Abstract
Many iron-sulfur proteins involved in cluster trafficking form [2Fe-2S]-cluster-bridged complexes that are often challenging to characterize because of the inherent instability of the cluster at the interface. In this work, we illustrate the use of fast, online buffer exchange coupled to a native mass spectrometry (OBE nMS) method to characterize [2Fe-2S]-cluster-bridged proteins and their transient cluster-transfer reaction intermediates. The use of this mechanistic and protein characterization tool is demonstrated with holo glutaredoxin 5 (GLRX5) homodimer and holo GLRX5:BolA-like protein 3 (BOLA3) heterodimer. Using the OBE nMS method, cluster transfer reactions between the holo-dimers and apo-ferredoxin (FDX2) are monitored, and intermediate [2Fe-2S] species such as (FDX2:GLRX5:[2Fe-2S]:GSH) and (FDX2:BOLA3:GLRX5:[2Fe-2S]: GSH) are detected. In summary, we demonstrate the OBE nMS method to be a robust technique for characterizing iron-sulfur cluster bridged protein complexes and transient iron-sulfur cluster transfer intermediates.
Keywords: native mass spectrometry, [2Fe-2S]-cluster-bridged protein complexes, online buffer exchange, cluster transfer reaction, intermediates
Graphical Abstract
[2Fe-2S] cluster-bridged complexes are characterized by use of online buffer exchange coupled to native mass spectrometry (OBE nMS). Cluster-transfer reactions between holo complexes and apo ferredoxin (FDX2) are monitored by use of OBE nMS, and intermediate Fe-S species are detected.

Iron-sulfur (Fe-S) cluster proteins are a versatile class of metalloproteins, being involved in electron transfer, catalysis, regulation of gene expression, and DNA processing.[1-3] Iron-sulfur cluster cofactors are often sensitive to oxygen and prone to decomposition,[4] making characterization of cluster-bound proteins challenging. Typical identification and characterization methods include UV-Vis, circular dichroism (CD), Mössbauer, resonance Raman, and electron paramagnetic resonance (EPR) spectroscopies.[5] However, there are limitations of these techniques that include challenges in discriminating cluster types (often inconclusive with UV and CD, while EPR often requires additional experiments such as power saturation and temperature dependence[6]) and high concentration requirements (Mössbauer and Resonance Raman).[7-9] These techniques provide spectral data that are an average of the whole ensemble and cannot distinguish between sub-populations. Notably, all of these methods utilize unique properties of the metal cofactor and provide limited information concerning the protein component. Therefore, a characterization tool that requires smaller quantities of protein and provides insight into both protein and cluster components is desirable. Native mass spectrometry (nMS), which preserves non-covalent interactions during measurements, can overcome these challenges and serve as a robust technique for the identification and characterization of protein-bound Fe-S cofactors.
Native mass spectrometry has been applied to the study of Fe-S proteins. For example, Crack et al. first illustrated the application of nMS to study physiologically relevant cluster chemistry.[12-22] However, use of the application has been limited due to challenges from cluster instability and from the additional sample preparation steps required to spray intact cluster-bound proteins from mass spectrometric compatible buffers. The majority of such proteins characterized by mass spectrometry have Fe-S clusters buried inside the protein core and not at the protein-protein interface. Surface-accessible clusters that bridge protein monomers are challenging to characterize in the native holo dimeric form. NsrR from Streptomyces coelicolor binds a [4Fe-4S] cluster as a homodimer, but the native dimeric [4Fe-4S] species has not been characterized as the predominant product by MS.[22-24] The solution dimer form is susceptible to dissociation into monomers during ionization.[22] Herein, we attempted to overcome the challenges inherent to the characterization of such cluster-bridged complexes. Accordingly, there is a need for a robust technique to characterize them.
Online buffer exchange coupled to native MS does not require any extra sample preparation steps prior to MS analysis (Figure 1).[34,35] It can separate non-volatiles from proteins and/or protein complexes in the sample within a short time-frame, resulting[22] in clean protein and/or protein complex spectra generated directly from non-mass spectrometric compatible buffers. This method has low sample requirements, typically requiring 5 μL of 10 μM sample. Moreover, it minimizes the possibility of samples being exposed to oxygen and thereby preserves the native holo iron-sulfur cluster-containing protein complexes prior to detection. Using a home-packed PEEK tubing with P-6 Bio-Gel material as the stationary phase,[34,35] the protein samples typically elute in the time range from 0.55 to 0.9 minutes at a 100 μL/min flow rate (Figure S2a). This measurement time is faster than with commercial SEC columns.[35] The shorter time not only improves the efficiency of sample screening but also minimizes oxidation of unstable analytes, such as iron-sulfur cluster proteins, allowing rapid and robust characterization of these samples without a strict anaerobic environment. OBE nMS can serve as a simple screening method for these oxygen-sensitive iron-sulfur cluster samples without additional sample preparation steps.
Figure 1.
Experimental setup for OBE MS. The sample is injected and separated from non-volatile salts by a gel-filtration (OBE) column. The mobile phase is 200 mM ammonium acetate de-oxygenated by a constant stream of argon. The injection valve, sample syringe, and samples are kept inside a glove bag flushed with argon.
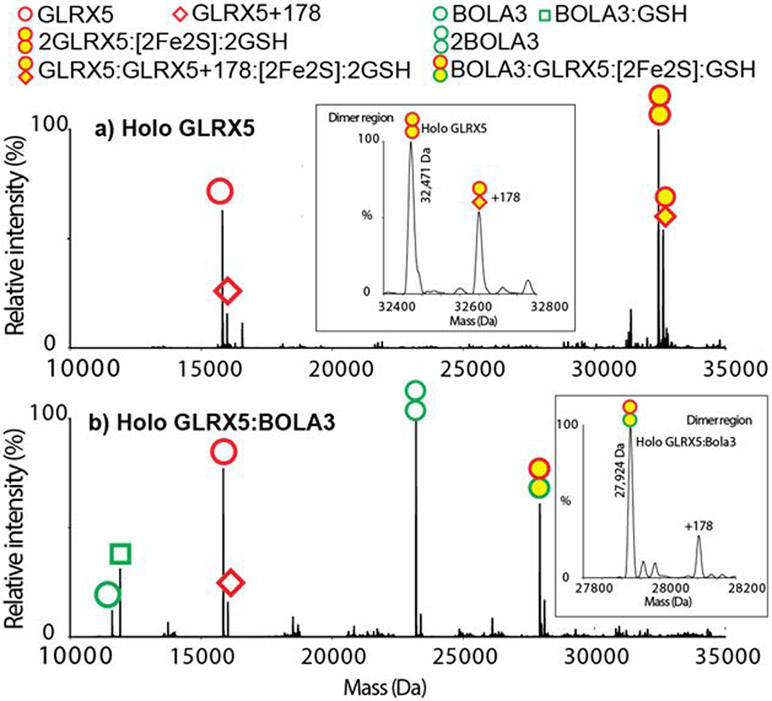
Glutaredoxin 5 (GLRX5), in its [2Fe-2S] bridged holo homodimeric form, is one of the core components of the cellular iron-sulfur cluster assembly machinery and serves as an intermediary cluster carrier, delivering cluster to several target proteins after synthesis of the [2Fe-2S] cluster by the scaffold protein ISU in the mitochondria.[36] [2Fe-2S]-Bridged holo heteromeric complexes of GLRX5 with other partner proteins have been implicated in important functions in cellular pathways. For example, [2Fe-2S] bridged heterodimeric complexes of GLRX5 with BolA type proteins BOLA1 and BOLA3 have been proposed to play important roles in redox regulation and Fe-S cluster trafficking, respectively.[37] Mutations in genes encoding the proteins mentioned above can result in disease conditions, often leading to death.[28] Homozygous mutation in the GLRX5 gene causes splicing errors and results in sideroblastic anemia and iron overload.[38] Genetic mutations in genes for BOLA3 result in fatal Multiple Mitochondrial Disease Syndrome. Though the exact molecular understanding has been lacking, these disease conditions are characterized by defective Fe-S cluster maturation and cluster trafficking pathways that could involve [2Fe-2S]-bridged heteromeric species. The [2Fe-2S]-bridged GLRX5 homodimer is a well-characterized protein complex with a crystallographically-determined structure in the Protein Data Bank (2WUL).[26] Mass spectra for GLRX5 were acquired using the OBE nMS method and deconvoluted using UniDec software[39]. The spectra of the holo GLRX5 (Figure 2a) show the 15,842 Da apo GLRX5 monomer and 32,471 Da holo GLRX5 dimers. The experimental and theoretical masses match within 1 Da (Table S2). The mass of the holo dimer contains two GLRX5, a 176 Da [2Fe-2S]2+ cluster and two 306 Da GS− ligands as predicted.[40] In addition to these peaks, we also observed the GLRX5 peaks with a post-translational modification of +178 Da, attributed to alpha-N-gluconoylation of the His tag (Figure S1).[41,42] Next we employed OBE nMS to characterize a [2Fe-2S]-bridged GLRX5:BOLA3 complex whose structural model has been proposed based on NMR experiments,[43] but there is no report of the complex being observed by MS. A mass of 27,925 Da is observed (Figure 2b). The experimental and theoretical masses match within 1 Da (Table S2). The mass of the holo heterodimer contains a GLRX5, a BOLA3, a 176 Da [2Fe-2S]2+ cluster and a 306 Da GS− ligands as predicted. Apo GLRX5 monomers and apo BOLA3 monomers and heterotetramers (Figure S2cd, Figure S3) are also observed. Comparing these results with the data collected using offline nanoelectrospray ionization (nESI) (Figure S4) and offline direct infusion with ESI (Figure S5), nESI shows that the dominant peaks are apo monomers and direct infusion showed unexpected additional iron and sulfur on the complex, indicating the advantage of the OBE nMS technique in properly retaining the O2-labile cluster.
Figure 2.
Deconvoluted OBE mass spectra of a) holo GLRX5 and b) holo GLRX5:BOLA3. The empty symbols indicate apo proteins, and the yellow filled symbols indicate holo proteins with a [2Fe-2S] cluster. The stoichiometry of 2GLRX5:[2Fe-2S]:2GSH and GLRX5:BOLA3:[2Fe-2S]:GSH are observed. Insets are deconvoluted spectra in the holo dimer region with holo 2GLRX5:[2Fe-2S]:2GSH at 32,471 Da and GLRX5:BOLA3:[2Fe-2S]:GSH at 27,924 Da.
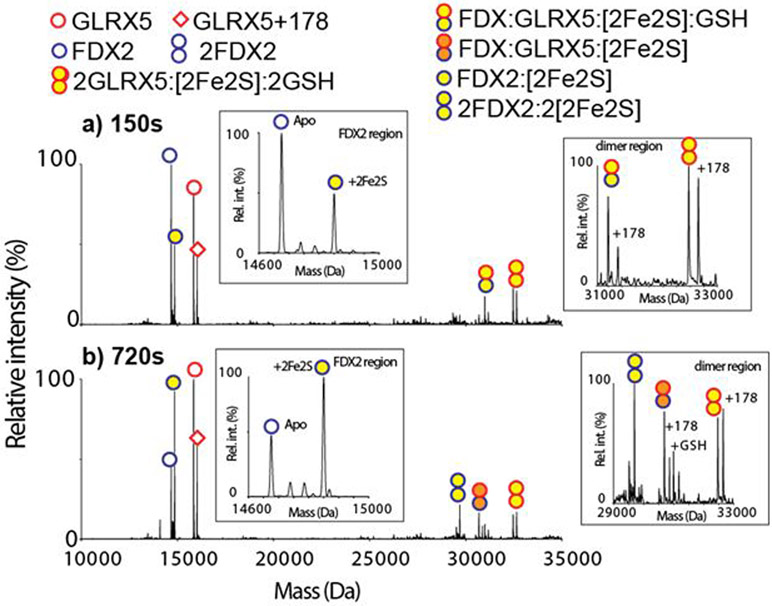
Previous work has demonstrated [2Fe-2S] cluster from both GLRX5 homodimer and GLRX5:BOLA3 heterodimer to be transferred to ferredoxin 2 (FDX2).[37,44] However, due to the challenge of characterizing the accurate stoichiometry of transient protein complexes in the reaction mixture by other techniques, the mechanistic details remain unclear. We monitored cluster transfer reactions using the OBE nMS method by mixing apo H. sapiens FDX2 with either holo GLRX5 or GLRX5:BOLA3 dimers in an approximately 1:1 molar ratio in reaction buffer (SI Cluster transfer reaction) and injected 5 μL aliquots into the OBE nMS at various reaction time points. Cluster transfer from holo GLRX5 homodimer to apo FDX2 was clearly observed (Figure 3, Figure S6). The relative ratio of holo GLRX5 homodimer to apo GLRX5 decreased over a period of 150 s of reaction time, while a peak for holo FDX2 emerged (Figure 3a). Spectra at intermediate times reveal the intermediate [2Fe-2S]-bridged cluster species that has not previously been observed. In the transfer from the GLRX5 homodimer, an intermediate complex of (FDX2:GLRX5:[2Fe-2S]:GSH) is captured at 150 s (Figure 3a). This intermediate loses the GSH molecule with longer reaction time and forms (FDX2:GLRX5:[2Fe-2S]) (Figure 3b). Formation of FDX2 holo dimer (2FDX2:2[2Fe-2S]) with increasing time may reflect non-specific dimerization due to increasing holo FDX2 concentration.
Figure 3.
Deconvoluted OBE mass spectra of the holo GLRX5 and apo FDX2 reaction mixture at a) 150 seconds and b) 720 seconds after mixing are shown. The empty symbols indicate apo proteins, and the yellow filled symbols indicate holo proteins with a [2Fe-2S] cluster. The [2Fe-2S] cluster transfers from the holo GLRX5 dimer to the apo FDX2 monomer. Intermediates (FDX2:GLRX5:[2Fe-2S]:GSH) and (FDX2:GLRX5:[2Fe-2S]) are observed. Insets are the zoom-in of the FDX2 and dimer regions.
The observation of (FDX2:GLRX5:[2Fe-2S]:GSH) provides mechanistic insight into the cluster transfer process. Cluster transfer requires the cleavage of Fe-cysteinyl bonds in the cluster-bound form of the homodimeric GLRX5 donor, accompanied by the formation of new Fe-cysteinyl bonds in the FDX2 acceptor with the release of two glutathione molecules. The detection of the (FDX2:GLRX5:[2Fe-2S]) species by mass spectroscopy suggests that during the reaction, one GLRX5 monomer is replaced by FDX2 in the complex as the reaction progresses, thereby allowing us to obtain an unprecedented mechanistic understanding of the individual steps in the cluster transfer process (Scheme 1).
Scheme 1.
The individual steps in the cluster transfer reaction from the holo GLRX5 homodimer to apo FDX2. Intermediates (FDX2:GLRX5:[2Fe-2S]:GSH) and (FDX2:GLRX5:[2Fe-2S]) are formed.
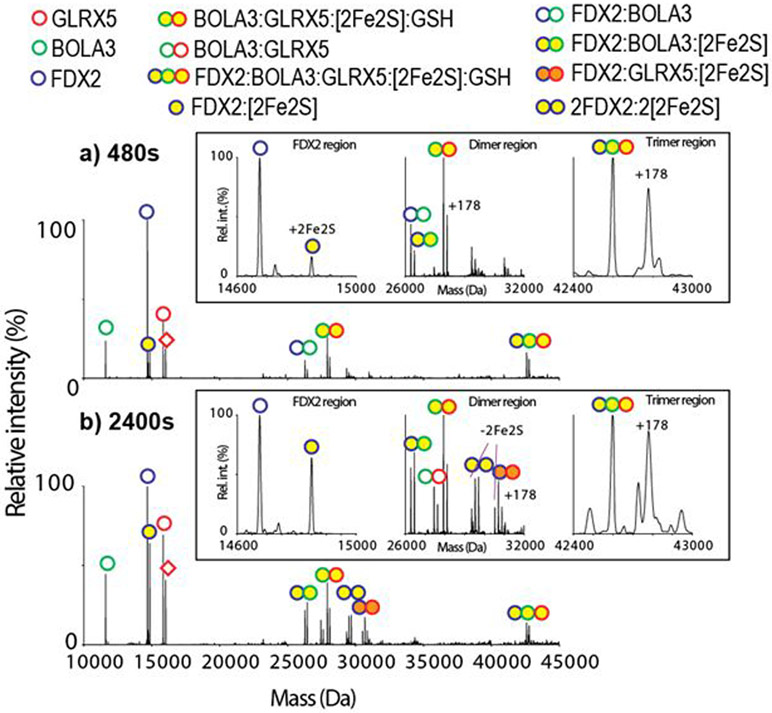
A similar cluster transfer reaction from holo heterodimer GLRX5:BOLA3 to the apo FDX2 was also monitored using OBE nMS (Figure 4, Figure S7). In the transfer reaction, intermediate complexes of (FDX2:BOLA3:[2Fe-2S]), (FDX2: GLRX5:BOLA3:[2Fe-2S]:GSH), and (FDX2:GLRX5:[2Fe-2S]) are observed. The formation of apo [FDX2:BOLA3] and the holo trimeric intermediate (FDX2: GLRX5:BOLA3:[2Fe-2S]:GSH) at the beginning of the reaction (Figure 4a), suggests that there are non-covalent interactions between FDX2 and BOLA3, bringing FDX2 to the holo GLRX5:BOLA3 heterodimer to form a trimeric intermediate. This initiates the cluster transfer process. The results indicate that there is no preference in cysteinyl bond cleavages from BOLA3 or GLRX5 in the holo BOLA3:GLRX5 heterocomplex when exposed to apo FDX2, as the formation of both intermediates (FDX2:BOLA3:[2Fe-2S]) and (FDX2:GLRX5:[2Fe-2S]) were detected (Figure 4b). Based on the MS data, a mechanism that demonstrates the steps in the cluster transfer from the holo BOLA3-GLRX5 heterodimer to apo FDX2 is shown in Scheme 2. The formation of [FDX2:GLRX5] after 2400 seconds most likely reflects slower kinetics of transfer in comparison to the transfer from the GLRX5 homodimer, which also resulted in detection of minor [FDX2:GLRX5].
Figure 4.
Deconvoluted OBE mass spectra of the holo GLRX5:BOLA3 and apo FDX2 reaction mixture at a) 480 seconds and b) 2400 seconds are shown. The empty symbols indicate apo proteins, and the yellow filled symbols indicate holo proteins with a [2Fe-2S] cluster. The [2Fe-2S] cluster transfers from the holo GLRX5:BOLA3 heterodimer to the apo FDX2 monomer. Intermediates [FDX2:BOLA3], (FDX2:BOLA3:[2Fe-2S]), (FDX2:BOLA3: GLRX5:[2Fe-2S]:GSH), [FDX2:GLRX5], and (FDX2:GLRX5:[2Fe-2S]) are observed. Insets are the zoom-in of the FDX2, dimer, and trimer regions.
Scheme 2.
The individual steps in the cluster transfer from the holo GLRX5:BOLA3 heterodimer to apo FDX2. Intermediates (FDX2:BOLA3:GLRX5:[2Fe-2S]:GSH), (FDX2:BOLA3:[2Fe-2S]), and (FDX2: GLRX5:[2Fe-2S]:GSH), (FDX2:GLRX5) are formed.
In vitro cluster transfer reactions have been routinely used to complement in vivo studies, and directionality of cluster transfer is often unique and well defined[40,46-48]. Many such reactions were demonstrated first in vitro and are now accepted as components of the current model for iron-sulfur cluster biosynthesis[28,49,50]. Though complexes between cluster donor and cluster acceptor proteins have been invoked as transient intermediates in mechanistic models of cluster delivery and assembly,[30,51,52] none of them have previously been observed experimentally. To our knowledge, this technique is currently the only method that can detect these intermediates as it provides information concerning both the cluster and protein components, thereby elucidating mechanistic details. The results confirm that this method is fast, robust, and of low sample consumption. OBE nMS has broad potential for characterizing iron-sulfur cluster-bridged protein complexes and transient iron-sulfur cluster intermediates.
Supplementary Material
Acknowledgments
We are grateful for the financial support from the National Institutes of Health (Grants P41GM128577 to VHW and AI072443 to JAC). We appreciate Zac VanAernum and Florian Busch for help in online buffer exchange and comments on this manuscript.
Footnotes
Supporting information including the experimental procedures for this article can be found under: https://doi.org/10.1002/anie.201915615
Contributor Information
Mengxuan Jia, Department of Chemistry and Biochemistry; Resource for Native Mass Spectrometry Guided Structural Biology, The Ohio State University, Columbus, OH 43210 (USA).
Sambuddha Sen, Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210 (USA).
Christine Wachnowsky, Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210 (USA).
Insiya Fidai, Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210 (USA).
J. A. Cowan, Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210 (USA)
Vicki H. Wysocki, Department of Chemistry and Biochemistry; Resource for Native Mass Spectrometry Guided Structural Biology, The Ohio State University, Columbus, OH 43210 (USA).
References
- [1].Beinert H, Science 1997, 277, 653–659. [DOI] [PubMed] [Google Scholar]
- [2].Lill R, Nature 2009, 460, 831–838. [DOI] [PubMed] [Google Scholar]
- [3].Fuss JO, Tsai C-L, Ishida JP, Tainer JA, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2015, 1853, 1253–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Imlay JA, Mol Microbiol 2006, 59, 1073–1082. [DOI] [PubMed] [Google Scholar]
- [5].Cowan JA, Inorganic Biochemistry: An Introduction, John Wiley & Sons, 1997. [Google Scholar]
- [6].Rupp H, Rao KK, Hall DO, Cammack R, Biochimica et Biophysica Acta (BBA) - Protein Structure 1978, 537, 255–269. [DOI] [PubMed] [Google Scholar]
- [7].Angove HC, Yoo SJ, Burgess BK, Münck E, J. Am. Chem. Soc 1997, 119, 8730–8731. [Google Scholar]
- [8].Berto P, Di Valentin M, Cendron L, Vallese F, Albertini M, Salvadori E, Giacometti GM, Carbonera D, Costantini P, Biochimica et Biophysica Acta (BBA) - Bioenergetics 2012, 1817, 2149–2157. [DOI] [PubMed] [Google Scholar]
- [9].Gao H, Subramanian S, Couturier J, Naik SG, Kim S-K, Leustek T, Knaff DB, Wu H-C, Vignols F, Huynh BH, et al. , Biochemistry 2013, 52, 6633–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Robinson CV, PNAS 2019, 116, 2814–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leney AC, Heck AJR, J. Am. Soc. Mass Spectrom 2017, 28, 5–13. [DOI] [PubMed] [Google Scholar]
- [12].Johnson KA, Verhagen MF, Brereton PS, Adams MW, Amster IJ, Anal. Chem 2000, 72, 1410–1418. [DOI] [PubMed] [Google Scholar]
- [13].Johnson KA, Amster IJ, J Am Soc Mass Spectrom 2001, 12, 819–825. [DOI] [PubMed] [Google Scholar]
- [14].Johnson KA, Shira BA, Anderson JL, Amster IJ, Anal. Chem 2001, 73, 803–808. [DOI] [PubMed] [Google Scholar]
- [15].Carroll KS, Gao H, Chen H, Leary JA, Bertozzi CR, Biochemistry 2005, 44, 14647–14657. [DOI] [PubMed] [Google Scholar]
- [16].Zeng J, Geng M, Jiang H, Liu Y, Liu J, Qiu G, Archives of Biochemistry and Biophysics 2007, 463, 237–244. [DOI] [PubMed] [Google Scholar]
- [17].Gao H, Leary J, Carroll KS, Bertozzi CR, Chen H, J Am Soc Mass Spectrom 2007, 18, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lipper CH, Paddock ML, Onuchic JN, Mittler R, Nechushtai R, Jennings PA, PLoS ONE 2015, 10, e0139699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Berkovitch F, Science 2004, 303, 76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Crack JC, Brun N. E. Le, Chemistry – A European Journal 2019, 25, 3675–3684. [DOI] [PubMed] [Google Scholar]
- [21].Crack JC, Thomson AJ, Brun NEL, PNAS 2017, 114, E3215–E3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Volbeda A, Dodd EL, Darnault C, Crack JC, Renoux O, Hutchings MI, Brun NEL, Fontecilla-Camps JC, Nat Commun 2017, 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Crack JC, Munnoch J, Dodd EL, Knowles F, Al Bassam MM, Kamali S, Holland AA, Cramer SP, Hamilton CJ, Johnson MK, et al. , J. Biol. Chem. 2015, 290, 12689–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Crack JC, Hamilton CJ, Brun NEL, Chemical Communications 2018, 54, 5992–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Iwema T, Picciocchi A, Traore DAK, Ferrer J-L, Chauvat F, Jacquamet L, Biochemistry 2009, 48, 6041–6043. [DOI] [PubMed] [Google Scholar]
- [26].Johansson C, Roos AK, Montano SJ, Sengupta R, Filippakopoulos P, Guo K, von Delft F, Holmgren A, Oppermann U, Kavanagh KL, Biochem. J. 2011, 433, 303–311. [DOI] [PubMed] [Google Scholar]
- [27].Yeung N, Gold B, Liu NL, Prathapam R, Sterling HJ, Willams ER, Butland G, Biochemistry 2011, 50, 8957–8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Maio N, Rouault TA, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2015, 1853, 1493–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li H, Outten CE, Biochemistry 2012, 51, 4377–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Melber A, Na U, Vashisht A, Weiler BD, Lill R, Wohlschlegel JA, Winge DR, eLife 2016, 5, e15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Uzarska MA, Nasta V, Weiler BD, Spantgar F, Ciofi-Baffoni S, Saviello MR, Gonnelli L, Mühlenhoff U, Banci L, Lill R, eLife 2016, 5, e16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cai K, Liu G, Frederick RO, Xiao R, Montelione GT, Markley JL, Structure 2016, 24, 2080–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gakh O, Ranatunga W, Smith DY, Ahlgren E-C, Al-Karadaghi S, Thompson JR, Isaya G, J. Biol. Chem 2016, 291, 21296–21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cavanagh J, Benson LM, Thompson R, Naylor S, Anal. Chem 2003, 75, 3281–3286. [DOI] [PubMed] [Google Scholar]
- [35].VanAernum Z, Busch F, Jones BJ, Jia M, Chen Z, Boyken SE, Sahasrabuddhe A, Baker D, Wysocki V, Rapid Online Buffer Exchange: A Method for Screening of Proteins, Protein Complexes, and Cell Lysates by Native Mass Spectrometry, 2019. [DOI] [PMC free article] [PubMed]
- [36].Couturier J, Przybyla-Toscano J, Roret T, Didierjean C, Rouhier N, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2015, 1853, 1513–1527. [DOI] [PubMed] [Google Scholar]
- [37].Sen S, Rao B, Wachnowsky C, Cowan JA, Metallomics 2018, 10, 1282–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, Rouault TA, J Clin Invest 2010, 120, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP, Robinson CV, Anal. Chem 2015, 87, 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Banci L, Brancaccio D, Ciofi-Baffoni S, Del Conte R, Gadepalli R, Mikolajczyk M, Neri S, Piccioli M, Winkelmann J, Proceedings of the National Academy of Sciences 2014, 111, 6203–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Geoghegan KF, Dixon HB, Rosner PJ, Hoth LR, Lanzetti AJ, Borzilleri KA, Marr ES, Pezzullo LH, Martin LB, LeMotte PK, et al. , Anal. Biochem 1999, 267, 169–184. [DOI] [PubMed] [Google Scholar]
- [42].Kim KM, Yi EC, Baker D, Zhang KY, Acta Crystallogr. D Biol. Crystallogr 2001, 57, 759–762. [DOI] [PubMed] [Google Scholar]
- [43].Nasta V, Giachetti A, Ciofi-Baffoni S, Banci L, Biochimica et Biophysica Acta (BBA) - General Subjects 2017, 1861, 2119–2131. [DOI] [PubMed] [Google Scholar]
- [44].Olive JA, Cowan JA, Journal of Inorganic Biochemistry 2018, 184, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bern M, Caval T, Kil YJ, Tang W, Becker C, Carlson E, Kletter D, Sen KI, Galy N, Hagemans D, et al. , J. Proteome Res 2018, 17, 1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wachnowsky C, Fidai I, Cowan JA, FEBS Lett. 2016, 590, 4531–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fidai I, Wachnowsky C, Cowan JA, Metallomics 2016, 8, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shakamuri P, Zhang B, Johnson MK, J. Am. Chem. Soc 2012, 134, 15213–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Stehling O, Wilbrecht C, Lill R, Biochimie 2014, 100, 61–77. [DOI] [PubMed] [Google Scholar]
- [50].Blanc B, Gerez C, Ollagnier de Choudens S, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2015, 1853, 1436–1447. [DOI] [PubMed] [Google Scholar]
- [51].Brancaccio D, Gallo A, Piccioli M, Novellino E, Ciofi-Baffoni S, Banci L, J. Am. Chem. Soc 2017, 139, 719–730. [DOI] [PubMed] [Google Scholar]
- [52].Sen S, Cowan JA, J Biol Inorg Chem 2017, 22, 1075–1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.