Abstract
The first total syntheses of the pro-resolving lipid mediators 7(S),13(R),20(S)-Resolvin T1 [7(S),13(R),20(S)-RvT1] and 7(S),13(R)-Resolvin T4 [7(S),13(R)-RvT4], derived from n-3 docosapentaenoic acid (n-3 DPA), are described. 7(S),13(R),20(S)-RvT1 was prepared from 7(S),13(R)-RvT4 via an enzymatic lipoxidase reaction. 7(S),13(R)-RvT4 was obtained by total synthesis where the chiral centers at C7 and C13 where introduced by a Noyori transfer hydrogenation and a chiral pool strategy respectively. Wittig reactions, Sonogashira coupling and Boland Zn(Cu/Ag) reduction were the key steps in the synthesis.
Keywords: 7(S),13(R),20(S)-RvT1; 7(S),13(R)-RvT4; Noyori transfer hydrogenation; Lipoxidase
Graphical Abstract
Inflammation is an immediate response to tissue injury and/or infection.1 The self-resolving acute inflammation is a protective mechanism, especially against infections, and it involves the biosynthesis of the specialized pro-resolving mediators (SPMs) which include the lipoxins,2 resolvins,3 maresins,4 protectins5 and their sulfide-conjugates.6–9 They are formed from poly-unsaturated fatty acids by lipoxidase (LOX), cytochrome P-450 monooxygenase and/or cyclooxygenase (COX) enzymes.3 In the situation that acute inflammation cannot be resolved chronic inflammatory diseases can develop including cancer, and autoimmune and neurological diseases.10–12
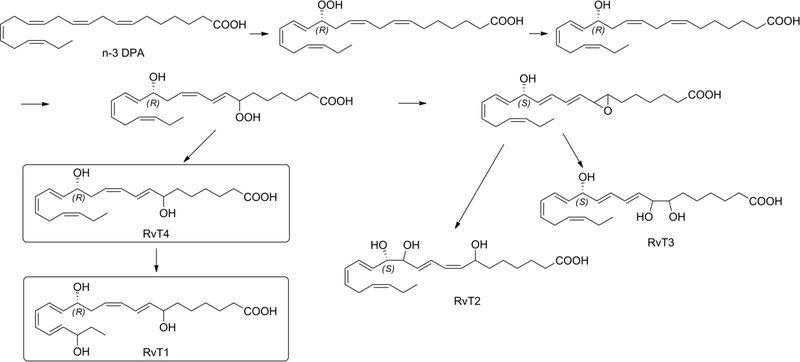
New SPMs derived from n-3 docosapentaenoic acid (n-3 DPA) have been identified.13–15 Recently Dalli, Chiang and Serhan discovered the 13-series Resolvin (RvT1, RvT2, RvT3 and RvT4) that are produced from n-3 DPA via an endothelial COX-2 mechanism combined with 5-lipoxidase from adjacent neutrophils to give the RvT1-RvT4.16 It should be noticed that neither eicosapentaenoic nor docosahexaenoic acid are substrates for this pathway. The RvT1, RvT2, RvT3 and RvT4 are formed during the early phase of bacterial infection. The RvTs protected animals against lethal doses of E-coli and therefore they could become an alternative to antibiotics.16
The proposed biosynthesis of RvT1-RvT4 is outlined in Figure 1.16,17 Dalli and collaborators showed that n-3 DPA is converted via endothelial COX-2 to the 13(R)-hydroperoxy-docosapentaenoic acid that after reduction gives 13(R)-hydroxy-docosapentaenoic acid. The synthesis of this precursor has been recently described by Hansen and collaborators.17 13(R)-hydroxy-docosapentaenoic acid is converted to 7-hydroperoxy13(R)-hydroxy-docosapentaenoic acid that after reduction of the hydroperoxy-group gives RvT4. Enzymatic lipoxidase reaction transforms RvT4 into RvT1. 18O2 incorporation experiments confirmed that the hydroxyl groups at C-7 and C-20 are derived from enzymatic lipoxidase reaction. The 7-hydroperoxy-13(R)-hydroxy-docosapentaenoic acid can form an allylic epoxide intermediate that gives rise to RvT2 and RvT3 as shown in Figure 1. It is worthwhile to note that due to the change in priority of the groups surrounding chiral center C13 in RvT2 and RvT3, according to the Cahn, Ingold, Prelog rules of nomenclature, this center has now to be assigned as S.
Figure 1.
Proposed biosynthesis of RvT1, RvT2, RvT3 and RvT4.
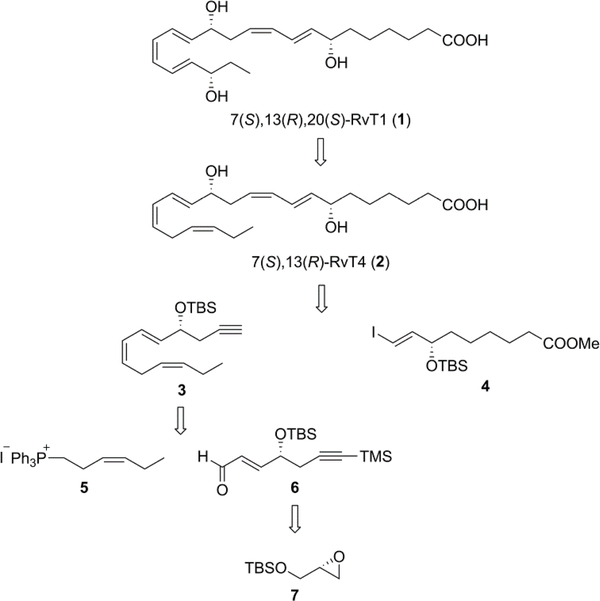
In order to explore the potent biological activities of these novel 13-series resolvins and due to the low abundance from natural sources they need to be prepared by total organic synthesis. Since lipoxidases in neutrophils introduce the hydroperoxy-groups with the (S)-chirality in poly-unsaturated fatty acids herein we describe the total synthesis of 7(S),13(R),20(S)-RvT1 [(7S,8E,10Z,13R,14E,16Z,18E,20S)-7,13,20-trihydroxy-8,10,14,16,18-docosapentaenoic acid (1)] and 7(S),13(R)-RvT4 [(7S,8E,10Z,13R,14E,16Z,19Z)-7,13-dihydroxy-8,10,14,16,19-docosapentaenoic acid (2)].
As shown in the retrosynthetic scheme (Figure 2) 7(S),13(R),20(S)-RvT1 (1) was synthesized from 7(S),13(R)-RvT4 (2) by enzymatic reaction with lipoxidase. Compound 2 was prepared by Sonogashira coupling of the key intermediates 3 and 4. The chiral center in 4 was generated by a Noyori Ru transfer hydrogenation. Intermediate 3 was synthesized from 5 and 6 via a Wittig reaction whereas the chiral center in 6 was obtained from optical pure glycidol derivative 7.
Figure 2.
Retrosynthetic approach to 7(S),13(R),20(S)-RvT1 and 7(S),13(R)-RvT4.
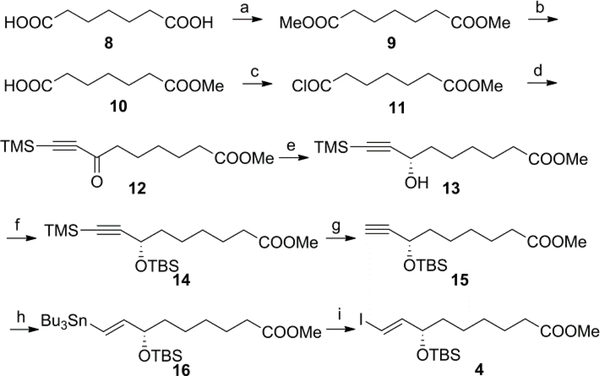
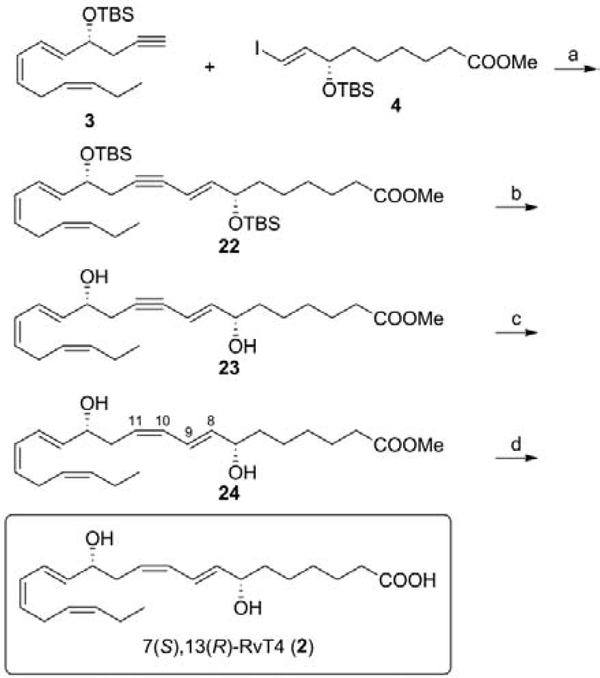
The C1–C9 fragment 4 was prepared in nine steps as outlined in Scheme 1. Pimelic acid was converted into its half ester 10 in two steps. Esterification at room temperature produced the diester 9 in quantitative yield.18 Enzymatic cleavage of 9 with porcine pancreatic lipase (PPL, Sigma) gave half ester 10 in 92% yield.19 Compound 10 was converted into the acid chloride 11 with oxalyl chloride in the presence of catalytic DMF in CH2Cl2.20 Crude compound 11 was reacted with bis(trimethylsilyl)acetylene in the presence of AlCl3 to afford ketone 12 in 65% yield over two steps. Asymmetric reduction of the ketone 12 in H2O/ethyl acetate in the presence of a catalytic amount of the phase transfer catalyst cetyltrimethylammonium bromide (CTABr) using the Noyori RuCl[(S,S)-TsDPEN](p-cymene) precatalyst (0.04 equiv) with sodium formate as a reducing agent, produced the chiral intermediate 13 with >94% ee as determined by chiral HPLC [Chiracel OD, hexane/i-PrOH 90:10, 0.6 mL/min, 210 nm, tR = 8.0 min (R-isomer) and tR = 8.5 min (S-isomer, 13)].21,22 Protection of 13 with TBSCl in the presence of imidazole and 4-DMAP in CH2Cl2 gave the silyl ether 14 in 93% yield. TMS desilylation of 14 with 1.2 equiv of K2CO3 in the presence of Na2SO4 in CH3OH gave cleanly the terminal acetylene 15. Hydrostannylation of 15 with 1.2 equiv tributyltin hydride in benzene at 80 °C in the presence of 10% of AIBN produced the trans-vinyl tin compound 16. The excess of tributyltin hydride was removed in high vacuo. The key intermediate 4 was obtained from 16 by reaction with iodine in CH2Cl2 at 0 °C in 83% yield over two steps.
Scheme 1.
Reagents and conditions: (a) TMSCl, 2,2-dimethoxypropane, CH3OH, rt, quantitative; (b) PPL, 0.05M phosphate buffer pH 7, 1 N NaOH, 92%; (c) (COCl)2, cat. DMF, CH2Cl2, rt; (d) bis(trimethylsilyl)acetylene, AlCl3, CH2Cl2, 0 °C, 65% (over two steps); (e) RuCl[(S,S)-TsDPEN](p-cymene), CTABr, NaCOOH, H2O, EtOAc, rt, 97%; (f) TBSCl, imidazole, 4-DMAP, CH2Cl2, 0 °C to rt, 93%; (g) K2CO3, Na2SO4, CH3OH, rt, 98%; (h) (Bu)3SnH, AIBN, benzene, reflux; (i) I2, CH2Cl2, 0 °C, 83% (over two steps).
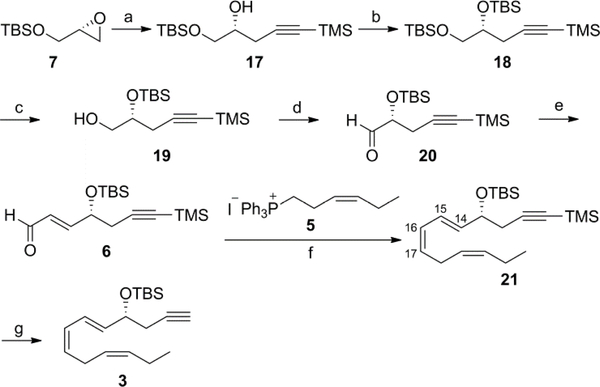
The C10-C22 intermediate 3 was obtained from commercial R(−)-TBS-glycidol (7) in seven steps (Scheme 2). Reaction of 7 with 2 equiv lithium trimethylsilylacetylene in the presence of BF3·Et2O gave the alcohol 17 in 90% yield that was converted to the di-TBS derivative 18 with 1.6 equiv TBSCl in CH2Cl2 in 89% yield.23 Selective deprotection of the primary TBS-ether was accomplished with 10-camphorsulfonic acid (CSA) in CH2Cl2/CH3OH at 0 °C to give 19 in 64% isolated yield. Dess-Martin periodinane oxidation of 19 in CH2Cl2 produced the aldehyde 20.24 Crude 20 was reacted with 1 equiv of (Triphenylphosphoranylidene)acetaldehyde in CH3CN at 30 °C for 15 h to give the α,β-unsaturated aldehyde 6 in 58% yield over two steps.25 Wittig reaction of 6 with 4 equiv of the ylide generated from the crystalline phosphonium salt 526,27 and n-BuLi in THF gave 21 in 82% yield. The geometry of the 14E,16Z-diene unit in 21 was confirmed by the 1H-1H coupling constants (J14,15 = 15.3 Hz and J16,17 = 11.1 Hz).28 Removal of the TMSgroup with K2CO3 in CH3OH gave the key intermediate 3 in quantitative yield.29
Scheme 2.
Reagents and conditions: (a) Trimethylsilylacetylene, n-BuLi, BF3·Et20, THF, –78 °C, 90%; (b) TBSCl, imidazole, 4-DMAP, CH2Cl2, 0 °C to rt, 89%; (c) CSA, CH2Cl2/CH3OH 1/1, 0 °C, 64%; (d) Dess-Martin periodinane, CH2Cl2, rt; (e) (Triphenylphosphoranylidene)acetaldehyde, CH3CN, 30 °C, 58% (over two steps); (f) 5, n-BuLi, THF, –78 °C to 0 °C, 82%; (g) K2CO3, CH3OH, rt, quantitative.
The skeleton of 7(S),13(R)-RvT4 was assembled from the key intermediates 3 and 4 as outlined in Scheme 3. Pd0/CuI Sonogashira coupling of 3 with 4 produced compound 22. The synthesis was completed via deprotection of the TBS groups of compound 22 with catalytic HCl, generated in situ from acetyl chloride in absolute CH3OH at 0 °C to rt, to give 23. Boland Zn(Cu/Ag) reduction30 of 23 in CH3OH/H2O at 40–45 °C (7 h) produced crude 7(S),13(R)-RvT4 methyl ester (24) that was purified by HPLC [Zorbax SB-C18 250 × 21.2 mm, 235 nm, CH3OH/H2O 76/24] to give 24 in 87% yield. The geometry of the 8E,10Z-diene unit in 24 was confirmed by the 1H-1H coupling constants (J8,9 = 15.3 Hz and J10,11 = 11.1 Hz).28 Mild alkaline hydrolysis of 24 with 1 N LiOH in CH3OH/H2O at 0 °C to rt gave after HPLC purification [Zorbax SB-C18 250 × 21.2 mm, 235 nm, CH3OH/H2O (0.1% NH4OAc, pH 5.6, 0.05% EDTA disodium) 70/30] and desalting 7(S),13(R)-RvT4 (2) in 80% yield. The 1H NMR, 13C NMR, UV, and HPLC/UV/MS analysis were consistent with the structure of 2.28
Scheme 3.
Reagents and conditions: (a) Pd(PPh3)4, CuI, piperidine, benzene, rt; (b) CH3COCl, CH3OH, 0 °C to rt, 31% (over two steps); (c) Zn(Cu/Ag), CH3OH, H2O, 40–45 °C, 87%; (d) 1 N LiOH, CH3OH/H2O 1/1, 0 °C to rt, 80%.
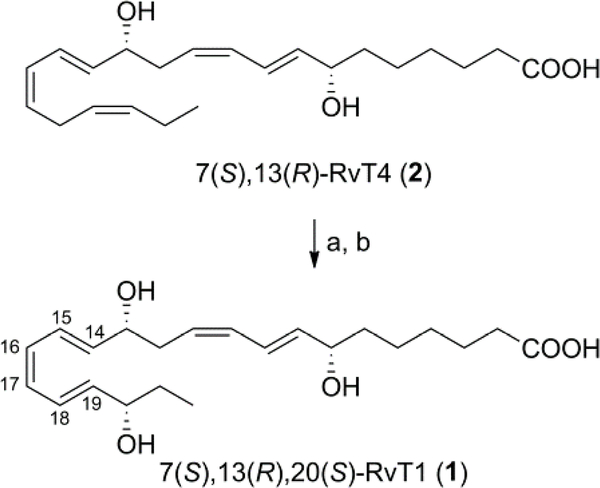
For the synthesis of 7(S),13(R),20(S)-RvT1 (1) we considered to use the enzymatic hydroxylation of 7(S),13(R)-RvT4 (2) (Scheme 4). This type of approach was successfully employed in the total syntheses of RCTRs,31 lipoxins and other lipid mediators.32–36 The reaction had to be optimized with respect to enzyme, pH and buffer. Compound 2 was reacted in borate buffer pH 10.7, with lipoxidase Type I-B to give after reduction with tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) directly 7(S),13(R),20(S)-RvT1 (1).37 Purification by HPLC and desalting gave pure 1. The geometry of the 14E,16Z,18E-triene unit in 1 was confirmed by the 1H-1H coupling constants (J14,15 = 15.3 Hz, J16,17 ~ 10.8 Hz and J18,19 = 15.3 Hz).28,38,39 The 1H NMR, 13C NMR, UV, and HPLC/UV/MS analysis were consistent with the structure of 1.28
Scheme 4.
Reagents and conditions: (a) lipoxidase type I-B from soybean, 0.01 M borate buffer pH 10.7, rt; (b) TCEP-HCl, rt, 22% (over two steps after HPLC purification and desalting).
In summary, the total syntheses of 7(S),13(R),20(S)-RvT1 and 7(S),13(R)-RvT4 have been achieved, making these pro-resolving lipid mediators from n-3 DPA available for further biological testing. The syntheses of RvT2 and RvT3, will be reported in due course.
Highlights.
First total syntheses 7(S),13(R),20(S)-Resolvin T1 and 7(S),13(R)-Resolvin T4 have been achieved.
RvT1 is synthesized from RvT4 via an enzymatic hydroxylation with lipoxidase.
RvT4 chiral centers at C7 and C13 were introduced by a Noyori transfer hydrogenation and a chiral pool strategy respectively.
Wittig reactions, Sonogashira coupling and Boland Zn(Cu/Ag) reduction were the key steps.
Acknowledgments
Financial support of this research by the NIH (R01AI128202) is gratefully acknowledged.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Serhan CN, Levy BD, J. Clin. Invest 128 (2018) 2657–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serhan CN, Hamberg M, Samuelsson B, Proc. Natl. Acad. Sci. USA 81 (1984) 5335–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serhan CN, Petasis NA, Chem. Rev 111 (2011) 5922–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serhan CN, Dalli J, Karamnov S, Choi A, Park CK, Xu ZZ, Ji RR, Zhu M, Petasis NA, FASEB J. 26 (2012) 1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serhan CN, Gotlinger K, Hong S, Lu Y, Siegelman J, Baer T, Yang R, Colgan SP, Petasis NA, J. Immunol 176 (2006) 1848–1859. [DOI] [PubMed] [Google Scholar]
- 6.Dalli J, Ramon S, Norris PC, Colas RA, Serhan CN, FASEB J. 29 (2015), 2120–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalli J, Chiang N, Serhan CN, Proc. Natl. Acad. Sci. USA 111 (2014) E4753–E4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalli J, Vlasakov I, Riley IR, Rodriguez AR, Spur BW, Petasis NA, Chiang N, Serhan CN, Proc. Natl. Acad. Sci. USA 113 (2016) 12232–12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Rosa X, Norris PC, Chiang N, Rodriguez AR, Spur BW, Serhan CN, Am. J. Pathol 188 (2018) 950–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serhan CN Nature 510 (2014) 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serhan CN, FASEB J. 31 (2017) 1273–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serhan CN, Chiang N, Dalli J, Mol. Aspects Med 64 (2018) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vik A, Dalli J, Hansen TV, Bioorg. Med. Chem. Lett 27 (2017) 2259–2266. [DOI] [PubMed] [Google Scholar]
- 14.Walker ME, Souza PR, Colas RA, Dalli J, FASEB J. 31 (2017) 3636–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen TV, Dalli J, Serhan CN, Prostaglandins Other Lipid Mediat. 133 (2017) 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalli J, Chiang N, Serhan CN, Nat. Med 21 (2015) 1071–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Primdahl KG, Aursnes M, Walker ME, Colas RA, Serhan CN, Dalli J, Hansen TV, Vik A, J. Nat. Prod 79 (2016) 2693–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez A, Nomen M, Spur BW, Godfroid J-J, Tetrahedron Lett. 39 (1998) 8563–8566. [Google Scholar]
- 19.Rodriguez A, Nomen M, Spur BW, Godfroid J-J, Eur. J. Org. Chem (1999) 2655–2662. [Google Scholar]
- 20.Meyners C, Mertens M, Wessig P, Meyer-Almes F-J, Chem. Eur. J 23 (2017) 3107–3116. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez AR, Spur BW, Tetrahedron Lett. 53 (2012) 1912–1915. [Google Scholar]
- 22.Rodriguez AR, Spur BW, Tetrahedron Lett. 53 (2012) 86–89. [Google Scholar]
- 23.Yahata K, Ye N, Iso K, Ai Y, Lee J, Kishi Y, J. Org. Chem 82 (2017) 8808–8830. [DOI] [PubMed] [Google Scholar]
- 24.Dess DB, Martin JC, J. Am. Chem. Soc 113 (1991) 7277–7287. [Google Scholar]
- 25.Allard M, Barnes K, Chen X, Cheung Y-Y, Duffy B, Heap C, Inthavongsay J, Johnson M, Krishnamoorthy R, Manley C, Steffke S, Varughese D, Wang R, Wang Y, Schwartz CE, Tetrahedron Lett. 52 (2011) 2623–2626. [Google Scholar]
- 26.Kojima K, Koyama K, Amemiya S, Tetrahedron 41 (1985) 4449–4462. [Google Scholar]
- 27.Oger C, Bultel-Ponce V, Guy A, Durand T, Galano J-M, Eur. J. Org. Chem (2012) 2621–2634. [Google Scholar]
- 28.Satisfactory spectroscopic data were obtained for all compounds. Selected physical data: Compound 10: 1H NMR (CDCl3, 300 MHz): δ 3.6 (s, 3H), 2.4–2.3 (t, J = 7.5 Hz, 2H), 2.3–2.2 (t, J = 7.5 Hz, 2H), 1.7–1.5 (m, 4H), 1.4–1.3 (m, 2H); 13C NMR (CDCl3, 75.5 MHz): δ 179.43, 174.12, 51.53, 33.77, 33.74, 28.44, 24.48, 24.23. Compound 11: 1H NMR (CDCl3, 300 MHz): δ 3.6 (s, 3H), 2.9–2.8 (t, J = 7.2 Hz, 2H), 2.3 (t, J = 7.5 Hz, 2H), 1.8–1.5 (m, 4H), 1.4–1.3 (m, 2H); 13C NMR (CDCl3, 75.5 MHz): δ 173.73, 173.62, 51.50, 46.77, 33.57, 27.78, 24.63, 24.27. Compound 12: 1H NMR (CDCl3, 300 MHz): δ 3.6 (s, 3H), 2.6–2.5 (t, J = 7.3 Hz, 2H), 2.3 (t, J = 7.3 Hz, 2H), 1.7–1.5 (m, 4H), 1.4–1.2 (m, 2H), 0.2 (s, 9H); 13C NMR (CDCl3, 75.5 MHz): δ 187.60, 173.96, 101.93, 97.73, 51.47, 44.96, 33.77, 28.33, 24.56, 23.45, –0.79 (3C). Compound 13: 1H NMR (CDCl3, 300 MHz): δ 4.4–4.3 (t, J = 6.6 Hz, 1H), 3.6 (s, 3H), 2.3 (t, J = 7.5 Hz, 2H), 1.9 (s, 1H), 1.7–1.6 (m, 4H), 1.5–1.3 (m, 4H), 0.14 (s, 9H); 13C NMR (CDCl3, 75.5 MHz): δ 174.18, 106.72, 89.37, 62.71, 51.46, 37.38, 33.91, 28.67, 24.75, 24.70, –0.15 (3C); [α]D20 = –0.5 (c 0.63, CHCl3). Compound 14: 1H NMR (CDCl3, 300 MHz): δ 4.3 (t, J = 6.6 Hz, 1H), 3.6 (s, 3H), 2.3 (t, J = 7.5 Hz, 2H), 1.7–1.6 (m, 4H), 1.5–1.3 (m, 4H), 0.87 (s, 9H), 0.12 (s, 9H), 0.10 (s, 3H), 0.08 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 174.21, 107.78, 88.41, 63.24, 51.44, 38.21, 33.99, 28.72, 25.80 (3C), 24.87, 24.84, 18.27, –0.18 (3C), –4.48, – 4.95; [α]D20 = –37 (c 0.58, CHCl3). Compound 15: 1H NMR (CDCl3, 300 MHz): δ 4.3 (td, J = 6.6, 2.1 Hz, 1H), 3.6 (s, 3H), 2.4–2.3 (d, J = 2.1 Hz, 1H), 2.3 (t, J = 7.5 Hz, 2H), 1.7–1.5 (m, 4H), 1.5–1.2 (m, 4H), 0.87 (s, 9H), 0.11 (s, 3H), 0.08 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 174.18, 85.58, 71.96, 62.61, 51.45, 38.30, 33.98, 28.75, 25.75 (3C), 24.85, 24.72, 18.20, –4.59, –5.09; [α]D20 = –36 (c 0.52, CHCl3). Compound 4: 1H NMR (CDCl3, 300 MHz): δ 6.5 (dd, J = 14.4, 6.0 Hz, 1H), 6.2–6.1 (dd, J = 14.4, 1.2 Hz, 1H), 4.2–4.0 (m, 1H), 3.6 (s, 3H), 2.3 (t, J = 7.5 Hz, 2H), 1.7– 1.2 (m, 8H), 0.86 (s, 9H), 0.02 (s, 3H), 0.00 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 174.17, 149.22, 75.58, 75.02, 51.47, 37.28, 33.99, 29.04, 25.80 (3C), 24.85, 24.48, 18.19, –4.53, –4.90; [α]D20 = –18.5 (c 0.35, CHCl3). Compound 18:1H NMR (CDCl3, 300 MHz): δ 3.9–3.7 (m, 1H), 3.6–3.4 (2 dd, J = 10.2, 5.4 Hz and J = 10.2, 6.3 Hz, 2H ABsystem), 2.5 (dd, J = 16.8, 5.4 Hz, 1H ABsystem), 2.3–2.2 (dd, J = 16.8, 6.6 Hz, 1H ABsystem), 0.88 (s, 18H), 0.12 (s, 9H), 0.09 (s, 3H), 0.06 (s, 3H), 0.03 (s, 3H), 0.03 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 104.71, 85.78, 72.08, 66.67, 25.96 (3C), 25.84 (3C), 25.76, 18.37, 18.13, 0.07 (3C), – 4.46, – 4.61, – 5.34, – 5.40; [α]D20 = +9.7 (c 0.23, CHCl3). Compound 19: 1H NMR (CDCl3, 300 MHz): δ 3.9–3.8 (m, 1H), 3.7–3.6 (dd, J = 11.1, 3.9 Hz, 1H ABsystem), 3.6–3.5 (dd, J = 11.1, 4.8 Hz, 1H ABsystem), 2.5–2.4 (dd, J = 16.8, 6.9 Hz, 1H ABsystem), 2.4–2.3 (dd, J = 16.8, 6.3 Hz, 1H ABsystem), 0.88 (s, 9H), 0.11 (s, 9H), 0.10 (s, 3H), 0.09 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 103.31, 86.73, 71.47, 65.87, 25.76 (3C), 25.35, 18.03, –0.02 (3C), –4.52, –4.77. Compound 20: 1C NMR (CDCl3, 300 MHz): δ 9.6 (d, J = 1.2 Hz 1H), 4.1 (ddd, J = 7.8, 5.1, 1.2 Hz, 1H), 2.7–2.6 (dd, J = 16.8, 5.1 Hz, 1H ABsystem), 2.6–2.4 (dd, J = 16.8, 7.8 Hz, 1H ABsystem), 0.94 (s, 9H), 0.15 (s, 9H), 0.13 (s, 3H), 0.07 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 202.10, 101.85, 87.36, 76.08, 25.70 (3C), 24.47, 18.21, 1.01, –0.09 (3C), –4.75. Compound 6: 1C NMR (CDCl3, 300 MHz): δ 9.6 (d, J = 7.8 Hz, 1H), 6.9 (dd, J = 15.6, 4.2 Hz, 1H), 6.3 (ddd, J = 15.6, 7.8, 1.8 Hz, 1H), 4.5 (dddd, J = 7.5, 6.3, 4.2, 1.8 Hz, 1H), 2.5 (dd, J = 16.5, 6.3 Hz, 1H ABsystem), 2.4 (dd, J = 16.5, 7.5 Hz, 1H ABsystem), 0.89 (s, 9H), 0.13 (s, 9H), 0.09 (s, 3H), 0.05 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 193.45, 157.82, 131.23, 102.05, 87.77, 70.49, 28.93, 25.70 (3C), 18.13, –0.06 (3C), –4.80, –4.87. Compound 21: 1C NMR (CDCl3, 300 MHz): δ 6.5 (dd, J = 15.3, 11.1 Hz, 1H), 6.0– 5.9 (t, J = 11.1 Hz, 1H), 5.7 (dd, J = 15.3, 6.0 Hz, 1H), 5.5–5.2 (m, 3H), 4.4–4.2 (m, 1H), 3.0–2.8 (m, 2H), 2.5–2.4 (dd, J = 16.8, 6.9 Hz, 1H ABsystem), 2.4–2.3 (dd, J = 16.8, 6.3 Hz, 1H ABsystem), 2.1–2.0 (m, 2H), 1.0–0.9 (t, J = 7.5 Hz, 3H), 0.90 (s, 9H), 0.12 (s, 9H), 0.09 (s, 3H), 0.05 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 135.34, 132.37, 130.32, 127.70, 126.60, 124.86, 104.17, 86.20, 71.92, 30.04, 25.96, 25.83 (3C), 20.55, 18.25, 14.22, 0.05 (3C), – 4.52, –4.71. Compound 3: 1C NMR (CDCl3, 300 MHz): δ 6.6–6.5 (ddt, J = 15.3, 11.1, 1.2 Hz, 1H), 6.0 (t, J = 11.1 Hz, 1H), 5.8–5.7 (dd, J = 15.3, 5.7 Hz, 1H), 5.5–5.2 (m, 3H), 4.4–4.3 (m, 1H), 3.0– 2.8 (m, 2H), 2.4 (ddd, J = 16.5, 6.0, 2.7 Hz, 1H ABsystem), 2.3 (ddd, J = 16.5, 6.9, 2.7 Hz, 1H ABsystem), 2.1–2.0 (m, 2H), 2.0– 1.9 (t, J = 2.7 Hz, 1H), 1.0–0.9 (t, J = 7.5 Hz, 3H), 0.90 (s, 9H), 0.08 (s, 3H), 0.05 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 134.97, 132.37, 130.46, 127.64, 126.56, 125.10, 81.29, 71.63, 69.97, 28.59, 25.98, 25.80 (3C), 20.55, 18.23, 14.22, –4.56, –4.80. Compound 22: 1C NMR (CDCl3, 300 MHz): δ 6.6–6.5 (br dd, J = 15.0, 11.1 Hz, 1H), 6.1–5.9 (m, 2H), 5.8–5.7 (dd, J = 15.0, 5.7 Hz, 1H), 5.6–5.5 (m, 1H), 5.5–5.2 (m, 3H), 4.4–4.3 (m, 1H), 4.2–4.0 (m, 1H), 3.6 (s, 3H), 3.0–2.8 (m, 2H), 2.5 (ddd, J = 16.5, 6.9, 2.1 Hz, 1H ABsystem), 2.4 (ddd, J = 16.5, 6.3, 2.1 Hz, 1H ABsystem), 2.3–2.2 (t, J = 7.5 Hz, 2H), 2.1–2.0 (m, 2H), 1.7–1.2 (m, 8H), 1.0–0.9 (t, J = 7.5 Hz, 3H), 0.89 (s, 9H), 0.86 (s, 9H), 0.08 (s, 3H), 0.05 (s, 3H), 0.01 (s, 3H), 0.00 (s, 3H). Compound 23: 1C NMR (CDCl3, 300 MHz): δ 6.6 (ddt, J = 15.3, 11.1, 1.2 Hz, 1H), 6.1–6.0 (dd, J = 15.9, 6.3 Hz, 1H), 6.0 (t, J = 11.1 Hz, 1H), 5.8–5.7 (dd, J = 15.3, 6.6 Hz, 1H), 5.7–5.6 (m, 1H), 5.5–5.2 (m, 3H), 4.4–4.3 (m, 1H), 4.2–4.0 (m, 1H), 3.6 (s, 3H), 3.0–2.8 (m, 2H), 2.6 (ddd, J = 16.8, 5.4, 2.1 Hz, 1H ABsystem), 2.5 (ddd, J = 16.8, 6.3, 2.1 Hz, 1H ABsystem), 2.3 (t, J = 7.5 Hz, 2H), 2.2–2.0 (m, 2H), 1.7–1.2 (m, 8H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 174.19, 145.16, 133.79, 132.48, 131.41, 127.53, 126.42, 126.37, 109.94, 86.39, 80.94, 72.17, 70.72, 51.48, 36.65, 33.94, 28.92, 28.70, 25.99, 24.86, 24.77, 20.56, 14.22. 7(S),13(R)-RvT4 methyl ester (24): 1C NMR (CD3OD, 300 MHz): δ 6.6–6.4 (2 dd, J = 15.3, 11.1Hz, 2H), 6.2–6.0 (t, J = 11.1 Hz, 1H), 6.0–5.9 (t, J = 11.1 Hz, 1H), 5.7–5.6 (m, 2H), 5.5–5.2 (m, 4H), 4.2–4.1 (m, 1H), 4.1–4.0 (m, 1H), 3.6 (s, 3H), 3.0–2.9 (m, 2H), 2.5–2.4 (m, 2H), 2.3 (t, J = 7.5 Hz, 2H), 2.2–2.0 (m, 2H), 1.7–1.3 (m, 8H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CD3OD, 75.5 MHz): δ 175.97, 138.02, 136.97, 133.13, 131.11, 131.08, 129.12, 128.13, 127.79, 126.55, 126.48, 73.22, 73.04, 51.97, 38.19, 36.80, 34.74, 30.11, 26.83, 26.22, 25.98, 21.47, 14.63. 7(S),13(R)-RvT4 (2): 1C NMR (CD3OD, 300 MHz): δ 6.6–6.4 (2 dd, J = 15.3, 11.1 Hz, 2H), 6.2–6.0 (t, J = 11.1 Hz, 1H), 6.0–5.9 (t, J = 11.1 Hz, 1H), 5.7–5.6 (2 dd, J = 15.3, 6.3 Hz and J = 15.3, 7.2 Hz, 2H), 5.5–5.2 (m, 4H), 4.2–4.1 (m, 1H), 4.1–4.0 (m, 1H), 3.0– 2.9 (m, 2H), 2.5–2.3 (m, 2H), 2.3–2.2 (t, J = 7.5 Hz, 2H), 2.2–2.0 (m, 2H), 1.7–1.3 (m, 8H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CD3OD, 75.5 MHz): δ 178.81, 138.07, 136.97, 133.12, 131.11 (2C), 129.13, 128.08, 127.79, 126.50 (2C), 73.24, 73.05, 38.29, 36.80, 35.86, 30.30, 26.83, 26.43, 26.32, 21.47, 14.63; UV (CD3OD) λmax 239 nm. HPLC/UV: Zorbax SB-C18, 1.8 μm, 50 × 2.1 mm, 237 nm, CD3OD/CD3OD (0.1% formic acid) 50:50–70:30, 0.2 mL/min, tR = 17.3 min; HPLC/MS (m/z): 361.2 [M-H] –. 7(S),13(R),20(S)-RvT1 (1): 1C NMR (CD3OD, 300 MHz): δ 6.8– 6.6 (m, 2H), 6.5 (dd, J = 15.3, 11.1 Hz, 1H), 6.2–6.0 (t, J = 11.1 Hz, 1H), 6.0–5.9 (m, 2H), 5.8–5.6 (3 dd, J = 15.3, 6.3 Hz, J = 15.3, 6.6 Hz and J = 15.3, 6.9 Hz, 3H), 5.5–5.4 (dt, J = 10.8, 7.5 Hz, 1H), 4.3–4.1 (m, 1H), 4.1–4.0 (m, 2H), 2.5–2.4 (m, 2H), 2.2 (t, J = 7.5 Hz, 2H), 1.7–1.3 (m, 10H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CD3OD, 75.5 MHz): δ C1 not observed, 138.42, 138.00, 137.74, 131.19, 130.11, 129.81, 127.95, 126.60, 126.53 (2C), 74.63, 73.27, 72.96, 38.19, 36.87, 36.70, 31.08, 30.32, 26.75, 26.27, 10.15. UV (EtOH) λmax 238, 260, 270, 280 nm. HPLC/UV: Zorbax SB-C18, 1.8 μm, 50 × 2.1 mm, 269 nm, CD3OD/CD3OD (0.1% formic acid) 50:50–70:30, 0.2 mL/min, tR = 12.4 min; HPLC/MS (m/z): 377.2 [M-H] –.
- 29.Cai C, Vasella A, Helv. Chim. Acta 78 (1995) 732–757. [Google Scholar]
- 30.Boland W, Schroer N, Sieler C, Feigel M, Helv. Chim. Acta 70 (1987) 1025–1040. [Google Scholar]
- 31.Rodriguez AR, Spur BW, Tetrahedron Lett. 58 (2017) 1662–1668. [Google Scholar]
- 32.Adams J, Fitzsimmons BJ, Girard Y, Leblanc Y, Evans JF, Rokach J, J. Am. Chem. Soc 107 (1985) 464–469. [Google Scholar]
- 33.Corey EJ, Mehrotra MM, Su W, Tetrahedron Lett. 26 (1985) 1919–1922. [Google Scholar]
- 34.Corey EJ, Su W, Cleaver MB, Tetrahedron Lett. 30 (1989) 4181–4184. [Google Scholar]
- 35.Itoh T, Saito T, Yamamoto Y, Ishida H, Yamamoto K, Bioorg. Med. Chem. Lett 26 (2016) 343–345. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez AR, Spur BW, Tetrahedron Lett. 59 (2018) 1143–1146. [Google Scholar]
- 37. To compound 2 (2.8 mg, 7.7×10−3 mmol) in 0.01 M solution of borate buffer pH 10.7 (10 ml) was added lipoxidase Type I-B from soybean (Sigma cat. # L7395–15MU, EC No. 232–853-1, 158000 U/mg) (9 mg) at rt and stirred for 1 h 45 min. The reaction was monitored by UV showing λmax at 271 nm. The solution was purged with argon and TCEP-HCl (4.6 mg, 0.016 mmol) was added. After stirring for 1 min the reaction mixture was passed through a reversed phase C–18 cartridge, washed with H2O and compound 1 eluted with CH3OH/H2O (8/2). Purification by HPLC [Zorbax SB-C18 250 × 21.2 mm, CH3OH/H2O (0.1% NH4OAc, pH 5.6, 0.05% EDTA disodium) 60/40] and desalting gave pure 7(S),13(R),20(S)-RvT1 (1) (0.64 mg, 22%).
- 38.Chen P, Fenet B, Michaud S, Tomczyk N, Vericel E, Lagarde M, Guichardant M, FEBS Lett. 583 (2009) 3478–3484. [DOI] [PubMed] [Google Scholar]
- 39.Liu M, Chen P, Vericel E, Lelli M, Beguin L, Lagarde M, Guichardant M, J. Lipid Res 54 (2013) 2083–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]