Dear Editor,
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has developed a global pandemic.1 The initial transmission of SARS-CoV-2 has been limited in the national wide of China during the first two month2, while a global spread is establishing with about 2 million laboratory confirmed infections and more than 126, 000 deaths from 185 countries by April 15, 2020.3
The genome of SARS-CoV-2 exhibited a relative high similarity among the early obtained strains.4 , 5 However, two key mutations were recently identified, potentially contributing to the sub-lineage classification of SARS-CoV-2.6 Although the genome structure of SARS-CoV-2 has been well documented, the temporal evolution and global transmission of the virus remains poorly investigated.
Here, we retrieved 313 SRAS-CoV-2 genomes from the GISAID (www.gisaid.org) database, from which 99 genomes with exact collection dates (before Feb 29, 2019) were selected to infer the origin time and global transmission of SARS-CoV-2 by Bayesian phylodynamic approaches.
To gain insight into the temporal evolutionary dynamics of SARS-CoV-2, we performed Markov Chain Monte Carlo (MCMC) algorithms implemented in BEAST 1.10.4 package with the 99 enrolled SARS-CoV-2 genomes. Generalized Time Reversible (GTR) with invariant sites as site heterogeneity model (GTR+I) was selected as the best-fit nucleotide substitution model by the Akaike Information Criterion (AIC) implemented in jModelTest. The estimated mean evolutionary rate of SARS-CoV-2 was estimated to be 6.14 × 10−6 subs/site/day (95% HPD: 3.61 × 10−6 –8.68 × 10−6 subs/site/day), corresponding to 2.24 × 10−3 subs/site/year (95% HPD: 1.32 × 10−3 –3.17 × 10−3 subs/site/year).
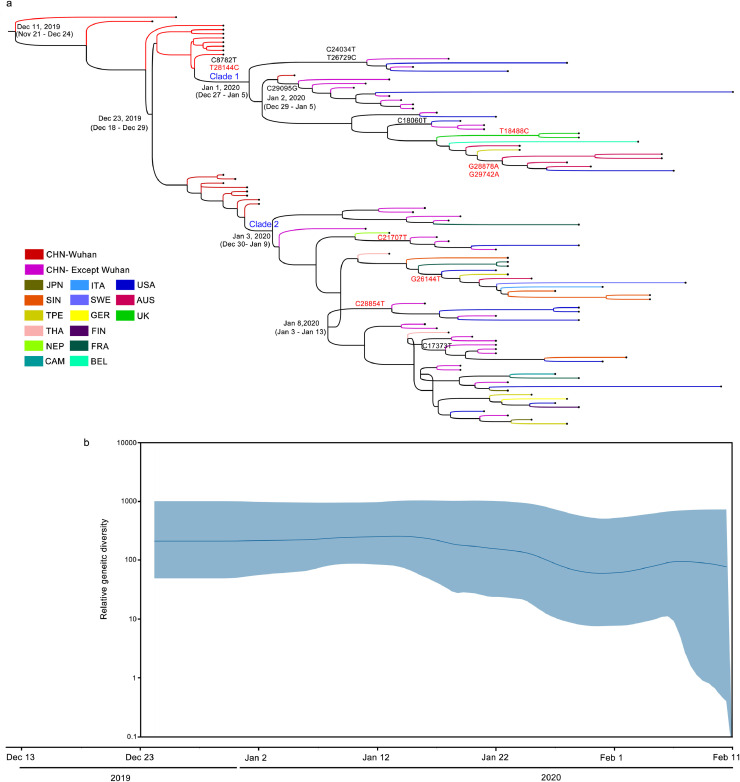
We recorded the information of MCMC reconstruction into a Maximum Clade Credibility (MCC) tree by using the program TreeAnnotator. From the MCC tree (Fig. 1 ), the tMRCA (Time to the Most Common Recent Ancestor) of SARS-CoV-2 was dated back to Dec 11, 2019 (95%HPD, Nov 21, 2019 – Dec 24, 2019). Two major clades were observed from the MCC tree, with a divergence time at Dec 23, 2019 (95%HPD, Dec 18, 2019 – Dec 29, 2019), both of which consist strains of SARS-CoV-2 from Wuhan and other regions of China.
Fig. 1.
Bayesian evolutionary dynamics of SARS-CoV-2. (a) Time-scaled Maximum Clade Credibility (MCC) tree based on MCMC analysis of the 99 SARS-CoV-2 genomes with an Exponential Growth tree prior. The time scale was set to the bottom of Fig. 1 shared by both (a) and (b). The tree branches were colored according to the collection countries with the color panel to the left lower part. Time to the Most Common Recent Ancestor (tMRCA) and the divergence time of clades and sub-clades were labelled on the corresponding nodes with 95% HPD in the following bracket. Clade and sub-clade specific mutations were labelled under the divergent time, with the non-synonymous mutations labelled in red. (b) population dynamics of SARS-CoV-2. The viral population dynamics was represented by the viral relative genetic diversity generated from Bayesian Skyline Plot reconstruction of the MCMC analysis. The Y-axis represents relative genetic diversity, which is equivalent to the product of the effective population size (Ne) and the generation length in days (τ). The color regions show 95% HPD limits, and the black line represent the median estimate of relative genetic diversity.
The circulating strains of SARS-CoV-2 could be separated into four sub-clades (Fig. 1a). The two sub-clades from Clade 1 was diverged at Jan 1, 2020 (95%HPD, Dec 27, 2019 – Jan 5, 2020), while the two sub-clades from Clade 2 was diverged at Jan 8, 2020 (95%HPD, Jan 3 – Jan 13). With respect to the country-specific strains of SARS-CoV-2, we observed that the circulating strains in USA were from both of the two clades, the UK and Australia circulating strains were from Clade 1, the circulating strains in Singapore, Japan, Germany, France and Italy seemed to be from Clade 2 (Fig. 1a, Table S1).
To infer the population growth dynamics of SARS-CoV-2, the viral relative genetic diversity was reconstructed by Bayesian Skyline Plot (BSP) analysis.7 BSP analysis suggested that SARS-CoV-2 possessed a relative stable effective population size (Ne) during the first month (Dec 23, 2019 to Jan 22, 2020) of the virus outbreak (Fig. 1b). A slow but accelerating reduction in the Ne was observed from Jan 22, 2020, with a sharp reduction of the lower 95% HPD of the Ne from Feb 5, 2020. A sharp reduction in the Ne suggests the initiation of a bottle-neck-effect in the virus population size. A bottle-neck-effect indicates that the current circulating virus strain was trapped, and more mutations in the virus genome will potentially occur to help the virus escape, resulting in a leap in the virus population. Despite the BSP was generated from a limited sample size, the results suggested a possible initiation of a bottle-neck-effect in the population size of SARS-CoV-2, indicating more infected cases will occur in the near future due to the increased mutations in the viral genome.
Despite SARS-CoV-2 remains relative stable, thirteen clade/sub-clade-specific mutations were observed in the present study (Fig. 1a). The mutations at nt 8782 and nt 28,144 were clade specific, i.e., C8782T and T28144C were only occurred in Clade 1, rather than in Clade 2. Only a viral strain (EPI_ISL_406,592 from Guangdong, China) in Clade 1 did not possess C8782T, while all strains in Clade 1 possess T28144C. Eleven out of the thirteen sub-clade specific mutations were also observed (Fig. 1a). Seven mutations were located in Clade 1, among which C29095G and C24034T/T26729C were observed in a sub-clade consisting of viral strains from China (outside Wuhan) and USA, respectively. G28878Aand G29742A were observed in a subclade of viral strains from Australia and USA. Four mutations were located in Clade 2, among which C21707T and C28854T were observed in a sub-clade consisting of viral strains from China (outside Wuhan) and USA. C17373T was observed in a sub-clade of viral strains from China (outside Wuhan), USA and Singapore. G26144T was observed in a sub-clade of viral strains from USA, Taiwan, Australia, Sweden, Italy, and Singapore.
Seven of the observed mutations resulted in non-synonymous mutations in the translated viral protein, including two mutations in nucleocapsid phosphoprotein (C28854T: Ser-Phe; G28878A: Ser-Asn), one mutation in ORF1ab polyprotein (T18488C: Ile-Thr), Surface glycoprotein (C21707T: His-Tyr), ORF3a protein (G26144T: Gly-Val), ORF8 protein (T28144C: Leu-Ser), and ORF10 protein (G29742A: Arg-His). Notably, all of the four sub-clades possessed at least one non-synonymous mutation (Fig. 1a, Table 1 ).
Table 1.
Clade-/Sub-clade specific mutations of SARS-CoV-2 observed in Maximum Clade Credibility tree.
| Mutation | Gene | Type | Amino acid mutation | Collection country/region of the viral strain |
|---|---|---|---|---|
| C8782T | ORF1a | synonymous | – | Clade 1 in Fig. 1a (detailed in Table S1) |
| C17373T | ORF1b | synonymous | – | China (outside Wuhan), USA and Singapore |
| C18060T | ORF1b | synonymous | – | China (outside Wuhan) and USA |
| C24034T | S | synonymous | – | |
| T26729C | M | synonymous | – | |
| C29095G | N | synonymous | – | |
| T18488C | ORF1b | non-synonymous | Ile-Thr | United Kingdom |
| C21707T | S | non-synonymous | His-Tyr | China (outside Wuhan) and USA |
| G26144T | ORF3 | non-synonymous | Gly-Val | USA, Taiwan, Australia, Sweden, Italy, and Singapore |
| T28144C | ORF8 | non-synonymous | Leu-Ser | Clade 1 in Fig. 1a (detailed in Table S1) |
| C28854T | N | non-synonymous | Ser-Phe | China (outside Wuhan) and USA |
| G28878A | N | non-synonymous | Ser-Asn | Australia and USA |
| G29742A | 3-UTR | non-synonymous | Arg-His (untranslated) |
Ile, Isoleucine; Thr, Threonine; His, Histidine; Tyr, Tyrosine; Gly, Glycine; Val, Valine; Leu, Leucine, Ser, Serine; Phe, Phenylalanine; Asn, Asparagine; Arg, Argnine. USA, United States of America.
In conclusion, continuous evolution occurred in almost all regions of the SARS-CoV-2 genome and potentially in a country-specific manner. Further efforts on monitoring the genomic mutations of SARS-CoV-2 from different countries are recommended.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Acknowledgements
The authors thank the researchers who share the genome data of SARS-CoV-2 to GISAID (http://www.gisaid.org/). This work was supported by a grant from Key R & D plan of Shanxi Province (No. 202003D31003/GZ to J. L.) and a grant from Major National Science and Technology Projects of the Ministry of Science and Technology of China (No. 2018ZX10305409-001–007 to J. L.).
Reference
- 1.Biondi-Zoccai G., Landoni G., Carnevale R., Cavarretta E., Sciarretta S., Frati G. SARS-CoV-2 and COVID-19: facing the pandemic together as citizens and cardiovascular practitioners. Minerva Cardioangiol. 2020 doi: 10.23736/S0026-4725.20.05250-0. Mar 9. PubMed PMID: 32150358. Epub 2020/03/10. [DOI] [PubMed] [Google Scholar]
- 2.Zhang S., Diao M.Y., Duan L., Lin Z., Chen D. The novel coronavirus (SARS-CoV-2) infections in China: prevention, control and challenges. Intensive Care Med. 2020 doi: 10.1007/s00134-020-05977-9. Mar 2. PubMed PMID: 32123989. Epub 2020/03/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30120-1. Feb 19. PubMed PMID: 32087114. Epub 2020/02/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. Feb 22. PubMed PMID: 32007145. Epub 2020/02/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. Mar 11. PubMed PMID: 32035028. Epub 2020/02/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilen C.B., Lee S., Hsieh L.L., Orchard R.C., Desai C., Hykes B.L., Jr. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science. 2018;360(6385):204–208. doi: 10.1126/science.aar3799. Apr 13. PubMed PMID: 29650672. Pubmed Central PMCID: PMC6039974. Epub 2018/04/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heled J., Drummond A.J. Bayesian inference of population size history from multiple loci. BMC Evol Biol. 2008;8:289. doi: 10.1186/1471-2148-8-289. Oct 23PubMed PMID: 18947398. Pubmed Central PMCID: PMC2636790. Epub 2008/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]