Summary
Bovine coronavirus (BCoV) is a causative agent of respiratory and enteric diseases in cattle and calves. BCoV infection was also evident in captive wild ruminants. Recently, water deer are recognized as the most common wildlife to approach farmhouses and livestock barns in Korea. Therefore, we investigated 77 nasal swab samples from non‐captive wild water deer (Hydropotes inermis) between November 2016 and September 2017 and identified three samples positive for coronavirus, indicating potential for respiratory shedding. The full genomic sequences of the water deer coronavirus were closely related to BCoV (>98%). Therefore, effective biosecurity system in bovine farms would be necessary to prevent contact between farm ruminants and free‐ranging wild water deer.
Keywords: bovine coronavirus, carrier, nasal swab, water deer
1. INTRODUCTION
Bovine coronavirus (BCoV) is a causative agent of respiratory and enteric diseases in calves and adult cattle. It belongs to the virus species Betacoronavirus 1 of the lineage A of the genus Betacoronavirus (Bidokhti et al., 2013).
BCoV has relatively recent common ancestors with human coronavirus OC43 (HCoV‐OC43) and porcine haemagglutinating encephalomyelitis virus (PHEV) (Vijgen et al., 2006). Due to their genetic closeness, the interspecies transmission potential of BCoV should not be ignored. In fact, human enteric coronavirus 4408 strain (HECV‐4408) was first isolated from a child's diarrhoeic stool and was genetically and antigenically more closely related to BCoV than HCoV‐OC43 (Zhang, Herbst, Kousoulas, & Storz, 1994).
In the late 1970s, bovine‐like CoVs were first detected in other ruminants by electron microscope (EM) and they have since been detected in the faecal samples of wild ruminants such as sambar deer (Cervus unicolor), waterbuck (Kobus ellipsiprymnus), sable antelope (Hippotragus niger), white‐tailed deer (Odocoileus virginianus), wisent (Bison bonasus), Himalayan tahr (Hemitragus jemlahicus), sitatunga (Tragelaphus spekii), nyala (Tragelaphus angasii) and giraffe (Giraffa camelopardalis) (Alekseev et al., 2008; Chung, Kim, Bae, Lee, & Oem, 2011; Hasoksuz et al., 2007). As no specific genetic markers were identified to discriminate the ruminant bovine‐like CoVs from BCoVs (Alekseev et al., 2008), interspecies transmission of BCoVs may be common among the ruminants.
However, the majority of cases were found in the captive wild ruminants rather than non‐captive ruminants and most of them were detected in the faecal samples. In Korea, no evidence of BCoV infection in water deer (Hydropotes inermis) was reported between 2010 and 2012 (Kim et al., 2014). Therefore, we investigated coronaviruses in water deer in Korea, using 77 nasal swabs collected between November 2016 and September 2017.
2. MATERIALS AND METHODS
2.1. Samples
In this study, 77 nasal swab samples from water deer (Hydropotes inermis) were obtained from Chungnam Wild Animal Rescue Center, between November 2016 and September 2017. The water deer in this study were rescued wildlife, and the nasal swabs were immediately collected as soon as they were delivered by wildlife veterinarians. The nasal swab samples were placed into universal viral transport medium (BD Diagnostics, USA) and transported to the laboratory for analysis.
2.2. Viral detection
RNA was extracted using TRIzol LS (Invitrogen, USA), and consensus primer‐based reverse transcriptase polymerase chain reaction (RT‐PCR), based on the RNA‐dependent RNA polymerase (RDRP) gene, was performed to detect coronaviruses (Poon et al., 2005). Among 77 samples, three samples appeared positive from RT‐PCR and the amplicons were sequenced using target‐specific forward and reverse primers synthesized by Cosmo Genetech Co., Ltd. The nucleotide sequences were further analysed against other coronavirus sequences from GenBank using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and MEGA version 6 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013).
2.3. Genome sequencing
The genome of water deer coronavirus was further sequenced by RT‐PCR using primers that were designed based on the genome of a related BCoV, strain Quebec (AF220295). Subsequently, 3ʹ and 5ʹ end sequences were obtained by rapid amplification of cDNA end (RACE) sequencing based on PCR using the adapter‐oligo dTVN primer from Cosmo Genetech Co., Ltd. The obtained partial viral genome was assembled and manually annotated by comparisons with the open reading frames (ORFs) of other CoVs in GenBank database and further schematized with Geneious R9 (Biomatters Ltd., Auckland, New Zealand). To compare the complete genome of the water deer coronavirus and other related CoVs, the orthologous average nucleotide identity (OrthoANI) value was preliminarily analysed using ANI calculator (http://www.ezbiocloud.net/tools/ani), and the nucleotide and deduced amino acid sequences of the viral ORFs were compared by multiple sequence alignment using CLUSTALW software (http://www.ebi.ac.uk/clustalw), respectively.
3. RESULTS AND DISCUSSION
The three positive samples (W17‐12, W17‐15 and W17‐18) were obtained in December 2016, January 2017, and February 2017, respectively, which covered the winter season in Korea (Table 1). Phylogenetic analysis, based on 256 bases of the partial RDRP gene, showed they were closely related to BCoV and HECV‐4408 (Figure 1a). The nucleotide identities of the viruses with BCoV and HECV‐4408 were 99.2% and 100%, respectively.
Table 1.
Information on nasal swab samples
| Month of Collection | Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| November 2016 | December 2016 | January 2017 | February 2017 | March 2017r | April 2017 | May 2017 | June 2017 | July 2017 | August 2017 | September 2017 | ||
| No. of nasal swabs | 1 | 8 | 5 | 2 | 5 | 10 | 11 | 16 | 7 | 7 | 5 | 77 |
| No. of CoV positivesa | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
Positives in RT‐PCR based on consensus primers targeting RNA‐dependent RNA polymerase region of coronaviruses (Poon et al., 2005).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 1.
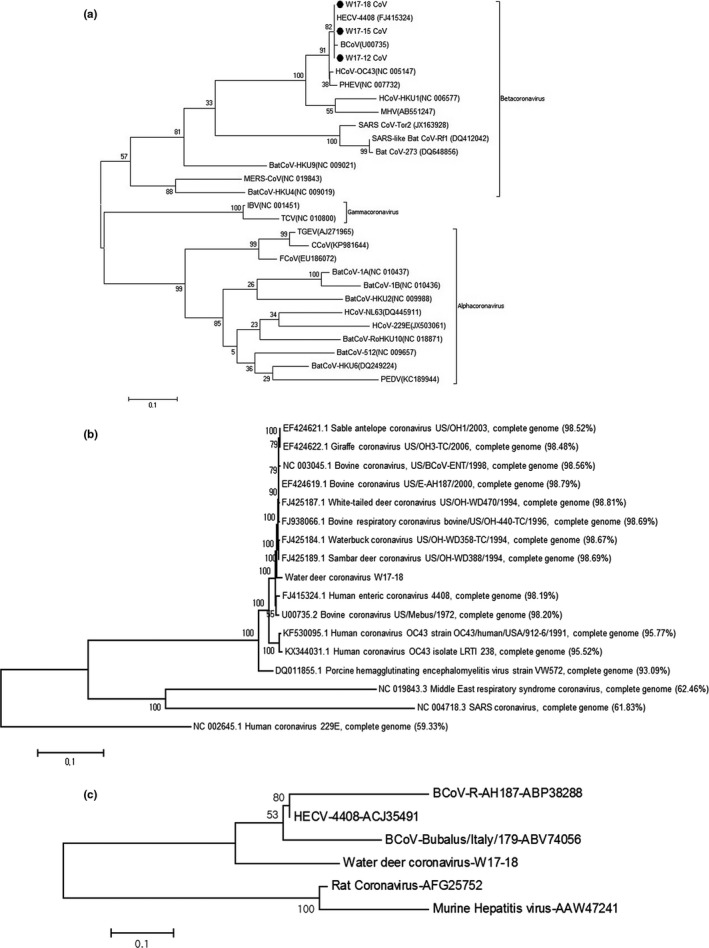
Phylogenetic analysis by maximum likelihood method based on the partial RNA‐dependent RNA polymerase (RDRP) gene (a), complete genomic sequence (b) and putative 4.8 kDa non‐structural protein (c). (a) The phylogenetic tree was drawn based on partial RDRP nucleotide sequences (256 bases) of the three water deer coronaviruses, (indicated by black circles) and other reference sequences; (b) The phylogenetic tree was drawn based on genomic sequence of water deer coronavirus W17‐18 and other reference sequences (sequence similarities enclosed in parentheses); (c) The phylogenetic tree was drawn based on amino acids sequences of the putative 4.8 kDa non‐structural protein of water deer coronavirus W17‐18 and other reference sequences
The fully sequenced water deer coronavirus W17‐18 genome (GenBank Accession No. MG518518) was 31,034 bp (37.1% G+C content); the overall gene content and arrangement in the strain were almost identical to those from its related BCoVs from captive wild ruminants in the GenBank database. Moreover, the OrthoANI value analysis suggested that the water deer coronavirus W17‐18 genome was very similar (>98%) to those BCoVs from bovine and wild animals and closely related to HCoV‐OC43 strains (>95%) and PHEV (>93%).
The full genome sequence‐based phylogenetic analysis by maximum likelihood method revealed that the virus was closely related to BCoV, bovine‐like coronavirus and HECV‐4408 (Figure 1b). Distinctively, the N2 protein (208 amino acids), which was solely found in the BCoVs, was detected in the sequenced genome, whereas the predicted 4.8‐kDa non‐structural protein in water deer coronavirus strain W17‐18 was elongated due to a 13‐nucleotide deletion compared to other BCoVs (Figure 2). Therefore, the phylogenetic analysis based on the 4.8 kDa‐ or 4.8 kDa‐like non‐structural protein amino acid sequences of other reference viruses infers that water deer coronavirus W17‐18 has created a unique clade within the BCoVs (Figure 1c). Collectively, the newly found water deer coronavirus in this study was closely related to BCoV showing minor unique genetic characteristic in the 4.8 kDa‐like non‐structural protein.
Figure 2.

Comparison of CoV genomes. The genome organization is schematically reported, grouping the viruses according to their genus or lineage. Grey‐coloured boxes represent accessory open reading frames (ORFs) of the CoV genomes [Colour figure can be viewed at http://wileyonlinelibrary.com]
As the water deer coronavirus was detected from nasal swabs during the winter season, it could be assumed that the water deer coronaviruses might also cause respiratory disease and circulate during the winter season like other BCoVs. The majority of BCoV cases previously detected in wild ruminants have been in captive animals. In this study, BCoV was detected in non‐captive wild water deer, which may provide further evidence that BCoV could be transmitted between freely ranging wild ruminants and farm ruminants. These findings may attract public attention for BCoV transmission between wild water deer and farms during winter season. In addition, BCoV‐like human coronavirus (HECV‐4408) was demonstrated to be able to infect gnotobiotic calves and provide cross‐protection against BCoV infection (Han, Cheon, Zhang, & Saif, 2006), and interspecies transmission of BCoV in turkey poults was also demonstrated under experimental condition (Ismail, Cho, Ward, Saif, & Saif, 2001). Therefore, interspecies transmission ecology of BCoV among livestock, wild animals and humans should be further investigated to control the virus.
We conclude that the water deer coronavirus is a new strain of BCoV, rather than a new species. This is the first report of BCoV detection in non‐captive wild water deer and presents a possible respiratory infection risk to water deer in Korea. Water deer (H. inermis) have recently been recognized as the most common wildlife to approach farmhouses and livestock barns in Korea (Kim et al., 2014). BCoV transmission between water deer and livestock farms should not be overlooked. Therefore, effective biosecurity systems in bovine farms should be investigated to prevent interspecies transmission of BCoV between farm ruminants and wild water deer.
ACKNOWLEDGEMENTS
This work was supported by grants from the KRIBB Initiative programme (Grant No. KGM4691713), the BioNano Health‐Guard Research Center, funded by the Ministry of Science and ICT (MSIT) of Korea as a Global Frontier Project (Grant number H‐GUARD_2013M3A6B2078954) and supported by Korea Ministry of Environment (MOE) as “Public Technology Program based on Environmental Policy (No. 2016000210002).”
Kim JH, Jang J‐H, Yoon S‐W, et al. Detection of bovine coronavirus in nasal swab of non‐captive wild water deer, Korea. Transbound Emerg Dis. 2018;65:627–631. 10.1111/tbed.12847
Contributor Information
D. G. Jeong, Email: dgjeong@kribb.re.kr.
H. K. Kim, Email: khk1329@kribb.re.kr.
REFERENCES
- Alekseev, K. P. , Vlasova, A. N. , Jung, K. , Hasoksuz, M. , Zhang, X. , Halpin, R. , … Saif, L. J. (2008). Bovine‐like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. Journal of Virology, 82, 12422–12431. 10.1128/JVI.01586-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidokhti, M. R. , Traven, M. , Krishna, N. K. , Munir, M. , Belak, S. , Alenius, S. , & Cortey, M. (2013). Evolutionary dynamics of bovine coronaviruses: Natural selection pattern of the spike gene implies adaptive evolution of the strains. Journal of General Virology, 94, 2036–2049. 10.1099/vir.0.054940-0 [DOI] [PubMed] [Google Scholar]
- Chung, J.‐Y. , Kim, H.‐R. , Bae, Y.‐C. , Lee, O. S. , & Oem, J.‐K. (2011). Detection and characterization of bovine‐like coronaviruses from four species of zoo ruminants. Veterinary Microbiology, 148, 396–401. 10.1016/j.vetmic.2010.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M. G. , Cheon, D. S. , Zhang, X. , & Saif, L. J. (2006). Cross‐protection against a human enteric coronavirus and a virulent bovine enteric coronavirus in gnotobiotic calves. Journal of Virology, 80, 12350–12356. 10.1128/JVI.00402-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasoksuz, M. , Alekseev, K. , Vlasova, A. , Zhang, X. , Spiro, D. , Halpin, R. , … Saif, L. J. (2007). Biologic, antigenic, and full‐length genomic characterization of a bovine‐like coronavirus isolated from a giraffe. Journal of Virology, 81, 4981–4990. 10.1128/JVI.02361-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, M. M. , Cho, K. O. , Ward, L. A. , Saif, L. J. , & Saif, Y. M. (2001). Experimental bovine coronavirus in turkey poults and young chickens. Avian Diseases, 45, 157–163. 10.2307/1593023 [DOI] [PubMed] [Google Scholar]
- Kim, S. H. , Choi, H. , Yoon, J. , Woo, C. , Chung, H. M. , Kim, J. T. , & Shin, J. H. (2014). Pathogens in water deer (Hydropotes inermis) in South Korea, 2010‐12. Journal of Wildlife Diseases, 50, 478–483. 10.7589/2013-06-137 [DOI] [PubMed] [Google Scholar]
- Poon, L. L. M. , Chu, D. K. W. , Chan, K. H. , Wong, O. K. , Ellis, T. M. , Leung, Y. H. C. , … Peiris, J. S. M. (2005). Identification of a novel coronavirus in bats. Journal of Virology, 79, 2001–2009. 10.1128/JVI.79.4.2001-2009.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. , & Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen, L. , Keyaerts, E. , Lemey, P. , Maes, P. , Van Reeth, K. , Nauwynck, H. , … Van Ranst, M. (2006). Evolutionary history of the closely related group 2 coronaviruses: Porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. Journal of Virology, 80, 7270–7274. 10.1128/JVI.02675-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. M. , Herbst, W. , Kousoulas, K. G. , & Storz, J. (1994). Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. Journal of Medical Virology, 44, 152–161. 10.1002/(ISSN)1096-9071 [DOI] [PMC free article] [PubMed] [Google Scholar]


