMP‐AMP synthase), its adaptor STING (stimulator of interferon genes) and the RNA sensor signaling adaptor MAVS (mitochondrial antiviral signaling protein) are required for the ERV‐induced intracellular activation of B cells and subsequent T‐cell‐independent (TI) antibody production. The ERV noncoding nucleic acids, including cytosolic ERV mRNA and reverse‐transcribed cDNA play as the TI antigens. The aberrant expression of ERV long noncoding nucleic acids is likely associated with the initiation of auto‐inflammatory response of autoimmune disease.64 Therefore, identification of these ERV lncRNAs might provide novel evidences for better understanding of the pathogenesis of autoimmune disease.
REGULATION OF lncRNAs BY PRR SIGNALING
Viral infection triggers innate immune response depending on the activation of PRR signaling and corresponding TFs. RIG‐I, MDA5, and TLRs pathways activate IRF3, IRF7 or NF‐κB.38 TLR2 and TLR4 are indicated capable to induce the differential expression of lncRNAs. TLR2, expressed on the surface of macrophages, monocytes, and other cells, recognizes viral, bacterial and fungal PAMPs and mediates their internalization. TLR2 signaling regulates the expression of various cytokines such as tumor necrosis factor α (TNF‐α). Pam3CSK4, a synthetic lipopeptide ligand of TLR2, promotes differential expression of 159 lincRNAs in human THP1‐derived macrophages, including suppressed lincRNA THRIL.27 It has been shown as well that 62 lncRNAs are induced in Pam3CSK4 stimulated mouse bone marrow‐derived macrophages (BMDMs), including the most significantly upregulated lincRNA‐Cox2.22 TLR4 is located on the membrane of endosome. It recognizes bacterial or viral pathogen and signals via adaptor MyD88 to activate NF‐κB. Using TLR4 agonist LPS, 20 lincRNAs in CD11C+ bone marrow‐derived DCs are found remarkably upregulated.54 Most (approximately 80%) of them are associated with NF‐κB signaling. Although the proportion of these PRR‐associated lncRNAs among all the differentially expressed lncRNAs in virus‐infected cells is unclear, it is very likely that they are important components of host antiviral innate immunity. Further characterization of these lncRNAs is needed to establish their roles in the host–virus interaction.
REGULATION OF lncRNAs BY IFNs AND TNF‐α
IFNs play a central role in innate immunity and mediate direct or indirect antiviral response.45, 65 Several groups simultaneously found that a variety of lncRNAs are regulated by type I, II, and III IFNs.16, 48, 59, 60 In addition, some IFN‐β and IFN‐α‐stimulated lncRNAs (ISRs) are identified in C57BL/6 J mice.46 IFN‐α‐induced lncRNAs are annotated and provided in database MONOCLdb.48 Over hundreds of human lncRNAs are differentially expressed in primate human hepatocytes treated by IFN‐α16 or in HuH7 cells treated by IFN‐α2,59 including ISRs and IFN‐downregulated lncRNAs (IDRs). These data strongly suggest a close relationship between lncRNAs and IFN‐mediated antiviral innate immunity.
Some of these ISRs are co‐expressed with ISGs that genomically locate as their neighboring genes. For example, lncRNA‐CMPK2 is 100‐fold induced by both IFN‐α and IFN‐γ, abrogated by repression of JAK‐STAT signaling pathway, and located near ISG gene CMPK2 in a nonoverlapping, head to tail orientation.16 Both lncRNA‐CMPK2 and CMPK2 mRNA are induced by IFNs simultaneously. lncRNA‐CMPK2 locates mainly in nucleus and acts as a negative regulator of IFN response (discussed below). In human hepatocytes, ISR2, ISR8, and ISR12 are induced by IFN‐β and IAV PR8ΔNS1, and the expression of these ISRs mimics that of adjacent genes GBP6, IRF1, and IL6, respectively.60 Increased expression of ISR2 is also detected in patients infected with HCV or HIV. However, their roles in antiviral response still remain unknown. lncISG15 and lncBST2 share the same promoter with their neighboring genes ISG15 and BST2, respectively.59 The expression of lncBST2 is significantly dependent on JAK/STAT signaling. It is thought that lncBST2 positively regulates BST2 transcription, as evidenced by gain‐ and loss‐of‐function analysis.66 However, the mechanism underlying the role of lncBST2 in BST2 transcription is unclear. Like ISG15, lncISG15 is regulated not only by IFN but also by other regulators, but its function in antiviral response needs to be clarified.
In addition to IFNs, the key proinflammatory cytokine TNF‐α also plays an important role in regulating the expression of lncRNAs. TNF‐α is mainly produced by macrophages and lymphocytes. It triggers fever and stimulates inflammatory response of immune cells by activating NF‐κB as well as MAPK pathway. Hundreds of lncRNAs were characterized by RNA‐seq as TNF‐α regulated transcripts, including NF‐κB activated pseudogene Lethe.17 Notably, TNF‐α‐induced expression of some lncRNAs is regulated by NF‐κB signaling. The relationship between these lncRNAs and virus‐induced inflammation remains to be further addressed.
FUNCTIONAL INVOLVEMENT OF lncRNAs IN ANTIVIRAL INNATE IMMUNITY
Roles of Host lncRNAs in Viral Infection
Hundreds of host lncRNAs are differentially expressed during viral infection, PRR stimulation, or IFN treatment, but only a small portion of these lncRNAs have been assayed in detail for their functioning. Importantly, current lines of evidences have strongly supported the functional involvement of lncRNAs in host antiviral response. Through gain‐ or loss‐of function experiments, some of these lncRNAs such as NeST, Lethe, lncRNA‐CMPK2, VIN, and NRON are found hijacked by virus to facilitate virus infection or susceptibility. For example, the expression of lincRNA VIN (virus‐inducible lincRNA) in human lung cells is upregulated by infection with IAV strains (H1N1, H3N2, and H7N7) and vesicular stomatitis virus (VSV).49 Although precise mechanism underlying the action of nuclear lncRNA VIN is still unknown, it is clear that virus takes advantage of the increased VIN to ensure its efficient replication and viral protein synthesis. On the other hand, other lncRNAs such as lincRNA‐Cox2, NRAV, NEAT1, 7SL, and lnc‐DC play critical roles in the antiviral response. For example, host lncRNA 7SL is selectively packaged into HIV‐1 particles and is a integral component of the viral ribonucleoprotein (RNP) complex.67 The packaged 7SL RNA is required for the antiviral activity of cytidine deaminase APOBEC3G. As a key scaffold, 7SL binds with APOBEC3G, viral protein Gag, viral genomic RNA and tRNA3Lys.68 Through the link of 7SL, APOBEC3G targets viral RNP and interferes with the HIV replication. Currently, most well‐characterized functional lncRNAs are involved in antiviral innate response through regulating activity of TFs, the transcription of cytokines or the expression of ISGs.
lncRNAs Function in Antiviral Immunity via Regulating TFs
During viral infection, host lncRNAs regulate various processes of antiviral response, including the TFs translocation and activity, IFN and cytokines production, ISGs transcription, and immune cell development (see Figure 1). For example, NFAT (Nuclear Factor of Activated T cells) is essential for immune response and cell development. lncRNA NRON (noncoding repressor of NFAT) is highly enriched in thymus, spleen, and lymph node and found to be a NFAT inhibitor.28, 50, 51 NRON exists in a scaffold RNA–protein complex via interaction with proteins KPNB1, CSE1L, and IQGAP1,28 which binds phosphorylated NFAT in cytoplasm and represses its nuclear trafficking (see Figure 2). When T cells are activated, dephosphorylated NFAT is released from the complex and enters the nucleus. A recent study suggests that downregulation of NRON by HIV infection enhances NFAT nuclear translocation and activity.51 Consequently, NFAT is hijacked by HIV to bind with the LTR of viral genome and facilitate viral protein synthesis and replication. Strikingly, HIV even utilizes NRON to control the balance between viral reproduction and cell death through HIV early expressed protein Neg and the late expressed protein Vpr to respectively decrease and increase NRON at different infection stage.51 The HIV‐induced upregulation of NFAT is simultaneously utilized by host as well to increase the adaptive immune response to HIV by activation of T cells.
Figure 2.
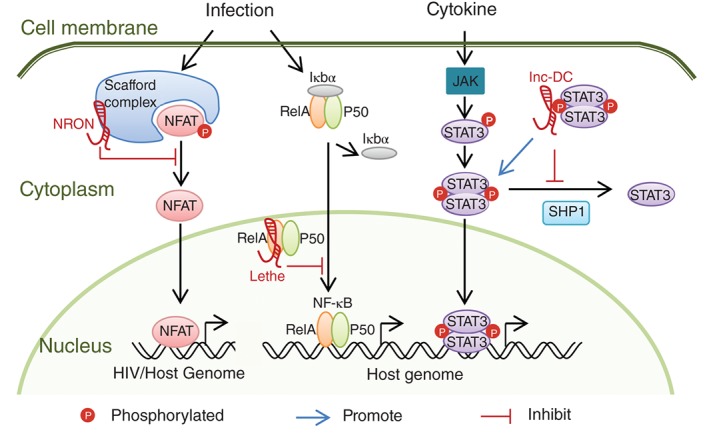
Host lncRNAs regulate the transcription factor activities as a scaffold, decoy or posttranslational modification shield. NRON inhibits the activation/dephosphorylation of NFAT as a component of the scaffold complex. Lethe interacts with RelA to repress the transcription activity of NF‐κB. lnc‐DC directly binds with STAT3, prevents the dephosphorylation of STAT3 by SHP1, and activates STAT3‐dependent transcription.
Another example is the pseudogene lncRNA Lethe. Lethe is upregulated directly by NF‐κB after stimulation with TNF‐α or glucocorticoid receptor agonist dexamethasone in mouse spleen,17 or induced by HCV infection or LIF‐mediated STAT3 activation in human liver Huh7 cells.55 In turn, Lethe acts as a negative feedback modulator of NF‐κB signaling through interaction with NF‐κB subunit RelA (see Figure 2). Like a decoy, Lethe blocks the binding site of RelA to target genes and subsequent transcription. In aged mice, increased NF‐κB activity might be associated with reduced Lethe abundance. The anti‐inflammatory therapeutic activity of dexamethasone might be mediated in part via NF‐κB repression by elevated Lethe. Overexpression of Lethe decreases the transcription of IL‐6, IL‐8, and NFKBia regulated by NF‐κB. The enhanced Lethe in Huh7 cells inhibits the expression of PKR, OAS, and IRF1, and promotes the HCV replication.55 However, the impact of Lethe on other antiviral genes driven by NF‐κB, such as IFNs, still requires additional investigation.
lncRNA NKILA (NF‐κB‐interacting lncRNA) is another lncRNA regulator of NF‐κB activity, but functions in posttranslational level.52 NKILA is highly induced by IL‐1β and TNF‐α treatment in breast cancer cells. The knockdown and overexpression analysis reveals that NKILA, similar to Lethe, is a negative regulator of the transcription of NF‐κB target genes. NKILA interacts with other molecules to form a cytoplasmic complex comprising NKILA, NF‐κB, and inhibitor of NF‐κB (IκB). In the complex, NKILA blocks the phosphorylation sites of IκB from IκB kinase (IKK) and protects IκB from subsequent degradation. Therefore, NF‐κB is sequestered in the cytoplasm and thereby NF‐κB driven transcription is inhibited. This mechanism underlying NKILA function is similar with that of cytoplasmic lnc‐DC (discussed below). In addition, lncRNA PACER (p50‐associated Cox2 extragenic RNA) acts as an extragenic positive regulator of NF‐κB to modulate the transcription of NF‐κB target gene Cox2, which is located on the antisense strand upstream of PACER. 58 Both PACER and Cox2 are significantly upregulated in PMA or LPS differentiated human U937 monocyte‐macrophages. PACER directly binds with the repressive NF‐κB subunit p50. This interaction promotes the formation of the active form of NF‐κB, i.e., RelA‐p50 heterodimer, leading to an increase in the transcription of Cox2. However, the precise role of NKILA and PACER in virus infection and replication remains to be further investigated.
DCs are the key antigen‐presenting cells in mammalian immune system. The lncRNA lnc‐DC is exclusively expressed in human conventional DCs (cDCs) with TF PU.1, the DC differentiation regulator, and is a specific and stable biomarker of the cDC differentiated from peripheral blood monocytes.29 Experiments using knockdown and overexpression have indicated that lnc‐DC is essential for T cell activation and antigen uptake. Further functional analysis has suggested that human or mouse lnc‐DC directly binds with the TF STAT3 in the cytoplasm and inhibits the dephosphorylation of STAT3 by phosphatase SHP1 (see Figure 2). The activated STAT3 mediates the transcription of genes necessary for DC differentiation. The mechanism governing action of lnc‐DC and NKILA is a new functional manner of cytoplasmic lncRNAs in addition to action as ceRNAs69 and STAU1‐ALU element mediated mRNA decay.70
The influences of these lncRNAs on the regulation of critical TFs, including NFAT, NF‐κB and STAT3, are solid evidences indicating the involvement of lncRNAs in antiviral response. It is reasonable to speculate that there exists more lncRNAs interacting with other important TFs, such as IRF3, IRF7, and STAT1.
Regulation of Cytokine Production by lncRNAs
It has been shown that some lncRNAs play important roles in cytokine production. IFN‐γ is the first IFN reported to be regulated by lncRNA. It is an important cytokine for cell self‐activation and secreted by many immune cells such as Th1 cells, CTL cells, NK cells and DCs.53 A mouse lncRNA gene NeST (nettoie Salmonella pas Theiler's [cleanup Salmonella not Theiler's], also known as Tmevpg1) is found in Tmevp3 (Theiler's murine encephalitis virus persistence 3) locus, which is adjacent to the IFN‐γ coding gene ifng.8 But more importantly, lncRNA NeST induces IFN‐γ transcription specifically in activated CD8+ T cells, but not in activated CD4+ T cells. This leads to increased Theiler's virus persistence but decreased Salmonella enterica pathogenesis. NeST epigenetically modulates the ifng locus through interaction with a protein partner WDR5, the component of H3K4 methyltransferase complex (see Figure 3). Nuclear lncRNA NeST recruits WDR5 in trans to increase the histone 3 methylation of the IFN‐γ‐encoding DNA, and then promotes the transcription of its neighboring gene. Nevertheless, to date little information is available about regulation of type I and III IFN expression by lncRNAs.
Figure 3.
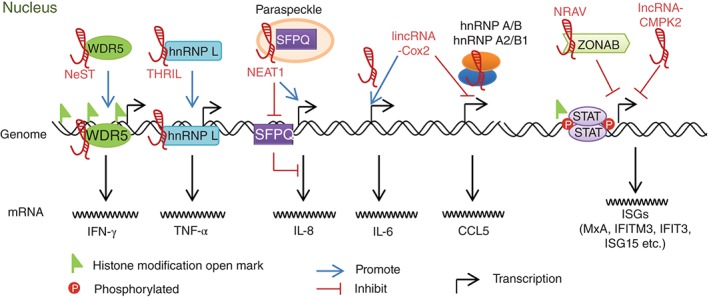
Host lncRNAs regulate the transcription of cytokines and IFN‐stimulated genes (ISGs) as a modulator of histone modification, promoter activation or repression. NeST interacts with WDR5 to increase the chromatin open modifications at the ifng loci. THRIL increases TNF‐α transcription via binding with hnRNP L on the TNF‐α promoter. NEAT1 sequesters the repressor SFPQ in the paraspeckle and thus increases the IL‐8 transcription. lincRNA‐Cox2 increases IL‐6 via an unknown mechanism and decreases the CCL5 transcription by interaction with hnRNP A/B and hnRNP A2/B1. NRAV as a repressive factor decrease the expression of several critical ISGs through modulation of histone modification. lncRNA‐CMPK2 functions as a repressor via an IFN‐independent unknown mechanism.
TNF‐α is a crucial cytokine activating NF‐κB signaling in immune cells and involving in inflammatory response. lincRNA THRIL (linc‐1992), named as TNFα and hnRNP L related immunoregulatory lincRNA, is required for TNF‐α expression.27 THRIL and hnRNP L form a specific complex, which binds with the promoter of TNF‐α to activate its transcription (see Figure 3). The high levels of TNF‐α, in turn feedback and negatively regulate the THRIL expression. The significantly lower THRIL levels are detected in TLR2 ligand Pam3CSK4‐stimulated human THP1 macrophages and the blood of Kawasaki disease patients at acute phase with elevated TNF‐α level. Therefore, THRIL is proposed as a biomarker for high TNF‐α level and immune activation. Other cytokines and chemokines, e.g., IL‐8, CXCL10, CCL1, and CSF1, are also induced by THRIL, but the mechanisms involved remain to be elucidated.
Mouse lincRNA‐Cox2 is another key lncRNA involved in regulation of cytokine expression. It is a central regulator of immune response since it mediates both activation and repression of many distinct immune genes.22 Its neighboring gene encodes protein Cox2 (also known as Ptgs2), which is a key mediator of inflammation and regulated by NF‐κB after TLR4 stimulation. lincRNA‐Cox2 is also highly upregulated by TLR4 agonist LPS or TLR2 agonist Pam3CSK4.22, 54 Previous study reveals that lincRNA‐Cox2 regulates expression of various genes, including PRRs (Tlr1), chemokines (Il‐6, Il‐23, Ccl5), chemokine receptors (Ccrl), and ISGs (Irf7, Ifi204, Isg15). For example, lincRNA‐Cox2 directly decreases CCL5 (Rantes) level and enhances IL‐6 expression (see Figure 3). The inhibition of CCL5 by lincRNA‐Cox2 is mediated through an RNA‐protein complex comprising hnRNP A/B and hnRNP A2/B1. While both lincRNA‐Cox2 and its adjacent gene Cox2 are important regulators of immune response, the expression of Cox2 is not affected by lincRNA‐Cox2. Whether there exists a functional relationship between them still needs to be addressed.
lncRNA NEAT1 (nuclear‐enriched abundant transcript)/VINC (virus‐inducible ncRNA) is induced by several viruses and acts as an important positive regulator of antiviral response. The antiviral function of NEAT1 depends on its essential role in nuclear paraspeckle formation and protein sequestration.71 The expression of NEAT1 is significantly enhanced by HIV‐1, JEV, RV, IAV, and HSV infection, TLR3 agonist (poly I:C) treatment, and the stresses from cancers and ALS.7, 56, 57 In addition, viral infections also induce excessive formation of paraspeckles in human or mouse immune cells. Loss‐of‐function analysis has revealed that NEAT1 inhibits the HIV protein production and the translocation of HIV unspliced INS‐containing mRNA (e.g., gag/pol and env RNAs) from nucleus to cytoplasm.56 Further investigations suggest that virus‐elevated NEAT1 increases the transcription of IL‐8 and other immune genes (e.g., CCL5, ISG20, IFIT3, RIG‐I). Upon viral infection, abundant NEAT1 relocates the repressor SFPQs from the IL‐8 gene promoters to paraspeckles, and then triggers the transcription of IL‐8 (see Figure 3). The most recent finding reveals that NEAT1 interacts with SWI/SNF chromatin‐remodeling complexes, which are newly identified components of paraspeckles.72 Therefore, NEAT1 probably regulates some gene expressions via epigenetic modulation.
Implication of lncRNAs in Regulating the Expression of ISGs
lncRNAs also participate in tight regulation of the expression of important antiviral effectors ISGs. Our previous studies reveal that lncRNA NRAV, named as negative regulator of antiviral response, suppresses the transcription of several critical ISGs through epigenetic regulation of these genes.9 Several viruses, such as ‐ssRNA virus [IAV and sendai virus (SeV)], dsRNA virus [Muscovy duck reovirus (MDRV)] and DNA virus [herpes simplex virus (HSV)], dramatically downregulate the NRAV expression in human cells. The reduction of NRAV causes robust expression of a variety of ISGs, i.e., MxA, IFITM3, IFIT2, IFIT3, and OASL, and significantly impairs the IAV replication in vivo and in vitro (see Figure 3). Therefore, the viral infection induced decrease of NRAV is suggested to be part of the host antiviral response. The histone modification ‘OPEN’ mark H3K4me3 and ‘CLOSE’ mark H3K27me3 of mxa and ifitm3 are shown to be modulated by NRAV. The NRAV‐associated protein ZONAB is a multifunctional TF and might be involved in the NRAV‐dependent regulation of the ISGs. However, further studies are required to determine the mechanism underlying the epigenetic regulation of NRAV during viral infection.
lncRNA‐CMPK2 is significantly upregulated in liver tissues of HCV‐infected patients.16 The elevated expression of lncRNA‐CMPK2 might benefit the HCV replication. lncRNA‐CMPK2 acts as a negative regulator of various ISGs, such as ISG15, CXCL10, IFIT3, IFITM1, Viperin, and CMPK2, but not that of Mx1 and IFIT1. The repression of ISGs by lncRNA‐CMPK2 seems to be independent of IFN. lncRNA‐CMPK2 might play a role in epigenetic modulation of a subset of ISG loci. However, its protein partner(s) and the mechanism underlying its function are still unknown.
VIRAL lncRNAs ANTAGONIZE THE INNATE IMMUNITY
Interestingly, several recent studies have revealed that the viral pathogens encode and express ncRNAs and these novel viral components are critically involved in host–virus interaction.73, 74, 75 For example, in Kaposi's sarcoma‐associated herpesvirus (KSHV) infected cells, viral polyadenylated nuclear (PAN) RNA facilitates lytic‐phase viral transcription and late viral protein production.73 Furthermore, PAN globally influences viral and cellular gene expression through binding with host proteins such as poly(A) binding protein C1 (PABPC1), IRF4, demethylases UTX, and JMJD3, histone methyltransferase MLL2, and PRC2 proteins (SUZ12 and EZH2).73 It is worth to note that PAN mediates the epigenetic downregulation of inflammatory cytokines including γ‐IFN, α‐IFN‐16 and IL‐18.73 Another example of viral lncRNA is the noncoding subgenomic flaviviral RNA (sfRNA) encoded by dengue virus 2 (DENV‐2). sfRNA targets and antagonizes a set of host RNA‐binding proteins G3BP1, G3BP2, and CAPRIN1 to interfere with the translation of critical ISGs, such as PKR and IFITM2.74 Thus, sfRNA functions as an inhibitory regulator of these antiviral effectors. In addition, a human immunodeficiency virus expresses an antisense RNA that plays a vital role in epigenetic regulation of viral genes through recruiting and guiding a chromatin‐remodeling complex consisting of proteins such as DNMT3a, EZH2, and HDAC‐1.75 However, to date little information is available about viral lncRNAs. Further identification and functional analysis of these RNAs will provide novel insights into complicated mechanisms underlying interaction between host and virus.
CONCLUSION
As an important new class of RNA regulators, lncRNAs have been shown to be involved in multiple processes of antiviral innate immunity. Expression of host lncRNAs during viral infection can be regulated by viral components, or host PRR signaling‐activated TFs, virus‐induced cytokines, IFNs and chemokines, or other stimulations. Differentially expressed lncRNAs function as negative or positive regulators in various critical steps of antiviral response. For example, expression of lncRNAs has profound effects on activation of TFs, the production of IFN‐γ, cytokines and chemokines, activation of JAK‐STAT signaling, and the transcription of ISGs. The altered expression or activity of these pivotal innate immune molecules significantly influences the host antiviral response and thereby affects the viral infection and replication. Interestingly, some lncRNAs acting as negative regulators of innate immunity can be hijacked by virus to inhibit the antiviral response, and lncRNAs functioning as positive regulators can be suppressed by virus during the infection. These findings provide strong evidences supporting the key role played by the ubiquitous and versatile lncRNAs in antiviral innate immunity. However, although thousands of lncRNAs are associated with viral infection, the number of lncRNAs with experimentally verified function is limited. Therefore, intensive studies are still needed to define the expression, regulation and functioning of lncRNAs during the viral pathogenesis. The future discoveries related to lncRNAs would provide a comprehensive understanding of the elaborate antiviral innate immunity.
ACKNOWLEDGMENTS
This work is supported by National Basic Research Program (973) of China (2015CB910502), Natural Science Foundation of China (U1305212), and National Key Technologies Research and Development Program of China (2013ZX10004‐611) to J.‐L.C. and Natural Science Foundation of China (81590765) to J. O.
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
FURTHER READING
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol 2014, 14:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Schneider WM, Rice CM. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol 2015, 36:124–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non‐coding RNAs: insights into functions. Nat Rev Genet 2009, 10:155–159. [DOI] [PubMed] [Google Scholar]
REFERENCES
- 1. Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity 2015, 42:792–804. [DOI] [PubMed] [Google Scholar]
- 2. Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol 2014, 26:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heward JA, Lindsay MA. Long non‐coding RNAs in the regulation of the immune response. Trends Immunol 2014, 35:408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atianand MK, Fitzgerald KA. Long non‐coding RNAs and control of gene expression in the immune system. Trends Mol Med 2014, 20:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011, 21:354–361. [DOI] [PubMed] [Google Scholar]
- 6. Zhang Q, Jeang KT. Long non‐coding RNAs (lncRNAs) and viral infections. Biomed Pharmacother 2013, 3:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al. Long noncoding RNA NEAT1‐dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 2014, 53:393–406. [DOI] [PubMed] [Google Scholar]
- 8. Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon‐gamma locus. Cell 2013, 152:743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon‐stimulated gene transcription. Cell Host Microbe 2014, 16:616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013, 20:300–307. [DOI] [PubMed] [Google Scholar]
- 11. Fatica A, Bozzoni I. Long non‐coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014, 15:7–21. [DOI] [PubMed] [Google Scholar]
- 12. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013, 152:1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012, 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 2011, 39:D146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, Carnero E, Barriocanal M, Fortes P, Anthony DD, et al. Negative regulation of the interferon response by an interferon‐induced long non‐coding RNA. Nucleic Acids Res 2014, 42:10668–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti‐inflammatory therapeutics. eLife 2013, 2:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya‐Jones A, Blanco M, Burghard C, Moradian A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP, Pauler FM. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development 2013, 140:1184–1195. [DOI] [PubMed] [Google Scholar]
- 21. Yul W, Yang RAF, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife 2014, 3:e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry‐Bezy M, Lawrence JB, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341:789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009, 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011, 43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guttman M, Rinn JL. Modular regulatory principles of large non‐coding RNAs. Nature 2012, 482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geisler S, Coller J. RNA in unexpected places: long non‐coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013, 14:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci USA 2014, 111:1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, Schmidt V, Hogan PG, Sacks DB, Rao A. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA‐protein scaffold complex. Proc Natl Acad Sci USA 2011, 108:11381–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3‐binding long noncoding RNA lnc‐DC controls human dendritic cell differentiation. Science 2014, 344:310–313. [DOI] [PubMed] [Google Scholar]
- 30. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nat Med 2008, 14:723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA‐p21 suppresses target mRNA translation. Mol Cell 2012, 47:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y, Ouyang J, Zhang L, Tan H, Chen R, et al. A long noncoding RNA critically regulates Bcr‐Abl‐mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 2015, 34:1768–1779. [DOI] [PubMed] [Google Scholar]
- 33. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel‐Duby R, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ingolia NT, Brar GA, Stern‐Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein‐coding genes. Cell Rep 2014, 8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al. The imprinted H19 lncRNA antagonizes let‐7 microRNAs. Mol Cell 2013, 52:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo G, Kang Q, Chen Q, Chen Z, Wang J, Tan L, Chen JL. High expression of long non‐coding RNA H19 is required for efficient tumorigenesis induced by Bcr‐Abl oncogene. FEBS Lett 2014, 588:1780–1786. [DOI] [PubMed] [Google Scholar]
- 37. Schneider WM, Chevillotte MD, Rice CM. Interferon‐stimulated genes: a complex web of host defenses. Annu Rev Immunol 2014, 32:513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol 2014, 14:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramirez‐Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 2009, 138:114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Menachery VD, Eisfeld AJ, Schafer A, Josset L, Sims AC, Proll S, Fan S, Li C, Neumann G, Tilton SC, et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon‐stimulated gene responses. MBio 2014, 5:e01174–01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang S, Chi X, Wei H, Chen Y, Chen Z, Huang S, Chen JL. Influenza A virus‐induced degradation of eukaryotic translation initiation factor 4B contributes to viral replication by suppressing IFITM3 protein expression. J Virol 2014, 88:8375–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Porritt RA, Hertzog PJ. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol 2015, 36:150–160. [DOI] [PubMed] [Google Scholar]
- 43. Wei H, Wang S, Chen Q, Chen Y, Chi X, Zhang L, Huang S, Gao GF, Chen JL. Suppression of interferon lambda signaling by SOCS‐1 results in their excessive production during influenza virus infection. PLoS Pathog 2014, 10:e1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Das A, Dinh PX, Panda D, Pattnaik AK. Interferon‐inducible protein IFI35 negatively regulates RIG‐I antiviral signaling and supports vesicular stomatitis virus replication. J Virol 2014, 88:3103–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol 2015, 15:231–242. [DOI] [PubMed] [Google Scholar]
- 46. Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel‐Tretheway BG, Korth MJ, Castle JC, Biery MC, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio 2010, 1:e00206–e00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yin Z, Guan D, Fan Q, Su J, Zheng W, Ma W, Ke C. lncRNA expression signatures in response to enterovirus 71 infection. Biochem Biophys Res Commun 2013, 430:629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Josset L, Tchitchek N, Gralinski LE, Ferris MT, Eisfeld AJ, Green RR, Thomas MJ, Tisoncik‐Go J, Schroth GP, Kawaoka Y, et al. Annotation of long non‐coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon‐stimulated transcripts. RNA Biol 2014, 11:875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Winterling C, Koch M, Koeppel M, Garcia‐Alcalde F, Karlas A, Meyer TF. Evidence for a crucial role of a host non‐coding RNA in influenza A virus replication. RNA Biol 2014, 11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza‐Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309:1570–1573. [DOI] [PubMed] [Google Scholar]
- 51. Imam H, Bano AS, Patel P, Holla P, Jameel S. The lncRNA NRON modulates HIV‐1 replication in a NFAT‐dependent manner and is differentially regulated by early and late viral proteins. Sci Rep 2015, 5:8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, et al. A cytoplasmic NF‐kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015, 27:370–381. [DOI] [PubMed] [Google Scholar]
- 53. Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol 2012, 189:2084–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non‐coding RNAs in mammals. Nature 2009, 458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiong Y, Yuan J, Zhang C, Zhu Y, Kuang X, Lan L, Wang X. The STAT3‐regulated long non‐coding RNA Lethe promote the HCV replication. Biomed Pharmacother 2015, 72:165–171. [DOI] [PubMed] [Google Scholar]
- 56. Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV‐1 posttranscriptional expression. MBio 2013, 4:e00596–e00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus‐inducible non‐coding RNA in mouse brain. J Gen Virol 2006, 87:1991–1995. [DOI] [PubMed] [Google Scholar]
- 58. Krawczyk M, Emerson BM. p50‐associated COX‐2 extragenic RNA (PACER) activates COX‐2 gene expression by occluding repressive NF‐kappaB complexes. eLife 2014, 3:e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barriocanal M, Carnero E, Segura V, Fortes P. Long non‐coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Front Immunol 2015, 5:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carnero E, Barriocanal M, Segura V, Guruceaga E, Prior C, Borner K, Grimm D, Fortes P. Type I interferon regulates the expression of long non‐coding RNAs. Front Immunol 2014, 5:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011, 54:1679–1689. [DOI] [PubMed] [Google Scholar]
- 62. Lu J, Xie F, Geng L, Shen W, Sui C, Yang J. Investigation of serum lncRNA‐uc003wbd and lncRNA‐AF085935 expression profile in patients with hepatocellular carcinoma and HBV. Tumour Biol 2015, 36:3231–3236. [DOI] [PubMed] [Google Scholar]
- 63. Zeng M, Hu Z, Shi X, Li X, Zhan X, Li XD, Wang J, Choi JH, Wang KW, Purrington T, et al. MAVS, cGAS, and endogenous retroviruses in T‐independent B cell responses. Science 2014, 346:1486–1492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64. Tugnet N, Rylance P, Roden D, Trela M, Nelson P. Human endogenous retroviruses (HERVs) and autoimmune rheumatic disease: is there a link? Open Rheumatol J 2013, 7:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McNab F, Mayer‐Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol 2015, 15:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kambara H, Gunawardane L, Zebrowski E, Kostadinova L, Jobava R, Krokowski D, Hatzoglou M, Anthony DD, Valadkhan S. Regulation of interferon‐stimulated gene BST2 by a lncRNA transcribed from a shared bidirectional promoter. Front Immunol 2014, 5:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol 2007, 81:13112–13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang W, Du J, Yu K, Wang T, Yong X, Yu XF. Association of potent human antiviral cytidine deaminases with 7SL RNA and viral RNP in HIV‐1 virions. J Virol 2010, 84:12903–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non‐coding RNA TINCR. Nature 2013, 493:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009, 33:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kawaguchi T, Tanigawa A, Naganuma T, Ohkawa Y, Souquere S, Pierron G, Hirose T. SWI/SNF chromatin‐remodeling complexes function in noncoding RNA‐dependent assembly of nuclear bodies. Proc Natl Acad Sci USA 2015, 112:4304–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rossetto CC, Tarrant‐Elorza M, Verma S, Purushothaman P, Pari GS. Regulation of viral and cellular gene expression by Kaposi's sarcoma‐associated herpesvirus polyadenylated nuclear RNA. J Virol 2013, 87:5540–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bidet K, Dadlani D, Garcia‐Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non‐coding RNA. PLoS Pathog 2014, 10:e1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saayman S, Ackley A, Turner AM, Famiglietti M, Bosque A, Clemson M, Planelles V, Morris KV. An HIV‐encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther 2014, 22:1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]


