Summary
Microarray technology can be useful for pathogen detection as it allows simultaneous interrogation of the presence or absence of a large number of genetic signatures. However, most microarray assays are labour‐intensive and time‐consuming to perform. This study describes the development and initial evaluation of a multiplex reverse transcription (RT)‐PCR and novel accompanying automated electronic microarray assay for simultaneous detection and differentiation of seven important viruses that affect swine (foot‐and‐mouth disease virus [FMDV], swine vesicular disease virus [SVDV], vesicular exanthema of swine virus [VESV], African swine fever virus [ASFV], classical swine fever virus [CSFV], porcine respiratory and reproductive syndrome virus [PRRSV] and porcine circovirus type 2 [PCV2]). The novel electronic microarray assay utilizes a single, user‐friendly instrument that integrates and automates capture probe printing, hybridization, washing and reporting on a disposable electronic microarray cartridge with 400 features. This assay accurately detected and identified a total of 68 isolates of the seven targeted virus species including 23 samples of FMDV, representing all seven serotypes, and 10 CSFV strains, representing all three genotypes. The assay successfully detected viruses in clinical samples from the field, experimentally infected animals (as early as 1 day post‐infection (dpi) for FMDV and SVDV, 4 dpi for ASFV, 5 dpi for CSFV), as well as in biological material that were spiked with target viruses. The limit of detection was 10 copies/μl for ASFV, PCV2 and PRRSV, 100 copies/μl for SVDV, CSFV, VESV and 1,000 copies/μl for FMDV. The electronic microarray component had reduced analytical sensitivity for several of the target viruses when compared with the multiplex RT‐PCR. The integration of capture probe printing allows custom onsite array printing as needed, while electrophoretically driven hybridization generates results faster than conventional microarrays that rely on passive hybridization. With further refinement, this novel, rapid, highly automated microarray technology has potential applications in multipathogen surveillance of livestock diseases.
Keywords: African swine fever, classical swine fever, foot‐and‐mouth disease, microarray, multiplex PCR, swine vesicular disease
1. INTRODUCTION
The swine industry is a major part of the global livestock industry and an important part of the food supply chain. The global pig industry produced approximately 963 million pigs and 109 million metric tons of pork in 2011 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Infectious diseases such as foot‐and‐mouth disease (FMD), classical swine fever (CSF) and African swine fever (ASF) are highly contagious viral diseases reportable to the OIE (World Organization for Animal Health) that can have a severe economic impact on affected areas due to production losses and the impact on the international trade of animals and animal products with disease‐free countries. For example, the 2001 FMD outbreak in the United Kingdom had an estimated cost of $13 billion US (Thompson et al., 2002). The direct costs of the 1997–1998 CSF epizootic in the Netherlands, excluding loss of exports, amounted to $2 billion US and the slaughter of approximately 10 million pigs (Terpstra & de Smit, 2000).
FMD affects over 70 species of cloven‐hoofed animals including cattle, pigs, sheep, goats and wild ruminants causing vesicular lesions on the mouth and hoof (Hedger, Condy, & Gradwell, 1980). The disease is endemic in many parts of South America, Asia and Africa. The aetiological agent is the FMD virus (FMDV, Picornaviridae, Aphthovirus). Non‐FMD viruses that can cause clinically indistinguishable vesicular lesions in swine include swine vesicular disease virus (SVDV, Picornaviridae, Enterovirus) and vesicular exanthema of swine virus (VESV, Caliciviridae), a virus derived from feeding pigs seal meat contaminated with San Miguel sea lion virus (SMSV; Zimmerman, Karriker, Ramirez, Schwartz, & Stevenson, 2012). CSF virus (CSFV, Flaviviridae) causes haemorrhagic disease in pigs (Giammarioli, Pellegrini, Casciari, & Mia, 2008) and lesions in less severe cases (Zimmerman et al., 2012). African swine fever virus (ASFV, Asfarviridae) also causes haemorrhagic fever (Costard et al., 2009; Giammarioli et al., 2008) that is clinically indistinguishable from infection with CSFV.
For the laboratory diagnosis of vesicular diseases, the OIE currently recommends methods such as virus isolation, antigen capture enzyme‐linked immunosorbent assay (ELISA) and PCR, including real‐time PCR. Virus isolation is often considered the gold standard, but ELISA and real‐time reverse transcriptase (RT)‐PCR are faster and thus more suitable for use as screening tests. Conventional and real‐time RT‐PCR assays have been developed for CSFV (Wernike, Hoffmann, & Beer, 2013), ASFV (Fernández‐Pinero et al., 2013) and FMDV (Hole, Clavijo, & Pineda, 2006; King et al., 2006; Reid et al., 2009) and its differentials that include vesicular stomatitis virus (VSV; Hole et al., 2006; Rasmussen, Uttenthal, & Agüero, 2006), SVDV (Fernández et al., 2008; Núñez et al., 1998), and VESV (Reid et al., 2007). User‐friendly reverse transcription insulated isothermal PCR (RT‐iiPCR) assays performed on compact, field‐deployable instruments with automatic display of “+” or “−” results have also been described for CSFV (Lung et al., 2015) and FMDV (Ambagala et al., 2016). Real‐time multiplex PCR assays have been reported for the detection of FMDV and CSFV (Shi et al., 2016; Wernike et al., 2013) and other targets (Haines, Hofmann, King, Drew, & Crooke, 2013; Li et al., 2013; Rasmussen et al., 2006; Wernike et al., 2012). Although RT‐iiPCR and real‐time RT‐PCR are rapid and highly sensitive, separate single‐target reactions are normally performed by the Canadian Food Inspection Agency's National Centre for Foreign Animal Disease when testing for the presence of foreign animal disease pathogens. Thus, a user‐friendly, multiplex diagnostic test for differential diagnosis of high‐consequence foreign animal diseases could be a useful tool.
In addition to the major vesicular diseases of swine, CSFV and ASFV, two viruses indigenous to North America; porcine circovirus type 2 (PCV2, Circoviridae) and porcine respiratory and reproductive syndrome virus (PRRSV, Arteriviridae), which are responsible for most of the production losses to the North American pig industry (Nicholson et al., 2011; Zimmerman et al., 2012), were also tested with available resources to evaluate the potential of this technology for testing of indigenous diseases. Depending on the strain of the virus and the immune status of the host, there is considerable variation in clinical signs of PRRS. Typically, PRRSV infection causes mild to severe respiratory disease in newborn and growing pigs and reproductive failure in pregnant sows (Lunney et al., 2016) and can also cause apoptosis in organs such as the lungs, testes, lymph nodes and thymus (Karniychuk et al., 2011). PCV2 infection can lead to lymphoid depletion and immunosuppression in pigs and is the primary causative agent of porcine circovirus‐associated disease (PCVAD). Infections can also cause wasting and increased mortality, as well as lesions in the lymphatic area (Meng, 2013).
Microarrays can accommodate multiple probes for each target that can provide redundancy and broader coverage of viral variants when compared with single probe assays. The development and evaluation of traditional microarrays for subtyping and multiplex detection of viruses that affect livestock have been previously described (Banér et al., 2007; Hindson et al., 2008; Jack et al., 2009; Lenhoff et al., 2008; Lung et al., 2011, 2016). The assay described by Banér et al. (2007) detected FMDV, SVDV, VSV, and differentiated between the two serotypes of VSV. Lung et al. (2011) described an assay that detected four vesicular disease viruses (FMDV, VSV, SVDV and VESV) and differentiated between the seven serotypes of FMDV (A, O, C, Asia 1, SAT1, SAT2 and SAT3) and the two serotypes of VSV (New Jersey & Indiana). These methods are laborious to perform, utilize conventional slide microarrays that rely on slower passive hybridization and require multiple pieces of equipment for capture probe printing, hybridization, washing, and slide scanning. In this study, the development and initial validation of an electronic microarray platform that integrates and automates capture probe printing with microarray hybridization, washing, and reporting on a single instrument and allows for onsite updating of probe sequences and custom printing of capture probes by the user for the detection of multiple viruses is described.
2. MATERIALS AND METHODS
2.1. Viruses
Sixty‐eight laboratory and field isolates of the seven targeted swine viruses were used in this study (Tables 1, 5). The strains tested represent viruses from all seven FMDV serotypes, three CSFV genotypes, and both North American (NA) and European (EU) genotypes of PRRSV that were available for testing at the Canadian Food Inspection Agency (CFIA). Strains of FMDV, SVDV, VESV, CSFV and ASFV were propagated and titered according to previously published methods (Moniwa, Clavijo, Li, Collignon, & Kitching, 2007; Senthilkumaran et al., 2016). PRRSV RNA and PCV2 DNA were kindly provided by Dr. Markus Czub (University of Calgary, Calgary, Alberta, Canada). PRRSV‐positive field serum samples were kindly provided by Dr. Davor Ojkic (University of Guelph). A panel of 11 samples from animals negative for the target viruses along with 11 non‐target bacteria and viruses that affect livestock were used to evaluate the specificity of the assay: swine influenza virus (SIV), porcine respiratory coronavirus (PRCV) KIVA, transmissible gastroenteritis virus (TGEV) TC1998, porcine circovirus 1 (PCV1), vesicular stomatitis virus (VSV) 89 GAS, bovine viral diarrhoea virus (BVDV) type 1 Singer, Border disease virus (BDV) CoosBay, Streptococcus suis, Pasteurella multocida, Salmonella enterica choleraesuis, and Mycoplasma hyopneumoniae.
Table 1.
Laboratory samples representing the seven targeted swine viruses used in this study
| Virus | Genotype/Serotype | Strain name | Country of origin |
|---|---|---|---|
| FMDV | A | Iran 1/96 (FMD 1) | Iran |
| ARG 2/2001 (FMD 2) | Argentina | ||
| ARG/87 (FMD 3) | Argentina | ||
| COL/85 (FMD 4) | Columbia | ||
| Iran/99 (FMD 5) | Iran | ||
| Iraq 24/64 (FMD 6) | Iraq | ||
| Cruzeiro/Bra/55 (FMD 7) | Brazil | ||
| O | Manisa (FMD 8) | Turkey | |
| TAW 10/97 (FMD 9) | Taiwan | ||
| UKG 11/2001 (FMD 10) | UK | ||
| BFS/1860 (FMD 11) | UK | ||
| C | Noville (FMD 12) | Switzerland | |
| Resende (FMD 13) | Brazil | ||
| Asia | PAK 1/54 (FMD 14) | Pakistan | |
| Shamir (FMD 15) | Israel | ||
| SAT1 | KEN 4/98 (FMD 16) | Kenya | |
| BOT 1/68 (FMD 17) | Botswana | ||
| SAT2 | ZIM 5/81 (FMD 18) | Zimbabwe | |
| SWA 1/69 (FMD 19) | Swaziland | ||
| SAU 1/2000 (FMD 20) | Saudi Arabia | ||
| ZIM 10/91 (FMD 21) | Zimbabwe | ||
| SAT3 | BEC 1/65 (FMD 22) | Botswana | |
| ZIM 4/81 (FMD 23) | Zimbabwe | ||
| SVDV | GRE 1/79 | Greece | |
| FRA 1/73 | France | ||
| HKN 1/80 | Hong Kong | ||
| HKN 3/89 | Hong Kong | ||
| ITL 1/66 | Italy | ||
| ITL 1/97 | Italy | ||
| JAP 1/74 | Japan | ||
| NET 3/92 | Netherlands | ||
| PORT 1/2003 | Portugal | ||
| SWI 1/74 | Switzerland | ||
| UK 27/72 | UK | ||
| ITL 19/92 | Italy | ||
| CSFV | 1.1 | Alfort/187 | France |
| 1.2 | Brescia | Italy | |
| 1.3 | VRI 4167 | Malaysia | |
| 2.1 | NL B64 | Spain | |
| 2.2 | Vi 3295/4/89 | Germany | |
| 2.3 | Diepholz 1/Han94 | Germany | |
| 3.1 | Congenital Tremor | UK | |
| 3.2 | 3.2a | USA | |
| 3.3 | 3.3a | USA | |
| 3.4 | Kanagawa (Tap 3) | Japan | |
| ASFV | 1 | Lisbon‐61 | Portugal |
| 1 | Lillie | France | |
| 2 | Georgia 2007 | Georgia | |
| VESV | VESV‐CAL | USA | |
| PCV2 | Type 2 | PCV2 B | Canada |
| PRRSV | 1 | PRRS LV | Netherlands |
| 2 | PRRSV 2.5 | USA | |
| 2 | PRRS Vaccine 2.5 | USA | |
| 2 | PRRS 1.4 | USA | |
| 2 | PRRS MLV June 2004 | USA | |
| 2 | PRRSV Vac 2,5 | USA | |
| 2 | PRRSV P‐YNL | USA | |
| 2 | PRRSV 93 44927 | USA |
Synthetic DNA construct.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 5.
Testing of assay with clinical and spiked biological samples
| Virus | Isolate | No. of samples | Sample type | Microarray earliest detection (dpi) | Real‐time PCR earliest detection (dpi) |
|---|---|---|---|---|---|
| FMDV | A IRN 1/2009, O UKG 1/2011, SAT1 ZAM 9/2008 | 14 | Serum | 1 | 2 |
| O UKG 1/2011, SAT1 ZAM 9/2008 | 10 | Nasal | 2 | 1 | |
| O UKG 1/2011 | 6 | Oral | 2 | 1 | |
| SVDV | PORT 1/2003, UK27/72 | 36 | Nasal | 1 | 1 |
| CSFV | Diepholz, Honduras | 36 | Serum | 5 | 5 |
| ASFV | Malawi | 24 | Whole Blood | 4 | 4 |
| VESVa | Cal | 1 | Oral | Detected | N/A |
| 1 | Nasal | Detected | N/A | ||
| PCV2a | Weirrenga | 1 | Oral | Detected | N/A |
| 1 | Nasal | Detected | N/A | ||
| PRRSV | 2 (17‐001377‐0001_1‐57‐1) | 1 | Serum | Detected | N/A |
| 7 (17‐007044‐2208_1‐3‐2) | 1 | Serum | Detected | N/A | |
| 11 (17‐010599‐0006_1‐30‐2) | 1 | Serum | Detected | N/A | |
| 20 (17‐019861‐0006_1‐1‐1) | 1 | Serum | Detected | N/A | |
| 21 (17‐020077‐0002_1‐8‐4) | 1 | Serum | Detected | N/A | |
| 22 (17‐020084‐0001_1‐3‐2) | 1 | Serum | Detected | N/A | |
| 23 (17‐020347‐0011_1‐8‐2) | 1 | Serum | Detected | N/A | |
| 33 (17‐035‐986_1‐4‐1) | 1 | Serum | Detected | N/A | |
| 36 (17‐039858‐0022_2‐5‐2) | 1 | Serum | Detected | N/A | |
| 43 (17‐052259‐0023_1‐57‐1) | 1 | Pooled | Detected | N/A | |
| YNL (NA), LV (EU) | 2a | Oral | Detected | N/A | |
| YNL (NA), LV (EU) | 2a | Nasal | Detected | N/A |
dpi, days post‐infection.
Biological samples spiked with virus.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.2. Samples
The details of the inoculation studies in animals and clinical samples were as previously described (Lung et al., 2011) or will be described elsewhere for other viruses. All animal studies complied with the guidelines of the Canadian Council on Animal Care.
Clinical samples containing VESV and PCV2 were not available, and thus, oral and nasal material obtained from healthy animals (Prairie Swine Centre, Saskatoon, Saskachewan, Canada) was spiked with cell culture‐amplified viruses for these two viruses and PRRSV.
2.3. Primers and probes
PCR primer and capture probe sequences were either identified from the literature, then used with or without modifications or were newly designed (Tables 2 and 3). Primer and probe design were performed as described previously (Lung et al., 2011, 2012) using AlleleID® v. 7.7 (Premier BioSoft International, Palo Alto, CA, USA), Clone Manager v 9.0 software (Scientific and Educational Software, Cary, NC, USA) and BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) using all publicly available sequences from NCBI. Primers were tested in silico using IDT's OligoAnalyzer 3.1 (http://www.idtdna.com/calc/analyzer), and any primers with self‐ and hetero‐dimers with a ΔG ≥ −11 kcal/mole were modified or screened out. The reverse primers were synthesized with the reverse complementary sequence of the NGEN™ Red Universal Reporter Probe (Nexogen, San Diego, CA) at the 5′ end (Lung et al., 2011). The primers were also evaluated in silico as previously described (Lung et al., 2017) against non‐target viruses using NCBI's electronic PCR (e‐PCR, Rotmistrovsky, Jang, & Schuler, 2004; Schuler, 1997) to evaluate potential cross‐reactivities. Furthermore, all unique primer pairs used in the multiplex PCR were tested against all non‐redundant nucleotide sequences of non‐FMDV picornaviruses (n = 61,995 unique sequences). This was achieved using a Python script developed in‐house to generate a multi‐FASTA file with all the unique primer pairs in the multiplex PCR (e.g., the forward primer of FMDV and the reverse primer of ASFV) that target viruses other than the viruses for which the primers were originally designed. The table was used as the input for the e‐PCR program to test the primers in silico. Hits that were returned with less than four mismatches were evaluated with ThermoBLAST's in silico simulations under reaction conditions to predict possible hybridizations based on thermodynamic parameters as well as sequence complementarity (SantaLucia, 2007). Candidate probes were screened for predicted specificity using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Virus‐specific candidate probes that did not show significant matches to the other target viruses (probes exhibiting <25% overall identity and cross‐homologous regions of less than eight nucleotides) were then screened on the NanoChip400 microarray (Nexogen) against a reference panel of viruses. All capture probes were modified with 5′‐biotinylation to allow attachment to the streptavidin‐containing hydrogel of the microarray. All primers and probes were synthesized by IDT (Integrated DNA Technologies, Coralville, IA, USA).
Table 2.
PCR primers used in this study
| Virus | Genomic region | Primer | Sequence (5′‐3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| FMDV | VP3/VP1/ 2A/2B | VP3com980(1) | GCT GAT TAC GCG TAC AC | 971 | This study |
| VP3com980(2) | GCT GAC TAC GCG TAC AC | This study | |||
| VP3com980(3) | GCG GAT TAC GCC TAC AC | This study | |||
| VP3com980(4) | GCG GAT TAC GCG TAC AC | This study | |||
| VP3com980(5) | GCA GAT TAC GCG TAT AC | This study | |||
| VP3com980(6) | GCA GAC TTT GCA TAC AC | This study | |||
| VP3com980(7) | AGT GAC TTC TCC TAC AC | This study | |||
| VP3com980(8) | GCT GAC TAT GCT TAC AC | This study | |||
| VP3com980(9) | GCA GAC TTT GCC TAY AC | This study | |||
| FMDV 2B Rev 4026‐S‐Deg2a | GCG GAC ACC ARC CGG TTR AAG TC | This study | |||
| SVDV | 3C/3D | SVDVCV‐3C‐17a‐F‐(5875bp) | CAG CGG CAC TCC TCA GAC ACT AC | 791 | Lung et al. (2011) |
| SVDVCV‐3D‐3a‐R‐(6642bp)a | GAG TTT CAG GCA CGT AAA CCA CAC | Lung et al. (2011) | |||
| CSFV | E1/E2 | KBH12‐5 E1 Ext FWD | AGR CCA GAC TGG TGG CCN TAY GA | 671 | Paton et al. (2000) |
| KBH12‐6 E2 Ext REVa | TTY ACC ACT TCT GTT CTC A | Paton et al. (2000) | |||
| ASFV | VP72 | King Long ‐ Fwd Primer | ATA GGA TTA AAA CCT ACC TGG AAC ATC TCC G | 537 | King et al. (2003) |
| King Long ‐ Rev Primera | GGT ACT GTA ACG CAG CAC AGC TGA ACC GTT CTG | King et al. (2003) | |||
| VESV | Polymerase | VESVSM‐2‐F‐(5101bp) | CGA CTC GAT GGA CCT GTT CAC ATA CG | 649 | Lung et al. (2011) |
| VESVSM‐5‐R‐(5749bp)a | CGT AGA GGT CGG TTA GGT CCT TTC TG | Lung et al. (2011) | |||
| PCV2 | Capsid | CircoV‐1222F | GTA ATC AAT AGT GGA ATC TAG GAC | 534 | Lung et al. (2017) |
| CircoV ‐1760Ra | TTC GTT TTC AGA TAT GAC GTA TC | Lung et al. (2017) | |||
| PRRSV | Matrix | PRRS‐Mtrx‐ F2 | AAG GTA AGT CGC GGC CGA C | 379 | Lung et al. (2017) |
| PRRS‐Mtrx ‐R2a | TGC CRC CCA ACA CGA GGC | Lung et al. (2017) |
Reverse primers contain a complimentary tag sequence for Red Universal Reporter probe at the 5′ end (Lung et al., 2012).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 3.
Capture probes used in this study
| Virus/Genomic region | Probe name | Sequence (5′ biotinylationa ‐ 3′) | Reference |
|---|---|---|---|
| FMDV ‐ VP3/VP1/2A/2B | FMD Common A | AAG TTG GCN GGA GAC GTB GAG TCC AAC CC | This study |
| FMD Common B | AAC TTY GAC CTG TTA AAG TTG GCB GGA GAC GTT GAG TC | This study | |
| FMD Common C | AAC TTC GAC CTG TTA AAG TTG GCY GGA GAC GTT GAG TCC AAC CCT | This study | |
| FMD Common D | GAG TCC AAC CCT GGG CCY TTC TTC TTC | This study | |
| FMD Common E | GAG AYG TBG AGT CCA ACC CTG GGC CYT T | This study | |
| SVDV ‐ 3C‐3D | SVDV‐020‐deg | AAG AGA CAT YCT ATC CAA GAA GAC CAG AGA CCT TRC CA | Lung et al. (2011) |
| SVDV‐014a | GGG TAG CGC CGT TGG GTG TGA CC | Lung et al. (2011) | |
| SVDV‐008‐deg | GTG GCY YTG GGT ATC AAG AAA AGA GAC AT | Lung et al. (2011) | |
| SVDV‐013 | GCA ATG AGG CAG ACA TTT GGA AAC CTA TA | Lung et al. (2011) | |
| SVDV‐016 | AAA GAG ACA TCC TAT CCA AGA AGA CCA GAG ACC T | Lung et al. (2011) | |
| SVDV‐010‐deg | TAT GGT CTA AAC YTR CCA ATG GTA ACC TA | Lung et al. (2011) | |
| SVDV‐019‐deg | ACA ACT AGC CAC ACT RGA CAT YAG CAC KGA RC | Lung et al. (2011) | |
| CSFV ‐E1/E2 | CSFV Common 1 | CTT AAK GTG GTY AGT AGG AGG TAY | This study |
| CSFV Common 4a | CTG RAY GAC GGR ACY GTY AR | This study | |
| bioKBH12‐8 E2 Int FWD | TCR WCA ACC AAY GAG ATA GGG | Paton et al. (2000) | |
| bioKBH12‐9 E2 Int REV | GAT GAC TTY GGR TTY GGR CTG TG | Paton et al. (2000) | |
| ASFV ‐ VP72 | bioASFV‐VP72‐1668 | CTG CTC ATG GTA TCA ATC TTA TCG A | King et al. (2003) |
| bioASFV‐VP72‐1898 | ACG GCY GAT CTT GTG GTA TC | King et al. (2003) | |
| VESV ‐ Polymerase | VESV‐SM‐010‐deg | CCA CYA TGG CTA CTA CTC AYA CGC TTC TGT CGT TTG AC | Lung et al. (2011) |
| VESV‐SM‐003a‐deg | CGG ATG CTG ARA TAA CGC CTA TCC C | Lung et al. (2011) | |
| PCV2 ‐ Capsid | CircoV‐1576 | ATA TCC GAA GGT GCG GGA T | Lung et al. (2017) |
| CircoV‐1657 | GAC GAG CCA GGG GCG GCG GC | Lung et al. (2017) | |
| PRRSV ‐ Matrix | PRRS 322‐346 COM | TAC ATT CTG GCC CCT GCC CAT CAC G | Lung et al. (2017) |
| PRRSV‐M‐378 | GGC AAA TGA TAA CCA CGC ATT TG | Lung et al. (2017) | |
| PRRSV‐M‐361 | GGC TTT CAT CCG ATT GCG GCA AAT G | Lung et al. (2017) | |
| PRRS 233‐252 EU | TTG TCA CCC TTC TGT GGG GC | Lung et al. (2017) | |
| PRRS 380‐402 EU | CGT CTG GTA ACC GAG CAT ACG CT | Lung et al. (2017) | |
| Non‐Specific Binding | NSBP | CAA AGT GGG AGA CGT CGT TG | Hindson et al. (2008) |
Probes modified with 5′ biotinylation for binding to streptavidin pad on NanoChip 400 microarray (Takahashi, Norman, Mather, & Patterson, 2008).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.4. Nucleic acid extraction
Nucleic acid was extracted from vesicular viruses (FMDV, SVDV and VESV) and clinical samples as described previously (Lung et al., 2011), while all other viruses were extracted using the QIAamp Viral RNA Kit (Deregt et al., 2006) or the Ambion Mag Max Kit (Thermofisher Scientific‐ Ambion, Burlington, ON, Canada) according to manufacturer's specification. Nucleic acid was eluted in 50 μl QIAamp or 90 μl MagMax elution buffer. For PCV2 and PRRSV, 14 μl of the neat laboratory‐propagated virus was spiked into 126 μl of biological material and extracted with the QIAmp viral RNA Kit (Qiagen, Toronto, ON, Canada). For VESV, 20 μl of virus culture was spiked into 120 μl of clinical material and extracted with the QIAamp viral RNA kit. All extracted spiked samples were tested for the presence of target viruses with the seven‐plex assay.
2.5. Multiplex reverse transcription PCR
A multiplex RT‐PCR consisting of 22 primers was developed to amplify genomic targets of seven swine viruses (FMDV, SVDV, VESV, CSFV, ASFV, PRRSV and PCV2) in a single reaction. The RT‐PCR was performed using the SuperScript® III (SSIII) One‐Step RT‐PCR with Platinum Taq kit (Life Technologies, Burlington, ON, Canada) in a 50‐μl reaction. The RT‐PCR consisted of FMDV, CSFV and PCV2 primers at 1 μM final concentration each, SVDV, VESV, ASFV and PRRSV primers at 0.5 μM final concentration each, 25 μl of 2 × RT buffer mix, 2 μl of SSIII enzyme mix, 15 μl of ultrapure water and 1 μl of sample. The RT‐PCRs were run on a Veriti thermal cycler (Life Technologies‐Applied Biosystems, Burlington, ON, Canada) in Lethbridge during initial assay development and switched to 9700 thermal cycler (Life Technologies) at the National Centre for Foreign Animal Disease. The cycling parameters were as follows: 55°C for 15 min for reverse transcription (RT), followed by 94°C for 2 min and then 35 cycles of: 94°C for 30 s, 50°C for 1 min and 68°C for 1 min. This was followed by a final 5‐min extension at 68°C. Post‐amplification reactions were visualized using either the QIAxcel (QIAGEN) instrument or agarose gel electrophoresis with SYBR® safe DNA gel stain (Life Technologies).
2.6. Electronic microarray
All buffers used in the computer‐controlled automated electronic microarray assay were from Nexogen, Inc. or prepared in‐house from chemicals purchased from Sigma‐Aldrich (Oakville, ON, Canada). Biotinylated capture probes (250 nM) were prepared in 50 mM L‐histidine/0.05% Proclin® 300 buffer (His/Proclin® buffer, Nexogen Inc.) and electronically addressed to selected electrodes on the NanoChip400 cartridge through the application of a positive 350 nA current for 30 s. After five washes of the cartridge with a mixture of His/Proclin® buffer containing 20% Triton X‐100, and water with 0.05% Proclin® solution, 8.75 μl of unpurified PCR amplicons, diluted 1 of 8 in 61.25 μl Cap‐Down A Buffer were electronically addressed at 800 nA for 60 s to selected electrode pads containing bound capture probes. The cartridge was washed five more times with His/Proclin® buffer, incubated at 40°C for 60 s and washed once with high salt buffer (HSB, Nexogen Inc.). Hybridized amplicons were detected using a fluoresceinated reporter probe resuspended in HSB at 1 μM final concentration and a touchdown passive reporting protocol as described previously (Lung et al., 2012). All electronic microarray hybridizations were performed in duplicate, and a non‐template PCR control (NTC) was included in all experiments. Raw fluorescent intensity (FI) data from all utilized pads on each cartridge were obtained and analysed using Microsoft Excel. Positive‐to‐negative (P/N) ratios were calculated by dividing the averaged FI value for each amplicon by the FI value produced by the average NTC for each probe. Samples that produced P/N ratios greater than an empirically determined cut‐off of 2 for both replicates of one or more virus‐specific capture probe were considered positive. P/N data derived from multiple microarrays were visualized through a heat map generated using TreeView (Eisen, Spellman, Brown, & Botstein, 1998).
2.7. Analytical sensitivity
RNA transcribed from a plasmid (CSFV, PRRSV, SVDV, VESV) or PCR amplicon template (FMDV) was used to determine the limit of detection of the RT‐PCR and the electronic microarray for the RNA viruses. In vitro transcribed RNA was generated from plasmid templates containing the targeted genomic regions of the swine viruses, blunt ligated into either the pJET1.2 vector (ThermoScientific‐Fermentas, Ottawa, ON) or the pCR™II‐blunt‐TOPO® vector (Life Technologies ‐ Invitrogen, Burlington, ON, Canada). Quantified plasmids were restriction digested with HindIII (10 μ/μl) or other appropriate restriction enzymes when HindIII sites were found within the amplicon (Thermo Scientific‐Fermentas, Ottawa, ON), purified using the Zymo DNA Clean & Concentrator™‐25 column kit (Zymo Research, Irvine, CA) and transcribed using the MEGAscript® kit (Life Technologies). For FMDV, the desired gene regions were synthesized by IDT, cloned into the pGEM‐3Zf(+) vector, digested with EcoRI (Life Technologies, Thermo Scientific‐Fermentas, Ottawa, ON), amplified under the same cycling conditions by the described RT‐PCR in single‐plex and transcribed as described previously (Lung et al., 2015). RNA was quantified using the Qubit® 2.0 Fluorometer and RNA broad range assay kit (Life Technologies). For the DNA viruses ASFV and PCV2, plasmids containing the targeted genes were extracted from overnight cultures of Escherichia coli transformed with the appropriate plasmid (Sambrook & Russell, 2001). Copy number was calculated using the following equation:
After quantification, in vitro transcribed RNA and plasmid DNA were diluted serially 10‐fold from neat stock to 10−12 in ultrapure dH2O. The dilution series were amplified using the standard seven‐plex multiplex, a five‐plex exotic virus multiplex (all viruses except PRRSV and PCV2), two‐plex indigenous virus multiplex (PRRSV and PCV2) and single‐plex RT‐PCRs and imaged using the QIAxcel Advanced instrument (QIAGEN). RT‐PCR endpoints were determined by the last reaction in the dilution series to give a visually detectable band. In duplicate, a panel of six samples from each dilution series, two samples above the RT‐PCR endpoint, one sample at the endpoint and three samples below the endpoint, were selected and ran on the electronic microarray with a positive control and NTC (n = 8). The eight samples were run against the specific target probes (i.e., FMDV samples run on five FMDV probes) as well as the non‐specific binding probe (NSBP) as a negative control. A P/N ratio cut‐off of 2 for positivity was used to determine the analytical sensitivity on the electronic microarray. P/N ratios ≥1.7 and ≤2 were designated as suspect reactors.
3. RESULTS
3.1. RT‐PCR amplification
The multiplex RT‐PCR utilized 22 primers to amplify selected genomic regions of the seven target viruses (Table 2). The RT‐PCR generated amplicons of the expected size for all 68 isolates of the seven target viruses (23 FMDV, 12 SVDV, 10 CSFV, one VESV, three ASFV, one PCV2 and 18 PRRSV; Figures 1a and 3a). The specificity of the amplification was evaluated using a total of 22 samples, including 11 oral clinical materials from healthy pigs, and 11 non‐target swine bacteria and viruses associated with livestock. Nucleic acid extracted from non‐target pathogens and clinical material from healthy animals either did not generate detectable RT‐PCR products or generated weak, non‐specific products in the absence of templates from target viruses (Figure 1b). The non‐specific amplifications were reduced substantially when the same samples were spiked with PRRSV RNA as an exogenous control (Figure 1c).
Figure 1.
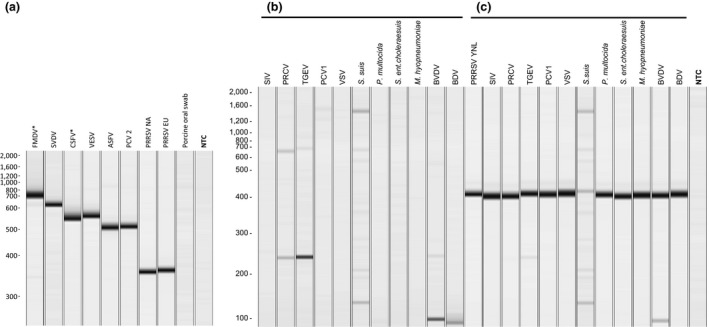
Amplification of target and non‐targets using the seven‐plex RT‐PCR. (a) QIAxcel gel image of representative amplified products after RT‐PCR amplification of representative strains of the seven targeted swine viruses and oral swab from healthy pigs. Asterisk denotes in vitro transcribed RNA was used as a template. (b) QIAxcel gel image of amplified products from amplification of 11 non‐target virus and bacteria that affect livestock: swine influenza virus (SIV), porcine respiratory coronavirus (PRCV) KIVA, transmissible gastroenteritis virus (TGEV) TC1998, porcine circovirus 1 (PCV1), vesicular stomatitis virus (VSV) 89 GAS, Streptococcus suis, Pasteurella multocida, Salmonella enterica choleraesuis, Mycoplasma hyopneumoniae, bovine viral diarrhoea virus (BVDV) type 1 Singer, and Border disease virus (BDV) CoosBay. (c) Amplification of nucleic acid from non‐target bacteria and viruses spiked with PRRSV YNL RNA as an exogenous control. NTC: no template control
Figure 3.
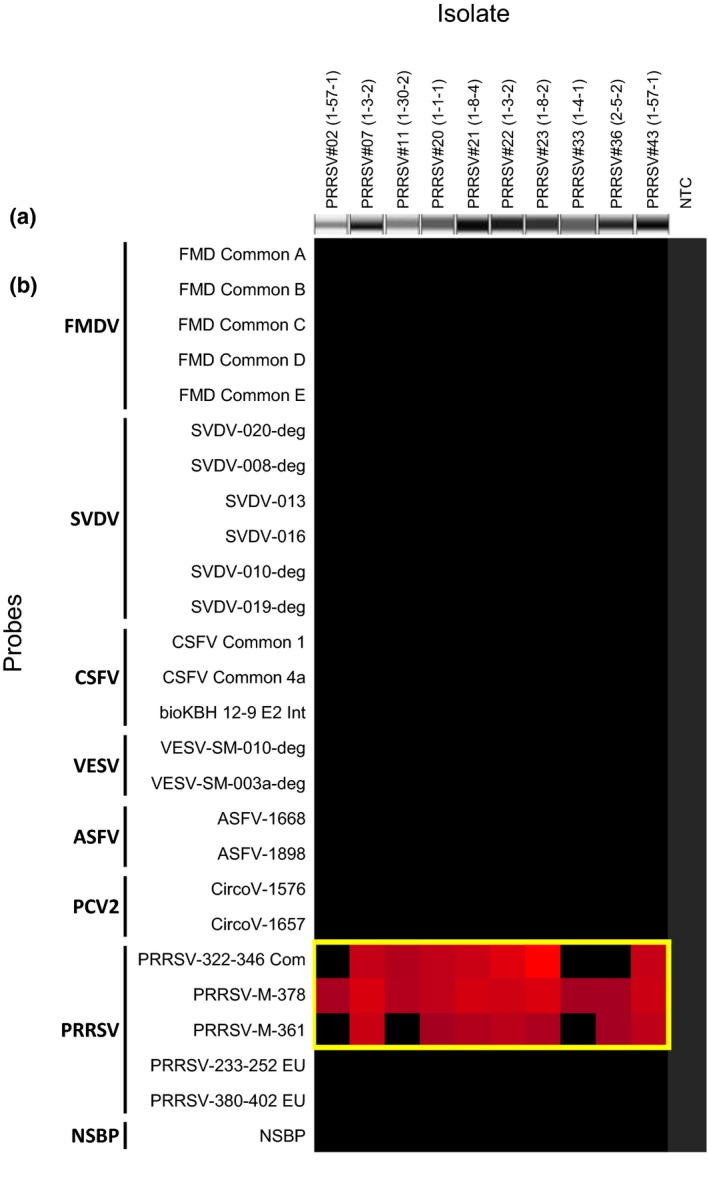
Multiplex RT‐PCR amplicons visualized using QIAxcel System (a) and heat map depicting the reactivity of 10 genetically diverse Canadian PRRSV‐positive serum samples from diagnostic submissions against 27 virus‐specific capture probes and one non‐specific binding probe (NSBP) (b). The panel of samples includes the following: #2 (17‐001377‐0001_1‐57‐1), #7 (17‐007044‐2208_1‐3‐2), #11 (17‐010599‐0006_1‐30‐2), #20 (17‐019861‐0006_1‐1‐1), #21 (17‐020077‐0002_1‐8‐4), #22 (17‐020084‐0001_1‐3‐2),‐#23 (17‐020347‐0011_1‐8‐2), #33 (17‐035‐986_1‐4‐1), #36 (17‐039858‐0022_2‐5‐2), #43 (17‐052259‐0023_1‐57‐1). A positive signal in red represents a positive‐to‐negative ratio (P/N) of >2, while negative results in black represent any P/N ≤ 2
3.2. Microarray for detection and differentiation of seven swine viruses
A total of 27 candidate capture probes for the seven targeted viruses and 1 non‐specific capture probe were tested on the electronic microarray with amplicons derived from the panel of 68 target viruses and 22 clinical negatives, non‐target bacteria and viruses. All amplified target virus samples reacted to their target‐specific capture probes. No cross‐reactivity with materials from healthy swine, non‐target bacteria or viruses was observed (Figure 2b).
Figure 2.
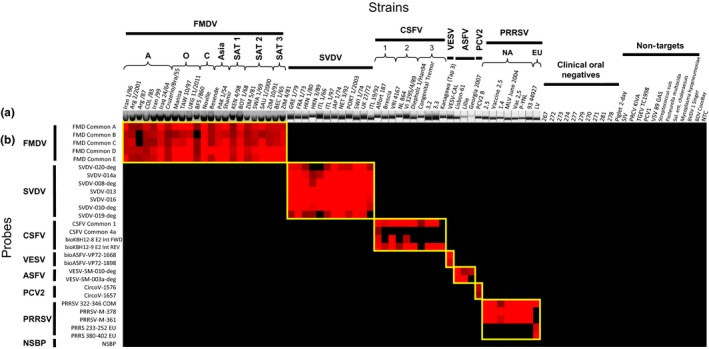
Multiplex RT‐PCR amplicons visualized using agarose gel and QIAxcel System (a) and heat map depicting the reactivity of samples against 27 virus‐specific capture probes and one non‐specific binding probe (NSBP) (b). The panel of samples includes 58 strains of the seven targeted swine viruses, 11 oral swab samples: 267, 270, 271, 272, 273, 274, 277, 278, 279, 281, pooled 2‐day piglet, taken from healthy pigs and 11 non‐target virus and bacteria that are associated with livestock: swine influenza virus (SIV), porcine respiratory coronavirus (PRCV) KIVA, transmissible gastroenteritis virus (TGEV) TC1998, porcine circovirus 1 (PCV1), vesicular stomatitis virus (VSV) 89 GAS, Streptococcus suis, Pasteurella multocida, Salmonella enterica choleraesuis, Mycoplasma hyopneumoniae, bovine viral diarrhoea virus (BVDV) type 1 Singer, and Border disease virus (BDV) CoosBay. The viral strains tested include representatives of each of the seven FMDV serotypes, three CSFV genotypes, two ASFV genotypes and genotypes NA and EU of PRRSV. A positive signal in red represents a positive‐to‐negative ratio (P/N) of >2, while negative results in black represent any P/N ≤ 2
3.3. Limits of detection
Nucleic acid from 10‐fold serial dilutions of transcribed RNA of known copy number were amplified by single‐plex RT‐PCR, a duplex RT‐PCR consisting of primers for PRRSV and PCV2, a five‐plex RT‐PCR for viruses exotic to Canada (FMDV, SVDV, VESV, CSFV and ASFV) or the seven‐plex RT‐PCR. The RT‐PCR amplicons were then tested on the microarray, to determine the limit of detection of the RT‐PCRs and microarray for each virus. The assay showed similar efficiencies for the single‐plex, duplex and five‐plex RT‐PCRs and microarray for SVDV, VESV, CSFV, ASFV, PCV2 and PRRSV, but the PCR had higher sensitivity for the smaller amplicons: ASFV, PCV2 and PRRSV (Table 4). The seven‐plex RT‐PCR showed the best analytical sensitivity with ASFV, PCV2 and PRRSV at a range between 1 copy in PCR and 10 copies on the electronic microarray, followed by SVDV, CSFV and VESV at around 10 and 100 copies in PCR and microarray, respectively. The detection limit was lowest for FMDV at hundreds of copies for both seven‐plex and five‐plex PCRs, and one (single‐plex, five‐plex) or two (seven‐plex) orders of magnitude lower on the microarray (Table 4).
Table 4.
Analytical sensitivity of RT‐PCR and electronic microarray assay
| Virus | Copy number | Assay detection | |||||||
|---|---|---|---|---|---|---|---|---|---|
| RT‐PCR | Microarray | ||||||||
| Single‐plex | 7‐plex | 5‐plex | 2‐plex | Single‐plex | 7‐plex | 5‐plex | 2‐plex | ||
| FMDV Asia 1 PAK | 47204 | + | + | + | + | + | + | ||
| 4720 | + | + | + | + | − | + | |||
| 472 | + | + | + | − | − | − | |||
| 47 | S | − | S | − | − | − | |||
| 4 | − | − | − | − | − | − | |||
| SVDV ITL 19/99 | 2333 | + | + | + | + | + | + | ||
| 233 | + | + | + | + | − | + | |||
| 23 | + | S | + | − | − | − | |||
| 2 | S | − | S | − | − | − | |||
| 0.2 | − | − | − | − | − | − | |||
| CSFV Alfort/187 | 7031 | + | + | + | + | + | + | ||
| 703 | + | + | + | + | − | + | |||
| 70 | + | − | + | − | − | − | |||
| 7 | S | − | S | − | − | − | |||
| 0.7 | − | − | − | − | − | − | |||
| VESV‐Cal | 1305 | + | + | + | + | + | + | ||
| 130 | + | + | + | + | − | + | |||
| 13 | + | + | + | − | − | − | |||
| 1 | − | − | − | − | − | − | |||
| ASFV Lisbon 61a | 300 | + | + | + | + | + | + | ||
| 30 | + | + | + | + | + | + | |||
| 3 | + | + | + | − | − | − | |||
| 0.3 | S | − | S | − | − | − | |||
| 0.03 | − | − | − | − | − | − | |||
| PCV2 Ba | 800 | + | + | + | + | + | + | ||
| 80 | + | + | + | + | + | + | |||
| 8 | + | + | + | − | − | − | |||
| 0.8 | S | − | S | − | − | − | |||
| 0.08 | − | − | − | − | − | − | |||
| PRRSV LV | 222 | + | + | + | + | + | + | ||
| 22 | + | + | + | + | S | + | |||
| 2 | + | + | + | − | − | − | |||
| 0.2 | S | − | S | − | − | − | |||
| 0.02 | − | − | − | − | − | − | |||
S, suspect reactor.
Plasmid DNA.
Dilutions that gave positive signals are shaded grey.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.4. Testing of clinical and spiked samples
Extracted nucleic acid from clinical samples was amplified using the seven‐plex multiplex assay. FMDV nucleic acid was detected in serum samples as early as 1 day post‐infection (dpi). All available targets were detected from nasal, oral, serum or whole blood with detection ranging from 1 dpi for SVDV, 4 dpi for ASFV and 5 dpi for CSFV (Table 5). PRRSV was detected in positive field serum samples from swine (n = 10, Table 5, Figure 3). All samples from oral and/or nasal swab material that were spiked with VESV, PRRSV and PCV2 (n = 8) produced amplicons of the expected size after the multiplex PCR and the viruses were accurately detected by the microarray (data not shown).
In addition to samples taken prior to experimental infection at 0 dpi, nucleic acid extracted from oral swabs from 11 healthy swine did not produce any amplicons or produced weak detectable bands on agarose gels after the multiplex RT‐PCR and no reactivity above background was observed subsequently on the microarray, demonstrating 100% specificity for these samples (Figure 2b).
4. DISCUSSION
This study describes the development and initial laboratory evaluation of a novel user‐friendly microarray primarily designed for the detection and differentiation of FMDV, SVDV, VESV, CSFV, and ASFV that are exotic to North America. Due to the ability of the electronic microarray assay to print capture probes on site, RT‐PCR primers and probes for PRRSV and PCV2 were tested with available samples to evaluate the potential use of electronic microarray assays for detection of indigenous diseases. The seven‐plex RT‐PCR and electronic microarray assay successfully amplified and accurately detected laboratory amplified (n = 58), experimentally inoculated (spiked, n = 8), and PRRSV‐positive field samples (n = 10). All capture probes were highly specific and showed no cross‐reactivity with heterologous viruses, non‐target bacteria and viruses, or samples from healthy animals. PRRSV was originally detected in North America and Europe almost simultaneously (Christianson & Joo, 1994; Lunney et al., 2016). The two genotypes were genetically diverse with about 55%–70% nucleotide identity (Lunney et al., 2016). Due to the high genetic diversity of RNA viruses and the lack of more PRRSV (especially genotype 2) isolates available for testing, regular evaluation of the primers and probe sequences against newly available sequences and additional validation will be needed. The large number of capture probes (up to 400 for the electronic microarray used here) should allow better coverage of genetically diverse strains compared with real‐time RT‐PCRs, which can typically accommodate only a few probes. The ability to print custom capture probes on site without additional instrumentation allows the assay to be updated conveniently.
Conventional testing methods, such as virus isolation, ELISA and PCR, have been advocated by the OIE as gold standard tests for the detection of many viral infections. Although these methods can be highly sensitive to specific viral serotypes, methods like virus isolation are time‐consuming, requiring multiple blind passages of a sample to generate usable amounts of infectious material. Additionally, multiple tests are required to detect coinfections or to differentiate between species with clinically indistinguishable signs. Although multiplex real‐time RT‐PCRs have been described for many swine pathogens (Haines et al., 2013; Li et al., 2013; Rasmussen et al., 2006; Shi et al., 2016; Wernike et al., 2012, 2013), the diagnosis of FMDV, SVDV, CSFV and ASFV still requires the performance of multiple single tests. Automated multiplex assays can potentially reduce labour cost and handling time. The highest analytical sensitivities observed for the 7‐plex RT‐PCR were as follows: PRRSV, PCV2 and ASFV which have the smallest amplicons (379–537 bp). Although the microarray assay successfully detected FMDV in dpi 1 samples from experimentally infected animals, the multiplex RT‐PCR was less efficient for amplification of FMDV, which had the largest amplicon size in the assay (971 bp). This is most likely due to reduced PCR efficiencies for large amplicons (Cheng, Fockler, Barnes, & Higuchi, 1994; Huang, Arnheim, & Goodman, 1992). The FMDV primers used in this study were specifically designed to amplify the highly variable VP1 capsid protein coding region to allow subtyping into the seven known FMDV serotypes either in the same assay or in a separate assay (unpublished results) or by sequencing. The choice of RT‐PCR primers that generates a smaller FMDV amplicon could (e.g., Moniwa et al., 2007) potentially further increase the detection limit of the assay for FMDV. With 22 primers in the described multiplex RT‐PCR, there may also be interactions between the different primers that reduce the efficiency of the amplification. When the PCR primers for PRRSV and PCV2 were removed and placed in a separate RT‐PCR, an increase in sensitivity was observed for FMDV, SVDV, VESV and CSFV. The electronic microarray component of the assay showed a reduction in sensitivity for some target pathogens both in this and other studies (Lung et al., 2016, 2017 and unpublished results) when compared with the RT‐PCR component of the assay. Possible explanations for this reduction in sensitivity at the microarray stage could be the formation of secondary structure of certain capture probes, the amplicon or the annealing of the two strands of the amplicons which could prevent efficient hybridization between the capture probes and the amplicon under the experimental conditions. The use of asymmetric PCR to generate single‐stranded products, increased amount of capture probe and/or amplicon may also further increase the sensitivity of the assay. Although not tested in this study due to resource constraints, previously published electronic microarray assays for detection of avian, swine and bovine viruses (Lung et al., 2012, 2016, 2017), were able to detect the presence of more than one target. Due to potentially high loads of viruses such as PCV2 in many countries, the splitting of the seven‐plex RT‐PCR into a duplex RT‐PCR for PRRSV and PCV2 and a five‐plex RT‐PCR for the other viruses, as described in this study, will improve the sensitivity of the assay for the detection of reportable diseases.
Although an RT step was not required for DNA viruses, previous studies have shown better amplification of the DNA targets when a RT step was included (Lung et al., 2017). This could be due to the utilization of messenger RNAs as well as genomic DNA as template (Lung et al., 2017). Further improvements in assay sensitivity and integration of the PCR with the electronic microarray in the new NanoChip400 XL electronic microarray platform could make this novel automated microarray technology a useful tool for multipathogen detection and subtyping.
ACKNOWLEDGEMENTS
This work was funded by Defence Research and Development Canada, Centre for Security Science's Chemical, Biological, Radiological‐Nuclear, and Explosives Research and Technology Initiative (CRTI) project 09‐403TA. The authors acknowledge Dr. Noriko Goji, Mr. Sandor Dudas, and two anonymous reviewers for suggestions regarding the manuscript.
Erickson A, Fisher M, Furukawa‐Stoffer T, et al. A multiplex reverse transcription PCR and automated electronic microarray assay for detection and differentiation of seven viruses affecting swine. Transbound Emerg Dis. 2018;65:e272–e283. 10.1111/tbed.12749
Reproduced with the permission of the Minister of Health, Canadian Food Inspection Agency
REFERENCES
- Ambagala, A. , Fisher, M. , Goolia, M. , Nfon, C. , Furukawa‐Stoffer, T. , Ortega Polo, R. , & Lung, O. (2016). Field‐deployable reverse transcription‐insulated isothermal PCR (RT‐iiPCR) assay for rapid and sensitive detection of foot‐and‐mouth disease virus. Transboundary and Emerging Diseases, 64, 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banér, J. , Gyarmati, P. , Yacoub, A. , Hakhverdyan, M. , Stenberg, J. , Ericsson, O. , … Belák, S. (2007). Microarray‐based molecular detection of foot‐and‐mouth disease, vesicular stomatitis and swine vesicular disease viruses, using padlock probes. Journal of Virological Methods, 143, 200–206. [DOI] [PubMed] [Google Scholar]
- Cheng, S. , Fockler, C. , Barnes, W. M. , & Higuchi, R. (1994). Effective amplification of long targets from cloned inserts and human genomic DNA. Proceedings of the National Academy of Sciences of the USA, 91, 5695–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, W. T. , & Joo, H. (1994). Porcine reproductive and respiratory syndrome: A review. Swine Health and Production, 2, 10–28. [Google Scholar]
- Costard, S. , Wieland, B. , de Glanville, W. , Jori, F. , Rowlands, R. , Vosloo, W. , … Dixon, L. K. (2009). African swine fever: How can global spread be prevented? Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 364, 2683–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deregt, D. , Gilbert, S. A. , Dudas, S. , Pasick, J. , Baxi, S. , Burton, K. M. , & Baxi, M. K. (2006). A multiplex DNA suspension microarray for simultaneous detection and differentiation of classical swine fever virus and other pestiviruses. Journal of Virological Methods, 136, 17–23. [DOI] [PubMed] [Google Scholar]
- Eisen, M. B. , Spellman, P. T. , Brown, P. O. , & Botstein, D. (1998). Cluster analysis and display of genome‐wide expression patterns. Proceedings of the National Academy of Sciences of the USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, J. , Agüero, M. , Romero, L. , Sánchez, C. , Belák, S. , Arias, M. , & Sánchez‐Vizcaíno, J. M. (2008). Rapid and differential diagnosis of foot‐and‐mouth disease, swine vesicular disease, and vesicular stomatitis by a new multiplex RT‐PCR assay. Journal of Virological Methods, 147, 301–311. [DOI] [PubMed] [Google Scholar]
- Fernández‐Pinero, J. , Gallardo, C. , Elizalde, M. , Robles, A. , Gómez, C. , Bishop, R. , … Arias, M. (2013). Molecular diagnosis of African swine fever by a new real‐time PCR using universal probe library. Transboundary and Emerging Diseases, 60, 48–58. [DOI] [PubMed] [Google Scholar]
- Giammarioli, M. , Pellegrini, C. , Casciari, C. , & Mia, G. (2008). Development of a novel hot‐start multiplex PCR for simultaneous detection of classical swine fever virus, African swine fever virus, porcine circovirus type 2, porcine reproductive and respiratory syndrome virus and porcine parvovirus. Veterinary Research Communications, 32, 255–262. [DOI] [PubMed] [Google Scholar]
- Haines, F. J. , Hofmann, M. A. , King, D. P. , Drew, T. W. , & Crooke, H. R. (2013). Development and validation of a multiplex, real‐time RT PCR assay for the simultaneous detection of classical and African swine fever viruses. PLoS One, 8, e71019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedger, R. S. , Condy, J. B. , & Gradwell, D. V. (1980). The response of some African wildlife species to foot‐and‐mouth disease vaccination. Journal of Wildlife Diseases, 16, 431–438. [DOI] [PubMed] [Google Scholar]
- Hindson, B. J. , Reid, S. M. , Baker, B. R. , Ebert, K. , Ferris, N. P. , Tammero, L. F. B. , … King, D. P. (2008). Diagnostic evaluation of multiplexed reverse transcription‐PCR microsphere array assay for detection of foot‐and‐mouth and look‐alike disease viruses. Journal of Clinical Microbiology, 46, 1081–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hole, K. , Clavijo, A. , & Pineda, L. A. (2006). Detection and serotype‐specific differentiation of vesicular stomatitis virus using a multiplex, real‐time, reverse transcription‐polymerase chain reaction assay. Journal of Veterinary Diagnostic Investigation, 18, 139–146. [DOI] [PubMed] [Google Scholar]
- Huang, M. M. , Arnheim, N. , & Goodman, M. F. (1992). Extension of base mispairs by Taq DNA polymerase: Implications for single nucleotide discrimination in PCR. Nucleic Acids Research, 20, 4567–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, P. J. M. , Amos‐Ritchie, R. N. , Reverter, A. , Palacios, G. , Quan, P.‐L. , Jabado, O. , … Boyle, D. B. (2009). Microarray‐based detection of viruses causing vesicular or vesicular‐like lesions in livestock animals. Veterinary Microbiology, 133, 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniychuk, U. U. , Saha, D. , Geldhof, M. , Vanhee, M. , Cornillie, P. , Van den Broeck, W. , & Nauwynck, H. J. (2011). Porcine reproductive and respiratory syndrome virus (PRRSV) causes apoptosis during its replication in fetal implantation sites. Microbial Pathogenesis, 51, 194–202. [DOI] [PubMed] [Google Scholar]
- King, D. P. , Ferris, N. P. , Shaw, A. E. , Reid, S. M. , Hutchings, G. H. , Giuffre, A. C. , … Beckham, T. R. (2006). Detection of foot‐and‐mouth disease virus: Comparative diagnostic sensitivity of two independent real‐time reverse transcription‐polymerase chain reaction assays. Journal of Veterinary Diagnostic Investigation: Official Publication of the American Association of Veterinary Laboratory Diagnosticians, Inc, 18, 93–97. [DOI] [PubMed] [Google Scholar]
- King, D. P. , Reid, S. M. , Hutchings, G. H. , Grierson, S. S. , Wilkinson, P. J. , Dixon, L. K. , … Drew, T. W. (2003). Development of a TaqMan® PCR assay with internal amplification control for the detection of African swine fever virus. Journal of Virological Methods, 107, 53–61. [DOI] [PubMed] [Google Scholar]
- Lenhoff, R. J. , Naraghi‐Arani, P. , Thissen, J. B. , Olivas, J. , Carillo, A. C. , Chinn, C. , … McBride, M. T. (2008). Multiplexed molecular assay for rapid exclusion of foot‐and‐mouth disease. Journal of Virological Methods, 153, 61–69. [DOI] [PubMed] [Google Scholar]
- Li, J. , Shi, J. , Wu, X. , Cong, X. , Xu, S. , Yuan, X. , … Huang, B. H. (2013). Differentiation of PCV1 and PCV2 by a multiplex real‐time PCR assay. The Veterinary Record, 173, 346. [DOI] [PubMed] [Google Scholar]
- Lung, O. , Beeston, A. , Ohene‐Adjei, S. , Pasick, J. , Hodko, D. , Hughes, K. B. , … Deregt, D. (2012). Electronic microarray assays for avian influenza and Newcastle disease virus. Journal of Virological Methods, 185, 244–253. [DOI] [PubMed] [Google Scholar]
- Lung, O. , Fisher, M. , Beeston, A. , Hughes, K. B. , Clavijo, A. , Goolia, M. , … Deregt, D. (2011). Multiplex RT‐PCR detection and microarray typing of vesicular disease viruses. Journal of Virological Methods, 175, 236–245. [DOI] [PubMed] [Google Scholar]
- Lung, O. , Furukawa‐Stoffer, T. , Burton Hughes, K. , Pasick, J. , King, D. P. , & Hodko, D. (2016). Multiplex RT‐PCR and automated microarray for detection of eight bovine viruses. Transboundary and Emerging Diseases, 10.1111/tbed.12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, O. , Ohene‐Adjei, S. , Buchanan, C. , Joseph, T. , King, R. , Erickson, A. , … Ambagala, A. (2017). Multiplex PCR and microarray for detection of swine respiratory pathogens. Transboundary and Emerging Diseases, 64, 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, O. , Pasick, J. , Fisher, M. , Buchanan, C. , Erickson, A. , & Ambagala, A. (2015). Insulated isothermal reverse transcriptase PCR (iiRT‐PCR) for rapid and sensitive detection of classical swine fever virus. Transboundary and Emerging Diseases, 63, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney, J. K. , Fang, Y. , Ladinig, A. , Chen, N. , Li, Y. , Rowland, B. , & Renukaradhya, G. J. (2016). Porcine reproductive and respiratory syndrome virus (PRRSV): Pathogenesis and interaction with the immune system. Annual Review of Animal Biosciences, 4, 129–154. [DOI] [PubMed] [Google Scholar]
- Meng, X.‐J. (2013). Porcine circovirus type 2 (PCV2): Pathogenesis and interaction with the immune system. Annual Review of Animal Biosciences, 1, 43–64. [DOI] [PubMed] [Google Scholar]
- Moniwa, M. , Clavijo, A. , Li, M. , Collignon, B. , & Kitching, R. P. (2007). Performance of a foot‐and‐mouth disease virus reverse transcription‐polymerase chain reaction with amplification controls between three real‐time instruments. Journal of Veterinary Diagnostic Investigation, 19, 9–20. [DOI] [PubMed] [Google Scholar]
- Nicholson, T. L. , Kukielka, D. , Vincent, A. L. , Brockmeier, S. L. , Miller, L. C. , & Faaberg, K. S. (2011). Utility of a panviral microarray for detection of swine respiratory viruses in clinical samples. Journal of Clinical Microbiology, 49, 1542–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez, J. I. , Blanco, E. , Hernández, T. , Gómez‐Tejedor, C. , Martín, M. J. , Dopazo, J. , & Sobrino, F. (1998). A RT‐PCR assay for the differential diagnosis of vesicular viral diseases of swine. Journal of Virological Methods, 72, 227–235. [DOI] [PubMed] [Google Scholar]
- Paton, D. , McGoldrick, A. , Greiser‐Wilke, I. , Parchariyanon, S. , Song, J.‐Y. , Liou, P. , … Belák, S. (2000). Genetic typing of classical swine fever virus. Veterinary Microbiology, 73, 137–157. [DOI] [PubMed] [Google Scholar]
- Rasmussen, T. B. , Uttenthal, A. , & Agüero, M. (2006). Detection of three porcine vesicular viruses using multiplex real‐time primer‐probe energy transfer. Journal of Virological Methods, 134, 176–182. [DOI] [PubMed] [Google Scholar]
- Reid, S. M. , Ebert, K. , Bachanek‐Bankowska, K. , Batten, C. , Sanders, A. , Wright, C. , … King, D. P. (2009). Performance of real‐time reverse transcription polymerase chain reaction for the detection of foot‐and‐mouth disease virus during field outbreaks in the United Kingdom in 2007. Journal of Veterinary Diagnostic Investigation: Official Publication of the American Association of Veterinary Laboratory Diagnosticians, Inc, 21, 321–330. [DOI] [PubMed] [Google Scholar]
- Reid, S. M. , King, D. P. , Shaw, A. E. , Knowles, N. J. , Hutchings, G. H. , Cooper, E. J. , … Ferris, N. P. (2007). Development of a real‐time reverse transcription polymerase chain reaction assay for detection of marine caliciviruses (genus Vesivirus). Journal of Virological Methods, 140, 166–173. [DOI] [PubMed] [Google Scholar]
- Rotmistrovsky, K. , Jang, W. , & Schuler, G. D. (2004). A web server for performing electronic PCR. Nucleic Acids Research, 32, W108–W112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , & Russell, D. W. (2001). Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- SantaLucia, J. (2007). Physical principles and visual‐OMP software for optimal PCR design In Yuryev A. (Ed.), PCR primer design (pp. 3–33). Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Schuler, G. D. (1997). Sequence mapping by electronic PCR. Genome Research, 7, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthilkumaran, C. , Bittner, H. , Ambagala, A. , Lung, O. , Babiuk, S. , Yang, M. , … Nfon, C. (2016). Use of oral fluids for detection of virus and antibodies in pigs infected with swine vesicular disease virus. Transboundary and Emerging Diseases, 10.1111/tbed.12563 [DOI] [PubMed] [Google Scholar]
- Shi, X. , Liu, X. , Wang, Q. , Das, A. , Ma, G. , Xu, L. , … Shi, J. (2016). A multiplex real‐time PCR panel assay for simultaneous detection and differentiation of 12 common swine viruses. Journal of Virological Methods, 236, 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H. , Norman, S. A. , Mather, E. L. , & Patterson, B. K. (2008). Evaluation of the NanoChip 400 system for detection of influenza A and B, respiratory syncytial, and parainfluenza viruses. Journal of Clinical Microbiology, 46, 1724–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpstra, C. , & de Smit, A. (2000). The 1997/1998 epizootic of swine fever in the Netherlands: Control strategies under a non‐vaccination regimen. Veterinary Microbiology, 77, 3–15. [DOI] [PubMed] [Google Scholar]
- Thompson, D. , Muriel, P. , Russell, D. , Osborne, P. , Bromley, A. , Rowland, M. , … Brown, C. (2002). Economic costs of the foot and mouth disease outbreak in the United Kingdom in 2001. Revue scientifique et technique (International Office of Epizootics), 21, 675–687. [DOI] [PubMed] [Google Scholar]
- Wernike, K. , Hoffmann, B. , & Beer, M. (2013). Single‐tube multiplexed molecular detection of endemic porcine viruses in combination with background screening for transboundary diseases. Journal of Clinical Microbiology, 51, 938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernike, K. , Hoffmann, B. , Dauber, M. , Lange, E. , Schirrmeier, H. , & Beer, M. (2012). Detection and typing of highly pathogenic porcine reproductive and respiratory syndrome virus by multiplex real‐time RT‐PCR. PLoS One, 7, e38251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, J. J. , Karriker, L. A. , Ramirez, A. , Schwartz, K. J. , & Stevenson, G. W. (2012). Diseases of swine. Chichester, West Sussex; Ames, Iowa: Wiley‐Blackwell. [Google Scholar]


