Summary
Porcine epidemic diarrhoea virus (PEDV) and porcine deltacoronavirus (PDCoV) were first identified in Canada in 2014. Surveillance efforts have been instrumental in controlling both diseases. In this study, we provide an overview of surveillance components for the two diseases in Ontario (Canada), as well as PEDV and PDCoV incidence and prevalence measures. Swine herds located in the Province of Ontario, of any type, whose owners agreed to participate in a voluntary industry‐led disease control programme (DCP) and with associated diagnostic or epidemiological information about the two swine coronaviruses, were eligible to be included for calculation of disease frequency at the provincial level. PEDV and PDCoV data stored in the industry DCP database were imported into the R statistical software and analysed to produce weekly frequency of incidence counts and prevalence counts, in addition to yearly herd‐level incidence risk and prevalence between 2014 and 2016. The yearly herd‐level incidence risk of PEDV, based on industry data, was 13.5%, 3.0% and 1.4% (95% CI: 11.1–16.2, 2.0–4.2, 0.8–2.3), while the yearly herd‐level incidence risk of PDCoV was 1.1%, 0.3%, and 0.1% (95% CI: 0.5–2.2, 0.1–0.9, 0.0–0.5), for 2014, 2015 and 2016, respectively. Herd‐level prevalence estimates for PEDV in the last week of 2014, 2015 and 2016 were 4.4%, 2.3% and 1.4%, respectively (95% CI: 3.1–6.0, 1.5–3.3, 0.8–2.2), while herd‐level prevalence estimates for PDCoV in the last week of 2014, 2015 and 2016 were 0.5%, 0.2% and 0.2%, respectively (95% CI: 0.1–1.2, 0.0–0.6, 0.0–0.6). Collectively, our results point to low and decreasing incidence risk and prevalence for PEDV and PDCoV in Ontario, making both diseases possible candidates for disease elimination at the provincial level.
Keywords: disease surveillance, herd‐level incidence, herd‐level prevalence, porcine deltacoronavirus (PDCoV), porcine epidemic diarrhoea virus (PEDV)
1. INTRODUCTION
Porcine epidemic diarrhoea (PED) was first described in England in 1971, and its causative agent, porcine epidemic diarrhoea virus (PEDV), was identified in 1978 (Chen et al., 2014). PEDV causes anorexia, vomiting, diarrhoea and dehydration in pigs, resulting in near 100% mortality for piglets during the first few days of life (Hill et al., 2014) and low mortality in older pigs. The virus spreads via the faecal–oral route, either through direct contact with an infected pig or through indirect contact with contaminated fomites. Widespread outbreaks were reported in Europe during the 1970s and 1990s, while epidemics in Asia have caused significant disruption to Asian pig production since 2008 (Williamson et al., 2013). Porcine deltacoronavirus (PDCoV, also known as swine deltacoronavirus, SDCV) was first identified in Hong Kong in 2012. The transmission modes and clinical signs due to PDCoV infections are similar to PEDV; however, the mortality rate is generally lower after PDCoV infections (Carvajal et al., 2015).
PEDV emerged in North America in May 2013, while PDCoV was first confirmed in February 2014, both in the United States (Chen et al., 2014; Ma et al., 2015). These novel viruses rapidly disseminated throughout the US swine population, resulting in the mortality of an estimated seven million animals by May 2014 (Jung & Saif, 2015). Due to the faecal–oral route of transmission, the infection spread through various mechanisms, including contaminated transportation vehicles (Lowe et al., 2014). In Canada, PEDV emerged in January 2014 when a swine herd in Ontario tested positive for the virus (Kochhar, 2014). Imported spray‐dried porcine plasma contaminated with PEDV was the likely pathway of introduction, as established through descriptive studies (Pasma, Furness, Alves, & Aubry, 2016), analytical epidemiological studies (Aubry, Thompson, Pasma, Furness, & Tataryn, 2017; O'Sullivan, 2015) and experimental investigations (Pasick et al., 2014). By July 2014, only 62 cases of PEDV had been reported in Ontario and the outbreak was largely under control (Pasma et al., 2016).
Despite considerable impact on animal health in completely susceptible populations, the diseases caused by the two emerging porcine coronaviruses were not notifiable globally, according to the World Organization for Animal Health (OIE, 2017), and were not federally reportable or notifiable in Canada (Canadian Food Inspection Agency, 2014). At the provincial level, animal health regulations enabled consideration of PEDV and PDCoV as emerging hazards (Government of Ontario, 2009), which allowed a measured and appropriate response to the outbreak by the Ontario Ministry of Agriculture, Food and Rural Affairs (OMAFRA) via close collaboration with swine industry organizations.
Initial emergence of the two novel porcine coronaviruses was followed by their successful elimination from several initial case farms (Misener, 2015). The high rate of successful PEDV elimination from individual herds, and effective measures that seemed to have minimized widespread viral dissemination, resulted in the current position of industry organizations that both infectious agents can and should be eliminated at the provincial level (Ontario Swine Health Advisory Board, 2017).
A disease control programme (DCP) involves disease monitoring, surveillance, intervention and control strategies (Salman, 2003). The DCP considered here has been voluntary in nature as defined elsewhere (Christensen, 2003). Furthermore, an important component of any disease control programme is measuring trends in incidence and prevalence, particularly when the disease of interest moves into the phase of possible elimination (Salman, 2003). With the infrastructure built for management of endemic diseases in Ontario, the data to support estimation of disease trends are available. Thus, the primary objective of this study was to estimate herd‐level incidence and prevalence measures for PEDV and PDCoV in swine herds in Ontario (Canada) between January 2014 and December 2016, based on industry data. The secondary objective was to describe relevant surveillance components that were used for identification of new PEDV cases.
2. MATERIALS AND METHODS
2.1. Data sources
The Ontario Swine Health Advisory Board (OSHAB) maintains a database which contains premises information and PEDV/PDCoV herd status for producers enrolled in its voluntary regional disease control programmes. The program, and a database originally designed for management of porcine reproductive and respiratory syndrome virus (PRRSV) (Arruda, Poljak, Friendship, Carpenter, & Hand, 2015), was expanded to PEDV and PDCoV after their emergence. The data relevant for this work included unique identifiers, herd type, date of herd enrolment into the database, PEDV and PDCoV status of individual premises, and the date that individual premises changed their PEDV and PDCoV status. For inclusion into the study, swine herds could be of any herd type, but had to meet the following criteria: (i) be located in the Province of Ontario, (ii) participate in the voluntary industry‐led disease control programme and (iii) have diagnostic or epidemiological information about the infection status of porcine epidemic diarrhoea virus (PEDV) or porcine deltacoronavirus (PDCoV). The industry organization (i.e., OSHAB) provided relevant data to researchers for calculation of disease frequencies under a separate data transfer agreement.
Due to the voluntary nature of the DCP, the enrolment of herds into the database has been an ongoing process. This could have resulted in the date of enrolment being later than the date the infection was originally detected in a specific herd. In rare instances where a herd's enrolment date was not specified or occurred later than the first reported case of PEDV or PDCoV for that herd, the disease status date was entered as the herd enrolment date for the purposes of this report. This was done so that herd‐level prevalence on a weekly basis could be properly calculated. An Open Database Connectivity (ODBC) connection to this database was established, and relevant tables were imported into R (R Core Team, 2016) using the rodbc library (Ripley & Lapsley, 2016).
2.2. Herd‐level PEDV and PDCoV infection status definition
The industry database maintained by OSHAB traces infection status of participating individual premises over time, which includes detection of infection, as well as any subsequent infection, elimination or other change in infection status with the associated dates. In this database, premises can have four possible values for their infection status, as defined below: (i) confirmed positive—premises with herds that tested positive for PEDV/PDCoV based on laboratory test results from the Animal Health Laboratory (AHL) at the University of Guelph (Guelph, ON, Canada). For a premise to be classified as confirmed positive, it had to have an associated diagnostic submission number that includes at least one positive test for PEDV/PDCoV using RT‐PCR, regardless of the number of specimens that were submitted. An AHL reference number (also known as a G Number) was not available for some confirmed positive cases in the database, as the attending veterinarian obtained test results, but did not provide OSHAB with the AHL reference number; (ii) presumed positive—premises which housed animals that were moved from positive sites at a prior stage in the production system (i.e., defined as positive due to pig movement). This information was obtained from attending veterinarians based on their knowledge of pig flow and movement, and was not based on diagnostic testing conducted on the premises of interest. For premises to be classified as presumed positive, the herd veterinarian simply needed to indicate that a specific site received pigs from PEDV/PDCoV‐positive sites; (iii) presumed negative—previously positive premises, either confirmed or presumed, that were tested using PCR tests according to industry guidelines and had all test results negative. The sampling requirements for declaring premises to be PEDV/PDCoV presumed negative varied based on the combination of herd type and the type of animal flow (i.e., all‐in/all‐out by barn, or continuous flow nursery and finisher herds). Complete criteria were, at the time of publishing, available on the website of the industry organization (Ontario Swine Health Advisory Board, 2015). Briefly, sampling strategy for farrow‐wean, nursery and finisher sites aimed to detect prevalence of virus‐positive animals of at least 10%, with expected herd sensitivity of 95%, assumed test sensitivity of 98%, test specificity of 100%. In instances where the sampling material was oral fluid collected through cotton ropes, the assumption was that five pigs contributed oral fluids to one rope, and such fluids were considered a pooled sample. For farrow‐finish or farrow‐feeder sow sites, the same assumptions were made, except that the sampling strategy was required to detect prevalence of 5% with 95% herd sensitivity. Because of alternative strategies, the required sample size varied but a minimum sample size was four oral fluids. In addition, the testing time in sow herds was prescribed to be a minimum of 10 weeks post‐infection and was required to be repeated three times in the case of farrow‐wean (FW) sites, or two times in the case of farrow‐finish (FF) or farrow‐feeder (FG) sites. The recommended specimen type could be swab, Swiffer (for covering larger areas in a pen), or oral fluids, depending on the target age group. In FW herds, individual farrowing crates were the target population for each individual sampling occasion, in particular if diarrhoea was evident. The minimum recommendation for one sampling occasion in FW herds was to sample four Swiffer samples, at least eight farrowing crates per one sample. Alternatively, individual swabs of 30 farrowing crates were deemed as acceptable sample after pooling 5:1. In FG farms, the recommended sample type was oral fluid, with recommendation to collect 12 oral fluids from nursery pigs. Similarly, in FF farms, six oral fluids were recommended for collection from nursery pigs and six for collection from finisher pigs. For all‐in/all‐out nursery and finisher farms, the recommendation was to sample six oral fluids, with added requirement that these herds should be supplied from sow herds with a confirmed negative status. All testing has been assumed to be performed using RT‐PCR tests. Full description is available elsewhere (Ontario Swine Health Advisory Board (OSHAB), 2015); (iv) confirmed negative—premises which have had no clinical signs or diagnostic evidence of PEDV/PDCoV for at least 6 months after the presumed negative status date. In addition, herds that were part of the Ontario voluntary DCP but were not tested for emerging porcine coronaviruses—due to lack of clinical or other types of diagnostic or epidemiological triggers—had assigned status of NA (not available).
2.3. Weekly measures of disease frequency
Weekly time series were then obtained for the three sets of statistics for both viruses: (i) number of premises with specific infection status in each week, (ii) number of new positive and new negative premises and (iii) number of premises in the database.
For each week where a herd's PEDV/PDCoV status was not reported, the status was set to the last‐reported status using the zoo package (Zeileis & Grothendieck, 2005). For example, if a status is not reported for the current week, and “confirmed positive” was reported for the prior week, then the current week's status is “confirmed positive.” The individual premises data were then aggregated to counts of premises on a weekly basis. Based on the former time series, prevalence area plots were generated, providing a visual assessment of “confirmed positive,” “presumed positive,” “presumed negative” and “confirmed negative” herds over time. The prevalence numerator was the positive herd count (sum of confirmed positive and presumed positive herds) for a specific week, while the denominator was the herd count in the premises table for that week (calculated previously).
Subsequently, the following disease status changes were tracked by week for each premise: (1) “Not Available” to “Presumed Positive,” (2) “Not Available” to “Confirmed Positive,” (3) “Presumed Positive” to “Presumed Negative,” (4) “Presumed Positive” to “Confirmed Negative,” (5) “Confirmed Positive” to “Presumed Negative,” (6) “Confirmed Positive” to “Confirmed Negative,” (7) “Presumed Negative” to “Presumed Positive,” (8) “Presumed Negative” to “Confirmed Positive,” (9) “Confirmed Negative” to “Presumed Positive,” (10) “Confirmed Negative” to “Confirmed Positive.”
Any status change leading to new presumed or confirmed positive status (i.e., status changes 1, 2, 7, 8, 9 and 10) was classified as new positive. Similarly, status changes 3, 4, 5 and 6 were classified as new negative. The number of new positives and new negatives was then aggregated to the weekly level throughout the study period. The latter time series were then used to construct a chart of cumulative incidence counts for each year, and epidemic curves were constructed for positive herds and herds which became negative. In addition, for each week, the incidence risk was calculated by dividing the number of cases that occurred in a specific week and by the number of herds that were eligible to become cases at the beginning of the week.
2.4. Yearly measures of disease frequency
Yearly herd‐level incidence risk and prevalence at the end of the year were calculated in the following manner: the numerator for the yearly incidence risk calculation was defined as the cumulative number of incident cases at the end of each year, while the denominator was defined as the number of herds in the DCP at the end of each year, minus the number of confirmed positive and presumed positive herds at the beginning of the specified year, minus half the number of herd additions to the database during the year (the number of additions was treated as the number of withdrawals so this inherently open population could be converted to a closed population, and equations for calculation of risk used) (Eq. (1) (adapted from Dohoo, Martin, & Stryhn, 2003)):
where N = cumulative incident cases at yearend and
| (1) |
For yearly herd‐level prevalence, the numerator was defined as the total number of confirmed positive and presumed positive herds at the end of the year, while the denominator was defined as the number of herds in the DCP at the end of each year. Both the yearly incidence risk and prevalence, originally calculated as proportions, were then multiplied by 100 to get the percentage values, with exact 95% confidence intervals obtained from the binom.test function in R.
For the yearly incidence rate, we calculated total number of herd‐years at risk for each year from the number of herds under risk in each week and used this as a denominator. The incidence rate was then expressed as number of cases per one herd‐year. Exact 95% confidence intervals on the incidence rate were obtained via the poisson.test function in R.
2.5. Surveillance components for PEDV
Three surveillance components for identification of new cases were considered in this study. The cumulative number of presumed and confirmed cases between January 2014 and December 2016 maintained in the OSHAB voluntary DCP database represents surveillance component 1. Surveillance component 2 is the official count of new cases maintained by OMAFRA and publicly available when this report was written (Ontario Pork, 2017). OMAFRA does not monitor PDCoV and defines a PEDV case as a herd which tests positive based on AHL's RT‐PCR test (Pasma et al., 2016), and is the first such case in the production system. In other words, the PEDV case count pertains to primary cases only; subsequent secondary cases due to animal movement in the production system are not included in OMAFRA reporting, although they could be confirmed as PEDV‐positive in the diagnostic laboratory. Surveillance component 3 represents cumulative number of positive submissions, due to any reason, maintained by the Animal Health Laboratory (AHL, University of Guelph), the largest diagnostic laboratory in Ontario for testing livestock diseases. The overlap between the three surveillance components was calculated on the basis of demographic data.
3. RESULTS
3.1. Surveillance components for PEDV
Figure 1 depicts three surveillance components considered in this study and their overlap. Briefly, during the 3‐year period (January 2014–December 2016), AHL reported a total of 974 positive submissions for PEDV (Fairles, personal communication 2017). Of the 974 positive submissions, a cumulative total of 118 cases are reported in OMAFRA and OSHAB as new cases. The remaining 856 positive submissions were not part of any surveillance component considered in this study. Of the 118 cases, 99 are captured by OMAFRA as primary PEDV cases; however, 70 of these 99 primary PEDV cases are also reported in the OSHAB database, leaving 29 primary PEDV cases (99–70) reported in OMAFRA but not OSHAB. The remaining 19 cases (974–856–99) are non‐primary PEDV‐positive cases (i.e., not the first outbreak in the associated production system); hence, they are captured in AHL and reported in the OSHAB voluntary DCP database, but not OMAFRA. Lastly, 52 PEDV‐positive cases are strictly in OSHAB and are not reported in any other surveillance component. It should be noted that these 52 cases are “presumed positive” due to pig movement, and as such have no supporting AHL test result, an attribute which also makes them ineligible for OMAFRA reporting. The OSHAB total for PEDV cases is 52 + 19 + 70 = 141, which is also the cumulative number of incident cases over the 3‐year period.
Figure 1.
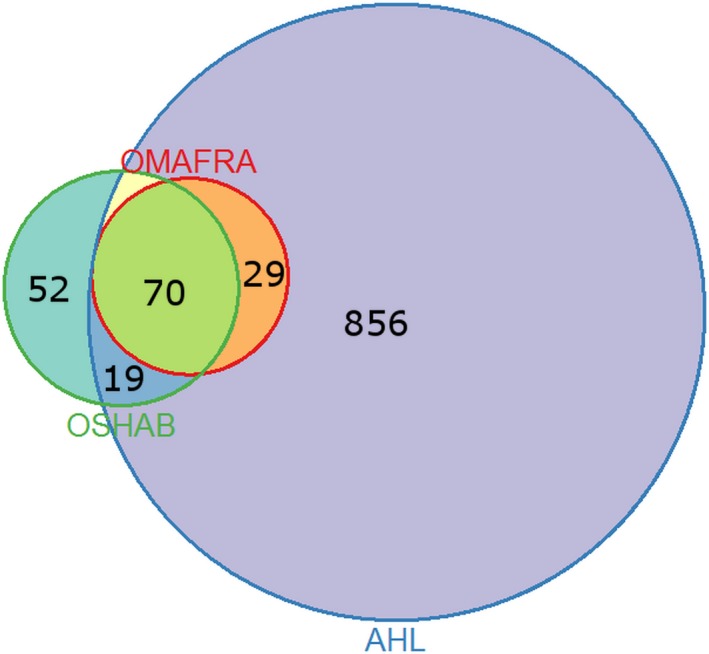
Venn diagram representing overlap among the three surveillance components for identification of new cases of the porcine epidemic diarrhoea virus (PEDV) in Ontario (Canada) between 2014 and 2016—The Animal Health Laboratory (AHL), Ontario Ministry of Agriculture Food and Rural Affairs (OMAFRA) and the Ontario Swine Health Advisory Board (OSHAB). The size of individual circles should be proportional to the number of positive cases recorded by a specific surveillance component. The overlapping areas should represent cases that were identified by as many surveillance components as are being overlapped. The numbers listed in a specific area represent number of cases that are identified by a specific surveillance component or combination thereof
3.2. Yearly incidence and prevalence measures
Table 1 contains number of new infected premises for the two viruses in each year based on the industry's voluntary DCP database, number of positive cases in the last week of each year, as well as incidence risk and prevalence estimates. In addition, 95% confidence intervals are provided, recognizing that this surveillance component was a census of herds participating in the voluntary DCP, but does not contain all herds from the source population. In brief, annual incidence risk was the highest in 2014 for PEDV (13.49%) and PDCoV (1.14%), and the lowest in 2016 for PEDV (1.42%) and PDCoV (0.08%). The end of year prevalence for PEDV ranged between 4.4% and 1.4% in 2014 and 2016, respectively. A similar trend can be seen for PDCoV (Table 1). Furthermore, similar values have been observed for incidence rates in each year (Table 1).
Table 1.
Herd‐level incidence risk and rate of two novel porcine coronaviruses (PEDV and PDCoV) in Ontario swine herds between 2014 and 2016, and estimated prevalence of positive cases at the end of each year based on data provided in the Ontario Swine Health Advisory Board (OSHAB) Disease Control Program (DCP) database (average number of herds for 2014–2016 = 1093)
| Year | Cumulative n of new cases | Incidence risk (%) | 95% CI (%) | Incidence rate (cases per herd‐year) | 95% CI (cases per herd‐year) | Number of cases at year‐end | Prevalence at year‐end (%) | 95% CI (%) |
|---|---|---|---|---|---|---|---|---|
| Porcine epidemic diarrhoea virus | ||||||||
| 2014 | 95 | 13.49 | (11.06–16.24) | 0.14 | (0.12–0.18) | 36 | 4.36 | (3.07–5.99) |
| 2015 | 29 | 2.97 | (2.00–4.24) | 0.03 | (0.02–0.05) | 27 | 2.25 | (1.49–3.26) |
| 2016 | 17 | 1.42 | (0.83–2.26) | 0.02 | (0.01–0.03) | 17 | 1.35 | (0.79–2.16) |
| Porcine deltacoronavirus | ||||||||
| 2014 | 8 | 1.14 | (0.49–2.23) | 0.011 | (0.005–0.022) | 4 | 0.48 | (0.13–1.24) |
| 2015 | 3 | 0.30 | (0.06–0.87) | 0.003 | (0.001–0.009) | 2 | 0.17 | (0.02–0.60) |
| 2016 | 1 | 0.08 | (0.00–0.45) | 0.001 | (0.000–0.005) | 2 | 0.16 | (0.02–0.57) |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.3. Weekly incidence and prevalence measures
The cumulative number of new PEDV and PDCoV cases per week for the 3 years is provided in Figures 2 and 3, respectively. Notably, there were many new PEDV cases detected in the winter of 2014 when the disease was first introduced to Ontario and Canada.
Figure 2.

Cumulative weekly confirmed and presumed positive cases for porcine epidemic diarrhoea virus (PEDV) in Ontario (Canada) for 2014–2016, based on data provided in the Ontario Swine Health Advisory Board (OSHAB) Disease Control Program (DCP) database (average number of herds for 2014–2016 = 1093)
Figure 3.
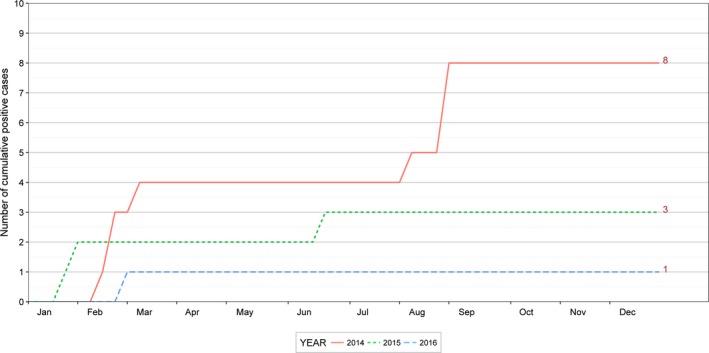
Cumulative weekly confirmed and presumed positive cases for porcine deltacoronavirus (PDCoV) in Ontario (Canada) for 2014–2016, based on data provided in the Ontario Swine Health Advisory Board (OSHAB) Disease Control Program (DCP) database (average number of herds for 2014–2016 = 1093)
Figure 4 depicts the number of herds categorized into one of the four groups with respect to PEDV infection. From this figure, it could be observed that PEDV had a rapid increase in the number of prevalent cases until March 2014, which then peaked in the summer of the same year, only to be followed by a decline. Figure 5 displays the weekly development of infection status as a proportion. Note that in the latter figure, the denominator changed on a weekly basis as the number of herds in the voluntary DCP changed. Figures 6 and 7 display the number of premises in a distinct PDCoV status as a count (Figure 6) and proportion (Figure 7). The weekly incidence risk of PEDV and PDCoV in 2014–2016 is provided in the supplementary material (Figures S1 and S2, respectively).
Figure 4.
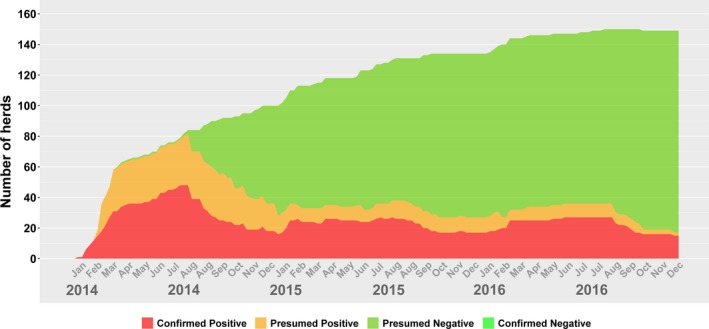
Number of Herds with confirmed positive, presumed positive, presumed negative or confirmed negative status for porcine epidemic diarrhoea virus (PEDV) in Ontario (Canada) for 2014–2016, based on data provided in the Ontario Swine Health Advisory Board (OSHAB) Disease Control Program (DCP) database (average number of herds for 2014–2016 = 1093)
Figure 5.
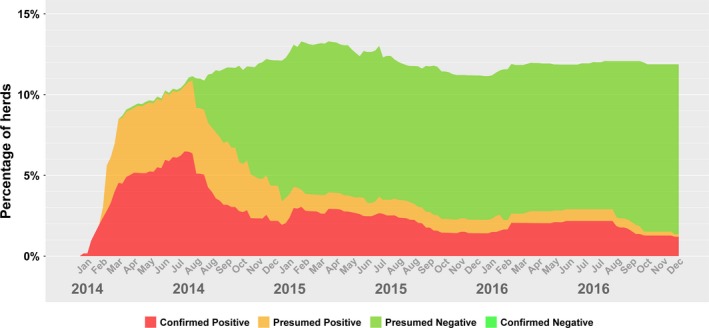
Percentage of Herds with confirmed positive, presumed positive, presumed negative or confirmed negative status for porcine epidemic diarrhoea virus (PEDV) in Ontario (Canada) for 2014–2016, based on data provided in the Ontario Swine Health Advisory Board (OSHAB) Disease Control Program (DCP) database (average number of herds for 2014–2016 = 1093)
Figure 6.
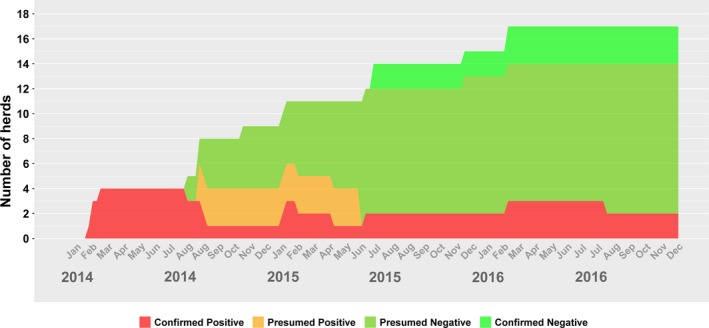
Number of Herds with confirmed positive, presumed positive, presumed negative or confirmed negative status for porcine deltacoronavirus (PDCoV) in Ontario (Canada) for 2014–2016, based on data provided in the Ontario Swine Health Advisory Board (OSHAB) Disease Control Program (DCP) database (average number of herds for 2014–2016 = 1093)
Figure 7.
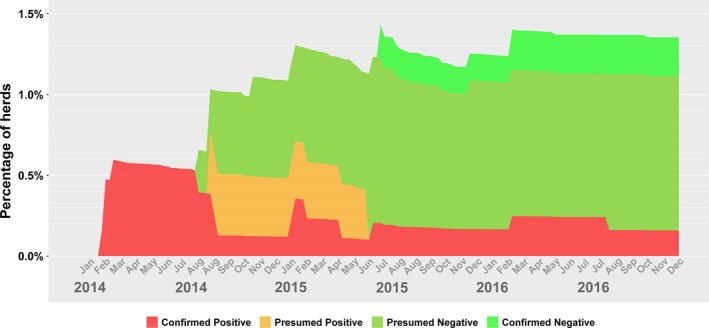
Percentage of Herds with confirmed positive, presumed positive, presumed negative or confirmed negative status for porcine deltacoronavirus (PDCoV) in Ontario (Canada) for 2014–2016, based on data provided in the Ontario Swine Health Advisory Board (OSHAB) Disease Control Program (DCP) database (average number of herds for 2014–2016 = 1093)
4. DISCUSSION
Emerging infections considered in this work are examples of production‐limiting diseases that are not federally reportable (Canadian Food Inspection Agency, 2017) or listed in an OIE list of diseases (World Organization for Animal Health, 2017). Consequently, resources available through response by the regulatory veterinary authorities could be limited. In the case of the Ontario outbreak, the regulatory framework existed at the provincial level because PEDV was considered an emerging hazard under the provincial Animal Health Act (Government of Ontario, 2009) and because of suspicion of feed involvement, which is under the jurisdiction of national veterinary authorities. The response to the outbreak was collaborative in nature and is best depicted by the evaluation of surveillance components for PEDV.
The OMAFRA and industry approaches were different with respect to surveillance coverage, case definition and follow‐up time. OMAFRA surveillance was mostly concerned about identification of new cases and their investigation in the early phase of the outbreak. It contained a census of all PEDV cases, and all herds from the source population were eligible to be listed as a case, at least during the initial phase of the outbreak when PEDV could be considered an emerging hazard. However, only the first case in a given production system was counted as a case and secondary cases due to planned animal movement to other premises were not counted as additional cases. For example, if PEDV was detected in a sow herd in a multisite production system, the sow herd would be counted as a case, but not the nursery or finisher sites supplied from the sow herd. For this surveillance system, all cases had to have diagnostic confirmation through laboratory submissions. For this reason, the OMAFRA PEDV surveillance system is completely nested within the diagnostic laboratory. It is notable that ~17.1% of PEDV cases were identified through this surveillance system only, which is reflective of the reportable nature (provincially) of the emerging hazard.
The OSHAB surveillance system was concerned with identification of new cases, reducing dissemination of infection through animal movement, and following the infection status over time for the purposes of monitoring disease trends over time. As a voluntary programme, the source population consisted of premises that volunteered to participate. At the end of 2016, the number of sites in the OSHAB database was 1255, which represents approximately 49% of 2,556 active swine sites in Ontario (Brisson, 2014). Thus, the OSHAB surveillance coverage was lower than for the OMAFRA surveillance, which had 100% coverage—by law, PEDV‐infected herds were reported to OMAFRA during the phase when the hazard was still considered as emerging.
The OSHAB case definition also included secondary sites that were confirmed positive due to animal movement and had laboratory confirmation, as well as secondary sites where movement of PEDV‐positive animals occurred although testing was not carried out. This approach allowed identification of ~41.8% of cases that would otherwise be unaccounted for in the OMAFRA system (n = 71), which included 30.6% of total cases (n = 52) that were not confirmed through diagnostic testing (Figure 1). Given the purpose of the industry's surveillance system, recording infection in the secondary sites was a logical choice as they could still contribute to disease dissemination through multiple pathways. Such premises had to be declared as presumed or confirmed negative, based on diagnostic testing that met pre‐determined criteria conducted on the premises.
The Venn diagram of surveillance components (Figure 1) also shows a large volume of positive test results for PEDV in a laboratory that did not contribute to the identification of new cases. Such testing serves different purposes and likely contains additional surveillance components which are not considered here. An example is testing trailers in abattoirs for the purposes of case identification through risk‐based approaches. Surveillance systems for emerging diseases could benefit from a clearer definition of such surveillance components, so that diagnostic tests could be easily aggregated into appropriate streams.
The collaborative efforts at controlling PEDV are most clearly seen in the incidence counts for 2014. As expected at the onset of any outbreak, particularly due to massive contamination through a common source, there was a rapid climb in the number of infected farms. However, this outbreak was brought under control relatively quickly. By May 2014, the worst of the outbreak was over, and there were only single digit weekly infections (no more than 5) for the remainder of the year. In addition, the initially positive herds which contributed to the rapid climb in infections between January and May 2014 became negative within a short period and contributed to the rapid increase in negative herds between June and December 2014.
The incidence counts for 2015 reveal a more proactive approach to OSHAB PEDV surveillance and disease control. Not only were there substantially fewer infections (infections were only reported for 15 weeks out of the year, and there were at most 5 infections per week), the number of negative herds also kept up with the number of positive herds (for 9 of the 15 weeks where infections were reported, there were also negative herds). Overall, the number of negative herds in 2015 outpaced the number of positive herds, indicating proactive steps to minimize the number of new infections while aggressively taking steps to eradicate existing ones. This stands in stark contrast to 2014, where disease eradication efforts only seemed to take on momentum towards the latter half of the year.
The trend in incidence counts from 2015 continued into 2016, with lower numbers for positive herds and the number of negative herds outpacing positive ones. However, in contrast to 2015, roughly 75% of new infections occurred in the first quarter of the year (January–March), while 80% of new negative herds were reported in the last quarter of the year (late September–December). The cluster of negative herds at the end of the year suggests that with new infections under control, emphasis was placed on the management of prevalent cases, taking into consideration lower viral transmission during warmer periods of the year.
The prevalence plots reveal ebbs and inclines in PEDV prevalence, in what appears to be a cyclical pattern: a peak in July 2014, a low in January 2015, a peak in July 2015, a low in October 2015 and a peak in February 2016 (Figure 4). The numbers were relatively constant from late February to July 2016, declined in late July and slowly declined until the end of the year. Given the prevalence fluctuations between 1% and 2% for September 2015–December 2016 (Figure 5), and the repeated patterns, there is a danger that PED may become an endemic disease with low prevalence and limited pockets of infection in Ontario. As such, further investigation and action are needed to ensure that the peaks are replaced by a steady decline.
As the same disease prevention and control measures for PEDV also apply to PDCoV (Ontario Ministry of Agriculture Food and Rural Affairs, 2014), there were few reported infections for PDCoV (essentially, controlling for PEDV also controls for PDCoV). It is therefore not surprising that the incidence count patterns are similar: in 2014, there were more PDCoV‐positive herds than negative herds, while in 2015, there were more negative herds than positive ones. However, in 2016 there was just one new PDCoV infection, with a single herd also becoming negative, an indication that if current disease control efforts continue, there may be no new infections in subsequent years.
The prevalence figures for PDCoV are quite different from PEDV—rather than a cyclical pattern as previously observed, the numbers are more erratic, with sudden peaks followed by constants over prolonged periods, and then declines. The relatively static and low prevalence (0.1%–0.2%) observed between July 2015 and December 2016 suggests that PDCoV is a candidate for disease elimination. There is also a possibility that PDCoV cases are underreported by the industry, either because of potentially lower clinical impact in swine herds or because of perceived lower importance than PEDV. Nonetheless, when any of the three porcine coronaviruses (PEDV, PDCoV, TGEV—transmissible gastroenteritis virus) is suspected in a herd and diagnostic material is submitted to the AHL, the diagnostic testing is automatically conducted for all three viruses.
Some limitations of the current study include the fact that PEDV and PDCoV cases are actively pursued for inclusion into the voluntary DCP by industry organizations. Such strategy is likely to result in estimates of incidence and prevalence measures that are higher than in the source population. Because of the inherently open nature of the voluntary DCP, we had to modify the formulae for calculation of incidence risk. It could be argued that presumed cases are not diagnostically confirmed and are therefore subject to misclassification. However, the reality of a voluntary DCP for production‐limiting diseases is that resources to conduct large‐scale testing are scarce and, as such, need to be carefully deployed. Furthermore, in order to confirm premises as presumed negative, diagnostic testing to confirm absence of infection at the design prevalence level is still required. Also, the criteria to declare confirmed negative status is arguably open‐ended and could be further improved.
In conclusion, this study provides estimates of incidence and prevalence measures in Ontario based on industry data collected through voluntary disease control programmes. The data suggest that annual incidence risk and prevalence estimates are low and have been steadily decreasing between 2014 and 2016 for PEDV and PDCoV. Current estimates of disease frequency support planning of disease elimination at the provincial level, but much information should be available about factors that led to time to elimination in individual herds. In addition, our evaluation of surveillance components indicates that the two surveillance components were complementary and focused on different aspects of surveillance. OMAFRA surveillance was mostly focused on identification of primary cases aimed at quick disease investigations and traceability in the face of the outbreak, whereas OSHAB surveillance has the added benefit of having sufficient data that allow long‐term evaluation of disease trends, long‐term disease management and tracing disease status of individual herds over time. The OSHAB voluntary DCP database also provides a good tool for calculating weekly prevalence and incidence measures, which is a valuable statistic for producers and animal health experts during all phases of disease outbreak and control.
Supporting information
ACKNOWLEDGEMENTS
Funding for this study was provided by the NSERC Discovery Grants Program. Authors are grateful to participating industry organizations and producers for providing access to data.
Ajayi T, Dara R, Misener M, Pasma T, Moser L, Poljak Z. Herd‐level prevalence and incidence of porcine epidemic diarrhoea virus (PEDV) and porcine deltacoronavirus (PDCoV) in swineherds in Ontario, Canada. Transbound Emerg Dis. 2018;65:1197–1207. 10.1111/tbed.12858
REFERENCES
- Arruda, A. G. , Poljak, Z. , Friendship, R. , Carpenter, J. , & Hand, K. (2015). Descriptive analysis and spatial epidemiology of porcine reproductive and respiratory syndrome (PRRS) for swine sites participating in area regional control and elimination programs from 3 regions of Ontario. Canadian Journal of Veterinary Research, 79, 268. [PMC free article] [PubMed] [Google Scholar]
- Aubry, P. , Thompson, J. L. , Pasma, T. , Furness, M. C. , & Tataryn, J. (2017). Weight of the evidence linking feed to an outbreak of porcine epidemic diarrhea in Canadian swine herds. Journal of Swine Health and Production, 25(2), 69–72. https://www.aasv.org/shap/issues/v25n2/v25n2p69.pdf [Google Scholar]
- Brisson, Y . (2014). The changing face of the Canadian hog industry. Canadian Agriculture at a Glance. Retrieved from http://www.statcan.gc.ca/pub/96-325-x/2014001/article/14027-eng.pdf
- Canadian Food Inspection Agency . (2014, March 3). Porcine Epidemic Diarrhea (PED) situation in Canada ‐ Animals. Retrieved from http://www.inspection.gc.ca/animals/terrestrial-animals/diseases/other-diseases/ped/eng/1392762503272/1392762576176
- Canadian Food Inspection Agency . (2017). Reportable Diseases – Animals. Retrieved March 25, 2017, from http://www.inspection.gc.ca/animals/terrestrial-animals/diseases/reportable/eng/1303768471142/1303768544412
- Carvajal, A. , Argüello, H. , Martínez‐Lobo, F. J. , Costillas, S. , Miranda, R. , de Nova, P. J. , & Rubio, P. (2015). Porcine epidemic diarrhoea: New insights into an old disease. Porcine Health Management, 1(1), 12 10.1186/s40813-015-0007-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Li, G. , Stasko, J. , Thomas, J. T. , Stensland, W. R. , Pillatzki, A. E. , … Zhang, J. (2014). Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. Journal of Clinical Microbiology, 52(1), 234–243. 10.1128/jcm.02820-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, J. (2003). Danish swine salmonellosis control program: 1993 to 2001 In Salman M. D. (Ed.), Animal disease surveillance and survey systems: Methods and applications (pp. 185–207). Ames, Iowa: Iowa State Press. [Google Scholar]
- Core Team, R . (2016). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (Vol. 0). 10.1038/sj.hdy.6800737 [DOI] [Google Scholar]
- Dohoo, I. , Martin, W. , & Stryhn, H. (2003). Veterinary epidemiologic research (pp. 69–72). Charlottetown, PE, Canada: Atlantic Veterinary College Inc.. [Google Scholar]
- Government of Ontario . (2009). Animal Health Act, 2009, S.O. 2009, c. 31, Pub. L. No. 09a31. Government of Ontario. Retrieved from https://www.ontario.ca/laws/statute/09a31
- Hill, C. , Raizman, E. , Snider, T. , Goyal, S. , Torremorell, M. , & Perez, A. M. (2014). Emergence of porcine epidemic diarrhoea in North America. Focus on, 9(July), 1–8. [Google Scholar]
- Jung, K. , & Saif, L. J. (2015). Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Veterinary Journal, 204(2), 134–143. 10.1016/j.tvjl.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhar, H. S. (2014). Canada: Porcine epidemic diarrhea in Canada: an emerging disease case study. The Canadian Veterinary Journal, 55(11), 1048–1049. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/25392546 [PMC free article] [PubMed] [Google Scholar]
- Lowe, J. , Gauger, P. , Harmon, K. , Zhang, J. , Connor, J. , Yeske, P. , … Main, R. (2014). Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerging Infectious Diseases, 20(5), 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Zhang, Y. , Liang, X. , Lou, F. , Oglesbee, M. , Krakowka, S. , & Li, J. (2015). Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio, 6(2), e00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misener, M. (2015, April). PED – A Canadian Update. Paper presented at the 15th London Swine Conference, London, Ontario
- OIE ‐ World Organization for Animal Health (2017). OIE‐Listed diseases 2017. Retrieved March 25, 2017 from http://www.oie.int/animal-health-in-the-world/oie-listed-diseases-2017/
- Ontario Ministry of Agriculture Food and Rural Affairs . (2014, March 18). Swine DeltaCoronavirus Disease Advisory. Retrieved from https://www.opic.on.ca/images/pdfs/2014-03-18_Swinedeltacoronavirus_Advisory.pdf
- Ontario Pork . (2017). Herd Health: Porcine Epidemic Diarrhea Virus (PED) On‐Farm Testing ‐ Total Confirmed Cases. Retrieved March 25, 2017, from http://www.ontariopork.on.ca/Producers/Herd-Health#PED
- Ontario Swine Health Advisory Board (OSHAB) . (2015). OSHAB ARC&E Progress Update – August 15, 2015. Retrieved from http://opic.on.ca/images/ARCE_Interim_report_August_2015.pdf
- Ontario Swine Health Advisory Board (OSHAB) . (2017). The Ontario Swine Health Advisory Board (OSHAB), Porcine Epidemic Diarrhea Virus (PEDV) and Porcine Delta Coronavirus (PDCoV) Position Statement. Retrieved from https://www.opic.on.ca/images/OSHAB_PED_position_statement_final.pdf
- O'Sullivan, T. (2015, June). Investigation of Factors That Led to Emergence of PEDV through Feed During Early Phase of Canadian Outbreak. Paper presented at the 7th International Symposium on Emerging and Re‐Emerging Pig Diseases, Kyoto, Japan
- Pasick, J. , Berhane, Y. , Ojkic, D. , Maxie, G. , Embury‐Hyatt, C. , Swekla, K. , … Alexandersen, S. (2014). Investigation into the role of potentially contaminated feed as a source of the first‐detected outbreaks of porcine epidemic diarrhea in Canada. Transboundary and Emerging Diseases, 61(5), 397–410. 10.1111/tbed.12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasma, T. , Furness, M. C. , Alves, D. , & Aubry, P. (2016). Outbreak investigation of porcine epidemic diarrhea in swine in Ontario. The Canadian Veterinary Journal, 57(1), 84–89. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/26740705 [PMC free article] [PubMed] [Google Scholar]
- Ripley, B. , & Lapsley, M. (2016). RODBC: ODBC Database Access. R package version 1.3‐14. Retrieved from https://cran.r-project.org/package=RODBC
- Salman, M. (2003). Surveillance and monitoring systems for animal health programs and disease surveys In Salman M. D. (Ed.), Animal disease surveillance and survey systems: Methods and applications (pp. 3–13). Ames, Iowa: Iowa State Press. [Google Scholar]
- Williamson, S. , Strugnell, B. , Thomson, J. , Webster, G. , McOrist, S. , & Clarke, H. (2013). Emergence of severe porcine epidemic diarrhoea in pigs in the USA. Veterinary Record, 173(6), 146–148. 10.1136/vr.f4947 [DOI] [PubMed] [Google Scholar]
- Zeileis, A. , & Grothendieck, G. (2005). zoo: S3 infrastructure for regular and irregular time series. Journal of Statistical Software, 14(6), 1–27. 10.18637/jss.v014.i06 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


