Abstract
Fusicoccin A (FC) is a diterpene glycoside that stabilizes protein–protein interactions (PPIs) between 14–3–3 adapter proteins and their phosphoprotein interaction partners. Recently, FC has gained attention for its pro-apoptotic and neuroprotective properties in cell culture. Although the exact molecular mechanism(s) is (are) unresolved, 14–3–3 PPIs are central to this activity. With the goal of refining the pharmacology of this chemotype, we conducted a systematic analysis of the structural features that govern FC-induced stabilization of 14–3–3 PPIs utilizing a C-terminal phosphorylation recognition motif. This study confirmed that a C-terminal amino acid with a small alkyl group is required for the interaction of FC at canonical C-terminal 14–3–3 PPI interfaces. Using bioinformatics, this structural insight was leveraged to assemble a database of 119 candidate 14–3–3 PPIs that can serve as targets for FC. This group includes a subset of proteins with experimentally determined C-terminal phosphosites that have not been explored as potential targets of FC.
Graphical Abstract

Protein–protein interactions (PPIs) encompass an important contemporary class of drug targets.1,2 Existing small-molecule-based strategies for PPI modulation focus on inhibition. This approach has been effective in instances where a well-defined conformational binding motif (e.g., an α-helix) at the PPI interface supports the rational design of a peptidomimetic.3,4 In contrast, the strategy of using a stabilizer molecule to promote PPIs is comparatively underexplored.5 In principle, this mode of action provides an avenue to interrogate PPI networks where inhibition is intractable; however, the development of PPI stabilizers is intrinsically challenging because there is no clear starting point for initial ligand identification.
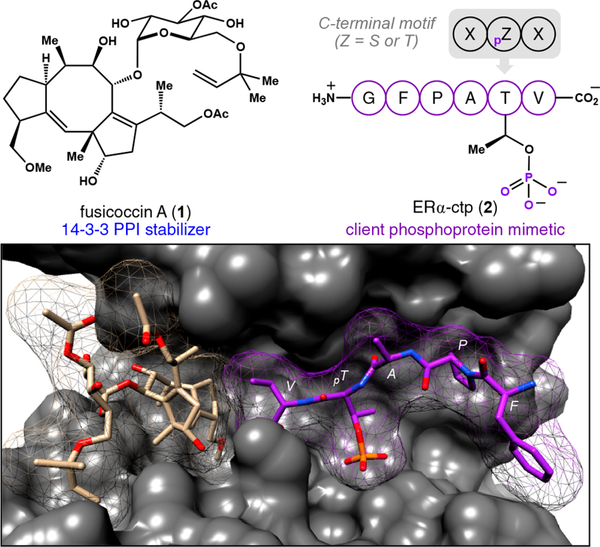
Natural products offer a rich source of molecular scaffolds that can be used to develop PPI stabilizers.6 With this idea in mind, we became interested in fusicoccin A (1, Figure 1), a complex diterpene glycoside that stabilizes PPIs involving 14–3–3 proteins.7,8 These adapter proteins are tightly integrated into phosphorylation-dependent cell signaling pathways, and their aberrant function is linked to the pathobiology of cancer and certain neurological disorders.9,10 Nevertheless, 14–3–3 is widely perceived as an intractable target because it forms PPIs with over 500 client proteins.11,12 In most cases, these interactions are mediated through a canonical “internal” (RXXpZXP, X = any residue, pZ = phosphorylated S or T) phosphorylation recognition motif presented on the surface of client proteins that locates to the amphipathic binding groove of 14–3–3.13 Intriguingly, 1 is not compatible with this group of 14–3–3 PPIs. Instead, the selectivity profile of 1 is largely constrained to a distinct part of the 14–3–3 interactome where binding is mediated though an atypical C-terminal phosphorylation recognition motif (XpZXCOOH).14–17
Figure 1.
A key contact between 1 (khaki) and the C-terminal valine of 2 (purple) in the 14–3–3 binding groove (gray) revealed by analysis of the 14–3–3σ·ERα-ctp·1 ternary complex (PDB 4JDD).
Recently, 1 has gained attention for its pro-apoptotic and neuroregenerative activity in human cell culture.18,19 Although the precise molecular mechanism(s) remain(s) undefined, 14–3–3 PPIs contribute to the pharmacology.20 This view is supported by characterized interactions between 1 and various systems that recapitulate C-terminal 14–3–3 PPIs in vitro.11 As a result, further development of this chemotype requires a systematic investigation of the structural features that govern stabilization of C-terminal 14–3–3 PPIs and a comprehensive profile of 14–3–3 client proteins that are compatible with 1. To this end, Aducci and Camoni reported that the stabilizing effect of 1 is primarily dependent on the identity of the C-terminal residue of the client phospholigand.21 Using in silico site-directed mutagenesis, this study predicted that C-terminal recognition motifs with hydrophobic, polar, or charged C-terminal residues could be targets for 1. Ohkanda subsequently showed that only 14–3–3 clients possessing the aliphatic residues I, V, and L at the C-terminus were strongly stabilized by 1.22 However, this study utilized a semisynthetic variant of 1 harboring a modified diterpene core attached to a non-natural furanose ring system. As a result, we set out to resolve this discrepancy and, in the process, develop a tool to predict C-terminal 14–3–3 PPIs that are compatible with biosynthetic 1.
Our investigation began by selecting a suitable C-terminal 14–3–3 PPI to benchmark the activity of 1. As shown in Figure 1, the complex formed between 14–3–3σ and estrogen receptor alpha (ERα) possessed several attractive features: (i) The 14–3–3 interaction site of ERα is located at the C-terminus of the F-domain, and binding to 14–3–3 is known to be dependent on phosphorylation of the penultimate threonine (T594) in vivo. (ii) The dynamics between 14–3–3σ and ERα have been assayed in vitro using a synthetic peptide (15 amino acids) harboring the 14–3–3 recognition sequence (590GFPApTVCOOH), referred to herein as the ERα C-terminal phospholigand (ERα-ctp, 2), and (iii) an X-ray structure of the 14–3–3σ·ERα-ctp·1 ternary complex (Protein Data Bank [PDB] ID: 4JDD, 2.1 A resolution) was available to guide our analysis.23 Notably, among the seven isoforms of 14–3–3 present in humans, 14–3–3σ is arguably the most important in cancer biology.9,24 Therefore, we focused on analyzing interactions between full-length, recombinant 14–3–3σ (see the SI for details) and the synthetic ERα-ctp mimetic 2 in the presence and absence of 1.
As shown in Figure 2, we started by establishing a uniform method to evaluate the stabilization induced by biosynthetic 1. Using isothermal titration calorimetry (ITC), we found that peptide 2 binds to 14–3–3σ with a Kd of 0.75 ± 0.14 μM. In contrast, the Kd of 2 for 14–3–3σ decreased to 0.02 ± 0.05 μM when 200 μM 1 was present. Thus, 1 stabilizes the 14–3–3σ·ERα-ctp complex by approximately 40-fold. This result was confirmed by fluorescence polarization (FP) experiments. Using this technique, we determined that the apparent affinity of fluorescein-labeled 2 (N-FAM-2) and 14–3–3σ was enhanced by approximately 70-fold, from an apparent Kd of 3.61 ± 0.41 μM to 0.05 ± 0.02 μM, when titrated in the presence of 80 μM 1. We also found that 1 stabilized the 14–3–3·N-FAM-2 complex with an EC50 of 3.16 ± 1.03 μM. Given that the apparent affinity determined by FP using N-FAM-2 was comparable to the Kd measured by ITC using 2, we concluded that the more convenient fluorescence polarization-based assay would be used to analyze 14–3–3/phospholigand interactions moving forward.
Figure 2.
Stabilization of the 14–3–3σ·ERα-ctp complex by fusicoccin A (1): (a) Isothermal titration calorimetry (ITC) of ERα-ctp (2) binding to 14–3–3σ in the absence (innate) or presence (stabilized) of 200 μM 1. (b) Fluorescence polarization (FP) measurements of fluorescein-labeled N-FAM-ERα-ctp (N-FAM-2) in the absence (innate) or presence (stabilized) of 80 μM 1 titrated with 14–3–3σ to obtain the apparent Kd of the 14–3–3σ·ERα-ctp complex. (c) Fluorescence polarization (FP) measurements of N-FAM-2 and 14–3–3σ titrated with 1 to obtain an EC50 value for its stabilizing activity toward the 14–3–3σ·ERα-ctp complex. (d) A comparison of ITC and FP methods for measuring the fold stabilization (SF) induced by 1. Error bars indicate the mean ± SD of at least two experiments.
Next, we chose to revisit a series of structurally distinct C-terminal 14–3–3 PPIs from the literature. The five targets we selected have been analyzed in the presence and absence of 1 previously;21 however, in most cases, existing studies relied on alternative analytical methods, different 14–3–3 isoforms, and/or longer synthetic phosphopeptides. As shown in Table 1, we focused on the interactions of 14–3–3σ and a series of fluorescein-labeled hexaphosphopeptides replicating the C-terminal recognition sequences of the potassium ion channel Task-3,25 the platelet adhesion receptor GpIBα,26 the G-protein coupled receptor GPR15,27 the interleukin 9 receptor α chain (IRL9),28 and the cyclin-dependent kinase inhibitor p27Kip1.29 Importantly, our analysis of the 14–3–3σ·Task3-ctp complex in the absence (Kd = 3.15 ± 1.12 μM) and presence of 80 μM 1 (Kd = 0.10 ± 0.05 μM) were in agreement with data reported by Ottmann using a similar FP assay and identical interaction partners.25 We also observed a stabilizing effect of 1 in combination with 14–3–3σ and either GpIBα-ctp (SF = 14 ± 2) or GPR15-ctp (SF = 7 ± 2). Notably, both sequences possess a C-terminal leucine, and the magnitude of increased affinity induced by 1 is consistent with literature reports.21 In contrast, the IRL9-ctp, which terminates in phenylalanine, was not strongly stabilized by 1 (SF = 3 ± 1). Similarly, 1 did not markedly enhance the affinity of 14–3–3σ for the p27Kip1-ctp, which possesses a C-terminal phosphothreonine (SF = 3 ± 2), and intriguingly, the innate affinity of the 14–3–3σ·p27Kip1-ctp complex itself was diminished (apparent Kd = 82.2 ± 31.0 μM). Taken together, these results reinforce the idea that the identity of the final residue within the C-terminal recognition motif is a key determinant of the activity of 1 at 14–3–3 PPI interfaces. These data also suggest that the C-terminal residue plays a direct role in innate binding to 14–3–3.
Table 1.
Apparent Kd and Fold Stabilization (SF) of Known C-Terminal 14–3–3 PPIs in the Absence and Presence of 80 μM 1a
| client protein | phospholigand sequenceb | innate Kd (μM) | Kd (μM) in presence of 1c | SF |
|---|---|---|---|---|
| Task3 | RKKRpSVCOOH | 3.15 ± 1.12 | 0.10 ± 0.05 | 14 ± 2 |
| GpIBα | YSGHpSLCOOH | 22.5 ± 9.71 | 1.69 ± 0.30 | 14 ± 2 |
| GPR15 | KRSVpSLCOOH | 7.36 ± 0.36 | 1.10 ± 0.09 | 7 ± 1 |
| IRL9 | ARSWpTFCOOH | 3.16 ± 0.32 | 1.54 ± 0.26 | 3 ± 1 |
| p27Kip1 | ALRRQpTCOOH | 82.2 ± 31.0 | 39.9 ± 8.09 | 3 ± 2 |
Reported apparent Kd is the mean ± standard deviation of at least two experiments.
Synthetic hexapeptides were labeled at the N-termini with the fluorophore fluorescein.
Titrations carried out in the presence of 80 μM 1. See the Supporting Information for details.
Analysis of the 14–3–3σ·ERα-ctp·1 ternary structure shown in Figure 1 shows that interactions between 1 and client phospholigand 2 in the 14–3–3σ binding groove are restricted to a hydrophobic contact between the 5–8–5 tricyclic core of 1 and the isopropyl side chain of the C-terminal valine within 2. On the basis of this observation, we hypothesized that amino acids with small alkyl side chains could make similar contacts with 1 and therefore support formation of a ternary complex. However, it was more challenging to predict which of these C-terminal residues might support binding to 14–3–3σ in the absence of 1, particularly given the intrinsic plasticity of the 14–3–3 binding groove that can accommodate hundreds of client phosphoproteins.12 Therefore, to interrogate these features systematically, we analyzed a focused library of N-FAM-2 variants with single amino acid changes at the C-terminus.
As summarized in Table 2, we observed a significant difference in the affinity of ERα-ctp variants for 14–3–3σ depending on the identity of the C-terminal residue. The results were divided into three groups: The first group contains hydrophobic residues I and V, which had micromolar affinities for 14–3–3σ (apparent Kd ~ 3.0–3.5 μM). A second group includes residues F, L, and A, which exhibited moderate affinities for 14–3–3σ (apparent Kd ~ 10.5–20.0 μM). The last group contained T, N, S, G, and D, which showed comparably weak affinity for 14–3–3σ (apparent Kd 50.0–200 μM). These titrations highlight a clear preference for docking of a hydrophobic C-terminal residue in the 14–3–3 binding groove.
Table 2.
Impact of the Modulation of the C-Terminal Residue within ERα-ctp on the Apparent Affinity of the 14–3–3σ·ERα-ctp Complex in Both the Absence and Presence of 1a
 | |||
|---|---|---|---|
| C-terminal residue (X) | innate Kd (μM) | Kd (μM) in the presence of 1b | ΔG stabilization (kcal·mol–1)c |
| I | 3.03 ± 0.31 | 0.06 ± 0.01 | 2.3 |
| V | 3.61 ± 0.41 | 0.05 ± 0.02 | 2.5 |
| F | 10.5 ± 3.36 | 4.03 ± 0.26 | 0.6 |
| L | 11.0 ± 2.72 | 0.17 ± 0.02 | 2.5 |
| A | 19.9 ± 6.99 | 0.70 ± 0.05 | 2.0 |
| T | 50.0 ± 13.3 | 1.32 ± 0.14 | 2.2 |
| S | 79.8 ± 9.62 | 3.54 ± 0.44 | 1.8 |
| N | 59.5 ± 13.5 | 32.4 ± 4.75 | 0.3 |
| G | 144 ± 60.0 | 36.5 ± 17.3 | 0.8 |
| D | 173 ± 82.3 | 48.2 ± 0.26 | 0.7 |
Apparent Kd reported as the mean ± standard deviation of two experiments.
Titrations carried out in the presence of 80 μM 1. See the SI for details.
ΔG stabilization was calculated using mean Kd values in the absence and presence of 1.
Next, we compared the affinities of our phospholigand library with 14–3–3σ in the presence of 80 μM 1. Consistent with our hypothesis, variants of N-FAM-2 bearing a C-terminal residue with an alkyl side chain were stabilized by 1. We found that residues I, V, L, A, T, and S supported a substantial decrease in apparent Kd in the presence of 1 (>20-fold, Table 2). The effect of 1 on the alanine variant was particularly striking. In this case, the apparent affinity to 14–3–3σ was enhanced by approximately 30-fold, from a Kd of 19.9 ± 6.99 μM to 700 ± 50 nM, in the presence of 80 μM 1. When interpreted in terms of ΔG of stabilization (kcal·mol–1), the relative ranking of C-terminal residues was found to be V or L (2.5) > I (2.3) > T (2.2) > A (2.0) > S (1.8). In contrast, variants of N-FAM-2 harboring F, N, G, and D at the C-terminus were not stabilized to a large extent by 1 (ΔG < 0.9 kcal/mol). These results point to an upper (N) and lower (G) constraint on the size of amino acid side chains that support interactions with 1 in the 14–3–3 binding groove. In this regard, hydrophobic residues L, V, and I with branched side chains were optimal, and the stabilizing effect of 1 decreased as the size of the C-terminal residue was decreased (i.e., V → A → G). A direct comparison of the V and T variants of N-FAM-2 indicated that polar functional groups were tolerated but diminished the stabilizing effect of 1. Consistent with this model, we found that the S variant of N-FAM-2 was stabilized by 22-fold in the presence of 1, whereas the corresponding A variant was stabilized by 29-fold under identical conditions. In contrast, slightly larger polar C-terminal residues (i.e., N) and C-terminal residues with small, charged side chains (i.e., D) disrupt hydrophobic contacts with 1.
With an improved picture of the structural criteria required for stabilization by 1, we employed bioinformatics to establish the scope of potential 14–3–3 client phosphoproteins that might serve as biological targets for 1 in vivo. Thus, using the C-terminal recognition motif as a search criterion, we performed a BLAST search to identify human proteins terminating in the motif -XZBCOOH (Z = T or S and B = V, L, I, T, A, or S).30 This inquiry returned over 800 unique FASTA sequences, which were further subjected to filtering algorithms. Using NetPhos 3.1,31 we compiled a list of proteins with predicted phosphorylation at the penultimate S or T residue. These putative client phosphoproteins were then filtered using the 14–3–3 Prediction Tool.32 This analysis returned a list of 119 C-terminal 14–3–3 PPIs that are candidate targets for 1 (the full list is reported in the SI). Next, utilizing the PhosphoSitePlus database, we evaluated each candidate phosphoprotein for experimentally validated phosphorylation at the predicted phosphosite.33 This search uncovered a subset of proteins within our initial list that have a reported C-terminal 14–3–3 phosphorylation recognition motif (Figure 3). These proteins are the most likely to be new targets for 1 in vivo. Importantly, our bioinformatics analysis also captured the C-terminal 14–3–3 PPIs previously reported with 1, including the sequences shown in Table 1.
Figure 3.
Select proteins that are putative targets for fusicoccin A (1): (a) FP measurements of fluorescein-labeled Nup160-ctp (3) in the absence (innate) or presence (stabilized) of 80 μM 1 titrated with 14–3–3σ to obtain the apparent Kd of the 14–3–3σ·Nup160-ctp complex. (b) The same FP experiment repeated with fluorescein-labeled TNFRSF19-ctp (4). (c) A comparison of the fold-stabilization (SF) induced by 1 using phospholigands 3 and 4. Error bars indicate the mean ± SD of at least two experiments.
As shown in Figure 3, this study identified several disease relevant proteins that have not been explored as interaction partners for 14–3–3 or as biological targets of 1. Thus, in an initial effort to validate the results of our search, we probed interactions between 14–3–3σ and phosphopeptides replicating the putative C-terminal 14–3–3 recognition domains of two structurally distinct candidate proteins from our list. The first was nucleoporin 160 (Nup160-ctp, 3),34 which binds to 14–3–3σ with an apparent Kd of 5.34 ± 0.31 μM in the absence of 1 and with an apparent Kd of 0.25 ± 0.02 μM when titrated in the presence of 80 μM 1 (Figure 3A). The affinity of the second candidate protein, TNF receptor 19 (TNFRSF19-ctp, 4),35 was also improved from an apparent Kd of 538 ± 141 μM to 38.0 ± 2.91 μM, when titrated in the presence of 80 μM 1 (Figure 3B). Importantly, the 80 μM concentration of 1 used in these experiments exceeds the concentrations of 1 reported to illicit phenotypic changes in cell culture (~2–20 μM) and is far beyond the EC50 recorded for the 14–3–3σ·ERα-cpt PPI in vitro (~3 μM).18,23 It is also noteworthy that the sequences of these phospholigands are different at every position except for the C-termini, and although 3 was a superior ligand for 14–3–3σ, the magnitude of stabilization afforded by 1 was comparable for 3 and 4 (20-fold vs 14-fold, Figure 3C). As such, these results demonstrate that the presence of a small alkyl residue at the C-terminus is the main requirement for stabilization of canonical C-terminal 14–3–3 PPIs by 1.
In conclusion, we have shown that for C-terminal 14–3–3 PPIs, 1 is best accommodated in the 14–3–3 biding groove when the client phosphoprotein terminates with V, L, I, T, A, or S. Using bioinformatics, we have leveraged this observation to assemble a database of 119 candidate 14–3–3 PPIs that can be targets for 1 in vivo. This group is notably enriched in proteins terminating with L, V, or I (68 out of 119), the residues found to be most compatible with 1 in Table 2. To further validate this analysis, we have demonstrated that interactions between 14–3–3σ and potential phospholigand targets identified from our search are enhanced by 1 in vitro. More broadly, this study points to an expanded family of potential molecular targets for 1. As a result, we contend that chemical synthesis will play a critical role unlocking the utility of 1 as a chemical probe of 14–3–3 functions.
Supplementary Material
Figure S1. Representative 12% SDS-PAGE gel of purified His-14–3-3σ (27 kD band). Lanes from right to left indicate: (1) ladder, (2) post-induction fraction, (3) whole cell lysate, (4) supernatant, (5) column flow-through, (6) wash, (7) 4 μg, (8) 8 μg, (9) 10 μg, (10) 12 μg.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01-GM125926 to J.H.F. and R01-GM115388 to B.G.M. B.G.M. acknowledges support from the Pfeiffer Endowed Professorship for Cancer Research. We thank E. Bienkiewicz (FSU) for assistance in developing fluorescence polarization assays. We are also grateful to B. Stefanovic (FSU) for access to his microplate reader.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.9b00795.
Experimental methods, supplementary fluorescence polarization data, and Table S1 listing a database of 119 candidate C-terminal 14–3–3 PPIs compatible with 1 (PDF)
Contributor Information
Ananya Sengupta, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, Florida 32306, United States.
Josue Liriano, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, Florida 32306, United States.
Brian G. Miller, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, Florida 32306, United States.
James H. Frederich, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, Florida 32306, United States.
REFERENCES
- (1).Wells JA, and McClendon CL (2007) Reaching for the highhanging fruit in drug discovery at protein–protein interfaces. Nature 450, 1001–1009. [DOI] [PubMed] [Google Scholar]
- (2).Scott DE, Bayly AR, Abell C, and Skidmore J (2016) Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat. Rev. Drug Discovery 15, 533–550. [DOI] [PubMed] [Google Scholar]
- (3).Arkin MR, Tang Y, and Wells JA (2014) Small-molecule inhibitors of protein–protein interactions: progressing toward the reality. Chem. Biol 21, 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Azzarito V, Long K, Murphy NS, and Wilson AJ (2013) Inhibition of α-helix-mediated protein–protein interactions using designed molecules. Nat. Chem 5, 161–173. [DOI] [PubMed] [Google Scholar]
- (5).Thiel P, Kaiser M, and Ottmann C (2012) Small-molecule stabilization of protein–protein interactions: an underestimated concept in drug discovery. Angew. Chem., Int. Ed 51, 2012–2018. [DOI] [PubMed] [Google Scholar]
- (6).Milroy L-G, Grossmann TN, Hennig S, Brunsveld L, and Ottmann C (2014) Modulators of protein–protein interactions. Chem. Rev 114, 4695–4748. [DOI] [PubMed] [Google Scholar]
- (7).Fullone MR, Visconti S, Marra M, Fogliano V, and Aducci P (1998) Fusicoccin effect on the in vitro interaction between plant 14–3–3 and plasma membrane H+ATPase. J. Biol. Chem 273, 7698–7702. [DOI] [PubMed] [Google Scholar]
- (8).Wurtele M, Jelich-Ottmann C, Wittinghofer A, and Oecking C (2003) Structural view of a fungal phytotoxin acting on a 14–3–3 regulatory complex. EMBO J. 22, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hermeking H (2003) The 14–3–3 cancer connection. Nat. Rev. Cancer 3, 931–943. [DOI] [PubMed] [Google Scholar]
- (10).Foote M, and Zhou Y (2012) 14–3–3 proteins in neurological disorders. Int. J. Biochem. Mol. Biol 3, 152–164. [PMC free article] [PubMed] [Google Scholar]
- (11).Stevers LM, Sijbesma E, Botta M, MacKintosh C, Obsil T, Landrieu I, Cau Y, Wilson AJ, Karawajczyk A, Eickhoff J, Davis J, Hann M, O’Mahony G, Doveston RG, Brunsveld L, and Ottmann C (2018) Modulators of 14–3–3 protein–protein interactions. J. Med. Chem 61, 3755–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Pennington KL, Chan TY, Torres MP, and Andersen JL (2018) The dynamic and stress-adaptive signaling hub of 14–3–3: emerging mechanisms of regulation and context-dependent protein–protein interactions. Oncogene 37, 5587–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, and Cantley LC (1997) The structural basis for 14–3–3:phosphopeptide binding specificity. Cell 91, 961–971. [DOI] [PubMed] [Google Scholar]
- (14).Coblitz B, Wu M, Shikano S, and Li M (2006) C-terminal binding: An expanded repertoire and function of 14–3–3 proteins. FEBS Lett. 580, 1531–1535. [DOI] [PubMed] [Google Scholar]
- (15).Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, and MacKintosh C (2010) Bioinformatic and experimental survey of 14–3–3 binding sites. Biochem. J 427, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ottmann C, Weyand M, Sassa T, Inoue I, Kato N, Wittinghofer A, and Oecking C (2009) A structural rationale for selective stabilization of antitumor interactions of 14–3–3 proteins by cotylenin. J. Mol. Biol 386, 913–919. [DOI] [PubMed] [Google Scholar]
- (17).Fusicoccin A also stabilizes 14–3–3 PPIs involving noncanonical recognition motifs: Stevers LM, Lam CV, Leysen SFR, Meijer F, vanScheppingen DS, de Vries RMJM, Carlile GW, Milroy LG, Thomas DY, Brunsveld L, and Ottmann C (2016) Characterization and small-molecule stabilization of the multisite tandem binding between 14–3–3 and the R domain of CFTR. Proc. Natl. Acad. Sci. U. S. A 113, 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).deVries-van Leeuwen IJ, Kortekaas-Thijssen C, NzigouMandouckou JA, Kas S, Evidente A, and de Boer AH (2010) Fusicoccin A selectively induces apoptosis in tumor cells after interferon-alpha priming. Cancer Lett. 293, 198–206. [DOI] [PubMed] [Google Scholar]
- (19).Kaplan A, Morquette B, Kroner A, Leong S, Madwar C, Sanz R, Banerjee SL, Antel J, Bisson N, David S, and Fournier AE (2017) Small-molecule stabilization of 14–3–3 protein–protein interactions stimulates axon regeneration. Neuron 93, 1082–1093. [DOI] [PubMed] [Google Scholar]
- (20).Variants of 1 with enhanced affinity for C-terminal 14–3–3 PPIs in vitro exhibit improved efficacy in cancer cell culture: Andrei SA, de Vink P, Sijbesma E, Han L, Brunsveld L, Kato N, Ottmann C, and Higuchi Y (2018) Rationally designed semisynthetic natural product analogues for stabilization of 14–3–3 protein–protein interactions. Angew. Chem., Int. Ed 57, 13470–13474. [DOI] [PubMed] [Google Scholar]
- (21).Paiardini A, Aducci P, Cervoni L, Cutruzzola F, Di Lucente C, Janson G, Pascarella S, Rinaldo S, Visconti S, and Camoni L (2014) The phytotoxin fusicoccin differentially regulates 14–3–3 proteins association to mode III targets. IUBMB Life 66, 52–62. [DOI] [PubMed] [Google Scholar]
- (22).Ohkanda J, Kusumoto A, Punzalan l., Masuda R, Wang C, Parvatkar P, Akase D, Aida M, Uesugi M, Higuchi Y, and Kato N (2018) Structural effects of fusicoccin upon upregulation of 14–3–3 phospholigand interaction and cytotoxic activity. Chem. - Eur. J 24, 16066–16071. [DOI] [PubMed] [Google Scholar]
- (23).De Vries-van Leeuwen IJ, da Costa Pereira D, Flach KD, Piersma SR, Haase C, Bier D, Yalcin Z, Michalides R, Feenstra KA, Jimenez CR, de Greef TFA, Brunsveld L, Ottmann C, Zwart W, and de Boer AH (2013) Interaction of 14–3–3 proteins with estrogen receptor alpha F domain provides a drug interface. Proc. Natl. Acad. Sci. U. S. A 110, 8894–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Li Z, Liu JY, and Zhang JT (2009) 14–3–3-sigma, the double-edged sword of human cancers. Am. J. Transl. Res 1, 326–340. [PMC free article] [PubMed] [Google Scholar]
- (25).Anders C, Higuchi Y, Koschinsky K, Bartel M, Schumacher B, Thiel P, Nitta H, Preisig-Muller R, Schlichthorl G, Renigunta V, Ohkanda J, Daut J, Kato N, and Ottmann C (2013) A semisynthetic fusicoccane stabilizes a protein–protein interaction and enhances the expression of K+ channels at the cell surface. Chem. Biol 20, 583–593. [DOI] [PubMed] [Google Scholar]
- (26).Camoni L, Di Lucente C, Visconti S, and Aducci P (2011) The phytotoxin fusicoccin promotes platelet aggregation via 14–3–3glycoprotein Ib-IX-V interaction. Biochem. J 436, 429–436. [DOI] [PubMed] [Google Scholar]
- (27).Okamoto Y, and Shikano S (2011) Phosphorylation-dependent C-terminal binding of 14–3–3 proteins promotes cell surface expression of HIV co-receptor GPR15. J. Biol. Chem 286, 7171–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sliva D, Gu M, Zhu XY, Chen J, Tsai S, Du X, and Yang YC (2000) 14–3–3 zeta interacts with the alpha-chain of human interleukin 9 receptor. Biochem. J 345, 741–747. [PMC free article] [PubMed] [Google Scholar]
- (29).Fujita N, Sato S, and Tsuruo T (2003) Phosphorylation of p27Kip1 at threonine 198 by p90 ribosomal protein S6 kinases promotes its binding to 14–3–3 and cytoplasmic localization. J. Biol. Chem 278, 49254–49260. [DOI] [PubMed] [Google Scholar]
- (30).Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990) Basic local alignment search tool. J. Mol. Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- (31).Blom N, Gammeltoft S, and Brunak S (1999) Sequenceand structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- (32).Madeira F, Tinti M, Murugesan G, Berrett E, Stafford M, Toth R, Cole C, MacKintosh C, and Barton GJ (2015) 14–3–3-Pred: Improved methods to predict 14–3–3 binding phosphopeptides. Bioinformatics 31, 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, and Sullivan M (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Vasu S, Shah S, Orjalo A, Park M, Fischer WH, and Forbes DJ (2001) Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol 155, 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Loftus JC, Dhruv H, Tuncali S, Kloss J, Yang Z, Schumacher CA, Cao B, Williams BO, Eschbacher JM, Ross JTD, and Tran NL (2013) TROY (TNFRSF19) promotes glioblastoma survival signaling and therapeutic resistance. Mol. Cancer Res. 11, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Representative 12% SDS-PAGE gel of purified His-14–3-3σ (27 kD band). Lanes from right to left indicate: (1) ladder, (2) post-induction fraction, (3) whole cell lysate, (4) supernatant, (5) column flow-through, (6) wash, (7) 4 μg, (8) 8 μg, (9) 10 μg, (10) 12 μg.