Summary
As one of the top pork producers in China, Shandong Province suffered frequent outbreaks of porcine epidemic diarrhoea virus (PEDV) on pig farms from January 2012 to July 2015, resulting in significant economic losses. To better understand the prevalence situation, we conducted molecular epidemiological analyses of 38 PEDV strains isolated from 13 cities in Shandong Province. The detection rate of PEDV was 71.2% (146/205) by reverse transcription polymerase chain reaction (RT‐PCR). The S genes of the 38 isolated samples were 4146 to 4161 nt in length and shared high levels of sequence identity (93.3–99.6% nt, 92.1–99.4% aa) with those of the 41 reference strains. Among the 38 strains, 31 strains that occupied 12 cities were classed into G3 genotype, while the other seven that only existed in four cities were classed into G2 genotype. In addition, the strains CH‐SDLY‐2‐2014 and CH‐SDLY‐3‐2014 isolated from Linyi were classed into the Gd subgenotype. Notably, there were multiple insertions or deletions in the S genes and several mutations in the neutralizing epitopes of the PEDV S protein. Overall, the results revealed that G2 and G3 are the predominant PEDV genotypes circulating in Shandong Province during 2012–2015, and Gd subgenotype in G3 group had already spread towards northern China in 2014.
Keywords: porcine epidemic diarrhoea virus, S gene, genotype, mutation, Gd subgenotype
Introduction
Porcine epidemic diarrhoea virus (PEDV) is an enveloped, single‐stranded RNA virus belonging to the genus Alphacoronavirus, subfamily Coronavirinae, family Coronaviridae (Pensaert and de Bouck, 1978). The PEDV genome with a 5′ cap and a 3′ polyadenylated tail is approximately 28 kb in length and contains seven open reading frames (ORFs), which encode replicase 1a and 1b, the spike (S) protein, ORF3 protein, envelope (E) protein, membrane (M) protein and nucleoprotein (N), arranged in the order 5′‐replicase 1a/1b‐S‐ORF3‐E‐M‐N‐3′ (Murphy et al., 1999). The PEDV S protein is a glycoprotein peplomer (surface antigen) on the viral surface and plays a pivotal role in induction of neutralizing antibodies in natural hosts (Chang et al., 2002; Temeeyasen et al., 2014).
Porcine epidemic diarrhoea virus was first reported in Belgium and the United Kingdom in 1978 (Pensaert and de Bouck, 1978). Within two decades, it swept over continents of Europe, Asia and North America (Song and Park, 2012; Huang et al., 2013; Jung et al., 2014; Vlasova et al., 2014; Sun et al., 2015).
In China, PED has been reported in many provinces, with mortality up to 100%, mainly involving breeding piglets within 10 days of age, usually 2–3 days after birth (Li et al., 2012). As a major swine‐producing province in China, Shandong has suffered great economic losses from outbreaks of PED, even with vaccination of the flocks. Since 2012, PED outbreaks have remarkably increased in Shandong Province (Zhao et al., 2012). To investigate the molecular diversity and epidemiology of the virus in Shandong, we sequenced the full‐length S gene of 38 PEDV‐positive samples collected from January 2012 to July 2015.
Materials and Methods
Specimens
A total of 205 intestinal and faecal samples were collected from 2‐day‐old to 7‐day‐old dead piglets with typical symptoms of PEDV infection from 13 cities in Shandong Province of China from 20 January 2012 to 15 July 2015. The morbidity within infected herds was near 100%, a total mortality rate was usually over 50% in suckling piglets, and the sick piglets died in 2–3 days with typical watery diarrhoea, dehydration and vomiting. According to the manufacturer's instructions, total RNA was extracted from the samples using TriZol reagent (Invitrogen, Carlsbad, CA, USA). Then, the samples were detected by reverse transcription polymerase chain reaction (RT‐PCR) with the primers targeted at PEDV M gene: Mf, 5′‐CCC CAG TAC TGT TAT TGA CGT ATA AAC‐3′ and Mr, 5′‐GTT TAG ACT AAA TGA AGC ACT TTC‐3′. A total of 38 PEDV‐positive samples were randomly selected for amplification of the S gene and sequence analysis (Table 1).
Table 1.
The porcine epidemic diarrhoea virus strains isolated from Shandong Province, China, in this study
| Strain | City | Isolated year | S gene (nt/aa) | Accession numbers |
|---|---|---|---|---|
| CH‐SDCQ‐2014 | Jinan | 2014 | 4149/1382 | KU133234 |
| CH‐SDQF‐1‐2014 | Jining | 2014 | 4152/1383 | KU133256 |
| CH‐SDQF‐2‐2014 | Jining | 2014 | 4152/1383 | KU133257 |
| CH‐SDQF‐3‐2014 | Jining | 2014 | 4152/1383 | KU133258 |
| CH‐SDRZ‐2013 | Rizhao | 2013 | 4152/1383 | KU133262 |
| CH‐SDZB‐2012 | Zibo | 2012 | 4146/1381 | KU133267 |
| CH‐SDZC‐2012 | Zibo | 2012 | 4161/1386 | KU133268 |
| CH‐SDBZ‐1‐2015 | Binzhou | 2015 | 4161/1386 | KU133232 |
| CH‐SDBZ‐2‐2015 | Binzhou | 2015 | 4161/1386 | KU133233 |
| CH‐SDCQ‐2015 | Jinan | 2015 | 4161/1386 | KU133235 |
| CH‐SDDP‐1‐2014 | Taian | 2014 | 4161/1386 | KU133236 |
| CH‐SDDP‐2‐2014 | Taian | 2014 | 4161/1386 | KU133237 |
| CH‐SDDP‐3‐2014 | Taian | 2014 | 4161/1386 | KU133238 |
| CH‐SDDY‐2012 | Dongying | 2012 | 4161/1386 | KU133239 |
| CH‐SDDZ‐2012 | Dezhou | 2012 | 4161/1386 | KU133240 |
| CH‐SDHZ‐1‐2013 | Heze | 2013 | 4161/1386 | KU133241 |
| CH‐SDHZ‐2‐2013 | Heze | 2013 | 4161/1386 | KU133242 |
| CH‐SDHZ‐3‐2013 | Heze | 2013 | 4161/1386 | KU133243 |
| CH‐SDHZ‐4‐2013 | Heze | 2013 | 4161/1386 | KU133244 |
| CH‐SDJY‐2012 | Jinan | 2012 | 4161/1386 | KU133245 |
| CH‐SDLQ‐2015 | Liaocheng | 2015 | 4161/1386 | KU133246 |
| CH‐SDLS‐1‐2014 | Jining | 2014 | 4161/1386 | KU133247 |
| CH‐SDLS‐2‐2014 | Jining | 2014 | 4161/1386 | KU133248 |
| CH‐SDLY‐1‐2012 | Linyi | 2012 | 4161/1386 | KU133249 |
| CH‐SDLY‐1‐2013 | Linyi | 2013 | 4161/1386 | KU133250 |
| CH‐SDLY‐1‐2014 | Linyi | 2014 | 4161/1386 | KU133251 |
| CH‐SDLY‐2‐2012 | Linyi | 2012 | 4161/1386 | KU133252 |
| CH‐SDLY‐2‐2013 | Linyi | 2013 | 4161/1386 | KU133253 |
| CH‐SDLY‐2‐2014 | Linyi | 2014 | 4158/1385 | KU133254 |
| CH‐SDLY‐3‐2014 | Linyi | 2014 | 4158/1385 | KU133255 |
| CH‐SDQF‐4‐2014 | Jining | 2014 | 4161/1386 | KU133259 |
| CH‐SDQF‐5‐2014 | Jining | 2014 | 4161/1386 | KU133260 |
| CH‐SDQH‐2014 | Dezhou | 2014 | 4161/1386 | KU133261 |
| CH‐SDTA‐2012 | Taian | 2012 | 4161/1386 | KU133263 |
| CH‐SDWF‐2012 | Weifang | 2012 | 4161/1386 | KU133264 |
| CH‐SDWF‐2015 | Weifang | 2015 | 4161/1386 | KU133265 |
| CH‐SDYT‐2012 | Yantai | 2012 | 4161/1386 | KU133266 |
| CH‐SDZC‐2015 | Zibo | 2015 | 4158/1385 | KU133269 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Amplification of S genes
The primers for PCR and sequencing the S genes were as follows: fragment A (S1f: 5′‐ATG AGG TCT TTA ACC T‐3′ and S1r: 5′‐CTG TTC GTG ACT CAG AAG‐3′), fragment B (S2f: 5′‐TAC CTT CTA CTG TCA GGG‐3′ and S2r: 5′‐CTC GCT TGA ACA GCA TAG‐3′) and fragment C (S3f: 5′‐ACT CTG GTG TCA TGG TAC‐3′ and S3r: 5′‐TCA CTG CAC GTG GAC TTT‐3′). According to the manufacturer's instructions, S genes were amplified from the PEDV‐positive samples with Quant One Step RT‐PCR Kit (Tiangen Biotech, Beijing, China). The Quant One Step reaction was carried out under the following conditions: reverse transcription at 50°C for 30 min, initial denaturation at 94°C for 2 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, extension at 65°C for 1 min and a final elongation step at 65°C for 10 min. Following purification by AxyPrep DNA Gel Extraction kit (Axygen, Union City, CA, USA), the PCR products were ligated into pGEM‐T easy vector (Promega, Madison, WI, USA). The nucleotide (nt) sequences of the positive clones were determined from a commercial service (Shanghai Sangon Biological Engineering Technology & Service Co., Ltd). At least five individual clones were sequenced for each sample.
Sequences analysis
The S gene sequences of 38 PEDV strains isolated from 13 cities in Shandong Province were obtained for phylogenetic analysis (Table 1). A total of 41 PEDV reference strains from other provinces of China and other countries were selected from the GenBank database for comparison (Table 2). Multiple protein sequences were aligned using the MegAlign program by the Clustal W method in the DNASTAR software package (Burland, 2000). A phylogenetic tree was constructed by the neighbour‐joining method using the Mega 6.0 software (Tamura et al., 2013).
Table 2.
The 41 reference porcine epidemic diarrhoea virus strains used in this study
| Field strain | Regions | Isolated year | S gene (nt/aa) | Accession numbers |
|---|---|---|---|---|
| JS‐HZ2012 | Jiangsu/China | 2012 | 4161/1386 | KC210147 |
| CH‐ZJJS‐2Z‐2012 | Zhejiang/China | 2012 | 4161/1386 | KF840546 |
| GD‐B | Guangdong/China | 2012 | 4161/1386 | JX088695 |
| SHOP‐YM‐2013 | Shanghai/China | 2013 | 4161/1386 | KJ196348 |
| CH‐ZJHZ‐1C‐2012 | Zhejiang/China | 2012 | 4161/1386 | KF840553 |
| BJ‐2011‐1 | Beijing/China | 2011 | 4146/1381 | JN825712 |
| AH2012 | Anhui/China | 2012 | 4161/1386 | KC210145 |
| CH‐SDZD‐1‐2012 | Shandong/China | 2012 | 4161/1386 | KF840550 |
| CH‐SDZD‐2‐2012 | Shandong/China | 2012 | 4161/1386 | KF840549 |
| CH‐JXZS‐3H‐2012 | Jiangxi/China | 2012 | 4161/1386 | KF840541 |
| LC | Guangdong/China | 2012 | 4158/1385 | JX489155 |
| CH‐GDGZ‐2012 | Guangdong/China | 2012 | 4158/1385 | KF384500 |
| GD‐1 | Guangdong/China | 2012 | 4158/1385 | JX647847 |
| GD‐A | Guangdong/China | 2012 | 4158/1385 | JX112709 |
| CH‐ZMDZY‐11 | Henan/China | 2011 | 4161/1386 | KC196276 |
| IA2 | USA | 2013 | 4161/1386 | KF468754 |
| IA1 | USA | 2013 | 4161/1386 | KF468753 |
| USA‐Ohio69‐2013 | USA | 2013 | 4161/1386 | KJ645665 |
| PC21A | USA | 2013 | 4161/1386 | KF272920 |
| KUIDL‐PED‐2014‐007 | Korea | 2014 | 4161/1386 | KJ588064 |
| KUN‐141112‐feces | Korea | 2014 | 4161/1386 | KR873431 |
| KUIDL‐PED‐2014‐002 | Korea | 2014 | 4161/1386 | KJ588063 |
| MN | USA | 2013 | 4161/1386 | KF468752 |
| CH‐HuBWHYQ‐2012 | Hubei/China | 2012 | 4161/1386 | KF840551 |
| CH‐FJND‐3‐2011 | Fujian/China | 2011 | 4161/1386 | JQ282909 |
| OH851 | USA | 2013 | 4152/1383 | KJ399978 |
| USA‐Lowa‐106‐2013 | USA | 2013 | 4152/1383 | KJ645695 |
| KUN‐1406‐01 | Korea | 2014 | 4152/1383 | KM403155 |
| USA‐M52‐2013 | USA | 2013 | 4152/1383 | KJ645704 |
| FR‐001‐2014 | France | 2014 | 4152/1383 | KR011756 |
| 15V010‐BEL‐2015 | Belgium | 2015 | 4152/1383 | KR003452 |
| GER‐L00719‐2014 | German | 2014 | 4152/1383 | LM645058 |
| CH‐S | Shanghai/China | 1986 | 4152/1383 | JN547228 |
| Attenuated Chinese PEDV | Hubei/China | 2012 | 4149/1382 | KC189944 |
| Attenuated DR13 | Korea | 2003 | 4149/1382 | JQ023162 |
| JS2008 | Jiangsu/China | 2008 | 4161/1386 | KC109141 |
| MK | Japan | 1996 | 4152/1383 | AB548624 |
| Virulent DR13 | Korea | 1999 | 4152/1383 | JQ023161 |
| SM98 | Korea | 1998 | 4143/1380 | GU937797 |
| CV777 | Belgium | 1978 | 4152/1383 | AF353511 |
| LZC | Gansu/China | 2006 | 4152/1383 | EF185992 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Results
Sequence properties of the S genes
Among the 205 intestinal and faecal samples from 13 cities in Shandong Province of China, 146 were positive for PEDV by RT‐PCR, and the detection rate was 71.2% (146/205). The S gene sequences of the 38 PEDV samples were submitted to GenBank and assigned accession Nos: KU133232–KU133269 (Table 1). The S genes of the 38 PEDVs were 4146 to 4161 nt in length, encoding the S proteins of 1381 to 1386 amino acids, respectively. Among the 38 samples, 29 were 4161 nt in length and encoded a protein of 1386 amino acids, while the other nine samples were 4152 nt (CH‐SDQF‐1‐2014, CH‐SDQF‐2‐2014, CH‐SDQF‐3‐2014 and CH‐SDRZ‐2013), 4158 nt (CH‐SDLY‐2‐2014, CH‐SDLY‐3‐2014 and CH‐SDZC‐2015), 4149 nt (CH‐SDCQ‐2014) and 4146 nt (CH‐SDZB‐2012) in length, respectively (Table 1). Multiple sequence alignment showed that the S genes above shared 95.0–99.9% nucleotide sequence identity and 94.3–99.8% amino acid sequence identity with each other, while shared 93.3–99.6% nucleotide similarity and 92.1–99.4% amino acid similarity with those of the 41 PEDV reference strains.
Phylogenetic analysis of the S gene
Phylogenetic analysis based on the Shandong PEDV field isolates (Table 1) and PEDV reference strains (Table 2) was performed. The results showed that the 79 PEDV strains could be divided into three genotypes: G1, G2 and G3 (Fig. 1). Nine reference strains belonged to G1, namely previous Chinese (JS2008, LZC and CH‐S), Korean (SM98 and virulent DR13), Japanese (MK) and European (CV777) strains and two vaccine strains (attenuated Chinese PEDV and attenuated DR13). None of the strains isolated in this study were included in G1.
Figure 1.
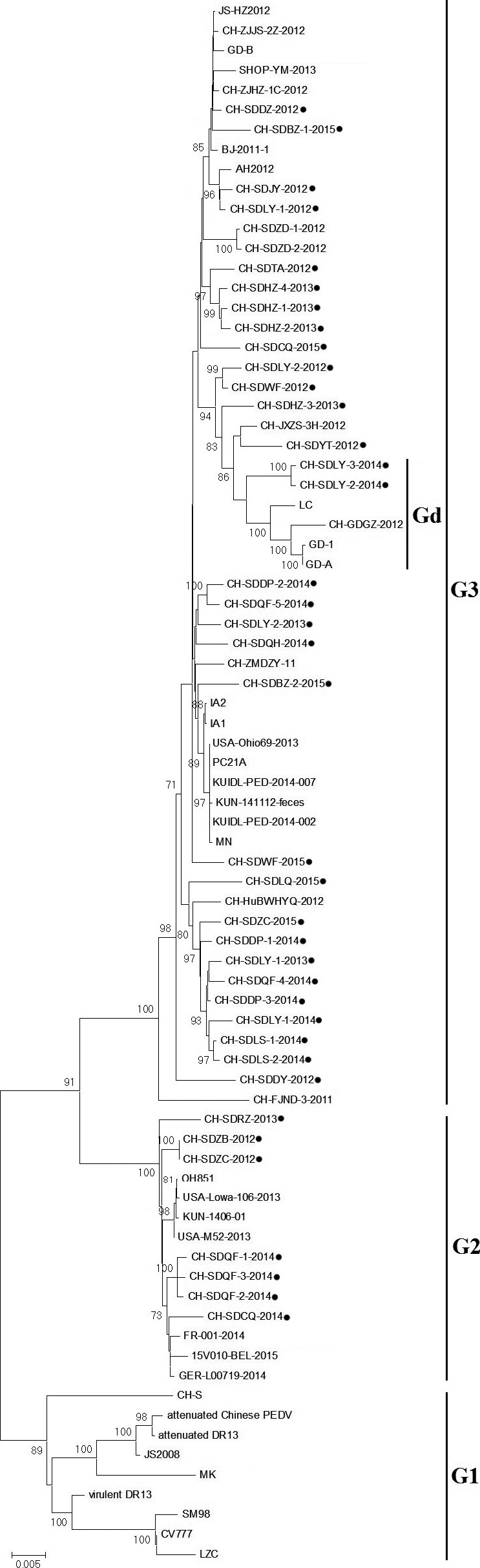
The rooted phylogenetic tree of 38 field strains homolog and other representative S gene constructed by the neighbour‐joining method within the Mega 6.0 software. Bootstrap majority consensus values on 1000 replicates are indicated at each branch point in per cent. Black solid circles indicate the 38 porcine epidemic diarrhoea virus strains isolated in this study. Bootstrap values are represented at key nodes. Scale bar represents nucleotide substitutions per site.
Seven of the 38 sequenced samples were clustered in G2, together with three current European strains (15V010‐BEL‐2015, GER‐L00719‐2014, FR‐001‐2014), three recent emergent S INDEL strains from the US (OH851, USA‐Iowa‐106‐2013, USA‐M52‐2013) and one current Korean strain (KUN‐1406‐01). The majority of the S genes were consistent in length in G2 (4152 nt), except for CH‐SDZB‐2012 (4146 nt) and CH‐SDCQ‐2014 (4149 nt). The seven strains were isolated from four cities in Shandong Province (Fig. 2), sharing 98.8–99.8% nucleotide identity with each other and 98.9–99.5% with the other reference strains from G2.
Figure 2.
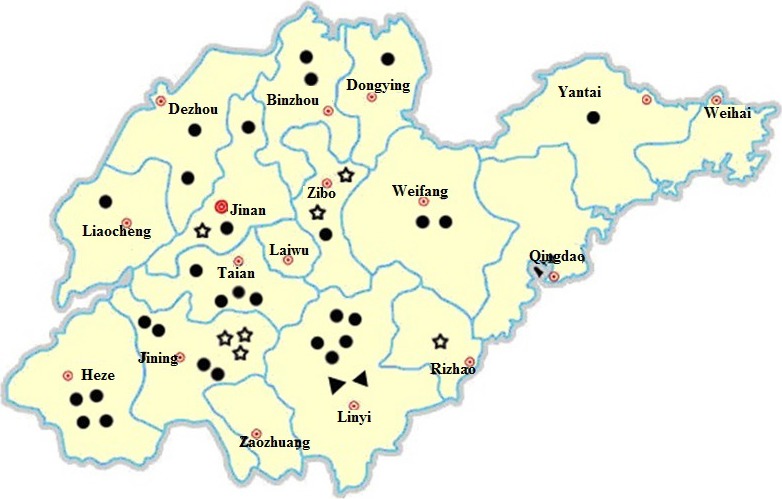
Map representing the 17 cities of Shandong Province in China. The 38 porcine epidemic diarrhoea virus (PEDV) strains were collected from 13 cities in Shandong. Black dot represents the PEDV strains in G3 genotype; black triangled dot represents the PEDV strains in Gd subgenotype; star represents the PEDV strains in G2 genotype.
Fifty‐six strains were classified to G3, including the rest 31 strains isolated in this study, 17 recent Chinese field strains, three Korean strains and five US strains. The 17 reference strains from China were originated from ten provinces in eastern China. The five US strains were reported to belong to North American clades I and II, following the PEDV outbreak in April 2013 (Vlasova et al., 2014). The three Korean strains are thought to be co‐circulating in Korea and showed high homology with the five US strains (Choi et al., 2014; Lee et al., 2015). The 31 isolated strains occupied all the 13 cities, suggesting that G3 was the most prevalent genotype in Shandong Province (Fig. 2). In group G3, the 31 strains isolated in this study shared 97.3–99.9% nucleotide identity with each other and 96.9–99.5% with other reference strains. Interestingly, phylogenetic analysis showed that G3 strains predominantly displayed a 4161‐nt S gene sequence with the exception of Gd subgenotype (4158 nt). CH‐SDLY‐2‐2014 and CH‐SDLY‐3‐2014 both belonged to the Gd subgenotype, which is a newly described variant subgenotype in 2012.
Till now, four neutralizing epitopes of the PEDV S protein have been reported, that is COE (CO‐26K equivalent) (499–638 aa), SS2 (748–755 aa), SS6 (764–771 aa) and 2C10 (1368–1374 aa) (Chang et al., 2002; Cruz et al., 2008; Sun et al., 2008). According to the alignment results of selected sequences from the phylogenetic tree, SS2 and 2C10 were found to be conserved in all field strains, while epitope SS6 displayed 2‐aa or 3‐aa mutations in most field strains. Compared with strain CV777, amino acid changes in COE domain existed in all of the aligned sequences (Fig. 3).
Figure 3.
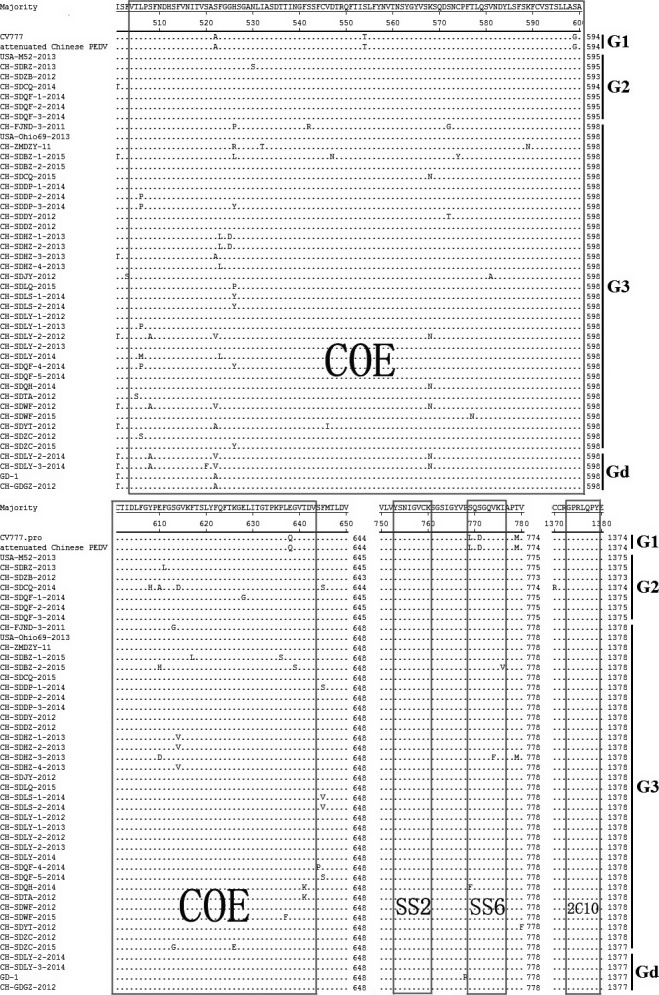
Comparison of the antigen epitopes of S proteins of strains and reference strains. Ellipses represent the consensus amino acids. COE, SS6, SS2 and 2C10 are in the box. Strains in this study are marked with asterisks (*).
Discussion
Since 2012, PEDV has re‐emerged and rapidly disseminated all over the world (Stevenson et al., 2013). In our study, the positive rate of suspected samples was 71.2% (146/205) in Shandong Province of China, indicating the high prevalence of PEDV among the pig populations. The S gene is important in the phylogenetic and epidemiological analysis of coronaviruses in the field (Stevenson et al., 2013). According to the phylogenetic analysis of S genes, the 38 representative isolates from this study were clustered into two distinct and evolutionarily distant groups (Fig. 1). Among the 38 strains from 13 cities, 31 (G3) occupied 12 cities and the other 7 (G2) only exist in four cities (Fig. 2), which revealed that the genotypes G2 and G3 are circulating in Shandong from 2012 to 2015, and G3 was the predominant genotype of PEDV.
Subgroup Gd strains were first identified in Guangdong Province during 2011 to 2013 and then rapidly spread to nearby areas (Chen et al., 2012; Fan et al., 2012; Wei et al., 2012; Tian et al., 2013). The CH‐SDLY‐2‐2014 and CH‐SDLY‐3‐2014 strains were first identified from Linyi, Shandong Province in 2014, which were further verified to be subgenotype Gd. This is the first report of Gd subgenotype stains in Northern China. Considering the frequent animal trades between Linyi and South China, the current data indicated that the transportation of animals might be a source of PEDV transmission, and this can provide instructive information for the prevention strategy of exotic PEDV genotype invasion.
The PEDV S protein is an important glycoprotein peplomer on the viral surface, and mutations related to virulence in the S gene antigenic regions may confer increased pathogenicity and antigenicity to the new PEDV variants, thereby reducing the protective efficacy of the vaccination (Park et al., 2007; Stevenson et al., 2013). Multiple alignment analysis showed that many amino acid mutations existed in the neutralizing epitopes of the PEDV S protein, which might be the most important reason for the frequent PEDV outbreaks in Shandong Province during 2012–2015.
Conflicts of Interest
The authors declare no conflict of interests.
Acknowledgements
This work was supported by the Science and Technology Commission of Shandong Province (No. 2013GC15003), China.
Contributor Information
F.‐L. Tian, Email: fulin_tian@163.com.
S.‐J. Jiang, Email: sjjing@sdau.edu.cn.
References
- Burland, T. G. , 2000: DNASTAR's Lasergene sequence analysis software. Methods Mol. Biol. 132, 71–91. [DOI] [PubMed] [Google Scholar]
- Chang, S. H. , Bae J. L., Kang T. J., Kim J., Chung G. H., Lim C. W., Laude H., Yang M. S., and Jang Y. S., 2002: Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells 14, 295–299. [PubMed] [Google Scholar]
- Chen, F. , Pan Y. F., Zhang X. B., Tian X. Y., Wang D. D., Zhou Q. F., Song Y. H., and Bi Y. Z., 2012: Complete genome sequence of a variant porcine epidemic diarrhea virus strain isolated in China. J. Virol. 86, 12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. C. , Lee K. K., Pi J. H., Park S. Y., Song C. S., Choi I. S., Lee J. B., Lee D. H., and Lee S. W., 2014: Comparative genome analysis and molecular epidemiology of the reemerging porcine epidemic diarrhea virus strains isolated in Korea. Infect. Genet. Evol. 26, 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, D. J. , Kim C. J., and Shin H. J., 2008: The GPRLQPY motif located at the carboxy‐terminal of the spike protein induces antibodies that neutralize porcine epidemic diarrhea virus. Virus Res. 132, 192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, H. Y. , Zhang J., Ye Y., Tong T. Z., Xie K. S., and Liao M., 2012: Complete genome sequence of a novel porcine epidemic diarrhea virus in south China. J. Virol. 86, 10248–10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. W. , Dickerman A. W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., and Meng X. J., 2013: Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio 4, e00737–e00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K. , Wang Q. H., Scheuer K. A., Lu Z. Y., Zhang Y., and Saif L. J., 2014: Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 20, 668–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Kim Y., and Lee C., 2015: Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU‐141112. Virus Res. 208, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. L. , Zhu L., Ma J. Y., Zhou Q. F., Song Y. H., Sun B. L., Chen R. A., Xie Q. M., and Bee Y. Z., 2012: Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field strains in south China. Virus Genes 45, 181–185. [DOI] [PubMed] [Google Scholar]
- Murphy, F. A. , Gibbs E. P., Horzinek M. C., and Studdert M. J., 1999: Veterinary Virology. 3rd edn Academic Press, San Diego. [Google Scholar]
- Park, S. J. , Song D. S., Ha G. W., and Park B. K., 2007: Cloning and further sequence analysis of the spike gene of attenuated porcine epidemic diarrhea virus DR13. Virus Genes 35, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert, M. B. , and de Bouck P., 1978: A new coronavirus‐like particle associated with diarrhea in swine. Arch. Virol. 58, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, D. , and Park B., 2012: Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 44, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, G. W. , Hoang H., Schwartz K. J., Burrough E. R., Sun D., Madson D., Cooper V. L., Pillatzki A., Gauger P., Schmitt B. J., Koster L. G., Killian M. L., and Yoon K. J., 2013: Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 25, 649–654. [DOI] [PubMed] [Google Scholar]
- Sun, D. , Feng L., Shi H., Chen J., Cui X., Chen H., Liu S., Tong Y., Wang Y., and Tong G., 2008: Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 131, 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, M. , Ma J. L., Wang Y. N., Wang M., Song W. C., Zhang W., Lu C. P., and Yao H. C., 2015: Genomic and epidemiological characteristics provide new insights into the phylogeographical and spatiotemporal spread of porcine epidemic diarrhea virus in Asia. J. Clin. Microbiol. 53, 1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher G., Peterson D., Filipski A., and Kumar S., 2013: MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temeeyasen, G. , Srijangwad A., Tripipat T., Tipsombatboon P., Piriyapongsa J., Phoolcharoen W., Chuanasa T., Tantituvanont A., and Nilubol D., 2014: Genetic diversity of ORF3 and spike genes of porcine epidemic diarrhea virus in Thailand. Infect. Genet. Evol. 21, 205–213. [DOI] [PubMed] [Google Scholar]
- Tian, Y. , Su D. P., Zhang H. M., Chen R. A., and He D. S., 2013: Complete genome sequence of a very virulent porcine epidemic diarrhea virus strain, CH/GDGZ/2012, Isolated in Southern China. Genome Announc. 1, e00645–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova, A. N. , Marthaler D., Wang Q., Culhane M. R., Rossow K. D., Rovira A., Collins J., and Saif L. J., 2014: Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 20, 1620–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z. Y. , Lu W. H., Li Z. L., Mo J. Y., Zeng X. D., Zeng Z. L., Sun B. L., Chen F., Xie Q. M., Bee Y. Z., and Ma J. Y., 2012: Complete genome sequence of novel porcine epidemic diarrhea virus strain GD‐1 in China. J. Virol. 86, 13824–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. J. , Sun Z., Zhang Y., Wang G. S., Wang H., Yang F. F., Tian F. L., and Jiang S. J., 2012: Complete genome sequence of a Vero cell‐adapted isolate of porcine epidemic diarrhea virus in eastern China. J. Virol. 86, 13858–13859. [DOI] [PMC free article] [PubMed] [Google Scholar]


