Abstract
In total, ~85% of malignant gastrointestinal stromal tumours (GISTs) harbour activating mutations in one of the genes KIT or PDGFRA, while 10-15% of all GISTs have no detectable KIT or PDGFRA mutations, but could have alterations in genes of the succinate dehydrogenase complex or in BRAF, PIK3CA or rarely RAS family genes. The clinical benefit of tyrosine kinase inhibitors, such as imatinib, depends on the GIST genotype, therefore molecular characterization of GIST has a crucial role in overall management of GIST. The aim of the present study was to molecularly characterize a cohort of 70 patients with metastatic GISTs from the Slovenian Cancer Registry (National Cancer Registry) treated between January 2002 and December 2011. Exons 9, 11, 13 and 17 of the KIT gene and exons 12, 14 and 18 of the PDGFRA gene were analysed by direct Sanger sequencing. All KIT/PDGFRA wild-type GISTs were tested for the presence of mutations in hot spot regions of KRAS, NRAS, BRAF, PIK3CA and AKT1 genes. Novel variants were characterized and classified using Cancer Genome Interpreter and according to The American College of Medical Genetics and Genomics/Association for Molecular Pathology guidelines. In total, 60 (85.7%) patients had mutations in KIT and 2 (2.9%) in PDGFRA. Whereas, 8 (11.4%) patients with GIST had no mutation in either of the analysed genes. The majority of GIST cases (n=52) had a mutation in KIT exon 11, where 40 different mutations were detected. Eight of the variants were novel: c.1652_1672del, c.1653_1660delinsAA, c.1665_1672delinsCC, c.1668_1686del, c.1676_1720del, c.1715_1756dup, c.1721_1765dup, and c.1722_1766dup. Mutation frequencies of KIT and PDGFRA genes observed in Slovenian patients are comparable with those in other European populations. In the present group of patients analysed, the most frequently mutated region was exon 11 in the KIT gene, responsible for coding juxtamembrane domain of KIT protein. In this region, eight novel mutations were identified and classified as likely pathogenic driver variants. In addition, the present study identified 6 patients with secondary KIT mutation and 1 patient with double mutant GIST, who had two different mutations in PDGFRA exon 14.
Keywords: gastrointestinal stromal tumour, KIT mutations, imatinib
Introduction
Gastrointestinal stromal tumours (GISTs) are the most common type of soft tissue sarcomas that arise from the mesenchymal cells of the gastrointestinal tract and have characteristic histological and molecular features (1). According to the literature, most GISTs are driven by activating mutations in the proto-oncogenes KIT and PDGFRA (2). In total, ~85% of malignant GISTs harbour activating mutations in one of the genes, KIT or PDGFRA, while 10-15% of all GISTs have no detectable KIT or PDGFRA mutations (2-4).
Receptors KIT and platelet derived growth factor receptor α (PDGFRA) have a similar structure and activation mechanism, as they belong to the same sub-family, the type III receptor tyrosine kinases (5). Their typical structure is composed of an extracellular (EC) domain, a transmembrane domain, an intracellular juxtamembrane (JM) domain and a cytoplasmic kinase domain, which is divided by a kinase insert into two catalytic parts, tyrosine kinase domain I (TK I) and tyrosine kinase domain II (TK II) (6).
Gain-of-function mutations in KIT or PDGFRA gene cause functional changes in KIT or PDGFRA receptor, which lead to constitutive, ligand-independent phosphorylation of the receptor and activation of downstream to KIT/PDGFRA signalling pathways (RAS/RAF/MAPK, PI3K/AKT/mTOR and JAK/STAT3), ultimately increasing cell proliferation and inhibiting apoptosis (6,7).
In GIST, 67% of KIT mutations occur in exon 11 (JM domain). The mutation frequency in other regions of the KIT receptor is markedly lower; mutations in exon 9 (EC domain) occur in 8-10% of GISTs, whilst exons 13 and 17 (TK I and TK II) are mutated in ~1% of all GISTs (8-10). PDGFRA mutations occur in ~8% of GISTs, of which exon 14 (TK I) and exon 18 (TK II) are mutated in 6-7% of the GISTs, and exon 12 (JM domain) in 1% (11,12).
GISTs without a mutation in the KIT or PDGFRA genes are historically known as wild-type (WT) GIST; however, their molecular biology is very heterogeneous. KIT/PDGFRA WT GISTs can have alterations in genes of the succinate dehydrogenase complex or in BRAF, PIK3CA, or rarely RAS family genes (13,14). BRAF V600E mutation has been reported in 7-15% of KIT/PDGFRA WT GISTs, meanwhile RAS and PIK3CA mutant GISTs are reported to be very rare (15-18). Even more rare are recently reported NTRK fusions in these patients, however they are important for treatment decisions (19).
Before the discovery of tyrosine kinase inhibitor (TKI) imatinib, no effective therapy was available for the treatment of unresectable, metastatic or recurrent GIST. The overall survival (OS) time of such patients was 9-20 months (20,21). With imatinib, the median OS time of patients with advanced GISTs is significantly longer and is reported to be ~55-57 months (21). Although the majority of KIT/PDGFRA-mutant GIST patients (90%) achieve clinical benefit from imatinib, primary resistance frequently occurs in GISTs that are KIT/PDGFRA WT, have KIT exon 9 mutation or have a substitution D842V in gene PDGFRA (22,23). Furthermore, secondary resistance develops in 40-50% of patients with GIST within 2 years of successful treatment due to secondary KIT/PDGFRA mutations or activating mutations of another downstream signalling pathway, such as the RAS/RAF/MAPK or PI3K/AKT/mTOR path-ways (18,22,23). Imatinib efficacy greatly depends on tumour genotype; therefore, molecular characterization of GIST is a very important step in optimizing GIST clinical treatment (21,23).
The aim of the present study was to molecularly characterize a cohort of 70 patients with metastatic GISTs from the Slovenian Cancer Registry (National Cancer Registry) treated between period 2002 and 2011. The spectrum of KIT and PDGFRA mutations was reported, as well as the spectrum of hot spot mutations in KRAS, NRAS, BRAF, PIK3CA and AKT1 genes in patients with GIST.
Materials and methods
Patients
In total, 70 patients with metastatic KIT-positive GIST, treated with TKIs at the Institute of Oncology Ljubljana (Ljubljana, Slovenia) between January 2002 and December 2011, were recruited and included in the present analysis.
Formalin-fixed, paraffin-embedded (FFPE) primary tissue samples were mainly obtained from the Department of Pathology, Institute of Oncology Ljubljana (72%) and also from pathology departments from other hospitals and clinics in Slovenia (University Medical Centre Ljubljana, Ljubljana; University Medical Centre Maribor, Maribor; Izola General Hospital, Izola; and Celje General Hospital, Celje). All patients were treated with TKIs at the Institute of Oncology Ljubljana. In total, 12 of the 70 samples were obtained from patients after the start of imatinib therapy. Clinicopathological characteristics of the patients, as well as prognostic risk groups according to Miettinen et al (4), are presented in Table I.
Table I.
Clinicopathological characteristics of 70 patients with gastrointestinal stromal tumour.
| Clinicopathological characteristic | n (%) |
|---|---|
| Sex | |
| Male | 32 (45.7) |
| Female | 38 (54.3) |
| Age | |
| Median, years | 65 |
| Range, years | 34-89 |
| ≤60 years | 28 (40.0) |
| >60 years | 42 (60.0) |
| Curative resection | |
| Yes | 13 (18.6) |
| No | 57 (81.4) |
| Imatinib therapy | |
| Yes | 70 (100.0) |
| No | 0 (0.0) |
| Outcome | |
| Alive | 21 (30.0) |
| Dead | 49 (70.0) |
| Primary tumour location | |
| Oesophagus | 2 (2.9) |
| Stomach | 33 (47.1) |
| Small intestine | 25 (35.7) |
| Rectum | 6 (8.6) |
| Extraluminal | 4 (5.7) |
| Mitotic index,/50 HPF | |
| ≤5 | 22 (31.4) |
| 5.1-10 | 11 (15.7) |
| >10 | 37 (52.9) |
| Primary tumour size, cm | |
| 0-5 | 6 (8.5) |
| 5.1-10 | 23 (32.9) |
| >10 | 37 (52.9) |
| Unknown | 4 (5.7) |
| Risk classificationa | |
| High | 49 (70.0) |
| Intermediate | 11 (15.7) |
| Low | 9 (12.9) |
| Very low | 1 (1.4) |
| Morphology | |
| Spindle cells | 47 (67.1) |
| Epithelioid cells | 8 (11.5) |
| Mixed cells | 15 (21.4) |
Risk groups for gastrointestinal tumours were adapted from Miettinen et al (4).
As controls for immunohistochemistry (IHC), normal appendiceal tissue samples were obtained from 5 patients (3 females, 2 males; age, 57-74 years; median, 63 years) undergoing various surgical procedures. In addition, tumour tissue samples were obtained from 10 patients (6 females, 4 males; age, 42-83 years; meadian, 60 years) with histopathologically confirmed CD117-positive non-metastatic GIST. All patients included as IHC controls were treated at the Institute of Oncology Ljubljana between January 2002 and December 2011.
All collected tumour samples were evaluated and were histopathologically examined under a microscope by pathology specialists, who confirmed the diagnosis using immunohistochemical staining for CD117 antigen expression. IHC analysis of CD117 was performed on 2-4 µm FFPE tissue sections, dried at 56°C for 2 h. Heat-mediated epitope retrieval was performed in a microwave oven using home-brew pH 9.0 Tris-EDTA buffer for 10 min at 100°C at high pressure. Detection was performed in the semi-automated IHC system LabVision™ Autostainer AS480. Endogenous peroxidases were blocked using Dako REAL™ Peroxidase blocking solution (cat. no. S2023; Dako; Agilent Technologies, Inc.) for 8 min at room temperature. CD117 was detected using a commercially available mouse polyclonal antibody CD117 (cat. no. A4502; Dako; Agilent Technologies, Inc.; 1:100), which was added for 30 min at room temperature. The antibody was diluted using Dako REAL™ antibody diluent (cat. no. S2022; Dako; Agilent Technologies, Inc.). Primary antibody was visualised using 2-step polymer detection system Dako REAL™ EnVision™ Detection system, Peroxidase/DAB+ Rabbit/Mouse (cat. no. K5007; Dako; Agilent Technologies, Inc.). Ready to-use secondary antibody Dako REAL™ EnVision™/HRP Rabbit/Mouse (cat. no. K5007; Dako; Agilent Technologies, Inc.) was added for 30 min at room temperature. Chromogene Dako REAL™ DAB+ Chromogen (cat. no. K5007; Dako; Agilent Technologies, Inc.) was diluted at 1:50, according to manufacturer's protocol and incubated twice for 7.5 min each at room temperature.
Tissue sections were viewed under a light microscope (magnification, ×10). The section of the FFPE slides that contained the highest number of tumour cells (always >70% in all GIST samples) was marked and used for DNA extraction.
The present study was approved by the Institutional Review Board of the Institute of Oncology Ljubljana (permission no. OIRIKE00036) and by the National Medical Ethics Committee of Republic of Slovenia (permission no. 1090216). Individual patient consent was waived for this study as it was a retrospective study, the research involved no risk to the subjects, and the institutional informed consent forms for treatment included consent for the use of patient's data, materials and/or test results for research purposes. All procedures followed in the present study were therefore in accordance with the ethical standards of the responsible committees on human experimentation (institutional and national) and the Helsinki Declaration of 1975, as revised in 2013.
Molecular analysis
Mutational analyses of KIT, PDGFRA, KRAS, NRAS and BRAF genes were performed by the Department of Molecular Diagnostic at the Institute of Oncology Ljubljana (Ljubljana, Slovenia). Genomic DNA of 70 sporadic GISTs was isolated from FFPE tissues, using macrodissection and standard procedures, according to the manufacturer's protocol of the QIAmp DNA FFPE Tissue kit (Qiagen GmbH). The quality and concentration of DNA extracts were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc.).
All GISTs (n=70) were examined for somatic mutations in KIT (exons 9, 11, 13 and 17) and PDGFRA (exons 12, 14 and 18) by polymerase chain reaction (PCR) amplification and direct Sanger sequencing. For PCR amplification, Invitrogen™EXPRESS SYBR™ GreenER™ qPCR Supermix, universal (Invitrogen; Thermo Fisher Scientific, Inc.), which includes Platinum™ Taq DNA Polymerase, was used. PCR reactions were run on an Applied Biosystems™ 7900HT Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the following thermocycling conditions: Activation at 50°C for 2 min; followed by denaturation at 95°C for 10 min; 20 cycles of denaturation at 95°C for 35 sec, annealing at 64°C for 45 sec and elongation at 72°C for 1 min; followed by another 30 cycles of denaturation at 95°C for 15 sec, annealing at 55°C for 45 sec and elongation at 72°C for 1 min. Direct Sanger sequencing reactions were performed using a Big Dye Terminator v3.1 (Applied Biosystems; Thermo Fisher Scientific, Inc.) and run on an ABI 3500 Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.). Specific primers for target exon amplification and sequencing reactions were designed using Primer3 web software (version 0.1g; http://biotools.nubic.northwestern.edu/Primer3.html) and are shown in Table SI.
Furthermore, all patients with KIT/PDGFRA WT GIST were tested for the presence of hot spot mutations in KRAS, NRAS, BRAF, PIK3CA and AKT1. Testing was performed using Colorectal Cancer Mutation Detection Panel v1.3 (EntroGen, Inc.), a quantitative PCR-based assay, according to the manufacturer's protocol. A list of all hot spot mutations that are included in the Colorectal Cancer Mutation Detection Panel are presented in Table SII.
Sequence analysis
DNA sequences were analysed using Sequencing Analysis software v5.4 software (Applied Biosystems; Thermo Fisher Scientific, Inc.), ChromasLite software v2.1.1 (Technelysium Pty Ltd.) and GeneRunner software (http://www.softpedia.com/get/Science-CAD/Gene-Runner.shtml). Mutation nomenclature followed the recommendations of the Human Genome Variation Society v19.01 (http://varnomen.hgvs.org/). Reference sequences used in the present study were taken from the National Centre for Biotechnology Information data-base (https://www.ncbi.nlm.nih.gov/): KIT (NM_000222.2), PDGFRA (NM_006206), KRAS (NM_004985), NRAS (NM_002524), BRAF (NM_004333), PIK3CA (NM_006218.2) and AKT1 (NM_005163.2).
Literature search of detected variants
A literature search for all detected variants found in the present study was performed using Alamut Visual v2.11 software (SOPHiA GENETICS). To perform a search for the references of every variant detected, a gene name and a nucleotide change (c.notation) were typed into Alamut Visual browser, which is linked to Google.
Online in silico analysis and bioinformatics tools
In silico mutation prediction analyses were performed using the online bioinformatics tools: Align GVGD (http://agvgd.hci.utah.edu/) MutationTaster2 (http://www.muta-tiontaster.org), and Cancer Genome Interpreter (https://www.cancergenomeinterpreter.org/analysis). Their pathogenicity was assessed according to The American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) criterions using Genetic Variant Interpretation Tool (http://www.medschool.umaryland.edu/Genetic_Variant_Interpretation_Tool1.html/).
Results
Genotype analysis KIT/PDGFRA mutant GISTs
Among 70 metastatic GIST tumours, 62 (88.6%) KIT/PDGFRA mutant GISTs were identified. Of the analysed GISTs, 60 (85.7%) had a mutation in KIT and only 2 (2.9%) had a mutation in PDGFRA. Of the 60 patients with KIT mutant GISTs, 6 (10%) developed a secondary mutation in exon 13 or 17 of the KIT gene. These secondary mutations were found in the patients whose tissue samples were obtained following imatinib treatment.
Among the 60 patients with KIT mutation, KIT exon 11 was mutated in 52 patients (86.7%), harbouring 40 different variants. KIT exon 9 was mutated in 6 cases (10%) and all carried a frequent six nucleotides duplication, c.1504_1509dup p.(Ala502_ Tyr503dup). KIT exon 13 was mutated in 5 cases (8.3%); 2 patients had a primary mutation and 3 patients had a secondary mutation. KIT exon 17 was mutated in 3 cases (5%), where all 3 patients had a secondary mutation (Table II).
Table II.
Mutational status and mutation location of all 70 patients with GIST from the Slovenian Cancer Registry.
| Mutation genotype | Number of GISTs |
|---|---|
| KIT/PDGFRA-mutant GIST | 62 |
| KIT (exon 9) | 6 |
| KIT (exon 11) | 52 |
| KIT (exon 13) | 5a |
| KIT (exon 17) | 3b |
| PDGFRA (exon 12) | 0 |
| PDGFRA (exon 14) | 1c |
| PDGFRA (exon 18) | 1 |
| Wild-typed GIST | 8 |
Two GISTs with primary mutation and three GISTs with secondary mutation.
Three GISTs with secondary mutation.
One GIST with two primary mutations.
Wild-type for KIT, PDGFRA, BRAF, KRAS, NRAS, PIK3CA and AKT1. GIST, gastrointestinal tumour.
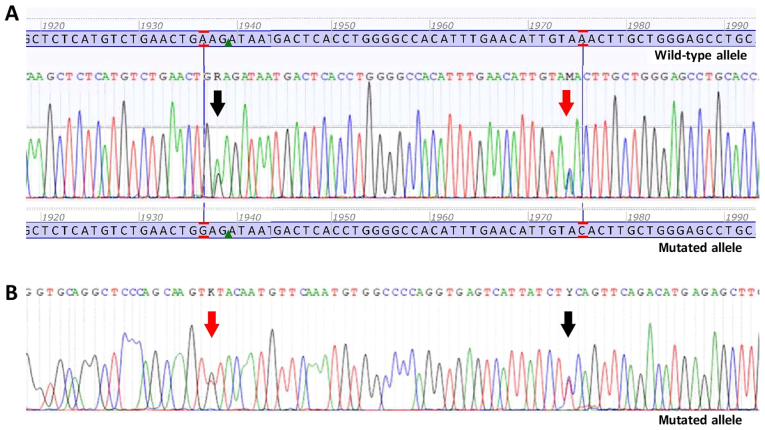
As aforementioned, mutations in PDGFRA were found in 2 cases (2.9%). The first patient had a single nucleotide change in exon 18, c.2525A>T p.(Asp842Val), while the second patient had two different single nucleotide changes, c.1936A>G p.(Lys627Glu) and c.1975A>C p.(Asn659His), both in exon 14. The latter case was a rare example of double mutant GIST (Fig. 1). Mutational status and mutation location of all 70 GISTs are presented in Table II. All detected alterations and their status of pathogenicity in 62 KIT/PDGFRA mutant GISTs from the Slovenian Cancer Registry are presented in Table SIII.
Figure 1.
Sanger sequencing electropherograms of double mutant gastrointestinal stromal tumour. (A) Forward sequence. Black arrow is indicating the nucleotide change c. 1936A>G p.(Lys627Glu) in PDGFRA exon 14. Red arrow is indicating the nucleotide change c.1975A>C p.(Asn659His) in PDGFRA exon 14. (B) Reverse sequence. Black arrow is indicating the nucleotide change c. 1936A>G p.(Lys627Glu) in PDGFRA exon 14. Red arrow is indicating the nucleotide change c.1975A>C p.(Asn659His) in PDGFRA exon 14.
KIT/PDGFRA wild-type GIST
All eight KIT/PDGFRA WT GISTs had no mutations in hot spot regions of KRAS (codons 12/13/59/61/117/146), NRAS (codons 12/13/59/61/117/146), BRAF (codon 600), PIK3CA (codons 542/545/1047) or AKT1 (codon 17).
Novel variants
The present study detected 49 different variants in total, of which eight were novel. A reference search for all detected variants (n=49) was performed using Google browser trough Alamut Visual software, on the 19th of January 2019.
All eight novel heterozygous variants, which had not been reported previously, were detected in exon 11 of gene KIT (Figs. 2 and 3). The precise effect of these eight novel variants on KIT protein function is unknown; however, based on Cancer Genome Interpreter and ACMG/AMP criterions, all of them were classified as likely pathogenic. The classification of all eight novel variants found in KIT exon 11 is presented in Table III and their descriptions are provided in the following paragraphs.
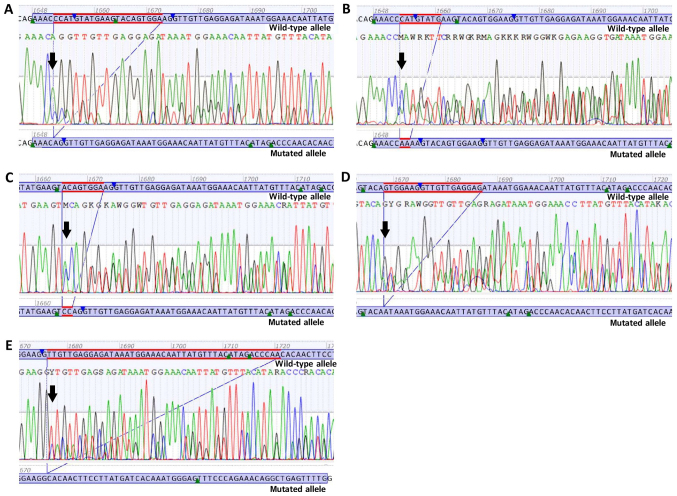
Figure 2.
Sanger sequencing electropherograms of novel deletions or deletion/insertions. (A) Novel deletion c.1652_1672del p.(Pro551_Lys558delinsGln). (B) Novel deletion/insertion c.1653_1660delinsAA p.(Met552_Glu554delinsLys). (C) Novel deletion/insertion c.1665_1672delinsCC p.(Trp557_Lys558del). (D) Novel deletion c.1668_1686del p.(Trp557*). (E) Novel deletion c.1676_1720del p.(Val559_Thr574delinsAla). Black arrows are indicating the start of deletions or deletion/insertions.
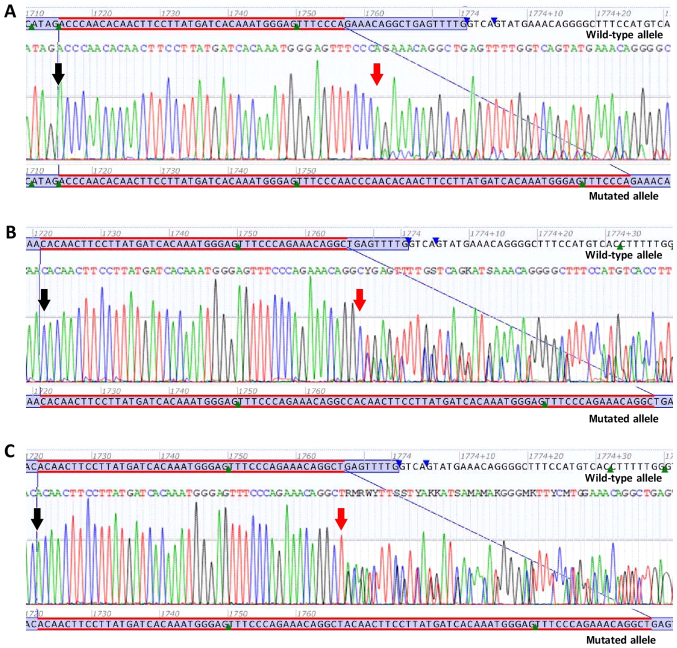
Figure 3.
Sanger sequencing electropherograms of novel duplications. (A) Novel duplication c.1715_1756dup p.(Pro585_Arg586ins14). (B) Novel duplication c.1721_1765dup p.(Arg588_Leu589ins15). (C) Novel duplication c.1722_1766dup p.(Gln575_Leu589dup). Black arrows are indicating the first duplicated nucleotide. Red arrows are indicating the last duplicated nucleotide.
Table III.
Classification of eight novel heterozygous variants in KIT exon 11.
| Location of die alteration
|
In silico prediction
|
ACMG/AMP Classification by Genetic Variant Interpretation Tool
|
Cancer Genome Interpreter
|
|||
|---|---|---|---|---|---|---|
| Nucleotide change (c .notation) | Amino ccid change (p.notation) | Align GVGD | MutadonTaster2 | Criterion | Pathogenicity | Oncogenic prediction |
| c.l652_1672del | p.(Pro55 l_Lys558dehnsGln) | Class C65 (GV: 0.00 - GD: 75.14) | Prediction disease causing - long InDel. Model: complex_aae. prob: 0.999999999999935 | PM1.PM2. PM4. PP3 | Likely pathogenic (IV) | Predicted driver: tier 1 |
| c.l653_16bOdelinsAA | p.(Met552_Ghi554delinsLys) | Class C65 (GV: 0.00 - GD: 94.49) | Prediction disease causing. Model: complex_aae. prob: 0.895033008598755 | PM1.PM2. PM4. PP3 | Likely pathogenic (IV) | Predicted driver: tier 1 |
| c.l665_1672delinsCC | p.(Trp557_Lys558del) | N/A | Prediction polymorphism. Model: complex_aae. prob: 0.981440878256269 | PM1.PM2. PM4 | Likely pathogenic (IV) | Predicted driver: tier 2 |
| c.l66S_16S6del | p.(Trp557') | N/A | Prediction disease causing. Model: complex_aae. prob. 1 | PM1.PM2. PM4. PP3 | Likely pathogenic (IV) | Predicted driver: tier 2 |
| c.l676_1720del | p .(Val559_Thr574dehnsAla) | Class C55 (GV: 0.00 - GD: 64.43) | Prediction disease causing - long InDel. Model: complex_aae. prob: 0. 99999994804898S | PM1.PM2. PM4. PP3 | Likely pathogenic (IV) | Predicted driver: tier 1 |
| c.l715_175odup | p.(Pro5S5_Arg5Soinsl4) | N/A | Prediction polymorphism Model: complex_aae. prob: 0.999999947411599 | PM1.PM2. PM4 | Likely pathogenic (IV) | Predicted passenger |
| c,1721_1765dup | p.(Arg5S8_Leu5S9insl5) | N/A | Prediction polymorphism. Model: complex_aae. prob: 0.999999998406041 | PM1.PM2. PM4 | Likely pathogenic (IV) | Predicted passenger |
| c,1722_1766dup | p .(Gln575_Leu5 89dup) | Class C65 (GV: 0.00 - GD: 112.44) | Prediction polymorphism Model: complex_aae. prob: 0.999999998406041 | PM1.PM2. PM4 | Likely pathogenic (IV) | Predicted driver: tier 2 |
Pathogenicity was determined using ACMG/AMP criterions. N/A, not avaliable.
c.1652_1672del p.(Pro551_Lys558delinsGln) is a deletion of 21 nucleotides in the exon 11 of the KIT gene, from position c.1652 to position c.1672. At the protein level, p.(Pro551_ Lys558delinsGln) results in the deletion of eight amino acids in the JM domain of the KIT protein from amino acids 551 to 558, combined with the insertion of a glutamine (Gln) at the same site (Fig. 2A).
c.1653_1660delinsAA p.(Met552_Glu554delinsLys) is a deletion of eight nucleotides in exon 11 of the KIT gene, from position c.1653 to position c.1660, and an insertion of two adenine bases at the same position. At the protein level, p.(Met552_Glu554delinsLys) results in the deletion of three amino acids in the JM domain of the KIT protein from amino acids 552 to 554, combined with the insertion of a lysine (Lys) at the same site (Fig. 2B).
c.1665_1672delinsCC p.(Trp557_Lys558del) is a deletion of eight nucleotides in exon 11 of the KIT gene, from position c.1665 to position c.1672, and an insertion of two cytosine bases at the same position. At the protein level, p.(Trp557_ Lys558del) results in the deletion of two amino acids in the JM domain of the KIT protein from amino acids 557 to 558 (Fig. 2C).
c.1668_1686del p.(Trp557*) is a deletion of 19 nucleotides in exon 11 of the KIT gene, from position c.1668 to position c.1686. At the protein level, p.(Trp557*) results in the premature termination of the KIT protein at amino acid 557. Due to the loss of the protein kinase domain, the mutation c.1668_1686del p.(Trp557*) is predicted to lead to a loss of KIT protein function (Fig. 2D).
c.1676_1720del p.(Val559_Thr574delinsAla) is a deletion of 45 nucleotides in exon 11 of the KIT gene, from position c.1676 to position c.1720. At the protein level, p.(Val559_Thr574delinsAla) results in the deletion of 16 amino acids in the JM domain of the KIT protein from amino acids 559 to 574, combined with the insertion of an Alanine (Ala) at the same site (Fig. 2E).
c.1715_1756dup p.(Pro585_Arg586ins14) is a duplication of 42 nucleotides in exon 11 of the KIT gene, from position c.1715 to position c.1756. At the protein level, p.(Pro585_Arg586ins14) indicates the insertion of 14 amino acids (Asn, Pro, Thr, Gln, Leu, Pro, Tyr, Asp, His, Lys, Trp, Glu, Phe and Pro) in the JM domain of the KIT protein, starting with Asparagine (Asn) at position 586 (Fig. 3A).
c.1721_1765dup p.(Arg588_Leu589ins15) is a duplication of 45 nucleotides in exon 11 of the KIT gene, from position c.1721 to position c.1765. At the protein level, p.(Arg588_Leu589ins15) indicates the insertion of 15 amino acids (Pro, Gln, Leu, Pro, Tyr, Asp, His, Lys, Trp, Glu, Phe, Pro, Arg, Asn and Arg) in the JM domain of the KIT protein, starting with Proline (Pro) at position 589 (Fig. 3B).
c.1722_1766dup p.(Gln575_Leu589dup) is a duplication of 45 nucleotides in exon 11 of the KIT gene, from position c.1722 to position c.1766. At the protein level, p.(Gln575_Leu589dup) indicates the duplication of 15 amino acids (Gln 575 through Leu 589) in the JM domain of the KIT protein, starting with Gln at position 590 (Fig. 3C).
Discussion
Genotype analysis of 70 patients from the Slovenian Cancer Registry was performed in the present study to determine the spectrum and frequency of KIT and PDGFRA alterations in Slovenian patients with metastatic GISTs, as well as the frequency of hot spot mutations in KRAS, NRAS, BRAF, PIK3CA and AKT1 in KIT/PDGFRA WT GISTs. Known mutation status of the aforementioned genes has been proven to be an important prognostic and predictive factor in the management and treatment of patients with GIST with TKIs (21,23).
Mutation analysis performed on Slovenian patients with GIST has confirmed a high degree of mutation heterogeneity in KIT and PDGFRA genes, as reported in previous studies (24-29). With direct Sanger sequencing of exons 9, 11, 13 and 17 of KIT gene and exons 12, 14 and 18 of PDGFRA gene, a total of 49 different alterations were detected. In the present study, the overall mutation rate was 88.5%, which was slightly higher than in a study from Italy (80%), yet comparable to frequencies observed in studies from Germany (86.1%) and Poland (85.1%), a French population-based study (KIT, 82.8%; PDGFRA, 2.1%), and in two phase III clinical trials that enrolled patients with metastatic GIST from the European Organisation for Research and Treatment of Cancer (KIT, 86.2%; PDGFRA, 1.6%) and Cancer and Leukemia Group B (KIT, 84.6%; PDGFRA, 2.65%) (24-29).
The mutation frequency in the patients analysed in the present study, was expectedly the highest in the KIT gene, with most mutations in exon 11 (86.7%). The highest mutation frequency in KIT exon 11 has also been reported in other studies (57, 66.1 and 87.3%, respectively) (14,24,30). In 8 patients with GIST in the present study (11.4%), no alterations in KIT/PDGFRA genes were found. According to the literature, ~10-15% of adult GISTs are KIT/PDGFRA WT (13,31). All 8 patients with no alterations in KIT or PDGFRA genes were also negative for hot spot mutations in KRAS, NRAS, BRAF, PIK3CA and AKT1 genes, which are included in the RAS/RAF/MAPK and PI3K/AKT/mTOR signalling pathways, suggesting that the frequency of these mutations is low (32). As reported by Shi et al (33), patients with KIT/PDGFRA WT GIST can harbour mutations in EGFR. Since in the present cohort, only 8 patients had KIT/PDGFRA WT GIST, and since the likelihood of EGFR mutations in these patients is <1%, the EGFR gene was not tested.
As aforementioned, the majority of identified KIT alterations in the present study were located in exon 11, between codons 550 and 580, which is similar to results from other studies (2,10,34-37). The most common type of KIT exon 11 alterations identified was an in-frame deletion of one to several codons. This is in line with the previously described frequencies of this change, representing 60-70% of all alterations in exon 11 (30,38,39). Furthermore, it was observed that a high presence of deletions or deletion/insertions in the present study was detected between codons 556 and 561 (38.5%). Of these, a specific and commonly reported deletion, p.(Trp557_Lys558), was identified in 9.6%, which is the same as that reported by Emile et al (27). Similar with the reports in the literature, the appearance of single nucleotide changes located around the same region of KIT exon 11, at codons 557, 559 and 560, such as p.(Trp557Arg), p.(Val559Asp) and p.(Val560Asp), was lower in the present study (5.8, 3.8 and 5.8%, respectively) (27,40).
In the present cohort of patients with metastatic GIST, there was a relatively high frequency (11.5%) of specific KIT exon 9 duplication, p.(Ala502_Tyr503dup), which was reported to be more frequent in metastatic GISTs (27,40).
In KIT exon 13, two single nucleotide changes, c.1924A>G p.(Lys642Glu) and c.1961T>C p.(Val654Ala), were identified. The substitution of amino acids Lys to Glu at codon 642, has been rarely described in literature and it has been suggested that this alteration disrupts normal auto-inhibitory function of the JM domain (13).
Single nucleotide changes in PDGFRA were identified in exon 18, c.2525A>T p.(Asp842Val), which is the most common PDGFRA mutation and accounts for 60-65% of all PDGFRA mutations seen in GIST (known to be resistant to the imatinib treatment) (12,41). Notably, 1 patient was identified with a rare example of double mutant GIST, which had two different primary mutations in the PDGFRA gene, c.1936A>G p.(Lys646Glu) and c.1975A>C p.(Asn659His), both located in exon 14. These patient samples were collected prior to the initiation of TKI treatment. According to the literature, double mutant GISTs, harbouring two primary mutations in the PDGFRA gene, found prior to TKI therapy, are rare (42,43).
Most of the patients who initially respond to imatinib develop resistance after a long period of treatment. Resistance to imatinib has been associated with secondary heterogeneous KIT receptor mutations in up to 90% of the patients. Secondary mutations most commonly arise in exons 13 and 14 (ATP-binding domain) or exons 17 and 18 (the activation loop), whereas primary KIT mutations predominantly affect the JM domain encoded by exon 11 (44). The present study identified 6 patients with KIT exon 13 or 17 secondary mutation. Primary tumour samples from these patients were obtained after imatinib treatment, suggesting secondary mutations most probably occurred as a response to the treatment, which has been reported in previous studies (10,41,45). In 3 out of 4 patients with secondary mutation in KIT exon 13 the same mutation was detected, a single nucleotide change c.1961T>C p.(Val654Ala), confirming that this is the most commonly described secondary mutation (10). Due to the existence of intra-tumour heterogeneity in GIST, with different clones being sensitive/resistant to different drugs, using a combination of different drugs could be a better approach for maximal clinical benefit (44). The most relevant drug combination in GIST aims to circumvent upstream KIT reactivation following imatinib failure by blocking KIT-downstream effectors, or bypass oncoproteins or KIT-mutated receptors themselves. If two or more not cross-resistant TKIs are used simultaneously in patients with KIT-receptor dependant secondary mutations, their cumulative toxic side effects are very high. Therefore, for clinical use their simultaneous combination is hardly feasible. However, their rapid alternation could improve the modest clinical benefit seen when they are used sequentially as second or third line treatment (44).
Eight novel likely pathogenic somatic variants were found in KIT exon 11 and they have been submitted to ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/), accession numbers SCV000902496 - SCV000902503. In order to characterize and classify the novel variants, the present study performed online in silico mutation prediction analysis using Align GVGD, MutationTaster2 and Cancer Genome Interpreter. Cancer Genome Interpreter categorises variants using OncodriveMUT bioinformatics method, which classifies variants into four classes: i) Predicted driver tier 1; ii) predicted driver tier 2 (where tier 1 and 2 represent higher and lower level of stringency of the driver prediction, respectively); iii) predicted passenger; and iv) polymorphism. Variants pathogenicity was determined according to ACMG/AMP guidelines using Genetic Variant Interpretation Tool (criterion selection PM1, PM2, PM4, PP3).
Exon 11 encodes the JM domain of KIT protein that prevents the kinase activation loop from shifting into the active state. Alterations in exon 11 predominantly disrupt the auto-inhibitory function and trigger ligand-independent receptor activation (46,47). Since all eight novel variants detected in the present study were located in a well-established functional domain and mutational hot spot, ACMG/AMP criterion PM1 was applied. None of the novel variants are described in public databases of human variation GnomAD (https://gnomad.broadinstitute.org/) or ExAC (https://www.re3data.org/repository/r3d100012122), hence ACMG/AMP criterion PM2 was applied. All of the novel variants cause protein length change due to deletion, insertion or duplication, therefore ACMG/AMP criterion PM4 was applied.
Five of the novel variants in KIT gene detected in the present study were deletions or deletions/insertions, [c.1652_1672del p.(Pro551_Lys558delinsGln), c.1653_1660delinsAA p.(Met552_Glu554delinsLys), c.1665_1672delinsCC p.(Trp557_Lys558del), c.1668_1686del p.(Trp557*), and c.1676_1720del p.(Val559_Thr574delinsAla)], localized within the most commonly mutated region of exon 11, between codons 550 and 580. Occurrence of deletions/insertions in this region of KIT receptor are described as activating alterations, which are according to literature, associated with malignant tumour behaviour (2,8,48,49). Therefore it could be predicted that newly identified variants in the present study, [c.1652_1672del p.(Pro551_Lys558delinsGln), c.1653_1660delinsAA p.(Met552_Glu554delinsLys), c.1676_1720del p.(Val559_Thr574delinsAla), and c.1665_1672delinsCC p.(Trp557_Lys558del)], are a gain-of-function mutations, leading to protein activation. Variant c.1665_1672delinsCC p.(Trp557_Lys558del) results in deletion of two amino acids Trp557 and Lys558. Deletion of codons 557-558 is frequently reported in other studies and has been described to cause constitutive activation of KIT and increased ERK phosphorylation (38,50,51). However, deletion of codons 557-558 as a result of this specific deletion of eight nucleotides from c.1665 to c.1672 coupled with insertion of two cytosines is firstly described in the present study. Deletion of codon 557 was detected also as a result of a novel deletion causing premature stop codon c.1668_1686del p.(Trp557*). Because of the premature termination of the KIT protein at amino acid 557, it was predicted that p.(Trp557*) leads to the loss of KIT protein function. Using Cancer Genome Interpreter, an evidence-based somatic variant classification tool, these five novel somatic variants were classified as predicted driver (tier 1 or tier 2), with high level of pathogenicity. Additionally, MutationTaster2 analysis predicted that four out of these five novel variants were disease-causing; based on their predicted oncogenic effect the ACMG/AMP criterion PP3 was applied.
Three of the novel variants were duplications of 17 to 45 nucleotides at the 3′-end of exon 11: c.1715_1756dup p.(Pro585_Arg586ins14), c.1721_1765dup p.(Arg588_ Leu589ins15) and c.1722_1766dup p.(Gln575_Leu589dup). Duplications in GISTs are known to almost exclusively appear at the 3′-end of KIT exon 11, they range from 3 to 57 nucleotides and they are reported to have a relatively good clinical outcome (52,53).
In conclusion, by identifying and classifying eight novel variants in KIT exon 11, and by detecting one double-mutant GIST with two mutations in PDGFRA exon 14, the present study contributes to broadening the spectrum of known mutations in GIST tumours. Mutation frequencies of KIT and PDGFRA gene observed in the present cohort (85.7 and 2.9%, respectively) are in accordance with previous literature. The current results also confirm the incidence of identified alterations in the previously reported and most frequently altered regions of KIT or PDGFRA, and suggest that the occurrence of alterations in other genes underlying GIST development, including KRAS, NRAS, BRAF, PIK3CA and AKT1, are very rare.
The present study is the first from Slovenia where a detailed characterization of gene abnormalities in patients with metastatic GIST from the Slovenian Cancer Registry has been performed. Knowing the accurate mutation status of patients with GIST is of great value for understanding GIST molecular biology, oncogenesis and GIST subtype classification, as well for the appropriate use of specific targeted therapy with KIT/PDGFRA TKIs.
Supplementary Data
Acknowledgements
Not applicable.
Funding
This study received funding from the Slovenian Research Agency (grant nos. P3-0352 and P3-0321).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
AB wrote the manuscript, collected the related literature, performed the majority of wet lab work, and contributed to the analysis and interpretation of sequencing data, bioinformatics analysis and classification of novel variants. BZ contributed to the conception of the study, acquired tumour samples and patient clinical data, and contributed to the revision of the manuscript. MB contributed to the wet lab experiments and contributed to the analysis of sequencing data. VSD contributed to the bioinformatics analysis, and interpretation and classification of novel variants. BG performed the pathological evaluation of tumour samples. VS contributed to the interpretation and classification of novel variants. GK created figures and contributed to the interpretation and classification of novel variants. SN contributed to the conception of this study and designed, reviewed, wrote and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institute of Oncology Ljubljana Ethic's Committee (approval no. OIRIKE00036) and the Republic of Slovenia National Medical Ethics Committee (approval no. 1090216). Individual patient consent was waived for this study as it was a retrospective study, the research involved no risk to the subjects, and the institutional informed consent forms for treatment included consent for the use of patient's data, materials and/or test results for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miettinen M, Lasota J. Gastrointestinal stromal tumors - definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch. 2001;438:1–12. doi: 10.1007/s004280000338. [DOI] [PubMed] [Google Scholar]
- 2.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, et al. Gain-of-Function Mutations of c-kit in Human Gastrointestinal Stromal Tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 3.Corless CL, Heinrich MC. Molecular Pathobiology of Gastrointestinal Stromal Sarcomas. Annu Rev Pathol Mech Dis. 2008;3:557–586. doi: 10.1146/annurev.pathmechdis.3.121806.151538. [DOI] [PubMed] [Google Scholar]
- 4.Miettinen M, Lasota J. Gastrointestinal stromal tumors: Pathology and prognosis at different sites. Semin Diagn Pathol. 2006;23:70–83. doi: 10.1053/j.semdp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Duensing A, Medeiros F, McConarty B, Joseph NE, Panigrahy D, Singer S, Fletcher CDM, Demetri GD, Fletcher JA. Mechanisms of oncogenic KIT signal transduction in primary gastrointestinal stromal tumors (GISTs) Oncogene. 2004;23:3999–4006. doi: 10.1038/sj.onc.1207525. [DOI] [PubMed] [Google Scholar]
- 6.Miettinen M, Lasota J. Gastrointestinal Stromal Tumors Review on Morphology, Molecular Pathology, Prognosis, and Differential Diagnosis. Arch Pathol Lab Med. 2006;130:1466–1478. doi: 10.5858/2006-130-1466-GSTROM. [DOI] [PubMed] [Google Scholar]
- 7.Heinrich MC, Rubin BP, Longley BJ, Fletcher JA. Biology and genetic aspects of gastrointestinal stromal tumors: KIT activation and cytogenetic alterations. Hum Pathol. 2002;33:484–495. doi: 10.1053/hupa.2002.124124. [DOI] [PubMed] [Google Scholar]
- 8.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 9.Miettinen M, El-Rifai W, H L Sobin L, Lasota J. Evaluation of malignancy and prognosis of gastrointestinal stromal tumors: A review. Hum Pathol. 2002;33:478–483. doi: 10.1053/hupa.2002.124123. [DOI] [PubMed] [Google Scholar]
- 10.Lasota J, Miettinen M. Clinical significance of oncogenic KIT and PDGFRA mutations in gastrointestinal stromal tumours. Histopathology. 2008;53:245–266. doi: 10.1111/j.1365-2559.2008.02977.x. [DOI] [PubMed] [Google Scholar]
- 11.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 12.Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P, Shiraga S, Bainbridge T, Morich J, Heinrich MC. PDGFRA mutations in gastrointestinal stromal tumors: Frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23:5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- 13.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: Origin and molecular oncology. Nat Rev Cancer. 2011;11:865–878. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 14.Joensuu H, Hohenberger P, Corless CL. Gastrointestinal stromal tumour. Lancet. 2013;382:973–983. doi: 10.1016/S0140-6736(13)60106-3. [DOI] [PubMed] [Google Scholar]
- 15.Daniels M, Lurkin I, Pauli R, Erbstösser E, Hildebrandt U, Hellwig K, Zschille U, Lüders P, Krüger G, Knolle J, et al. Spectr um of KIT/PDGF RA/BRAF mutations and Phosphatidylinositol-3-Kinase pathway gene alterations in gastro-intestinal stromal tumors (GIST) Cancer Lett. 2011;312:43–54. doi: 10.1016/j.canlet.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 16.Miranda C, Nucifora M, Molinari F, Conca E, Anania MC, Bordoni A, Saletti P, Mazzucchelli L, Pilotti S, Pierotti MA, et al. KRAS and BRAF mutations predict primary resistance to imatinib in gastrointestinal stromal tumors. Clin Cancer Res. 2012;18:1769–1776. doi: 10.1158/1078-0432.CCR-11-2230. [DOI] [PubMed] [Google Scholar]
- 17.Pantaleo MA, Nannini M, Corless CL, Heinrich MC. Quadruple wild-type (WT) GIST: Defining the subset of GIST that lacks abnormalities of KIT, PDGFRA, SDH, or RAS signaling pathways. Cancer Med. 2015;4:101–103. doi: 10.1002/cam4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasota J, Felisiak-Golabek A, Wasag B, Kowalik A, Zięba S, Chłopek M, Wang ZF, Coates T, Kopczynski J, Gozdz S, et al. Frequency and clinicopathologic profile of PIK3CA mutant GISTs: Molecular genetic study of 529 cases. Mod Pathol. 2016;29:275–282. doi: 10.1038/modpathol.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isozaki K, Hirota S. Gain-of-Function Mutations of Receptor Tyrosine Kinases in Gastrointestinal Stromal Tumors. Curr Genomics. 2006;7:469–475. doi: 10.2174/138920206779315755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishida T, Takahashi T, Miyazaki Y. Gastrointestinal stromal tumor: A bridge between bench and bedside. Gastric Cancer. 2009;12:175–188. doi: 10.1007/s10120-009-0525-8. [DOI] [PubMed] [Google Scholar]
- 22.Fletcher JA, Rubin BP. KIT mutations in GIST. Curr Opin Genet Dev. 2007;17:3–7. doi: 10.1016/j.gde.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Gramza AW, Corless CL, Heinrich MC. Resistance to tyrosine kinase inhibitors in gastrointestinal stromal tumors. Clin Cancer Res. 2009;15:7510–7518. doi: 10.1158/1078-0432.CCR-09-0190. [DOI] [PubMed] [Google Scholar]
- 24.Origone P, Gargiulo S, Mastracci L, Ballestrero A, Battistuzzi L, Casella C, Comandini D, Cusano R, Dei Tos AP, Fiocca R, et al. Liguria GIST Unit Molecular characterization of an Italian series of sporadic GISTs. Gastric Cancer. 2013;16:596–601. doi: 10.1007/s10120-012-0213-y. [DOI] [PubMed] [Google Scholar]
- 25.Penzel R, Aulmann S, Moock M, Schwarzbach M, Rieker RJ, Mechtersheimer G. The location of KIT and PDGFRA gene mutations in gastrointestinal stromal tumours is site and phenotype associated. J Clin Pathol. 2005;58:634–639. doi: 10.1136/jcp.2004.021766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wozniak A, Rutkowski P, Schöffski P, Ray-Coquard I, Hostein I, Schildhaus HU, Le Cesne A, Bylina E, Limon J, Blay JY, et al. Tumor genotype is an independent prognostic factor in primary gastrointestinal stromal tumors of gastric origin: A european multicenter analysis based on ConticaGIST. Clin Cancer Res. 2014;20:6105–6116. doi: 10.1158/1078-0432.CCR-14-1677. [DOI] [PubMed] [Google Scholar]
- 27.Emile JF, Brahimi S, Coindre JM, Bringuier PP, Monges G, Samb P, Doucet L, Hostein I, Landi B, Buisine MP, et al. Frequencies of KIT and PDGFRA mutations in the MolecGIST prospective population-based study differ from those of advanced GISTs. Med Oncol. 2012;29:1765–1772. doi: 10.1007/s12032-011-0074-y. [DOI] [PubMed] [Google Scholar]
- 28.Debiec-Rychter M, Sciot R, Le Cesne A, Schlemmer M, Hohenberger P, van Oosterom AT, Blay JY, Leyvraz S, Stul M, Casali PG, et al. EORTC Soft Tissue and Bone Sarcoma Group. Italian Sarcoma Group. Australasian GastroIntestinal Trials Group KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;42:1093–1103. doi: 10.1016/j.ejca.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 29.Heinrich MC, Owzar K, Corless CL, Hollis D, Borden EC, Fletcher CDM, Ryan CW, Von Mehren M, Blanke CD, Rankin C, et al. Correlation of kinase genotype and clinical outcome in the North American intergroup phase III trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 study by cancer and leukemia group B and southwest oncology gr. J Clin Oncol. 2008;26:5360–5367. doi: 10.1200/JCO.2008.17.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson J, Bümming P, Meis-Kindblom JM, Sihto H, Nupponen N, Joensuu H, Odén A, Gustavsson B, Kindblom L-G, Nilsson B. Gastrointestinal stromal tumors with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology. 2006;130:1573–1581. doi: 10.1053/j.gastro.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 31.Boikos SA, Pappo AS, Killian JK, LaQuaglia MP, Weldon CB, George S, Trent JC, von Mehren M, Wright JA, Schiffman JD, et al. Molecular Subtypes of KIT/PDGFRAWild-Type Gastrointestinal Stromal Tumors. JAMA Oncol. 2016;2:922. doi: 10.1001/jamaoncol.2016.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huss S, Pasternack H, Ihle MA, Merkelbach-Bruse S, Heitkötter B, Hartmann W, Trautmann M, Gevensleben H, Büttner R, Schildhaus HU, et al. Clinicopathological and molecular features of a large cohort of gastrointestinal stromal tumors (GISTs) and review of the literature: BRAF mutations in KIT/PDGFRA wild-type GISTs are rare events. Hum Pathol. 2017;62:206–214. doi: 10.1016/j.humpath.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Shi SS, Wu N, He Y, Wei X, Xia QY, Wang X, Ye S, Li R, Rao Q, Zhou X-J. Bin, Li R, Rao Q and Zhou XJ: EGFR gene mutation in gastrointestinal stromal tumours. Histopathology. 2017;71:553–561. doi: 10.1111/his.13251. [DOI] [PubMed] [Google Scholar]
- 34.Nakahara M, Isozaki K, Hirota S, Miyagawa J, Hase-Sawada N, Taniguchi M, Nishida T, Kanayama S, Kitamura Y, Shinomura Y. A novel gain-of-function mutation of c-kit gene in gastrointestinal stromal tumors. Gastroenterology. 1998;115:1090–1095. doi: 10.1016/S0016-5085(98)70079-4. [DOI] [PubMed] [Google Scholar]
- 35.Lasota J, Jasinski M, Sarlomo-Rikala M, Miettinen M. Mutations in exon 11 of c-Kit occur preferentially in malignant versus benign gastrointestinal stromal tumors and do not occur in leiomyomas or leiomyosarcomas. Am J Pathol. 1999;154:53–60. doi: 10.1016/S0002-9440(10)65250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res. 1999;59:4297–4300. [PubMed] [Google Scholar]
- 37.Moskaluk CA, Tian Q, Marshall CR, Rumpel CA, Franquemont DW, Frierson HF., Jr Mutations of c-kit JM domain are found in a minority of human gastrointestinal stromal tumors. Oncogene. 1999;18:1897–1902. doi: 10.1038/sj.onc.1202496. [DOI] [PubMed] [Google Scholar]
- 38.Wardelmann E, Losen I, Hans V, Neidt I, Speidel N, Bierhoff E, Heinicke T, Pietsch T, Büttner R, Merkelbach-Bruse S. Deletion of Trp-557 and Lys-558 in the juxtamembrane domain of the c-kit protooncogene is associated with metastatic behavior of gastrointestinal stromal tumors. Int J Cancer. 2003;106:887–895. doi: 10.1002/ijc.11323. [DOI] [PubMed] [Google Scholar]
- 39.Martín-Broto J, Rubio L, Alemany R, López-Guerrero JA. Clinical implications of KIT and PDGFRA genotyping in GIST. Clin Transl Oncol. 2010;12:670–676. doi: 10.1007/s12094-010-0576-7. [DOI] [PubMed] [Google Scholar]
- 40.Van Glabbeke M, Gastrointestinal Stromal Tumor Meta-Analysis Group (MetaGIST) Comparison of two doses of imatinib for the treatment of unresectable or metastatic gastrointestinal stromal tumors: A meta-analysis of 1,640 patients. J Clin Oncol. 2010;28:1247–1253. doi: 10.1200/JCO.2009.24.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lasota J, Miettinen M. KIT and PDGFRA mutations in gastrointestinal stromal tumors (GISTs) Semin Diagn Pathol. 2006;23:91–102. doi: 10.1053/j.semdp.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Conca E, Miranda C, Dal Col V, Fumagalli E, Pelosi G, Mazzoni M, Fermeglia M, Laurini E, Pierotti MA, Pilotti S, et al. Are two better than one? A novel double-mutant KIT in GIST that responds to Imatinib. Mol Oncol. 2013;7:756–762. doi: 10.1016/j.molonc.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pai T, Bal M, Shetty O, Gurav M, Ostwal V, Ramaswamy A, Ramadwar M, Desai S. Unraveling the spectrum of KIT mutations in gastrointestinal stromal tumors: An Indian Tertiary Cancer Center Experience. South Asian J Cancer. 2017;6:113–117. doi: 10.4103/sajc.sajc_275_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serrano C, Mariño-Enríquez A, Tao DL, Ketzer J, Eilers G, Zhu M, Yu C, Mannan AM, Rubin BP, Demetri GD, et al. Complementary activity of tyrosine kinase inhibitors against secondary kit mutations in imatinib-resistant gastrointestinal stromal tumours. Br J Cancer. 2019;120:612–620. doi: 10.1038/s41416-019-0389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang CM, Huang K, Zhou Y, Du CY, Ye YW, Fu H, Zhou XY, Shi YQ. Molecular mechanisms of secondary imatinib resistance in patients with gastrointestinal stromal tumors. J Cancer Res Clin Oncol. 2010;136:1065–1071. doi: 10.1007/s00432-009-0753-7. [DOI] [PubMed] [Google Scholar]
- 46.Gajiwala KS, Wu JC, Christensen J, Deshmukh GD, Diehl W, Dinitto JP, English JM, Greig MJ, He Y-A, Jacques SL, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc Natl Acad Sci U S A. 2009;106:1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN, Snell GP, Zou H, Sang BC, Wilson KP. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J Biol Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- 48.Roskoski R., Jr Structure and regulation of Kit protein-tyrosine kinase - the stem cell factor receptor. Biochem Biophys Res Commun. 2005;338:1307–1315. doi: 10.1016/j.bbrc.2005.09.150. [DOI] [PubMed] [Google Scholar]
- 49.Bachet JB, Hostein I, Le Cesne A, Brahimi S, Beauchet A, Tabone-Eglinger S, Subra F, Bui B, Duffaud F, Terrier P, et al. Prognosis and predictive value of KIT exon 11 deletion in GISTs. Br J Cancer. 2009;101:7–11. doi: 10.1038/sj.bjc.6605117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garner AP, Gozgit JM, Anjum R, Vodala S, Schrock A, Zhou T, Serrano C, Eilers G, Zhu M, Ketzer J, et al. Ponatinib inhibits polyclonal drug-resistant KIT oncoproteins and shows thera-peutic potential in heavily pretreated gastrointestinal stromal tumor (GIST) patients. Clin Cancer Res. 2014;20:5745–5755. doi: 10.1158/1078-0432.CCR-14-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang HC, Li TY, Chao YJ, Hou YC, Hsueh YS, Hsu KH, Shan YS. Kit exon 11 codons 557-558 deletion mutation promotes liver metastasis through the CXCL12/CXCR4 Axis in gastroin-/CXCR4 Axis in gastroin CXCR4 Axis in gastroin--testinal stromal tumors. Clin Cancer Res. 2016;22:3477–3487. doi: 10.1158/1078-0432.CCR-15-2748. [DOI] [PubMed] [Google Scholar]
- 52.Wozniak A, Rutkowski P, Piskorz A, Ciwoniuk M, Osuch C, Bylina E, Sygut J, Chosia M, Rys J, Urbanczyk K, et al. Polish Clinical GIST Registry Prognostic value of KIT/PDGFRA mutations in gastrointestinal stromal tumours (GIST): Polish Clinical GIST Registry experience. Ann Oncol. 2012;23:353–360. doi: 10.1093/annonc/mdr127. [DOI] [PubMed] [Google Scholar]
- 53.Lasota J, Wasag B, Steigen SE, Limon J, Miettinen M. Improved detection of KIT exon 11 duplications in formalin-fixed, paraffin-embedded gastrointestinal stromal tumors. J Mol Diagn. 2007;9:89–94. doi: 10.2353/jmoldx.2007.060104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.