Summary
The centriole, or basal body, is the center of attachment between the sperm head and tail. While the distal end of the centriole templates the cilia, the proximal end associates with the nucleus. Using Drosophila, we identify a centriole-centric mechanism that ensures proper proximal end docking to the nucleus. This mechanism relies on the restriction of Pericentrin-Like Protein (PLP) and the pericentriolar material (PCM) to the proximal end of the centriole. PLP is restricted proximally by limiting its mRNA and protein to the earliest stages of centriole elongation. Ectopic positioning of PLP to more distal portions of the centriole is sufficient to redistribute PCM and microtubules along the entire centriole length. This results in erroneous, lateral centriole docking to the nucleus, leading to spermatid decapitation as a result of a failure to form a stable head-tail linkage.
Keywords: Centriole, centrosome, cilia, sperm, pericentriolar material, spermatogenesis, fertility, PLP, Pericentrin, Drosophila
eTOC
Functional sperm require a stable linkage between the sperm head and tail, a linkage mediated by the centriole. Galletta et al. reveal how restriction of pericentriolar material to the proximal end of the centriole – achieved via restriction of the Pericentrin-like protein - ensures proximal end docking to the nucleus.
Introduction
An essential element of functional flagellated sperm is proper attachment between the head, which contains the genetic material, and the tail, which generates the force for swimming. A failure in this connection can result in decapitated, decaudated, or malformed sperm, ultimately leading to reduced fertility (Baccetti et al., 1989; Chemes et al., 1999; Chemes and Rawe, 2010). The most well documented human study investigated 10 infertile males with acephalic sperm (Chemes et al., 1999). These patients showed a variety of “abnormal head–neck configurations,” including breaks between the head and tail, and sperm with nuclei laterally attached to the midpiece near the centrioles. The head to tail linkage is centered around the centriole, referred to in this context as a basal body (we use the term centriole for simplicity). The ‘distal end’ of the centriole templates and anchors the axoneme, the core structural element of the tail, while the ‘proximal end’ of the centriole forms a connection with the nuclear surface (Fawcett, 1975; Tates, 1971). Thus, the proximal and distal ends of centrioles play distinct and critical roles in sperm assembly. Note that we are discussing the proximal-end and distal-end of an individual centriole, which are distinct from the “proximal centriole” and “distal centriole” terms used in mammalian systems to describe the two centrioles within each sperm. Except when specifically discussing mammalian spermiogenesis, the later terms will not be used or referenced in our study.
A prerequisite for a tight connection between the head and tail of the sperm is the relocation of the centriole to the nuclear envelope during early spermiogenesis in Drosophila and in mammals (Fawcett, 1981; Holstein and Roosen-Runge, 1981; Tates, 1971; Yuan et al., 2015). In Drosophila, after the exit from meiosis II, the centriole (Figure 1A) is repositioned against the reformed nuclear envelope (Figure 1B) and eventually becomes embedded in the nuclear envelope and surrounded by an electron dense material suggested to provide a tight connection (Figure 1B; (Fuller, 1993; Tates, 1971). Similarly in mammals, the ‘proximal centriole’ moves and attaches to the nucleus where an electron dense material accumulates and the ‘connecting piece’ assembles around the centriole pair (Chemes and Alvarez Sedo, 2012; Fawcett, 1981; Holstein and Roosen-Runge, 1981; Yuan et al., 2015). In both systems, the centriole templating the flagellar axoneme has its proximal end closest to the nucleus and is positioned perpendicular to the nuclear surface.
Figure 1. The centriole proximal end maintains MTOC activity throughout Nuclear Attachment.
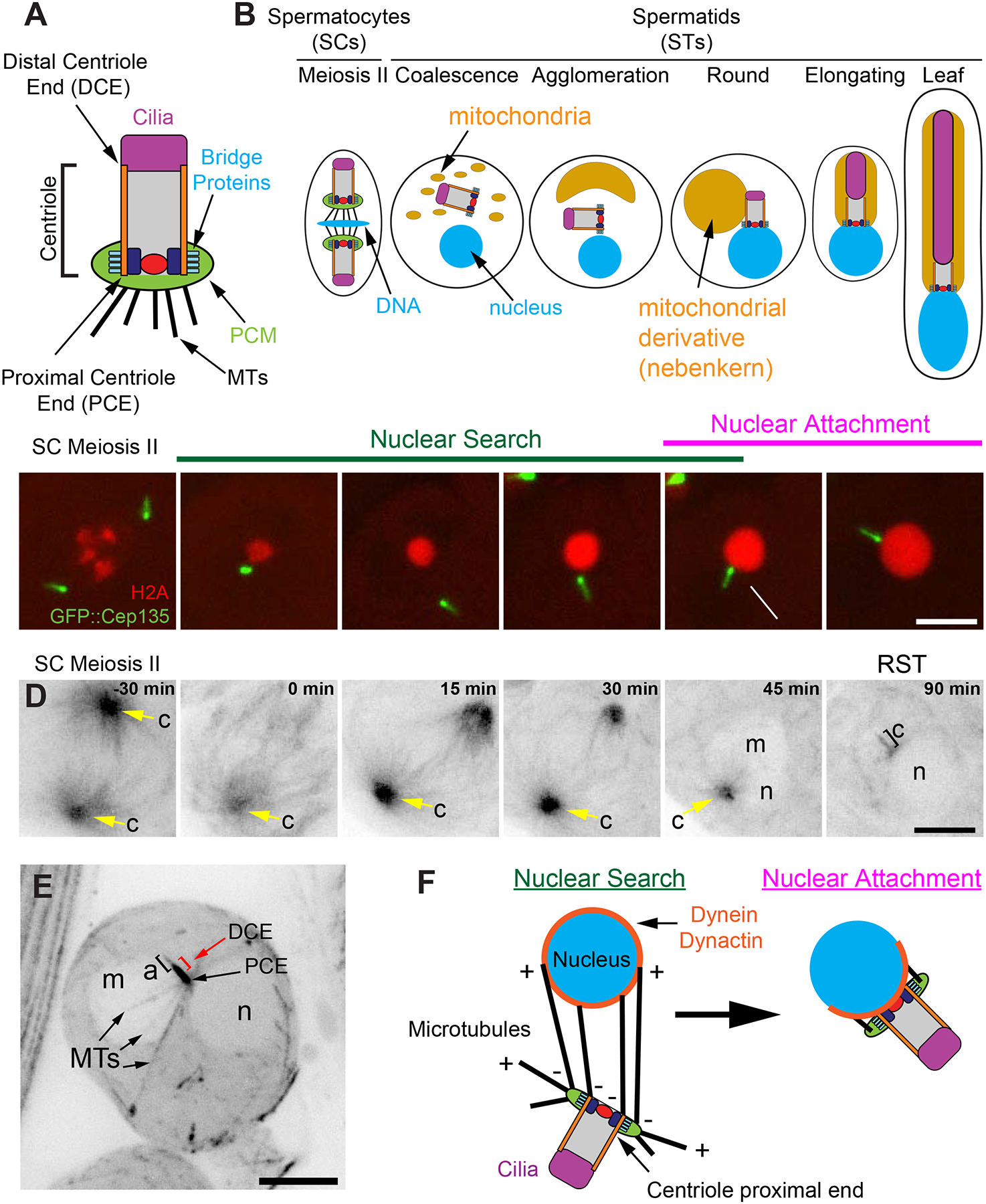
A) SC centriole showing the major components along the proximal-distal axis. Internal, proximal structures (navy and red). B) Centriole, nucleus (DNA), and mitochondria behavior during spermatogenesis from meiosis II through leaf stage STs. Development proceeds is left to right. C) A centriole (Cep135, green) and DNA (H2Av, red) in a SCs exiting meiosis through the RST stage showing the Nuclear Search and Nuclear Attachment stages (Video 1). Cep135 is brighter on the proximal end (Galletta et al., 2016a). Arrow indicates the arrival of the proximal end at the nucleus. Mean time of Nuclear Search = 40±19 minutes, N=20, 4 cysts, 2 testes. D) GFP::tubulin in a SC exiting meiosis through the RST stage (Video 2). c = centriole (yellow arrow, n = nucleus, m=mitochondrial derivative). E) Max intensity projection through a live RST expressing GFP::tubulin. Labels as in D, a = axoneme, PCE = proximal centriole end, DCE = distal centriole end (red bracket), MTs = microtubules. MTs emanate from the PCE, which is against the nucleus. F) Two step model of centriole docking: During Nuclear Search, MTs emanating from the proximal end of the centriole interact with dynein on the nuclear surface to bring the centriole and nucleus together. During Nuclear Attachment a stable connection is formed between the centriole and nucleus. Bars C,D,E = 5μm.
In mammalian model systems there are examples of mutations that cause the head-tail connection to fail, including mutations in centriole proteins Centrin 1 and Centrobin (Avasthi et al., 2013; Liska et al., 2009), but little is understood at the molecular level of how these centriole proteins are involved in establishing the head-tail connection. Mutant analysis in Drosophila has identified additional players such as Asunder, Lis-1, Spag4, Yuri gagarin, and Dynein/Dynactin components, indicating that positioning the microtubule (MT) motor dynein at the nuclear envelope is critical (Anderson et al., 2009; Fabian and Brill, 2012; Kracklauer et al., 2010; Sitaram et al., 2012; Texada et al., 2008; Wu et al., 2016). Finally, mutations in gamma tubulin ring complex proteins, which are required for proper MT formation, result in defective centriole-nuclear attachment in older developing spermatids (Vogt et al., 2006). We believe these studies in totality suggest a model whereby dynein on the nuclear surface binds and acts on centriolar MTs to reposition the centriole to the nucleus. However, this process has never been documented in live STs, the role of the centriole itself in this process has not been examined, and the molecular mechanism that ensures correct centriole orientation and docking has not been investigated in detail.
The highly stereotypical proximal end-on docking of centrioles to the nucleus suggests that the centriole proximal end is specialized. Many studies have carefully defined proteins uniquely positioned along the proximal-distal centriole axis (Brito et al., 2012; Fu et al., 2015; Galletta et al., 2016b; Loncarek and Bettencourt-Dias, 2018). This polarized localization can convey local functions (Figure 1A). For example, proteins that regulate centriole length such as Klp10A, Cep97, and Cp110 are positioned at the distal end of the centriole where centriole elongation is thought to exclusively occur (Delgehyr et al., 2012; Franz et al., 2013; Schmidt et al., 2009; Spektor et al., 2007). When a new centriole forms, proteins such as Ana2/STIL and Sas6, structural elements required for earliest steps daughter centriole formation, accumulate at the proximal end of the mother centriole (Reviewed in (Nigg and Holland, 2018). Additionally, pericentriolar material (PCM), which is critical for nucleating and organizing MTs appears to be restricted to the centriole proximal end in mammalian systems and the meiotic centrioles of Drosophila spermatocytes (Bornens et al., 1987; Fu and Glover, 2012; Galletta et al., 2016b; Giansanti et al., 2008; Paintrand et al., 1992; Roque et al., 2018). This suggests that MTs are predominantly produced and anchored at the proximal end of centrioles. However, unlike distal-end protein components, the importance of restricting PCM to the proximal-end and the mechanism by which this restriction is achieved are unknown. We sought to identify the mechanism by which PCM is proximally restricted and hoped, in turn, to gain insight into proximal-end nuclear docking during spermatogenesis.
Results
Centriole-Nuclear docking is achieved through a two-stage ‘Nuclear Search’ and ‘Nuclear Attachment’ mechanism
To investigate centriole-nuclear docking in early STs, we performed live cell imaging of SCs as they exited meiosis II through the initial steps of ST development. We also performed fixed cell analysis of the entirety of spermiogenesis on whole mount testes. From our live and fixed data, we identify all the stages of ST development previously described by EM (Tates, 1971) – Coalescence, Agglomeration, Round STs, Elongating STs, and the Leaf stage (Figure 1B). The Round ST (RST) stage includes the “Clew” and “Onion” stages (Tates, 1971) as they are indistinguishable by light microscopy. As predicted from EM, the centriole was not always immediately positioned near the reforming nuclear envelope (NE) following meiotic exit (Figure 1C). Instead, the centriole travelled through the cytoplasm for 40 ± 19 minutes before encountering the nucleus (Figure 1C, Video 1), we refer to this stage as the “Nuclear Search” stage, which occurs principally in the Coalescence and Agglomeration ST stages. Once the centriole reaches the NE, the proximal end of the centriole attaches to the nucleus and the two structures begin to move synchronously, we refer to this second stage as the “Nuclear Attachment” stage, which is completed in RSTs. An attractive hypothesis that emerges from our data and published mutant phenotypes (Anderson et al., 2009; Kracklauer et al., 2010; Sitaram et al., 2012; Wu et al., 2016) is that Nuclear Search requires MTs emanating from the centriole to bind (capture) dynein motors stably anchored on the nuclear surface, which walk to the MT minus ends effectively pulling the centriole to the nucleus. Nuclear Attachment is then achieved by forming a specialized molecular linkage between nuclear surface proteins and the proximal centriole end.
Centrosomal MTs and PCM are proximally restricted
To follow MT behavior during Nuclear Search and Attachment we imaged flies expressing GFP::Tubulin as SCs exited meiosis. The MTOC activity of the centriole rapidly declined (Figure 1D, Video 2), but continued to organize MTs throughout the early events of ST development, including the entirety of Nuclear Search. Despite attempts to image MTs during Nuclear Search, it was extremely challenging due to the high MT density. However, we did identify RSTs at the Nuclear Attachment stage with MTs specifically emanating from the proximal end of the centrioles (Figure 1E, PCE; Figure S1A, z-stack in Figure S6A), suggesting a model where proximal MTs capture the nuclear envelope and ensure proximal centriole end docking (Figure 1F).
The specific nucleation of MTs from the proximal end is consistent with previous findings in Drosophila male meiosis where PCM, as detected by gamma tubulin (γ-tub), is proximally restricted (Fu and Glover, 2012; Galletta et al., 2016b; Giansanti et al., 2008; Roque et al., 2018); Figure 2A, B). Proximal restriction of PCM has also been reported for mammalian centrosomes (Bornens et al., 1987; Paintrand et al., 1992). Thus, we set out to more thoroughly characterize the organization of the PCM during ST formation. As shown previously (Blachon et al., 2009), our immunostaining shows that γ-tub in RSTs (Nuclear-Attachment stage) is enriched on the proximal end of the centriole, similar to meiosis (Figure 2A–C). We note that centrioles that have completed docking frequently show a second, small pool of γ-tub at the distal end (Figure 2C), or additional low levels of γ-tub along the centriole length. It is likely that γ-tub at these non-proximal positions represent older RSTs beginning to reorganize their PCM for the next developmental stage (Elongating and leaf STs; Figure S2A–C). We conclude that both γ-tub and MTs remain proximally enriched on centrioles following meiosis through the RST stage, and hypothesize that sustained proximal localization of PCM and MTs in early RSTs is critical for centriole-nuclear docking.
Figure 2. PLP is required for γ-tub organization and centriole-nuclear docking.
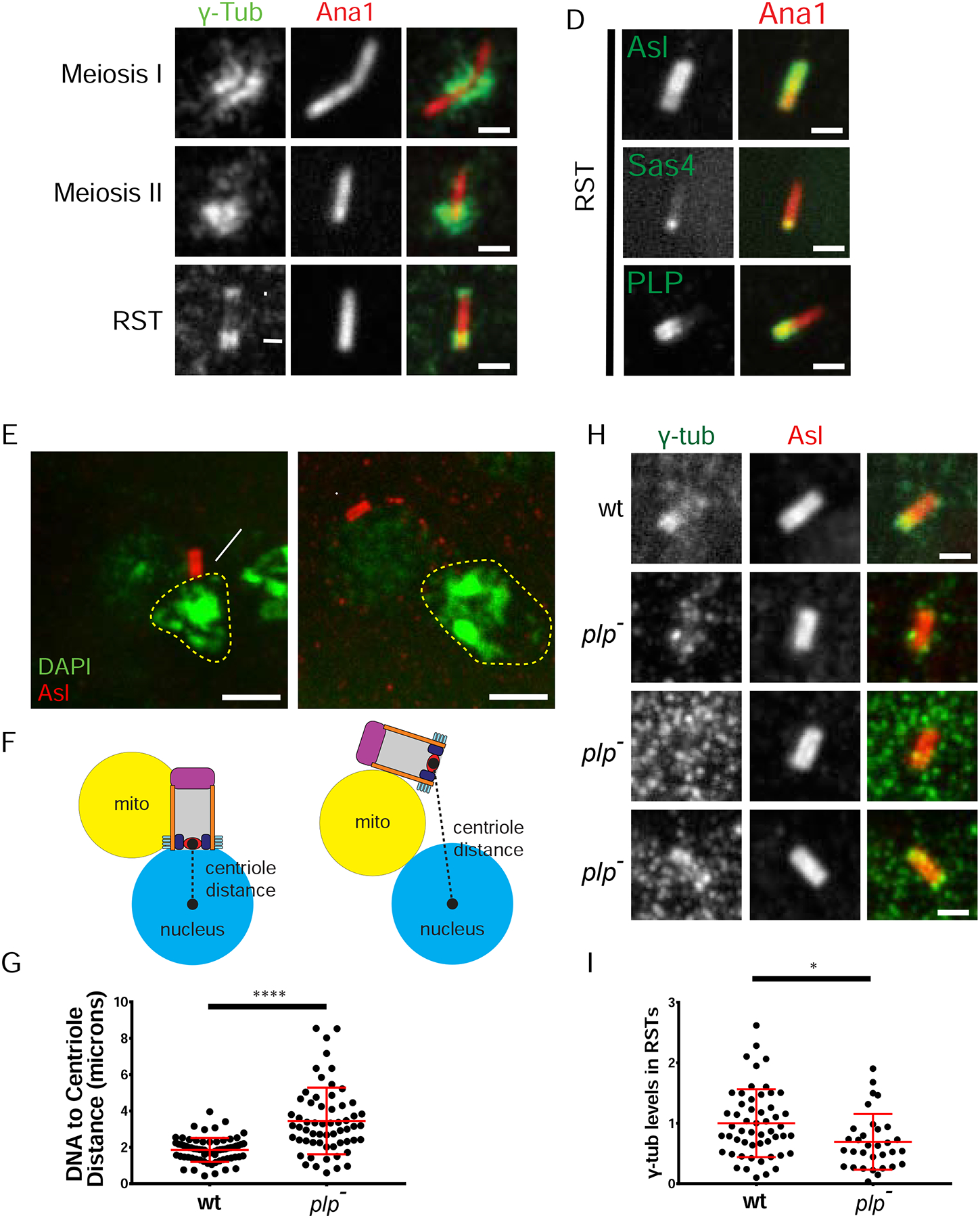
A-C) γ-tubulin (γ-tub; green) remains proximally enriched (arrow, N=46/56) on centrioles (Ana1, red) from meiosis through RSTs and the completion of Nuclear Attachment. RSTs also contain distal tip γ-tub in some cases (arrowhead, N=23/46). D) Bridge protein localization (green) along centrioles (Ana1, red) in RSTs. PLP and Sas4 remain proximal during Nuclear Search and Nuclear Attachment; Asl remains along the entire length. p = proximal end, d= distal end. E) RSTs showing centrioles (Asl, red) and DNA (DAPI, green). Approximate nucleus (n) boundary (dashed line), m = mitochondrial derivative. Wildtype RST shows proper perpendicular centriole attachment (arrow). plp- RST shows defective centriole positioning. F) Schematic of normal (left) and defective (right) centriole distance measurement. G) Centriole distance in wildtype (N=69) and plp- (N=60) RSTs. H) γ-tub (green) along centrioles (Asl, red) in wt and plp- RSTs (rows 2–4). plp- significantly disrupts localization of γ-tub (3 examples). Row 2, γ-tub on the centriole in a pattern different than wildtype (53%,17/32); row 3, little or no γ-tub (31%, 10/32); row 4, γ-tub enriched at one end (16%, 5/32). I) Relative amount of γ-tub on centrioles in wt (N=53) and plp- (N=32) RSTs normalized to the average wt signal. Bars A-D, H = 1μm, E = 2μm. (*) p=0.011, (****) p≤0.0001.
Pericentrin-Like-Protein is proximally restricted and critical for centriole docking
To understand how γ-tub is restricted to the proximal end, we focused on three proteins: Asterless (Asl), Pericentrin-Like-Protein (PLP), and Sas4. These ‘Bridge’ proteins are likely candidates to position the PCM based on their position at the centriole wall and functions in recruiting and organizing PCM (Varadarajan and Rusan, 2018). We used immunostaining (Asl, PLP) and CRISPR tagging (Sas4) to examine endogenous proximal-distal positions. Asl localized along the entire length of the centriole throughout meiosis and in RSTs (Figure 2D, Figure S2D), while PLP (Figure 2D, Figure S2E) and Sas4 (Figure 2D, Figure S2F; (Gopalakrishnan et al., 2011) were proximally restricted. PLP and Sas4 are therefore excellent candidates to control the proximal position of PCM during centriole docking.
If PLP and Sas4 are required for centriole docking, we predict that mutants in these proteins would disrupt docking. Since Sas4 is required for centriole duplication (Basto et al., 2006), centrioles are extremely rare in sas4 mutants, precluding a detailed analysis of centriole docking (Galletta et al., 2016b; Riparbelli and Callaini, 2011). In contrast, PLP is not required for centriole duplication and, therefore, we were able to examine plp- centrioles. plp- mutants have defects in meiosis and form dysfunctional STs, but form normal axonemes (Martinez-Campos et al., 2004). The meiotic defect has been attributed to a loss of the orthogonal arrangement of centrioles in SCs and to centriole fragmentation (Martinez-Campos et al., 2004); Roque et al., 2018). Upon reexamination of the same plp- mutant, we find defects in the orthogonal arrangement of centrioles, but centrioles do not fragment (Figure S2G, see legend for details). We also found that plp- mutants lead to 21% of RSTs with no centrioles or with more than one centriole per mitochondrial derivate (N = 143; 0% in wildtype, N = 154, methods; Castrillon et al., 1993), indicative of meiotic failure. Additionally, by the RST stage all centrioles have elongated to approximately 90% of wildtype length (Figure S2H). Slightly shorter centrioles have been observed in the sensory organ precursors of plp- mutants (Roque et al., 2018).
plp- RSTs have centrioles at much further distances from the nucleus (Figure 2E–G), suggesting that Nuclear Search is delayed or fails. To determine if loss of PLP results in loss or disorganization of PCM similar to other tissues (Galletta et al., 2014; Kawaguchi and Zheng, 2004; Martinez-Campos et al., 2004), we examined γ-tub, which was indeed significantly reduced and disorganized in RSTs (Figure 2H, I). Interestingly, PLP does not appear to be required for the recruitment and organization of γ-tub in slightly older STs (young elongating, Figure S2A’), indicating that PLP’s critical role in centriole-nuclear docking occurs between the completion of meiosis and the beginning of mitochondrial derivative elongation.
Together, our data support a centriole-centered model that ensures proper capture of the proximal end of the centriole by the nucleus in early STs. After meiosis, the centriole maintains MTOC activity through early spermiogenesis. This MTOC activity requires the bridge protein PLP to maintain its proper organization. MTs emanating from the proximal end of the centriole engage the nuclear surface, likely via dynein motors on that surface (Anderson et al., 2009; Fabian and Brill, 2012; Kracklauer et al., 2010; Sitaram et al., 2012; Texada et al., 2008; Wu et al., 2016), pulling the centriole towards the nucleus, proximal end first. This model raises two important questions. First, how is PLP specifically restricted to the proximal end of centrioles? Second, is the proximal restriction of PLP and PCM necessary for proper centriole docking?
PLP is proximally restricted by eliminating its availability during centriole elongation
To address the mechanism by which PLP is restricted to the proximal centriole end, we carefully followed PLP during the process of centriole elongation, which occurs throughout the four-day premeiotic G2 phase of SCs. Although centrioles elongate more than 10-fold to reach 1.8 μm by late SCs/meiosis (Figure 3A–C; (Tates, 1971), PLP length and position along the centriole changes only slightly from 0.6 ± 0.1 μm to 0.7 ± 0.1 μm (Figure 3B, D). Based on this observation, we proposed two models for PLP restriction. First, we hypothesized that sites of PLP binding to the centriole were restricted to the proximal end. However, we previously observed that when expressed using a ubiquitous promoter, PLP localized along the entire length of the centriole (Galletta et al., 2014), indicating that all positions along the centriole are competent to bind PLP. A second hypothesis was that, during centriole elongation, PLP protein is simply not readily available (absent from the cytoplasm), and thus cannot incorporate into more distal regions of the centriole. To determine if PLP protein is available during centriole elongation, we used CRISPR to C-terminally tag PLP with mNeonGreen (mNeon) at its endogenous locus. Since PLP is shown to have 12 protein isoforms (Flybase.org) that share a common C-terminal end, our CRISPR line should report on all possible PLP isoforms. We then used live imaging (no tissue fixation and extraction) of testes to measure cytoplasmic PLP levels using line-scan analysis along the long axis of the tissue (Figure S3A arrow, Methods). This revealed a significant decrease of PLP::mNeon signal from the tip containing the mitotic spermatogonia (SG) towards the SCs in the long premeiotic G2 (Figure S3B). In contrast, Asl remained at a constant level (Figure S3C,D), consistent with Asl loading along the entire centriole length (Figure 2D). We also imaged PLP::mNeon CRISPR testes live at higher magnification and performed single cell fluorescence intensity measurements of the cytoplasm (Figure 3E,F). There was significantly less PLP::mNeon signal in SCs compared to SG. Thus, by the time centriole elongation begins in SCs, the cytoplasmic PLP protein is less concentrated, possibly absent, resulting in restriction of PLP at the proximal end.
Figure 3. Proximal PLP is achieved by limiting PLP protein to early germline cells.
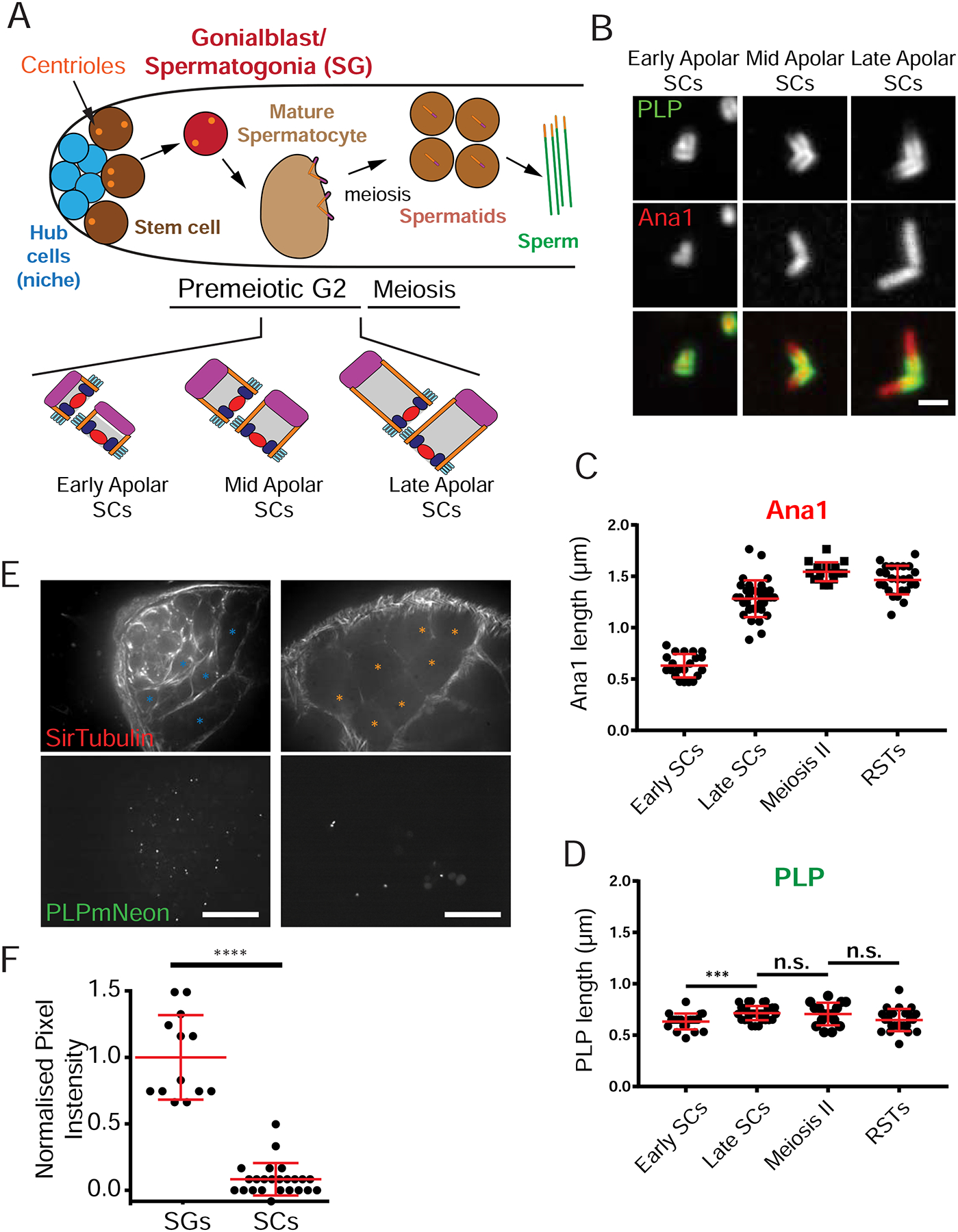
A) Stages and cell types of Drosophila male germ cell development. Centriole elongation (lower section) occurs during the long premeiotic G2. B) PLP (green) is present on SC centrioles (Ana1::tdtomato, red) in early premeiotic G2 (early apolar). As centrioles elongate (mid and late apolar SCs), PLP remains on the proximal ends. C) Centriole (Ana1) elongation. D) PLP length indicates a sustained proximal position with only a slight elongation (~100 nm). E) Live testes expressing PLP::mNeon at endogenous locus counterstained with SiR-Tubulin. Proliferating germ cells, or SG (blue asterisks) and SCs (premeiotic G2, orange asterisks) were identified based on the number of cells in a cyst, cell size and morphology. F) Normalized pixel intensity of PLP::mNeon signal (from E) showing significantly more PLP in the cytoplasm of SG (N = 13) vs SCs (N=24). Data normalized to the average intensity in SG. (****) p ≤ 0.0001, (***) p = 0.002, n.s. = not significant. Scale bars B = 1μm, C = 20μm.
PLP transcript is reduced during SC development
We next investigated if levels of the plp transcript were highest in SG and reduced during SCs development. Carefully monitoring mRNA levels in a specific cell within a complex, multicellular tissue such as the testis is challenging and demands examination at the individual cell level. To accomplish this, we utilized a recently generated single cell RNAseq dataset obtained from whole larval testes (Gene Expression Omnibus # GSE125947). The gene expression patterns of individual cells were used to group cells into clusters of similar lineage and developmental stage. This analysis separated cells of somatic origin from germline origin, which were then subdivided into SG and three clusters of SCs at different stages (Figure 4A). Thus, this dataset can reveal how mRNA levels change over time in the germline. We determined the change in mRNA levels of centriole, centrosome, cilia, and spermatogenesis genes in SCs compared to SG (Figure S4A). Interestingly, the mRNAs of centrosome bridge proteins show differing behaviors (Figure 4B; S4B–E). asl and spd-2 mRNAs remained relatively constant throughout germline development, suggesting that changes in their behavior or position along the centriole across germline development are not regulated at the mRNA level. In contrast, plp and sas4 mRNA levels were lower in later SCs than in SG (Figure 4B; S4B,C), consistent with their proximal localization.
Figure 4. PLP and PCM position is dictated by the timing of PLP expression.
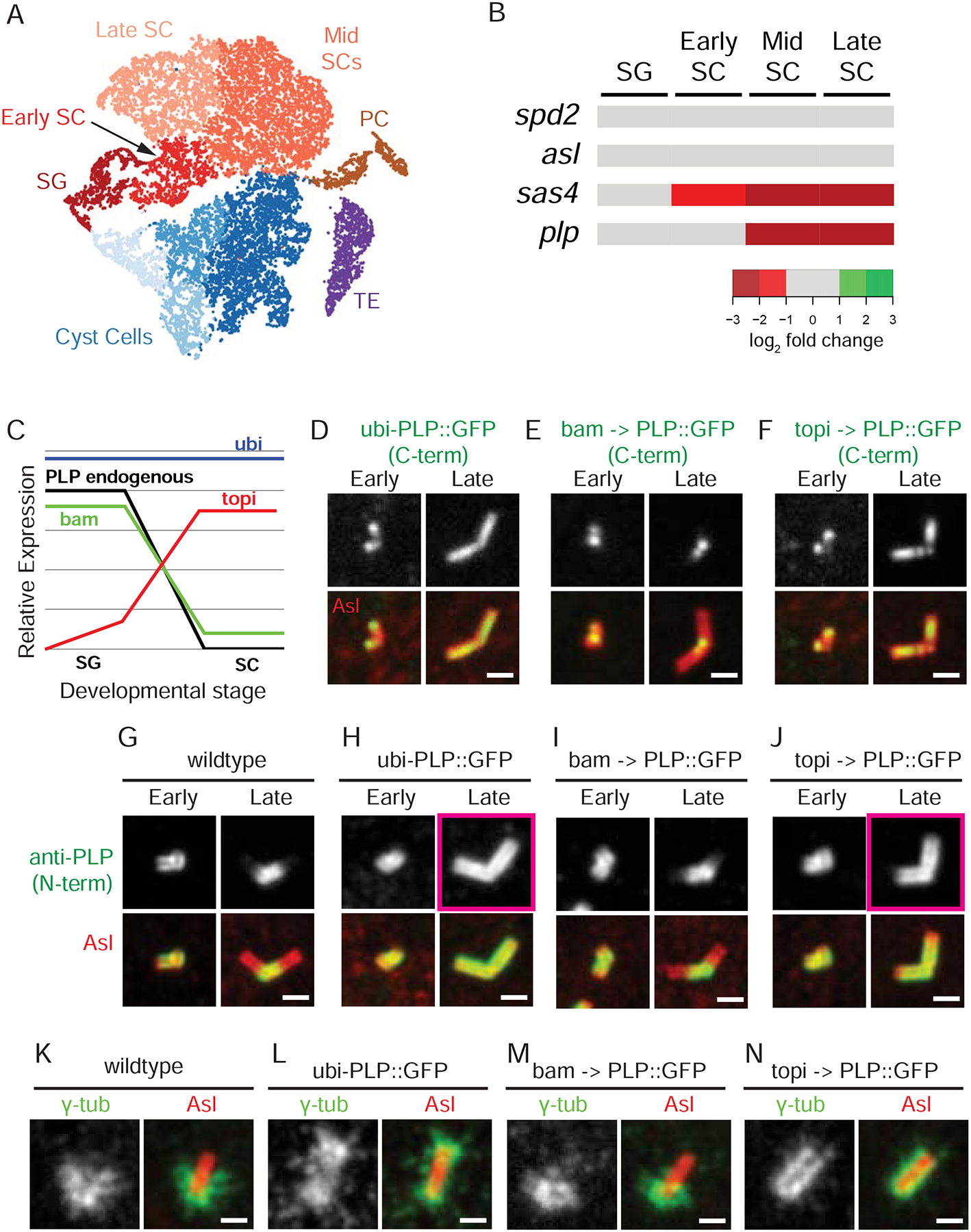
A) tSNE plot showing clusters of sequenced single cells color coded based on the cell type identified from known expression patterns. Somatic cells: Cyst Cells, Terminal Epithelia (TE), Pigment Cells (PC). Germline cells: SG, Early SCs, Mid SCs, and Late SCs. B) Expression of spd2, asl, sas4 and plp (rows) throughout germline developmental stages (columns). The RPKM for each gene was determined from all the cells assigned to a cluster. The log2 fold change, relative to SG at each stage (increase - green shades; decreased - red shades). Sas4 and PLP levels are lower in SC than in SG. C) Schematic of relative PLP levels driven by bam-Gal4 (green), topi-Gal4 (red), and ubi-PLP (blue) compared to endogenous (black) in SG vs SC. D-F) Position of exogenous PLP::GFP (green) determined by direct fluorescence along the centriole (Asl, red) under control of the ubi (D), bam (E) and topi (F) promoters in early and late SCs. G-J) Position of PLP (green = endogenous and exogenous) along the centriole (Asl, red) using N-terminal anti-PLP antisera in early and late SC in wildtype (G) and in flies expressing ubi (H), bam (I) and topi (J) driven PLP::GFP. Pink boxes in H and J highlight the elongation of PLP. Note: The N- and C-termini of PLP have distinct radial localizations with the N-terminus further from the center than the C-terminus (Lawo et al., 2012; Mennella et al., 2012). K-N) Position of γ-tub (green) along the centriole (Asl, red) in metaphase of meiosis II in wildtype (K) and in flies expressing ubi (H), bam (I) and topi (J) driven PLP::GFP. The proximal centriole end is positioned down in all panels. Bars = 1 μm
We also performed in situ hybridization on testes and showed that plp mRNA was significantly enriched in the most proximal region of the testis, where the youngest germline cells reside (Figure S4F, arrows). The signal from older germline cells that reside further down the testis is significantly reduced (Figure S4G), indicating a loss of plp mRNA in SC. In contrast, in situ hybridization of asl mRNA showed similar levels between young and old germline cells (Figure S4H,I). In total, our results indicate that plp mRNA is available to produce protein in the earliest stages of germline development but is no longer available at later stages (Figure 4C, black line). Thus, the proximal restriction of PLP is determined by restricting its availability at both the mRNA and protein level. Currently it is not clear if the decreased protein concentration is a result of increased degradation, decreased protein production, or simply protein dilution as a result of an increase in cell size.
Timing of PLP expression is critical for proximal positioning of PCM
If PLP is restricted to the proximal end of the centriole in SCs by restricting its expression to the earliest stages of germline development, then we predicted that ectopically providing PLP throughout germline development would extend PLP’s distribution along the centriole. We began by imaging ubi-PLP::GFP (expressed in a wildtype background) in live tissue and measuring GFP levels in the cytoplasm, which showed no decrease in protein levels from SG to SC (Figure S5A). Thus, ubi-PLP::GFP is readily available during centriole elongation, explaining why PLP is found on the entire length of centrioles in SCs (Figure 4D, H; Galletta et al., 2014).
We next utilized the GAL4/UAS system as a second method of modulating PLP mRNA and protein levels. We generated a UAS-PLP::GFP transgenic fly and crossed it with two Gal4 drivers. The first was bam-Gal4, which shows an expression pattern similar to endogenous PLP with high levels in SG and low/no levels in SCs (Figure 4C; S5A; White-Cooper, 2012). We found PLP was proximally restricted (Figure 4E, I), recapitulating the wildtype pattern (Figure 4G). The second was topi-Gal4, which expresses at higher levels in SCs than SG (Figure 4C; S5A; Raychaudhuri et al., 2012). This resulted in PLP localization along the entire centriole length (Figure 4F,J). Together, these observations support a model where the timing of high PLP protein concentration/levels determines its position along the centriole and shows that the binding site for PLP is not proximally restricted.
To determine if PLP position dictates PCM position, we used ubi-PLP::GFP or topi>PLP::GFP to localize PLP along the entire centriole length and analyze the position of the PCM in meiosis. We found that unlike wildtype and bam>PLP::GFP (Figure 4K,M), mispositioning PLP along the entire length of the centriole in meiosis resulted in γ-tub recruitment to the entire length (Figure 4L,N). Given the well-established role of Cnn and Spd2 in recruiting γ-tub, we investigated both proteins in meiosis to find that they are also positioned along the entire length of the centriole in ubi-PLP::GFP (Figure S5C). These results indicate that PLP is sufficient to instruct PCM position along the length of the centriole.
Proximal PCM restriction is critical for proper centriole-nuclear docking
With the ability to manipulate the position of PLP and γ-tub along the centriole, we could test our main hypothesis that PCM and MTOC restriction to the proximal end of the centriole is critical for centriole-nucleus docking. We first examined if centrioles with mispositioned PLP and γ-tub had a negative effect on meiosis (see methods; Castrillon et al., 1993). We find no evidence of meiotic defects when PLP was expressed under the control of the bam (100% normal, N = 175) or ubi promoter (99.4% normal, N = 175). When PLP was expressed under the control of topi, 18% (N = 383 RSTs) of RSTs showed evidence of meiotic defects. These meiotic defects, however, do not account for the downstream phenotypes we present below as they are found in both the ubi-PLP::GFP flies, which has no meiotic defect, and at greater penetrance than the rate of defective meiosis in topi>PLP:GFP.
To investigate post-meiotic stages, we first followed PLP and PCM localization in topi>PLP::GFP and ubi-PLP::GFP during early ST development. We found that both PLP and γ-tub positioning along the entire centriole length persisted through the RST stage (Figure 5A–H). In addition to γ-tub, we found that Cnn is retained on 85% of RST centrioles, in the position dictated by ubi-PLP::GFP along the entire length (Figure S5C–E, left). Similarly, the position of Spd2 along the length of the centriole is also dictated by the position of PLP, but it behaves differently than γ-tub and Cnn as ST development progress further. In RSTs, 74% of wildtype centrioles contain no detectable Spd2, and ubi-PLP::GFP does not alter the timing of Spd2 localization to the centriole (Figure S5C–E, right). Thus, ubi- and topi- driven PLP result in the persistence of γ-tub and Cnn at distal positions on the centriole through the RST stage.
Figure 5. Proximal PCM restriction is critical for normal centriole nuclear docking.
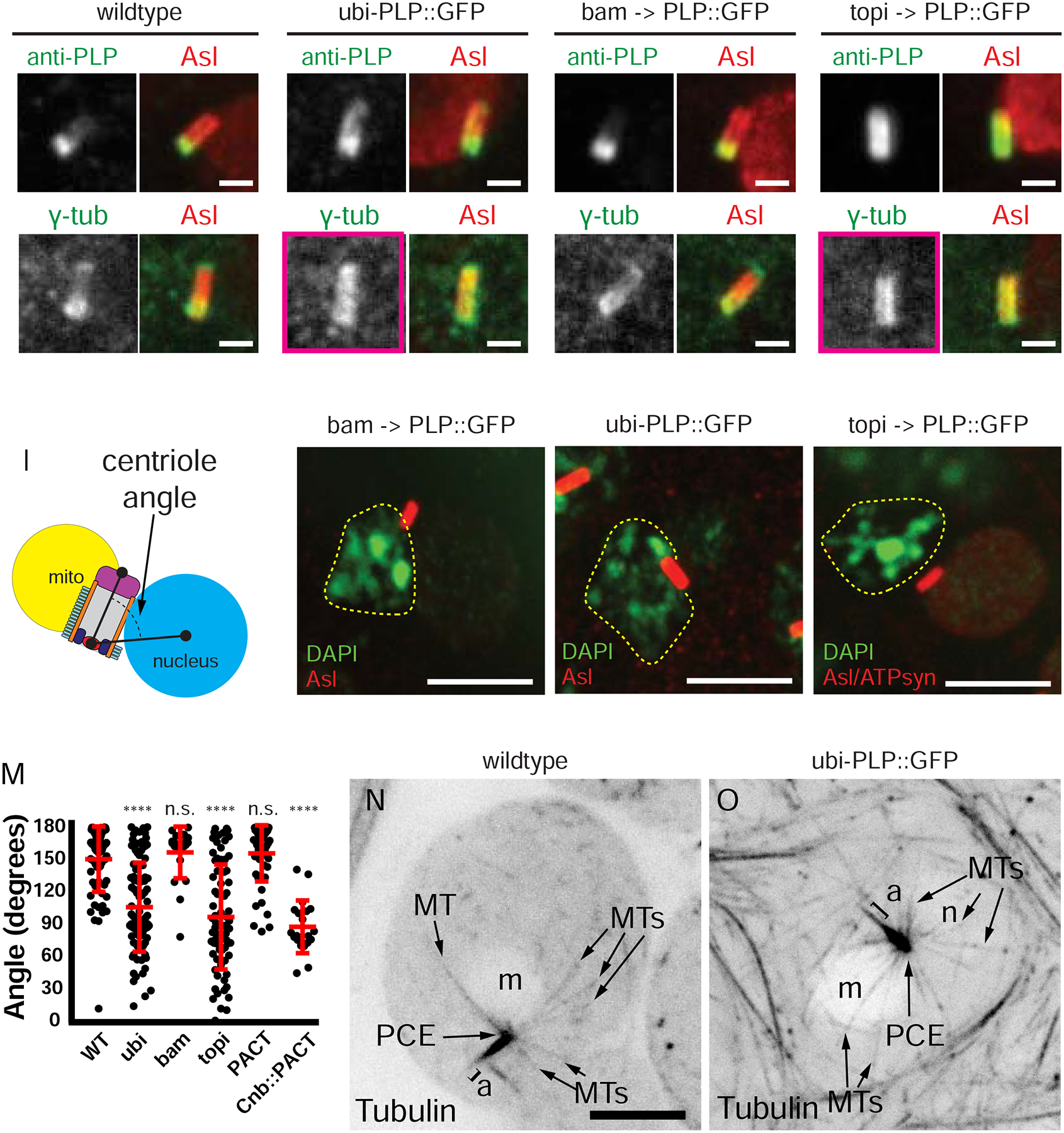
Position of PLP (top row, green) and PCM (bottom row, γ-tub, green) along centrioles (red, Asl) in RSTs: A, B) wildtype C,D) ubi-PLP::GFP E,F) bam-GAL4 driven UAS-PLP::GFP G,H) topi-GAL4 driven UAS-PLP::GFP. The position of PLP dictates the position of PCM along the centriole long axis. I) Centriole angle measurement in RSTs. J-L) Centriole (Asl, red) captured by the nucleus (n, dashed line, DAPI, green) in RSTs: J) bam-GAL4 driven UAS-PLP::GFP K) ubi-PLP::GFP L) topi-GAL4 driven UAS-PLP::GFP. Position of the mitochondrial derivative (m) determined from anti-ATP synthase (not shown in J, K; red in L). M) Angle of the centriole relative to the nucleus in RSTs: wildtype (n=69), ubi (n=98), bam (n=22), topi (n=74), ubi-GFP::PACT (Figure S7A, n=22), Centrobin (Cnb)::PACT (topi-GAL4 driven UAS-YFP::cnb::PACT, Figure S7C, n=20). N,O) MTs (black) in RSTs in wildtype (N) and ubi-PLP::GFP (O). n =nucleus, m = mitochondrial derivative, PCE, a = axoneme, MTs = microtubules. n.s. = not significant, (****) p ≤ 0.0001. Bars A-H,N,R = 1μm, I-L, M, O, Q = 5μm.
We then tested if mispositioning PLP affected centriole-nuclear docking. When exogenous PLP::GFP was expressed early in SG using bam-Gal4, which mimics the endogenous PLP expression pattern, normal Nuclear Search resulted in a normal docking angle of 156° ± 24, similar to wildtype 159 ° ± 30 (Figure 5I,J,M). In contrast, when PLP::GFP was expressed later in SCs, thus driving it along the centriole length (ubi-PLP::GFP and topi>PLP::GFP), Nuclear Search occurred normally, but centrioles adopted aberrant angles relative to the nucleus, 93° ± 41 and 88° ± 49 respectively (Figure 5K,L,M). To confirm that the presence of PCM along the length of centrioles during Nuclear Search resulted in MTs emanating from the entire length of the centriole, we examined MTs in ubi-PLP::GFP RSTs using a ubi-tubulin::GFP transgene. Unlike wildtype, where MTs emanate from the proximal end (Figure 5N, 1E; z-stack in Figure S6A; Figure S1A1–3), expression of ubi-PLP::GFP resulted in MTs emanating from the entire length of the centriole (Figure 5O, z-stack in Figure S6B; Figure S1B1–4). This is consistent with EM observations of MTs emanating from the entire length of the centriole much earlier in sperm development prior to meiosis in SCs from ubi-PLP::GFP (Roque et al., 2018). To further investigate the location of MT growth on centrioles, we performed MT regrowth following depolymerization assays. In wildtype cells treated on ice for 1 hour, the majority of MTs were depolymerized with the exception a small tuft of MTs at the proximal MT end (Figure S6C, left). Following a 1 minute release from cold, MTs predominantly nucleated from the proximal end (Figure S6C, right). In contrast, the ubi-PLP::GFP expressing RSTs showed MTs along the entire centriole length in both the iced and the 1 minute recovery conditions (Figure S6D).
To ensure that the docking defect was a consequence of mispositioned PCM and MTs, and not simply a result of overexpressing or mispositioned PLP leading to an indirect effect, we performed two controls. First, to test if the docking defect was due to the presence of PLP binding at distal sites on the centriole, rather than the distal position of PCM, we examined RSTs in flies expressing only the C-terminal portion of PLP which contains the centriole docking domain (PACT). Ubi-PACT::GFP localized along the entire length of centriole, but PCM remained proximally restricted and centrioles correctly docked to the nucleus (Figure S7A,B; Figure 5M). Second, to show that mispositioned PCM (not simply PLP) causes docking defects in STs, we mispositioned PCM along the entire centriole by expressing the chimeric protein Centrobin::PACT, which had been shown to be sufficient to recruit PCM to centrioles in neuroblasts (Januschke et al., 2013). In this case, PCM was recruited along the entire length and centrioles docked aberrantly to the nucleus, similar to PLP misexpression (Figure S7C,D, Figure 5M). Together these data strongly suggest that the docking defect arises from the failure to restrict PCM proximally and not simply a result of PLP occupying distal binding sites or of its overexpression.
Mispositioning PCM results in decapitated sperm
To determine the ultimate consequence of lateral centriole attachment to the nucleus, we examined near fully mature STs that have reshaped their nuclei. In wildtype cysts, all 64 nuclei from a single cyst cluster in a stereotypic configuration where the centrioles attach to the caudal side of the nucleus (Figure 6A). However, in mature STs where PLP and PCM were mispositioned, nuclei are not properly clustered with many lacking associated centrioles (Figure 6B,C). When combined with our earlier observations this suggests that lateral attachment of the centriole to the nucleus results in the formation of an unstable nuclear connection.
Figure 6. Abnormal centriole docking results in failed sperm head-tail attachment.
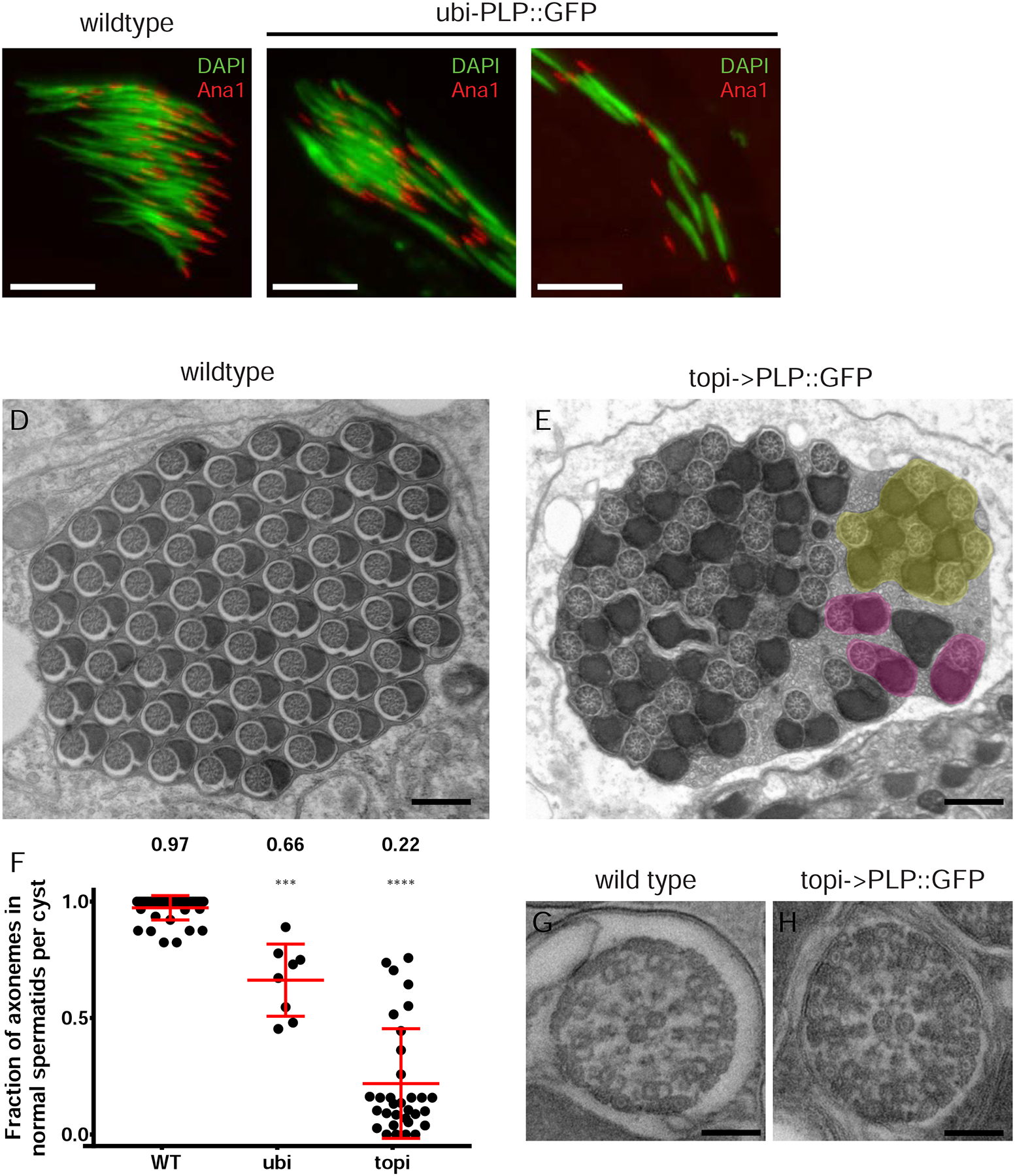
A,B) “Needle” shaped nuclei (DAPI, green) and centrioles (Ana1::tdtom, red) in STs from wildtype (A) and ubi-PLP::GFP (B,C) flies. ubi-PLP::GFP testes show a mix of clustered (B) and dispersed nuclei and centrioles in the same cyst (C) Many centriole-nucleus connections fail. D,E) TEM cross sections of mature ST cysts in wildtype (D) and topi-GAL4 driven UAS-PLP::GFP (E) testes where the later contain many STs that fail to properly individualized (yellow). Properly individualized STs with a normal complement of 1 axoneme and 2 mitochondrial derivates are in pink. F) The fraction of axonemes in each cyst that appear “normal”. Mislocalized PCM in early STs results in significant numbers of abnormal STs. Cysts analyzed: Wildtype (N=41), ubi (N=8), topi (N=32). G,H) Cross section TEM through a single axoneme from wildtype (G) and topi-GAL4 driving UAS-PLP::GFP (H) flies. Axoneme organization appears unaffected by PCM mislocalization. Bar A-C = 10μm. D,E = 500nm, G,H = 100nm.
Previous work has shown a link between failed centriole-nuclear docking in ST development and failure in ST individualization, when each sperm is encased in its own membrane (Fabian and Brill, 2012; Kracklauer et al., 2010; Texada et al., 2008). We performed TEM of testes from flies with mispositioned PCM and found clusters of axonemes and mitochondrial derivatives not enclosed in their own membrane, indicating a failure of individualization in the STs (Figure 6E). When PLP::GFP was expressed under the control of the ubi or topi promoter (Figure 6E,F), 34 ± 16 % or 78 ± 24 % of STs were defective, respectively (vs 3 ± 5 % in wildtype; Figure 6D, F). EM is an end-point readout of failed individualization, thus the mechanism by which nuclear attachment defects lead to individualization defect remains unknown. Reasonable hypotheses include the defect arising from the failed nuclear-clustering defect (Figure 6B,C), which also correlates with the failed centriole-nuclear docking, or an issue in assembling the ST individualization apparatus.
Closer examination shows that the axonemes themselves within defective clusters appear normal, indicating that the mispositioning of PCM did not affect ciliogenesis from the distal end (Figure 6G, H). Furthermore, we found ample sperm in the seminar vesicles (Figure S7E) and did not uncover any significant effects on fecundity (Figure S7F,G, see methods). Thus the number of sperm is sufficient to propagate the fly populations under standard laboratory conditions. A strong prediction would be that such a reduction in sperm production (as predicated by the TEM data, Figure 6E, F) would be outcompeted in nature. However, a comparable reduction in functional human sperm count is within the range of clinical subfertililty (Guzick et al., 2001).
Discussion
Functional sperm require a stable linkage between sperm head and tail, which is mediated by the centriole. To date, little is known about the molecular mechanisms underlying defective head-tail attachment, except for a few reports identifying mutations in genes such as Spata6, Sun5, BRDT, PMFBP1 and TSGA10 (Elkhatib et al., 2017; Fang et al., 2018; Li et al., 2017; Sha et al., 2018a; Sha et al., 2019; Sha et al., 2018b; Shang et al., 2018; Shang et al., 2017; Yuan et al., 2015; Zhu et al., 2018; Zhu et al., 2016). Directly relevant to our study on the role of the centriole in head-tail attachment, is TSGA10 (Cep135 paralog) and two other centriole proteins - Centrin1 (Avasthi et al., 2013) and Centrobin (Liska et al., 2009). Thus, the limited patient sequence analysis and mammalian model system mutants point to a critical role for the centriole in head-tail attachment.
Our live imaging of centrioles in Drosophila STs revealed that the docking of the centriole to the nucleus is a two-step process (Figure 7A). The first is ‘Nuclear Search’, where the centriole searches for its docking partner, the nucleus, immediately following meiotic exit. The second is ‘Nuclear Attachment’, where a stable connection of the centriole to the nuclear surface is formed. Work in Drosophila has also identified a number of mutations that ultimately result in decapitation, several of which affect the ability of the MT motor dynein to localize to the nuclear envelope, or to generate pulling forces on MTs (Anderson et al., 2009; Fabian and Brill, 2012; Kracklauer et al., 2010; Li et al., 2004; Sitaram et al., 2012; Texada et al., 2008; Wu et al., 2016). Interestingly, studies from human patients have also implicated the dynein adapter Hook1 (Mendoza-Lujambio et al., 2002), and the sun protein SPAG4 (Yang et al., 2018) in ensuring proper head-tail linkage. These proteins have been shown across species to position the centrosomes adjacent to the nucleus in interphase of normally cycling cells (Anderson et al., 2009; Gonczy et al., 1999; Malone et al., 2003; Robinson et al., 1999; Sitaram et al., 2012; Splinter et al., 2010; Zhang et al., 2009). Thus a simple model emerges where in developing STs a conserved dynein-based system on the surface of the nucleus interacts with MTs emanating specifically from the proximal end of the centriole to draw the proximal centriole end specifically to the nuclear surface. Our imaging of MTs suggests that this model is quite plausible, showing MTs specifically emerge from the proximal end of the centriole, which we then show is key to proper centriole-nuclear docking. One exciting future direction is to reexamine the known Drosophila decapitation mutants to separate players that affect Nuclear Search from those that affect only the later step of Nuclear Attachment.
Figure 7. Model for centriole-nuclear docking via proximal PCM restriction.
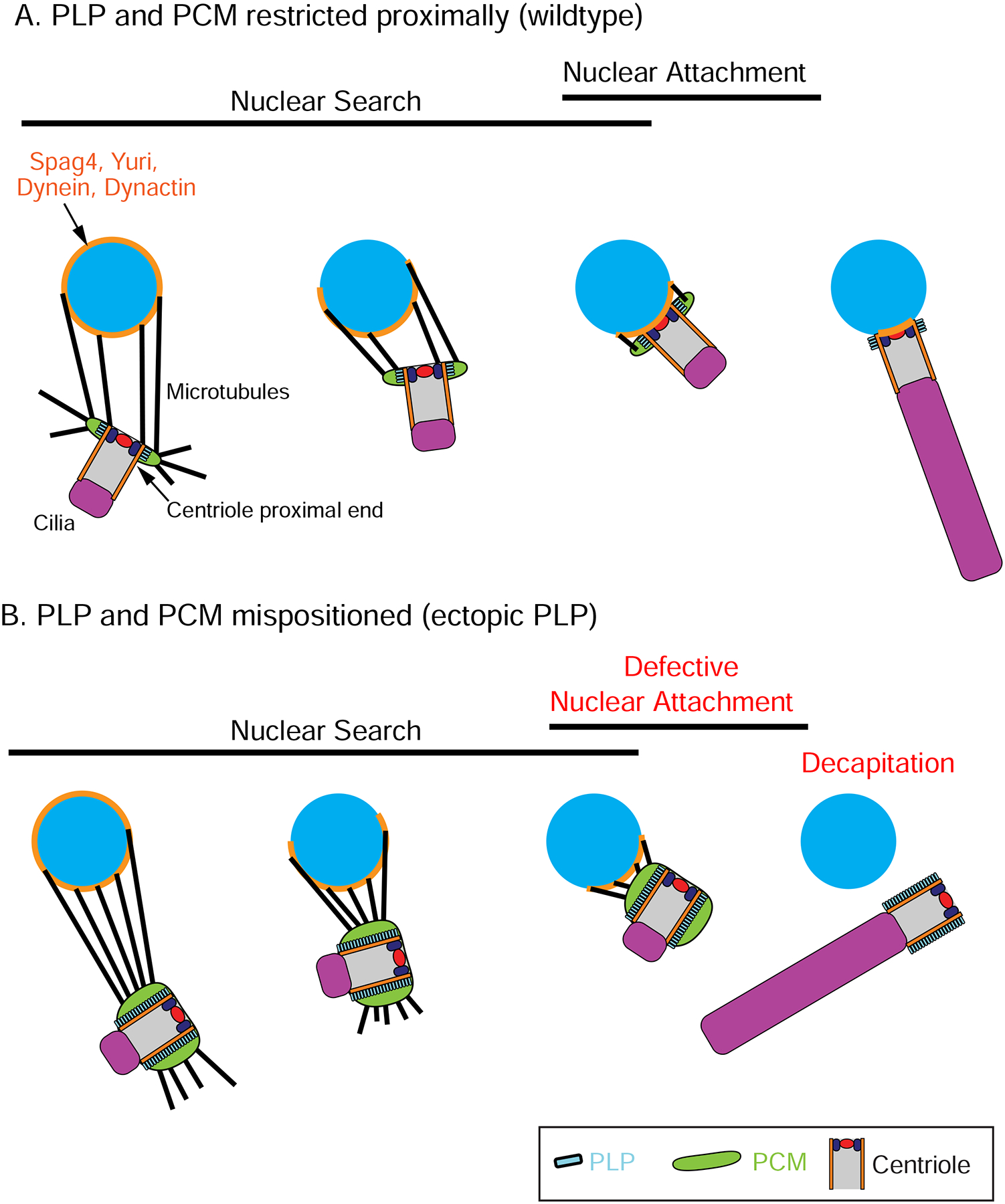
(A) During normal sperm development, proximal centriolar PCM (green) nucleates MTs (black) that capture the nucleus (blue). This capture is likely mediated by dynein motors tethered to the nuclear surface that drive centriole movement during Nuclear Search. Proper Nuclear Search then leads to Nuclear Attachment prior to axoneme (purple) elongation. (B) When PCM is not proximally restricted, MTs emanate from the entire length of the centriole. These centrioles undergo proper Nuclear Search, but adopt an improper angle relative to the nuclear surface and fail to properly undergo Nuclear Attachment (red). Subsequently, the centriole losses its connection with the nucleus, resulting in decapitation (red).
While our study focuses on the formation of functional sperm, it also provides insight into centrosome architecture, and how this architecture relates to function, in this case nuclear docking. In recent years, enormous progress has been made in identifying centrosome proteins and mapping their localization with nanometer precision. The challenge now is to link protein position with function; progress on this front has been most notably made at the centriole. For example Ana2/STIL, Sas6, and Cep135 form a cartwheel structure inside a new centriole at its proximal end, serving as a template for centriole symmetry (Reviewed in (Arquint and Nigg, 2016; Nigg and Holland, 2018). Another protein complex, Cep97,CP110 and Klp10A, localizes to the distal end and to control centriole length (Delgehyr et al., 2012; Franz et al., 2013; Schmidt et al., 2009; Spektor et al., 2007). Thus, subcentriolar localization and function are intimately linked. Despite PCM being documented at the proximal end of the centriole (Bornens et al., 1987; Fu and Glover, 2012; Galletta et al., 2016b; Paintrand et al., 1992), a link between the position and function of the PCM at the proximal end has not been established. One possible role for the PCM at the proximal end is to dictate the position of daughter centriole formation. Previous work has shown that new daughter centriole nucleation requires PCM (Dammermann et al., 2004; Loncarek et al., 2008; Zhu et al., 2008), and overexpression of PCM results in additional daughter centriole formation (Loncarek et al., 2008). However, the position of the ectopic centrioles along the proximal distal axis was not examined and the importance of the proximal position could not be inferred. Our study provides a clear demonstration of a proximal-specific function for PCM.
Through a series of wildtype, mutant and mis-expression experiments, we show that the bridge protein PLP is critical in proper centriole docking in STs. By examining plp mutant testes, we found that PLP is a major driver of PCM recruitment or retainment at centrioles in RSTs. In the absence of PLP, PCM is disorganized and the centriole is improperly positioned away from the nucleus. We then investigated how PLP itself is restricted to the proximal end. Using endogenously tagged PLP protein, single cell RNAseq and whole mount in situ, we show that proximal restriction of PLP is achieved through a reduction in PLP mRNA and protein concentration prior to centriole elongation. It will be important in future studies to determine precisely how PLP concentration is reduced; there is likely a delicate balance between decreased PLP translation, increased PLP degradation, and simple dilution of PLP as spermatocyte size increases.
While numerous studies have shown that PLP is necessary for PCM organization around centrioles and that PLP can interact with PCM components (Citron et al., 2018; Lerit et al., 2015; Martinez-Campos et al., 2004; Richens et al., 2015), the precise mechanism of how PLP acts to recruit or anchor γ-tub is not known. In addition to interacting with PCM components, both PLP, and its mammalian ortholog pericentrin, have been found in complexes with γ-tub itself (Dictenberg et al., 1998; Kawaguchi and Zheng, 2004). It remains to be determined precisely how the multiple possible pathways through which PLP can affect the PCM function at the centriole in different contexts. Whether direct or indirect, our study shows that mislocalizing PLP on the centriole is sufficient to dictate PCM position on the centriole. When PCM is present at a more distal position along the centriole in STs, MTs emanate from the entire centriole, which can result in lateral capture to the nucleus. This lateral capture appears unstable and firm nuclear attachment does not occur. We propose that these failed, or defective, centriole-nuclear attachments do not survive the forces applied as a result of axoneme and mitochondrial derivative elongation, nuclear clustering and/or ST individualization, ultimately resulting in sperm decapitation (Figure 7B). This is consistent with several studies linking a failure in individualization to a failure in centriole-nuclear attachment (Fabian and Brill, 2012; Kracklauer et al., 2010; Texada et al., 2008). Additional studies will be required to determine how the tight attachment between the nucleus and the centriole forms, precisely when the Nuclear Attachment fails in STs with laterally docked centrioles and how this connection relates to the machinery that drives the massive cellular reorganization required to build sperm.
The transcriptional mechanism to restrict protein localization we identified is one way to achieve sub-centriolar protein compartments. Other mechanisms include specific docking-site recognition, such as the LID domain of Sas4 recognizing the plus end of MTs at the centriole distal end (Sharma et al., 2016; Zheng et al., 2016) and protein symmetry-breaking and coalescence as seen with Plk4’s ability to concentrate into a single spot on the surface of the centriole (Dzhindzhev et al., 2017; Kim et al., 2013; Ohta et al., 2014; Ohta et al., 2018). While it is unknown if other proteins use a transcriptional mechanism like PLP for centriole position control, single cell RNA sequencing data could help identified such proteins.
One exciting finding from our work is that simply altering the timing of PLP expression can have major deleterious effects. This can be analogous to many human diseases that are frequently reported to correlate with higher levels of protein expression; understanding the underlying cell biology and physiology of the protein overexpression becomes quite critical. For example, overexpression of the master regulator of centriole duplication Plk4 is sufficient to promote tumorigenesis (Levine et al., 2017) and renal cysts (Dionne et al., 2018). Another example is seen in the case of having an extra copy of Pericentrin (the ortholog of PLP), which in humans is present on chromosome 21. An increase in Pericentrin protein levels by 50% can cause defects in ciliogenesis and cilia function (Galati et al., 2018). Therefore, understanding the role of the centriole in human disorders will not only require understanding the consequences of loss of protein function, but also the consequence of protein misexpression and misregulation.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Nasser M. Rusan (nasser@nih.gov). All unique/stable reagents generated in this study are available upon request without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
D. melanogaster
All fly stocks were maintained and genetic crosses were conducted at 25°C. Except when noted, all control flies in this study were yw. All plp mutants examined were plp2172/Df(3L)Brd15. UAS-Ana1::tdTomato contains the endogenous promoter; when used without a GAL4 line, Ana1 is likely expressed at endogenous levels ((Blachon et al., 2008). TAGRFP::Sas6 was previously generated using a BAC recombineering to introduce TAGRFP at the 5’ end of the sas6 locus (Lerit and Rusan, 2013). Sas6 is likely expressed at endogenous levels. All new transgenic animals were generated by BestGene (Chino Hills, CA).
METHOD DETAILS
Generating transgenic Drosophila
ubi-GFP::PACT was generated by mobilizing an existing ubi-GFP::PACT construct (Martinez-Campos et al., 2004) from the chromosome III to II using standard methods. UAS-PLP::GFP was generated using the Gateway cloning system to move the plpPFcDNA in pENTR/D (Galletta et al., 2014) into pPWG. GFP::Asl under endogenous control was generated using genomic DNA that contained 2322 bp upstream of the start codon (the asl promoter) and 605 bp downstream of the stop codon; GFP plus 54 bp of linker was inserted at the start codon. UAS-PLP::GFP and GFP::Asl transgenic flies were generated using standard P-element-mediated transformation. We confirmed that UAS-PLP::GFP produced a protein of the expected size by western blot (Figure S5B). PLP::mNeon was generated by CRISPR. y[1] M{vas-Cas9}ZH-2A w[1118]/FM7c flies were injected with pU6-Bbsl-chiRNA containing the PLP::mNeon gRNA and the repair template. The repair template included 1097 nucleotides (nts) upstream of the plp stop codon, 48 bp of linker sequence, the sequence of mNeon green, a stop codon and 1100 nts downstream of the plp stop codon. Recombinants were identified by PCR screening. The mNeon insertion recapitulates the endogenous PLP localization and expression pattern, is homozygous viable, and shows none of the known plp loss of function phenotypes, indicating it is fully functional. All transgenic animals were generated by BestGene (Chino Hills, CA).
Immunofluorescence methods
Whole mount samples were prepared as described (Galletta et al., 2016b). Testes from larvae, pupae and adults were dissected in Drosophila S2 media, fixed in 9% formaldehyde in PBS (from 16% Paraformaldehyde stock solution) for 20min and then washed three times in PBS + 0.3% Triton X-100 (PBST). Samples were either used immediately or stored for less than 2 weeks at 4°C in PBS. Samples were blocked for at least 2 hours at room temperature (RT) in PBST + 5% normal goat serum, incubated with primary antibodies in block overnight at 4°C a nd then washed three times in PBST for 10 minutes each. Secondary antibodies were incubated for 4–8 hours in block at RT then washed as above. Samples were mounted in Aquapolymount (Polysciences, Inc.) under No. 1.5 coverslips. Images of mature STs were acquired as described (Varadarajan et al., 2016) with modification. Testes were dissected in Schneider’s Drosophila (S2) Medium (Gibco and placed on a glass slide with a drop of S2 media. A small cut in the side of each testis was made and the sample was squashed under a coverslip to free cysts. Slides were flash frozen in liquid nitrogen, the cover slip removed and the slides stored in 95% ethanol. Fixation was in 4% formaldehyde in PBS for 20 min. Samples were then treated the same as other fixed samples. The following primary antibodies were used: Guinea pig anti-Asl (1:10,000, (Klebba et al., 2013)), affinity-purified rabbit anti-PLP (raised against the N-terminus, 1:4,000; (Rogers et al., 2008)), mouse anti-γ-tubulin (1:500, GTU88, T6556; Sigma-Aldrich), rabbit anti-Spd2 (1:7000; gift of M. Gatti; (Giansanti et al., 2008), rabbit anti-Cnn (1:10,000; (Galletta et al., 2016b), mouse anti-ATP5A (1:1000, 15H4C4, ab14748; Abcam). Secondary antibodies were Alexa Fluor 488, 568 or 647 conjugated (Thermo Fisher Scientific). DAPI (1:1000) was added to all samples during incubation with secondary antibodies.
Light microscopy
Confocal images of fixed samples were acquired on an Eclipse Ti2 (Nikon) with a 100X/1.4NA objective, a CSU-10 spinning disk confocal head (Visitech International), an interline-transfer charge coupled device camera (CoolSNAP HQ2; Photometrics) and 405, 491, 561 and 642 nm laser lines or a similar microscope with a 100X/1.49 NA objective, a CSU-22 confocal head, and an ORCA-Flash 4.0 CMOS camera (Hamamatsu Photonics). This second microscope was also used for live imaging using a 40x/1.3 NA objective and a 1.5X tube lens in the microscope body. MetaMorph (Molecular Devices) was used to control the microscope and acquire images. Movies of STs in testes were made as described (Galletta et al., 2016b). In brief, testes were dissected in Drosophila S2 media and the whole testis was mounted in a drop of media on a 50-mm lumox dish (Sarstedt). The media was surrounded by Halocarbon oil 700 (Sigma-Aldrich) and covered with a #1.5 coverslip. Stacks were acquired at 1 μm Z-intervals and at 5 – 15 minute time intervals. Images presented are projections of multiple confocal sections. Still images of live, disrupted testes were used to acquire images of microtubules emanating from centrioles in RSTs. These samples were prepared similar to samples for movies, except a glass slide was used instead of a lumox dish. Samples were then partially squashed to free STs from the testes and imaging was performed as for fixed images. Images of microtubule regrowth assays and images to quantify GFP levels in the cytoplasm of testes from transgenic flies were acquired on a Nikon W1 spinning disc confocal equipped with a Prime BSI cMOS camera (Photometrics) and a 40X/1.3 NA oil immersion objective. Images of in situs were acquired on a Nikon Eclipse Ni-E with a 20X/0.75 NA objective, DIC optics, 0.6X zoom and a Nikon DS-Fi2 camera. All image analysis was performed using ImageJ (National Institutes of Health) and data analysis was performed using Excel (Microsoft) and Prism (Graphpad).
Microtubule regrowth assay
Regrowth assays were performed as described (Chen et al., 2017) with modifications. Testes from adult flies were dissected in S2 media and transferred in 200 μl to a PCR tube. Media was exchanged with ice cold S2 media and the samples were placed in an ice/water bath for 1 hour. After 1 hour, samples were either immediately fixed as above or cold media was exchanged with media prewarmed at 25°C and samples were held at 25°C for 1 minute before fixing as above. Sample pr eparation for imaging is as described for whole tissue above.
Electron Microscopy
Drosophila testes were chemically fixed for one hour with 2.5% glutaraldehyde and 1% formaldehyde in 0.12 M sodium cacodylate buffer, pH 7.2, for conventional transmission electron microscopy (TEM). The specimens were then post fixed in 1% osmium tetroxide in cacodylate buffer, en bloc stained with 1 % uranyl acetate, dehydrated in an ethanol series/propylene oxide and embedded in EMbed 812 resin (Electron Microscopy Sciences, Hatfield PA). Ultrathin sections (50–60 nm) in both transverse and longitudinal orientations were obtained using an EM UC7 ultramicrotome (Leica, Vienna, Austria). Sections were post-stained with uranyl acetate and lead citrate and examined with either a JEM-1200EX (JEOL USA) TEM (accelerating voltage 80 keV) equipped with an AMT 6 megapixel digital camera (Advanced Microscopy Techniques Corp) or a JEM1400 (JEOL USA) TEM (accelerating voltage 120 keV) equipped with an AMT XR-111 digital camera (Advanced Microscopy Techniques Corp).
Single cell RNAseq
Testes from w1118 third instar larva, before the final prepupal defecation, were collected in Phosphate Buffered Saline, pH 7.4 (PBS) and fat body cells were removed with 0.075% (w/v) Porcine Powered Pancreas and 75U/μl Collagenase in PBS at 22° C for 2–3 min in 1.5ml Eppendorf tubes with gently agitation and decantation. Cleaned testes were then dissociated in 0.45% Porcine Powered Pancreas and 75U/μl Collagenase in PBS at 22° C for 30 min with a tungsten needle and pipetting under the dissecting scope. Collagenase dissociation was stopped with Fetal Bovine Serum (FBS, 1% final v/v) for 2 min. Samples were decanted and washed on a 35μm cell filter and cells were pelleted at 845 x g for 5 min, resuspended in 25 l PBS, 0.04% BSA and counted in a 1 l cell suspension to calculate density. Cell suspensions (6K for Replicates 1 and 2, 12K for replicate 3) were loaded onto the 10X Chromium system for barcoding and library preparation following the user guide for Single Cell 3’ Reagent Kits v2. Libraries were quantified with Quant-iT PicoGreen and 300–500 bp insert sizes were confirmed on a TapeStation 2200. scRNA-Seq profiles were generated in biological triplicate pools on the 10X Chromium System and sequenced (Read1 = 26 bp, Read2 = 98 bp and Readi7 = 8 bp) on a HiSeq Illumina 2500.
The single cell RNAseq data of whole Drosophila larval testes used in this study can be found at (Gene Expression Omnibus #GSE125947). Single-cells were clustered using K-nearest neighbors (KNN; Seruat v3.0.1) and germ cells were annotated as SG, early SC, mid SC and late SC using expression patterns of key marker genes from the literature including vas, bam, aub, p53, aly, sa, stg, twe, and bol. For a target list of 52 centriole, cilia and spermatogenesis genes, we compared expression patterns between SG and mid and late SCs using the FindMarkers function (Seurat v3.0.1) with no log fold change cutoff criteria. P-values were adjusted for multiple testing using false discovery rate (FDR) and considered a gene differentially expressed at an alpha level ≤ 0.05. Adjusted P value for plp = 3.8 × 10−59. For heatmaps, the read counts for each gene were summed for all cells assigned to each cluster. Data was normalized using reads per kilobase of transcript, per million mapped reads (RPKM). The RPKM for each gene at each stage was extracted and the log2 fold change relative to the SG cluster was calculated.
In situ hybridization
In situ hybridization on whole mount testes was performed as described (Morris et al., 2009), with one modification, the initial fixation was in 4% paraformaldehyde in PBS + 0.3% tween-20 for 1 hour. In brief, adult testes were dissected in Drosophila S2 media and fixed as above. Samples were then washed in PBS + 0.1% Tween 20 (PBSTw) and incubated in 50 μg/ml Proteinase K (Thermoscientific) in PBST for 5 – 7 minutes. The reaction was stopped by adding 2 mg/ml glycine and washed. Samples were then refixed, washed though a series of PBST, a 1:1 ratio of PBST : hybridization buffer (HB, 50% (vol/vol) Formamide, 5× SSC, 100 g/ml denatured sonicated salmon sperm DNA, 50 g ml−1 heparin, 0.1% Tween 20, pH to 4.5 with citric acid (100 mM final), and HB. Samples were prehybridized in HB buffer for 1 hour at 65°C. cDNAs encoding the last 1091 amino acids of PLP, which are shared by all predicted isoforms, and the entire Asl coding sequence were used as templates to generate sense and antisense RNA probes using T7, T3 or SP6 RNA polymerase (Roche and Thermo Scientific) labeled and DIG RNA Labeling mix (Roche). Probes were heat denatured at 80°C for 10 min then chilled on ice before adding to samples. Hybridization was overnight at 65°C. Samples were then washed in HB at 65°, followed by room temperat ure washes in sequential mixtures of HB:PBSTw at 4:1, 3:2, 2:3, 1:4, then PBST. Alkaline phosphatase conjugated sheep anti-DIG antibody (1:2000; Roche) was used for detection and incubated overnight at 4°C. Samples were washed 3X in HP (100 mM NaCl, 100 mM Tris (pH 9.5), 50 mM MgCl2, 0.1% Tween 20) then stained in HP with 7 – 10 μL NBT/BCIP premix (Roche). Samples were dehydrated through an ethanol series (30% - 100%), then incubated with 1:1 ethanol / methyl salicylate. Samples were then mounted in Aquapolymount (Polysciences, Inc.).
Western Blot
30 brains were dissected from wandering 3rd instar larvae of the indicated genotypes. Brains were homogenized in 50 μl 1X SDS running buffer (58 mM Tris pH 6.8, 5% glycerol, 1.95% SDS, 1.55 % DTT, ~0.05% Bromophenol Blue) then incubated at ~100°C for 5 min. Samples were run on a 7.5% gel an d transferred to nitrocellulose using the iBlot system (Thermo Fisher Scientific). PLP was detected using a 1:5000 dilution of the N-terminal antibody, followed by horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:1000, Thermo Fisher Scientific). Detection was performed using SuperSignal West Dura Extended Duration Substrate (Life Technologies) and visualized using a ChemiDoc MP Imaging System (Bio-Rad).
QUANTIFICATION AND STATISTICAL ANALYSIS
Docking angle measurement
To measure the docking angle of the centriole and distance from the nucleus, RSTs with a normal complement of nuclei and centrioles were selected. Only centrioles with their long axis almost parallel to the imaging plane were selected. If necessary, Z-projections to include the entire area covered by DAPI staining of the nucleus were generated. The perimeter of the DAPI area was selected by hand in ImageJ and the center of mass of the resulting region of interest (ROI) was measured. The points of the centriole closest to and furthest from the nucleus was then marked. The distance between the center of mass of the DAPI stain and the close end of the centriole was the “DNA to centriole distance”. The angle between the center of the DAPI, the close end of the centriole, and the far end was calculated as the “centriole angle. These results likely underestimate the severity of mispositioned centrioles as a result of measuring a 2D projection of the 3D arrangement of objects. Additionally, improperly docked centrioles might be in a different plane than the nucleus and might overlap with the nucleus in the 2D projection, thus underestimating the distance and the defect in the angle.
Centriole protein levels
Measurements of fluorescence intensity at the centriole were performed as described (Galletta et al., 2014). In brief, samples to be compared were dissected and processed the same day. Measurements are all relative to the control from the same day and performed in duplicate. Sum projections through the volume of the centriole were performed and the total integrated density within an (ROI) including the centriole associated signal was measured. An identically sized ROI was used for background subtraction. Measurements of cytoplasmic fluorescence of PLP::mNeon, ubi-PLP::GFP, bam->PLP::GFP and topi->PLP::GFP were conducted in live testes using SiR-Tubulin (Cytoskeleton, Inc.) to allow for morphological identification of cells. A ROI within the cytoplasm of cells of the indicated developmental stage was drawn, avoiding the centrioles. The mean pixel intensity of this ROI was determined. The mean pixel intensity of an ROI adjacent to the tissue sample was used for background subtraction. Data was normalized to the average signal in SG.
Centriole length measurement
Measurements of centriole length used the Asl or Ana1 signal. PLP length was measured along the long axis of the centriole. Length was determined using a line scan along the long axis of the centriole. The average intensity of the first and last 10 pixels of the line scan, not on the centriole, was used as the background and was subtracted from every point along the line scan. The maximum intensity along this scan was used to normalized all values. The number of pixels where the normalized intensity was >0.5 was determined and converted to microns using a conversion factor determined from a stage micrometer.
Determining meiotic failure
Meiotic failure was determined in RSTs. When there is a failure in meiosis in the male germline significant enough to disrupt cytokinesis, the mitochondria destined for more than one ST coalesce into a giant nebenkern in RSTs (Castrillon et al., 1993). When this occurs, more than one centriole is associated with this nebenkern. Defects in meiosis can also result in defects in centriole segregation leading to STs without centrioles. Confocal stacks through cysts of RSTs were acquired. The number of centrioles in a ST associated with each nebenkern was counted. The number of normal STs (1 centriole/nebenkern) and abnormal STs (0 or >1 centriole/nebenkern) was determined. The percentage of the total that were normal or abnormal was determined.
Normal Spermatid Fraction
A ST was considered “normal” if when examined by TEM the axoneme was enclosed in a membrane with 2 mitochondrial derivatives. The fraction of normal spermatids was determined by dividing the number axonemes in each cyst that were “normal”, as determined by the “normal” axoneme number by the total axoneme number, is presented.
Fertility assay
Specific details for each fertility assay performed can be found in the legend for Figure S7E, F. In general, fertility was assessed by mating 16 – 24 hour old naïve males with virgin yw female(s). After the specified mating period the male was removed and the female(s) were retained and allowed to lay eggs in the vial. Females were transferred to fresh vials at the indicated intervals. All adult progeny from all vials were counted and presented.
Statistical analysis
Except where specifically noted, unpaired t tests, with Welch’s correction when appropriate were used for statistical comparisons. Sample sizes and the number of experimental repeats are reported in the legends. Statistical analysis was performed with Prism (GraphPad). In all cases the mean ± the standard deviation (SD) is presented.
DATA AND CODE AVAILABILITY
The single cell RNAseq is reported in the Gene Expression Omnibus (GEO) repository accession #GSE125947)
Supplementary Material
Video 1. Nuclear Search and Nuclear Attachment (related to Figure 1C) Movie of a SC centriole exiting meiosis through “Nuclear Search” and “ Nuclear Attachment” in RST. GFP::Cep135 (green) and DNA (H2Av::RFP, red). Individual frames are a projection of multiple confocal slices. Frames were acquired at 15 minute intervals. Movie is presented at 3 frames / second. The centriole marker used is brighter on the proximal centriole end than on the distal centriole end (Galletta et al., 2016a) allowing for identification of the distinct ends of the centriole in the movie. Stills of the movie are presented in Figure 1C. Time stamp is relative to reformation of the nuclear envelope following meiosis.
Video 2. Centrioles organize microtubules throughout Nuclear Search and Nuclear Attachment (related to Figure 1D) Movie of a SC expressing GFP::α-tubulin exiting meiosis through “Nuclear Search” and “ Nuclear Attachment” in RST. Individual frames are a projection of multiple confocal slices. Frames were acquired at 5 minute intervals. Movie is presented at 5 frames / second. Stills of the movie are presented in Figure 1D. Time stamp is relative to reformation of the nuclear envelope following meiosis.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea pig anti-Asl | Klebba et al., 2013 | N/A |
| Rabbit anti-PLP | Rogers et al., 2008 | N/A |
| Mouse anti-γ-tubulin | Sigma Aldrich | GTU-88, Cat # T5326, AB_532292 |
| Rabbit anti-Spd2 | Giansanti et al., 2008 | AB_2567766 |
| Rabbit anti-Cnn | Galletta et al., 2016b | N/A |
| Mouse anti-ATP5A | Abcam | 15H4C4, Cat # ab14748, AB_301447 |
| Sheep anti-DIG-AP | Roche | Cat # 11093274910, AB_514497 |
| Mouse anti-GFP | Clonetech | JL-8, Cat # 632381 |
| Alexa Fluor 488 conjugated secondary antibodies | Thermo Fisher Scientific | Variable host and target species |
| Alexa Fluor 568 conjugated secondary antibodies | Thermo Fisher Scientific | Variable host and target species |
| Alexa Fluor 647 conjugated secondary antibodies | Thermo Fisher Scientific | Variable host and target species |
| Horseradish Peroxidase conjugated anti-mouse secondary antibody | Thermo Fisher Scientific | Cat # F1804 |
| Bacterial and Virus Strains | ||
| GC10 Competent Bacteria | Genesee Scientific | Cat # 42–659 or Cat # 42–661 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Paraformaldehyde 16% Solution | Electron Microscopy Sciences | Cat # 15710 |
| Triton X-100 | Thermo Fisher Scientific | Cat # BP151–500 |
| Tween-20 | Thermo Fisher Scientific | Cat # BP337–500 |
| Normal Goat Serum | Sigma Aldrich | Cat # G9023 |
| Aqua-Poly/Mount | Polysciences, Inc | Cat # 18606–20 |
| Schneider’s Drosophila (S2) Medium | Gibco, Thermo Fisher Scientific | Cat # 21720001 |
| DAPI | Thermo Fisher Scientific | Cat # D1306 |
| Sir-Tubulin | Cytoskeleton, Inc | Cat # CY-SC002 |
| Halocarbon oil 700 | Sigma Aldrich | Cat # H8898 |
| Glutaraldehyde | Electron Microscopy Sciences | Cat # 16120 |
| Osmium tetroxide | Electron Microscopy Sciences | Cat # 19152 |
| Uranyl Acetate| | Electron Microscopy Sciences | Cat # 22400 |
| EMbed 812 resin | Electron Microscopy Sciences | Cat # 14120 |
| Vectashield | Vector Laboratories | Cat # H-1000 |
| DIG RNA Labeling mix | Roche | Cat # 11277073910 |
| SP6 RNA Polymerase | Roche | Cat # 10810274001 |
| T3 RNA Polymerase | Thermo Fisher Scientific | Cat # EP0101 |
| T7 RNA Polymerase | Thermo Fisher Scientific | Cat # EP0111 |
| RNAse Inhibitor, Murine | New England Biolabs | Cat # M0314L |
| tRNA | Roche | Cat # 10109541001 |
| NBT/BCIP stock solution | Roche | Cat # 11681451001 |
| Methyl Salicylate | Acros Organics | Cat # 220495000 |
| Phusion High-Fidelity DNA Polymerase | Thermo Fisher Scientific | Cat # F530L |
| Taq DNA Ligase | New England Biolabs | Cat # M0208L |
| T5 Exonuclease | Epicentre | Cat # T5E4111K |
| T4 Polynucleotide Kinase | New England Biolabs | Cat # M0201S |
| T4 DNA Ligase | New England Biolabs | Cat # M0202L |
| Proteinase K | Thermo Fisher Scientific | E0491 |
| Collagenase | Sigma-Aldrich | C2674 |
| Fetal Bovine Serum | Gibco Life Technologies | 10438–034 |
| Bovine Serum Albumin | Millipore Sigma | 160069 |
| Critical Commercial Assays | ||
| GeneJet Gel Extraction Kit | Thermo Fisher Scientific | Cat # FERK0692 |
| GeneJet Plasmid Miniprep Kit | Thermo Fisher Scientific | Cat # FERK0503 |
| pENTR/D-TOPO Kit | Thermo Fisher Scientific | Cat # K2400–20 |
| Gateway LR Clonase | Thermo Fisher Scientific | Cat # 11791043 |
| SuperSignal West Dura Extended Duration Substrate | Life Technologies | Cat # 34076 |
| Chromium™ Single Cell 3’ Library & Gel Bead Kit v2 | 10X Genomics | 120237 |
| Quant-iT RiboGreen quantification kit | Life Technologies | R11490 |
| Deposited Data | ||
| Single cell RNAseq | GEO # GSE125947 | |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: yw | Peifer Lab (UNC-Chapel Hill) | N/A |
| D. melanogaster: plp2172 | Bloomington Drosophila Stock Center | BDSC: 12089 |
| D. melanogaster: Df(3L)Brd15 | Bloomington Drosophila Stock Center | BDSC: 5354 |
| D. melanogaster: H2Av::RFP | Bloomington Drosophila Stock Center | BDSC: 23651 |
| D. melanogaster: ubi-PLPPF::GFP | Galletta et al., 2014 | N/A |
| D. melanogaster: bam-Gal4 | Chen and McKearin, 2003 | N/A |
| D. melanogaster: topi-Gal4 | Raychaudhuri et al., 2012 | N/A |
| D. melanogaster: UAS-YFP::cnb::PACT | Januschke et al., 2013a | N/A |
| D. melanogaster: ubi-GFP::Cep135 | Galletta et al., 2016a | N/A |
| D. melanogaster: ubi-GFP::α-tubulin | Dr. Tomer Avidor-Reiss, University of Toledo | N/A |
| D. melanogaster: UAS-Ana1::tdTomato | Blachon et al., 2008 | N/A |
| D. melanogaster: ubi-GFP::PACT | This paper and Martinez-Campos et al., 2004 | N/A |
| D. melanogaster: UAS-PLP::GFP | This paper | N/A |
| D. melanogaster: GFP::Asl | This paper | N/A |
| D. melanogaster: PLP::mNeon | This paper | N/A |
| D. melanogaster: y[1] M{vas-Cas9}ZH-2A w[1118]/FM7c | Bloomington Drosophila Stock Center | BDSC: 51323 |
| Oligonucleotides | ||
| attB Forward GGTACCGTCGACGATGTAGGTCACGG |
IDTDNA | N/A |
| attB Reverse TATAGAATTCGTCGACATGCCCGCCGTG |
IDTDNA | N/A |
| Asl Genomic Upstream Forward TAATTTGCGAGTACGCAAAGCTTGGCTGCAAATGGTCTCGGTGCCGTTGGCAGTGGTAT |
IDTDNA | N/A |
| Asl Genomic Upstream Reverse GGTGAACAGCTCCTCGCCCTTGCTCACCATATTCGGCTAAGGGGACGCCACAAGCATCAA |
IDTDNA | N/A |
| GFP Forward TTGATGCTTGTGGCGTCCCCTTAGCCGAATATGGTGAGCAAGGGCGAGGAGCTGTTCACC |
IDTDNA | N/A |
| GFP Reverse CCCCTGAAAGAGGCTTATACCTGGCGTGTTGGTGAAGGGGGCGGCCGC |
IDTDNA | N/A |
| Asl Genomic Coding Forward AAAGCAGGCTCCGCGGCCGCCCCCTTCACCAACACGCCAGGTATAAGCCTCTTTCAGG |
IDTDNA | N/A |
| Asl Genomic Downstream Reverse GTTAACTCGAGGCCTCGAGGTCGACCTGCACAGAGCCACGCGGAGAGATTGTTATAGACA |
IDTDNA | N/A |
|
PLP::mNeon gRNA GGTGTTTTCGCTATTGTTAT |
IDTDNA | N/A |
| PLP 5’ Homology Arm Forward GTACTTCGCGAATGCGTCGAGATACCAATGTGGAAGGTTACCAGGCCAGTGAGCAATTGG |
IDTDNA | N/A |
| PLP 5’ Homology Arm Reverse GCCATCAACCACTTTGTACAAGAAAGCTGGGTCGGCGCGCCCACCCTTATGATGCCGCGCATGCGCTCTTTTTGG |
IDTDNA | N/A |
| mNeonGreen Forward AAGGGTGGGCGCGCCGACCCAGCTTTCTTGTACAAAGTGGTTGATGGCATGGTGAGCAAGGGCGAGGAGGATAACATG |
IDTDNA | N/A |
| mNeonGreen Reverse TTGATGGAACTCAACTCTGCTATCCTGGGCTTACTTGTACAGCTCGTCCATGCCCATCAC |
IDTDNA | N/A |
| PLP 3’ Homology Arm Forward GTGATGGGCATGGACGAGCTGTACAAGTAAGCCCAGGATAGCAGAGTTGAGTTCCATCAA |
IDTDNA | N/A |
| PLP 3’ Homology Arm Reverse ACTCGTCGGTCCCGGCATCCGATCCAATAGGGCTGGACGTGCAAAACATTGCAGCTTTTG |
IDTDNA | N/A |
| Recombinant DNA | ||
| pPWG | Drosophila Genomics Resource Center | DGRC: 1078 |
| pU6-BbsI-chiRNA | Addgene | Plasmid #45946 |
| pUC57 | GenScript | Cat # SD1176 |
| pCaSpeR4-attB | This paper | N/A |
| pOT2-PLP PC SD04227 | Drosophila Genomics Resource Center | DGRC:1078 |
| pBLuescript-Asl FL coding | This paper | N/A |
| Software and Algorithms | ||
| MetaMorph for CSU-10 and CSU-22 systems | Molecular Devices | N/A |
| Nikon Elements | Nikon | N/A |
| FIJI / Image J | NIH | http://fiji.sc/ |
| Excel | Microsoft | https://products.office.com/en-us/excel |
| Flycrispr design tool | http://flycrispr.molbio.wisc.edu/tools | |
| Seurat v3.0.1 | https://satijalab.org/seurat/install.html | |
| Prism 7 | GraphPad | www.graphpad.com/scientificsoftware/prism/ |
| Photoshop / Illustrator | Adobe | www.adobe.com/uk/products/ |
| Other | ||
| lumox dishes | Sarstedt | Cat # 94.6077.410 |
Highlights.
The proximal end of the centriole docks to the spermatid nucleus
Proper centriole docking to the nucleus requires proximal restriction of PCM
Pericentrin-Like-Protein (PLP) restricts PCM to the proximal centriole end
PLP is proximally restricted by eliminating its availability during centriole growth
Acknowledgements
We thank R. Guillen for cloning assistance. D. Lerit and K. Hasse-Plevock for fly genetics assistance. A. Sodeinde for brain dissections. R. O’Neil and M. Hannaford for critically reading the manuscript. T. Avidor-Reiss, C. Gonzalez, J. Raff and the Bloomington Drosophila stock center for flies. G. Rogers and M. Gatti for antibodies. We thank the NHLBI EM core facility for sample prep and discussion. We thank the NHLBI light microscopy core for use of equipment. This work is supported by the Division of Intramural Research at the NHLBI/NIH (1ZIAHL006126 to NMR) and NIDDK/NIH (1ZIADK015600 to BO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interests
The authors declare no competing interests.
References
- Anderson MA, Jodoin JN, Lee E, Hales KG, Hays TS, and Lee LA (2009). Asunder is a critical regulator of dynein-dynactin localization during Drosophila spermatogenesis. Mol Biol Cell 20, 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, and Nigg EA (2016). The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem Soc Trans 44, 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P, Scheel JF, Ying G, Frederick JM, Baehr W, and Wolfrum U (2013). Germline deletion of Cetn1 causes infertility in male mice. J Cell Sci 126, 3204–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccetti B, Burrini AG, Collodel G, Magnano AR, Piomboni P, Renieri T, and Sensini C (1989). Morphogenesis of the decapitated and decaudated sperm defect in two brothers. Gamete Res 23, 181–188. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, and Raff JW (2006). Flies without centrioles. Cell 125, 1375–1386. [DOI] [PubMed] [Google Scholar]
- Blachon S, Cai X, Roberts KA, Yang K, Polyanovsky A, Church A, and Avidor-Reiss T (2009). A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics 182, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, and Avidor-Reiss T (2008). Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics 180, 2081–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M, Paintrand M, Berges J, Marty MC, and Karsenti E (1987). Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskeleton 8, 238–249. [DOI] [PubMed] [Google Scholar]
- Brito DA, Gouveia SM, and Bettencourt-Dias M (2012). Deconstructing the centriole: structure and number control. Current opinion in cell biology. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Gonczy P, Alexander S, Rawson R, Eberhart CG, Viswanathan S, DiNardo S, and Wasserman SA (1993). Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135, 489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemes HE, and Alvarez Sedo C (2012). Tales of the tail and sperm head aches: changing concepts on the prognostic significance of sperm pathologies affecting the head, neck and tail. Asian J Androl 14, 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemes HE, Puigdomenech ET, Carizza C, Olmedo SB, Zanchetti F, and Hermes R (1999). Acephalic spermatozoa and abnormal development of the head-neck attachment: a human syndrome of genetic origin. Hum Reprod 14, 1811–1818. [DOI] [PubMed] [Google Scholar]
- Chemes HE, and Rawe VY (2010). The making of abnormal spermatozoa: cellular and molecular mechanisms underlying pathological spermiogenesis. Cell Tissue Res 341, 349–357. [DOI] [PubMed] [Google Scholar]
- Chen JV, Buchwalter RA, Kao LR, and Megraw TL (2017). A Splice Variant of Centrosomin Converts Mitochondria to Microtubule-Organizing Centers. Curr Biol 27, 1928–1940 e1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron YR, Fagerstrom CJ, Keszthelyi B, Huang B, Rusan NM, Kelly MJS, and Agard DA (2018). The centrosomin CM2 domain is a multifunctional binding domain with distinct cell cycle roles. Plos One 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, and Oegema K (2004). Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell 7, 815–829. [DOI] [PubMed] [Google Scholar]
- Delgehyr N, Rangone H, Fu J, Mao G, Tom B, Riparbelli MG, Callaini G, and Glover DM (2012). Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Current biology : CB 22, 502–509. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, and Doxsey SJ (1998). Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol 141, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne LK, Shim K, Hoshi M, Cheng T, Wang J, Marthiens V, Knoten A, Basto R, Jain S, and Mahjoub MR (2018). Centrosome amplification disrupts renal development and causes cystogenesis. J Cell Biol 217, 2485–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Abdelaziz M, Debski J, Dadlez M, and Glover DM (2017). Two-step phosphorylation of Ana2 by Plk4 is required for the sequential loading of Ana2 and Sas6 to initiate procentriole formation. Open Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhatib RA, Paci M, Longepied G, Saias-Magnan J, Courbiere B, Guichaoua MR, Levy N, Metzler-Guillemain C, and Mitchell MJ (2017). Homozygous deletion of SUN5 in three men with decapitated spermatozoa. Hum Mol Genet 26, 3167–3171. [DOI] [PubMed] [Google Scholar]
- Fabian L, and Brill JA (2012). Drosophila spermiogenesis: Big things come from little packages. Spermatogenesis 2, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Zhang J, Zhu F, Yang X, Cui Y, and Liu J (2018). Patients with acephalic spermatozoa syndrome linked to SUN5 mutations have a favorable pregnancy outcome from ICSI. Hum Reprod 33, 372–377. [DOI] [PubMed] [Google Scholar]
- Fawcett DW (1975). The mammalian spermatozoon. Dev Biol 44, 394–436. [DOI] [PubMed] [Google Scholar]
- Fawcett DW (1981). The cell, 2nd edn (Philadelphia: Saunders; ). [Google Scholar]
- Franz A, Roque H, Saurya S, Dobbelaere J, and Raff JW (2013). CP110 exhibits novel regulatory activities during centriole assembly in Drosophila. J Cell Biol 203, 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, and Glover DM (2012). Structured illumination of the interface between centriole and peri-centriolar material. Open biology 2, 120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Hagan IM, and Glover DM (2015). The centrosome and its duplication cycle. Cold Spring Harb Perspect Biol 7, a015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT (1993). Spermatogenesis In Development of Drosophila Martinez-Arias A, and Bate M, eds. (New York: Cold Spring Harbor Laboratory Press Cold Spring Harbor Laboratory Press; ), pp. 71–147. [Google Scholar]
- Galati DF, Sullivan KD, Pham AT, Espinosa JM, and Pearson CG (2018). Trisomy 21 Represses Cilia Formation and Function. Dev Cell 46, 641–650 e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Fagerstrom CJ, Schoborg TA, McLamarrah TA, Ryniawec JM, Buster DW, Slep KC, Rogers GC, and Rusan NM (2016a). A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat Commun 7, 12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Guillen RX, Fagerstrom CJ, Brownlee CW, Lerit DA, Megraw TL, Rogers GC, and Rusan NM (2014). Drosophila pericentrin requires interaction with calmodulin for its function at centrosomes and neuronal basal bodies but not at sperm basal bodies. Mol Biol Cell 25, 2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Jacobs KC, Fagerstrom CJ, and Rusan NM (2016b). Asterless is required for centriole length control and sperm development. J Cell Biol 213, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bucciarelli E, Bonaccorsi S, and Gatti M (2008). Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Current biology 18, 303–309. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, and Hyman AA (1999). Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol 147, 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J, Mennella V, Blachon S, Zhai B, Smith AH, Megraw TL, Nicastro D, Gygi SP, Agard DA, and Avidor-Reiss T (2011). Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun 2, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzick DS, Overstreet JW, Factor-Litvak P, Brazil CK, Nakajima ST, Coutifaris C, Carson SA, Cisneros P, Steinkampf MP, Hill JA, et al. (2001). Sperm morphology, motility, and concentration in fertile and infertile men. N Engl J Med 345, 1388–1393. [DOI] [PubMed] [Google Scholar]
- Holstein AF, and Roosen-Runge EC (1981). Atlas of human spermatogenesis (Berlin: Grosse; ). [Google Scholar]
- Januschke J, Reina J, Llamazares S, Bertran T, Rossi F, Roig J, and Gonzalez C (2013). Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat Cell Biol 15, 241–248. [DOI] [PubMed] [Google Scholar]
- Kawaguchi S, and Zheng Y (2004). Characterization of a Drosophila centrosome protein CP309 that shares homology with Kendrin and CG-NAP. Mol Biol Cell 15, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Park JE, Shukla A, Choi S, Murugan RN, Lee JH, Ahn M, Rhee K, Bang JK, Kim BY, et al. (2013). Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc Natl Acad Sci U S A 110, E4849–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba JE, Buster DW, Nguyen AL, Swatkoski S, Gucek M, Rusan NM, and Rogers GC (2013). Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr Biol 23, 2255–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracklauer MP, Wiora HM, Deery WJ, Chen X, Bolival B Jr., Romanowicz D, Simonette RA, Fuller MT, Fischer JA, and Beckingham KM (2010). The Drosophila SUN protein Spag4 cooperates with the coiled-coil protein Yuri Gagarin to maintain association of the basal body and spermatid nucleus. J Cell Sci 123, 2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, and Pelletier L (2012). Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol 14, 1148–1158. [DOI] [PubMed] [Google Scholar]
- Lerit DA, Jordan HA, Poulton JS, Fagerstrom CJ, Galletta BJ, Peifer M, and Rusan NM (2015). Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J Cell Biol 210, 79–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit DA, and Rusan NM (2013). PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J Cell Biol 202, 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, et al. (2017). Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev Cell 40, 313–322 e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sha Y, Wang X, Li P, Wang J, Kee K, and Wang B (2017). Whole-exome sequencing identified a homozygous BRDT mutation in a patient with acephalic spermatozoa. Oncotarget 8, 19914–19922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MG, Serr M, Newman EA, and Hays TS (2004). The Drosophila tctex-1 light chain is dispensable for essential cytoplasmic dynein functions but is required during spermatid differentiation. Mol Biol Cell 15, 3005–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liska F, Gosele C, Rivkin E, Tres L, Cardoso MC, Domaing P, Krejci E, Snajdr P, Lee-Kirsch MA, de Rooij DG, et al. (2009). Rat hd mutation reveals an essential role of centrobin in spermatid head shaping and assembly of the head-tail coupling apparatus. Biol Reprod 81, 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, and Bettencourt-Dias M (2018). Building the right centriole for each cell type. J Cell Biol 217, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, and Khodjakov A (2008). Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol 10, 322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, and White JG (2003). The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825–836. [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M, Basto R, Baker J, Kernan M, and Raff JW (2004). The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol 165, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Lujambio I, Burfeind P, Dixkens C, Meinhardt A, Hoyer-Fender S, Engel W, and Neesen J (2002). The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum Mol Genet 11, 1647–1658. [DOI] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, and Agard DA (2012). Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol 14, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CA, Benson E, and White-Cooper H (2009). Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nat Protoc 4, 1807–1819. [DOI] [PubMed] [Google Scholar]
- Nigg EA, and Holland AJ (2018). Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol 19, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, Oyama M, and Kitagawa D (2014). Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun 5, 5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Watanabe K, Ashikawa T, Nozaki Y, Yoshiba S, Kimura A, and Kitagawa D (2018). Bimodal Binding of STIL to Plk4 Controls Proper Centriole Copy Number. Cell Rep 23, 3160–3169 e3164. [DOI] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, and Bornens M (1992). Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol 108, 107–128. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, and Lehner CF (2012). Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol 10, e1001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richens JH, Barros TP, Lucas EP, Peel N, Pinto DM, Wainman A, and Raff JW (2015). The Drosophila Pericentrin-like-protein (PLP) cooperates with Cnn to maintain the integrity of the outer PCM. Biol Open 4, 1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli MG, and Callaini G (2011). Male gametogenesis without centrioles. Dev Biol 349, 427–439. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Wojcik EJ, Sanders MA, McGrail M, and Hays TS (1999). Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol 146, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Peifer M, and Rogers SL (2008). A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Molecular biology of the cell 19, 3163–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque H, Saurya S, Pratt MB, Johnson E, and Raff JW (2018). Drosophila PLP assembles pericentriolar clouds that promote centriole stability, cohesion and MT nucleation. PLoS Genet 14, e1007198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, and Nigg EA (2009). Control of centriole length by CPAP and CP110. Curr Biol 19, 1005–1011. [DOI] [PubMed] [Google Scholar]
- Sha YW, Sha YK, Ji ZY, Mei LB, Ding L, Zhang Q, Qiu PP, Lin SB, Wang X, Li P, et al. (2018a). TSGA10 is a novel candidate gene associated with acephalic spermatozoa. Clin Genet 93, 776–783. [DOI] [PubMed] [Google Scholar]
- Sha YW, Wang X, Xu X, Ding L, Liu WS, Li P, Su ZY, Chen J, Mei LB, Zheng LK, et al. (2019). Biallelic mutations in PMFBP1 cause acephalic spermatozoa. Clin Genet 95, 277–286. [DOI] [PubMed] [Google Scholar]
- Sha YW, Xu X, Ji ZY, Lin SB, Wang X, Qiu PP, Zhou Y, Mei LB, Su ZY, Li L, et al. (2018b). Genetic contribution of SUN5 mutations to acephalic spermatozoa in Fujian China. Gene 647, 221–225. [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan J, Tang W, Liu C, Xiao S, Guo Y, Yuan L, Chen L, Jiang H, Guo X, et al. (2018). Mechanistic insights into acephalic spermatozoa syndrome-associated mutations in the human SUN5 gene. J Biol Chem 293, 2395–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Zhu F, Wang L, Ouyang YC, Dong MZ, Liu C, Zhao H, Cui X, Ma D, Zhang Z, et al. (2017). Essential role for SUN5 in anchoring sperm head to the tail. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Aher A, Dynes NJ, Frey D, Katrukha EA, Jaussi R, Grigoriev I, Croisier M, Kammerer RA, Akhmanova A, et al. (2016). Centriolar CPAP/SAS-4 Imparts Slow Processive Microtubule Growth. Dev Cell 37, 362–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram P, Anderson MA, Jodoin JN, Lee E, and Lee LA (2012). Regulation of dynein localization and centrosome positioning by Lis-1 and asunder during Drosophila spermatogenesis. Development 139, 2945–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, and Dynlacht BD (2007). Cep97 and CP110 suppress a cilia assembly program. Cell 130, 678–690. [DOI] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, et al. (2010). Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8, e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tates AD (1971). Cytodifferentiation during spermatogenesis in Drosophila Melanogaster : an electron microscope study (S-Gravenhage,: J. H. Pasmans, Thesis-- Leiden.). [Google Scholar]
- Texada MJ, Simonette RA, Johnson CB, Deery WJ, and Beckingham KM (2008). Yuri gagarin is required for actin, tubulin and basal body functions in Drosophila spermatogenesis. J Cell Sci 121, 1926–1936. [DOI] [PubMed] [Google Scholar]
- Varadarajan R, Ayeni J, Jin Z, Homola E, and Campbell SD (2016). Myt1 inhibition of Cyclin A/Cdk1 is essential for fusome integrity and premeiotic centriole engagement in Drosophila spermatocytes. Mol Biol Cell 27, 2051–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan R, and Rusan NM (2018). Bridging centrioles and PCM in proper space and time. Essays Biochem 62, 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N, Koch I, Schwarz H, Schnorrer F, and Nusslein-Volhard C (2006). The gammaTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133, 3963–3972. [DOI] [PubMed] [Google Scholar]
- White-Cooper H (2012). Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis 2, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Zong Q, Du AL, Zhang W, Yao HC, Yu XQ, and Wang YF (2016). Knockdown of Dynamitin in testes significantly decreased male fertility in Drosophila melanogaster. Dev Biol 420, 79–89. [DOI] [PubMed] [Google Scholar]
- Yang K, Adham IM, Meinhardt A, and Hoyer-Fender S (2018). Ultra-structure of the sperm head-to-tail linkage complex in the absence of the spermatid-specific LINC component SPAG4. Histochem Cell Biol 150, 49–59. [DOI] [PubMed] [Google Scholar]
- Yuan S, Stratton CJ, Bao J, Zheng H, Bhetwal BP, Yanagimachi R, and Yan W (2015). Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc Natl Acad Sci U S A 112, E430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, and Han M (2009). SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Ramani A, Soni K, Gottardo M, Zheng S, Ming Gooi L, Li W, Feng S, Mariappan A, Wason A, et al. (2016). Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat Commun 7, 11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, Muller-Reichert T, Kittler R, Hyman AA, and Pelletier L (2008). The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol 18, 136–141. [DOI] [PubMed] [Google Scholar]
- Zhu F, Liu C, Wang F, Yang X, Zhang J, Wu H, Zhang Z, He X, Zhang Z, Zhou P, et al. (2018). Mutations in PMFBP1 Cause Acephalic Spermatozoa Syndrome. Am J Hum Genet 103, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Wang F, Yang X, Zhang J, Wu H, Zhang Z, Zhang Z, He X, Zhou P, Wei Z, et al. (2016). Biallelic SUN5 Mutations Cause Autosomal-Recessive Acephalic Spermatozoa Syndrome. Am J Hum Genet 99, 1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1. Nuclear Search and Nuclear Attachment (related to Figure 1C) Movie of a SC centriole exiting meiosis through “Nuclear Search” and “ Nuclear Attachment” in RST. GFP::Cep135 (green) and DNA (H2Av::RFP, red). Individual frames are a projection of multiple confocal slices. Frames were acquired at 15 minute intervals. Movie is presented at 3 frames / second. The centriole marker used is brighter on the proximal centriole end than on the distal centriole end (Galletta et al., 2016a) allowing for identification of the distinct ends of the centriole in the movie. Stills of the movie are presented in Figure 1C. Time stamp is relative to reformation of the nuclear envelope following meiosis.
Video 2. Centrioles organize microtubules throughout Nuclear Search and Nuclear Attachment (related to Figure 1D) Movie of a SC expressing GFP::α-tubulin exiting meiosis through “Nuclear Search” and “ Nuclear Attachment” in RST. Individual frames are a projection of multiple confocal slices. Frames were acquired at 5 minute intervals. Movie is presented at 5 frames / second. Stills of the movie are presented in Figure 1D. Time stamp is relative to reformation of the nuclear envelope following meiosis.
Data Availability Statement
The single cell RNAseq is reported in the Gene Expression Omnibus (GEO) repository accession #GSE125947)


