Abstract
It is estimated that ~50% of patients with melanoma harbour B-Raf (BRAF)V600 driver mutations, with the most common of these being BRAFV600E, which leads to the activation of mitogen-activated protein kinase proliferative and survival pathways. BRAF inhibitors are used extensively to treat BRAF-mutated metastatic melanoma; however, acquired resistance occurs in the majority of patients. The effects of long-term treatment with PLX4032 (BRAFV600 inhibitor) were studied in vitro on sensitive V600E BRAF-mutated melanoma cell lines. After several weeks of treatment with PLX4032, the majority of the melanoma cells died; however, a proportion of cells remained viable and quiescent, presenting senescent cancer stem cell-like characteristics. This surviving population was termed SUR cells, as discontinuing treatment allowed the population to regrow while retaining equal drug sensitivity to that of parental cells. RNA sequencing analysis revealed that SUR cells exhibit changes in the expression of 1,415 genes (P<0.05) compared with parental cells. Changes in the expression levels of a number of epigenetic regulators were also observed. These changes and the reversible nature of the senescence state were consistent with epigenetic regulation; thus, it was investigated as to whether the senescent state could be reversed by epigenetic inhibitors. It was found that both parental and SUR cells were sensitive to different histone deacetylase (HDAC) inhibitors, such as SAHA and MGCD0103, and to the cyclin-dependent kinase (CDK)9 inhibitor, CDKI-73, which induced apoptosis and reduced proliferation both in the parental and SUR populations. The results suggested that the combination of PLX4032 with HDAC and CDK9 inhibitors may achieve complete elimination of SUR cells that persist after BRAF inhibitor treatment, and reduce the development of resistance to BRAF inhibitors.
Keywords: melanoma, B-Raf, epigenetic inhibitors, plastic resistance, PLX4032
Introduction
It is estimated that ~50% of patients with melanoma harbour B-Raf (BRAF)V600 driver mutations, the most common of which being BRAFV600E; this mutation leads to the activation of mitogen-activated protein kinase (MAPK) proliferative and survival pathways (1). BRAF inhibitors, alone or in combination with MAPK kinase (MEK) inhibitors, are extensively used to treat BRAF-mutated metastatic melanoma; however, acquired resistance unfortunately occurs in the majority of patients (2). Resistance mechanisms involve mutations or changes in gene expression that result in the reactivation of MAPK signalling, or the activation of other proliferative and survival pathways, such as PI3K signaling (3-7). Increasing evidence suggests that these resistant states exist prior to treatment and may be selected by treatment with BRAF inhibitors (8,9). These subpopulations can contribute to tumour progression by presenting a slow-cycling behaviour or phenotype associated with epithelial-to-mesenchymal transition. This plasticity may allow a rapid and dynamic response, generating a phenotype that could more effectively tolerate drug treatment, underlying the limited cell death observed in vitro and in vivo during targeted therapy (8).
Acquired drug resistance could also be driven by epigenetic events; it has been shown that epigenetic alterations contribute to chemotherapy resistance in different types of tumours, including breast, colorectal and ovarian cancers (10-13). Recent evidence suggests that chromatin architecture reprogramming could be also implicated in drug resistance to MAPK inhibitors in melanoma cells (14,15). Several groups have reported that treatment with different epigenetic inhibitors, such as histone deacetylate (HDAC) inhibitors (16,17), bromodomain and extra-terminal motif (BET) inhibitors (18) and DNA methyltransferase (DNMT) inhibitors (19), in combination with BRAF inhibitors, could overcome resistance.
Aside from the resistant cells that are able to proliferate in the presence of MAPK inhibitors, our and other previous studies have shown that BRAF and MEK inhibitors can lead to the enrichment of a drug-tolerant tumour cell population that persists in a slow-cycling or quiescent state (9,20-22). This evidence now may be of greater clinical relevance, as combined BRAF and MEK inhibitor treatment has been recently approved in the adjuvant setting for patients with stage III recurrent BRAFV600-mutated melanoma, as it was reported to result in a significantly reduced risk of recurrence (23). If a population of persisting melanoma cells is present, once the treatment is discontinued, they could give rise to relapses.
Our previous study described a persistent melanoma cell population [surviving (SUR) cells], obtained following long-term PLX4032 treatment, of sensitive BRAFV600E-mutated melanoma cell lines (20). SUR cells express the cancer stem cell markers CD271 and ATP-binding cassette B5, and present senescence-associated characteristics, such as senescence-associated (SA) β-galactosidase activity. Discontinuing MAPK inhibitor treatment of SUR cells permits their regrowth, and they eventually regain drug sensitivity equal to parental cells, demonstrating the plasticity of the SUR phenotype. SUR cells exhibit an increased tumorigenicity compared with parental cells when injected subcutaneously in NOD/SCID-γ (NSG) mice, yet retain melanoma differentiation antigens (Ags) and human leukocyte Ag class I expression, and are therefore susceptible to Ag-specific cytotoxic T lymphocytes lysis (20).
It was hypothesized that the SUR phenotype may be determined by epigenetic changes. Thus, the aim of the present study was to determine if treatment with epigenetic inhibitors could efficiently eliminate the SUR population. SUR cell sensitivity to different epigenetic inhibitors was analysed, and it was found that both parental and SUR cells were sensitive to HDAC inhibitors. It is proposed that the combination of PLX4032 with epigenetic inhibitors could be efficacious to achieve complete elimination of SUR cells that persist after long-term BRAF inhibitor treatment.
Materials and methods
Cell lines and drugs
The MEL-XY3 and MEL-XY13 cell lines have already been described (20). MEL-XX12 and MEL-XX15 were obtained in-house from metastatic melanoma biopsies. Both cell lines present the BRAFV600E mutation. MEL-XX12 was established from a 58-year-old female diagnosed with cutaneous melanoma in the right side of the back. The patient developed a local recurrence in the back which was excised at the Hospital Naval Dr Pedro Mallo (Buenos Aires, Argentina). The patient provided oral consent (as the patient was hospitalised in a different institution at the time of collection, oral rather than written consent was obtained) for the collection of part of the fresh tumour to be used for the establishment of experimental cell lines for cancer research purposes. The MEL-XX15 cell line was established from a 38-year-old female diagnosed with cutaneous melanoma in the right forearm. The cell line was derived from a metastatic subcutaneous nodule excised from the breastbone site at 4 years after diagnosis. Surgery was conducted at the Instituto Alexander Fleming (Buenos Aires, Argentina), and the patient provided signed informed consent for surgery and the use of part of the tumour sample for scientific research. Ethical approval of the informed consent for surgery was granted by the Comité de Etica en Investigación del Instituto Alexander Fleming prior to the isolation and establishment of these two cell lines.
Melanoma cell lines were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.; MEL-XY3 cells only) or melanoma medium [DMEM:F-12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 2 mM glutamine, 20 nM sodium selenite, 100 µM ascorbic acid, 0.3 mg/ml galactose, 0.15 mg/ml sodium pyruvate, 5 µg/ml insulin; all cell lines] plus 10% fetal calf serum (AusGeneX), 100 IU/ml penicillin and 10 µg/ml streptomycin. All cells were maintained at 37°C in a 5% CO2 humidified atmosphere. Mycoplasma detection was regularly assayed by PCR. PLX4032, GDC-0973, MGCD0103, SAHA, S63845, panobinostat and tubacin were purchased from MedChemExpress LLC. IBET151, JQ-1, OTX-015, decitabine, EPZ-6438 and GSK126 were purchased from Selleck Chemicals. MC1568, sodium butyrate and trichostatin A (TSA) were purchased from Sigma-Aldrich (Merck KGaA). CDKI-73 was a gift from Dr Shudong Wang (University of South Australia).
Generation of SUR cells
SUR cells were generated by exposing parental cells to 10 µM PLX4032 or 10 µM PLX4032 + 1 µM GDC-0973 combined treatment for 5 weeks. Media was changed twice weekly.
RNA sequencing (RNA-seq)
RNA-seq was performed for MEL-XY3 and SUR MEL-XY3 cells following PLX4032 exposure using the Illumina Hiseq 4000 sequencing platform (Illumina, Inc.) with >20 million high-quality single-end reads per sample (sequencing was performed by BGI Americas). Quality control of reads was performed with FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and FastQC (http://www.bioinformatics.babraham.ac.uk/proj-ects/fastqc/). Reads were aligned to the latest human Hg38 reference genome using the STAR spliced read aligner (24). Fragment counts were derived using the HTSeq package (25). Differentially expressed genes were identified by a ranking based on adjusted P-values ≤0.005 and a false discovery rate (FDR) ≤0.1 using the R package edgeR (R version 3.2) (26,27). Gene set enrichment analysis (GSEA) (28) was performed using GSEA software from Broad Institute (https://www.gsea-msigdb.org/gsea/index.jsp). The present RNA-seq dataset was compared against a Molecular Signature Database gene set composed of a Gene Ontology (GO; http://geneontology.org/) gene set library (C5 collection), with an FDR <0.25 and P<0.01. The data discussed in this publication have been deposited in Gene Expression Omnibus (GEO) (29) and are accessible via GEO accession number GSE126960 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126960).
Normalized RNA-seq data were retrieved from GSE50509 dataset of the GEO database (29,30). Graphs were plotted using GraphPad Prism software (GraphPad Software, Inc.). A two-tailed paired t-test was performed to analyse the statistical significance of the given samples.
Reverse phase protein array (RPPA)
RPPA experiments were performed to analyze protein expression in MEL-XY3 cells, in MEL-XY3 cells treated for 7 days with PLX4032, in MEL-XY3 SUR cells and in MEL-XY3 SUR cells 7 days after drug removal, at which point the cells had resumed proliferating. RPPA was performed at the Victorian Centre for Functional Genomics RPPA platform at the Peter MacCallum Cancer Centre (Melbourne, Australia). Protein lysates were prepared in biological duplicates from cell lines freshly lysed in Zeptosens Cell Lysis Buffer 1 (CLB1; Bayer AG) and quantified using a Pierce™ Coomassie Plus Bradford Protein Assay Kit (Thermo Fisher Scientific, Inc.); triplicate measurements were performed for each sample. Employing a Sciclone/Caliper ALH3000 liquid handling robot (PerkinElmer, Inc.), samples were serially diluted in 10% CLB1:90% CSBL1 spotting buffer (Zeptosens AG; Bayer AG) and spotted onto ZeptoChips (Zeptosens AG; Bayer AG) in duplicate using a Nano-plotter-NP2.1 noncontact microarray system (GeSiM mbH). Chips were blocked under non-contact conditions for 1 h with BB1 buffer (Zeptosens AG; Bayer AG) and incubated with prevalidated primary antibodies (1:500) for 20 h (details of the antibodies used by the RPPA platform can be found at https://research.unimelb.edu.au/infrastructure/acrf-translational-reverse-phase-protein-array-platform), and Zenon™ Alexa Fluor® 647 anti-rabbit secondary antibody (1:1,000; cat. no. Z-25308; Thermo Fisher Scientific) for 4 h. Chips were read on a Zeptosens instrument (Zeptosens AG; Bayer AG), and ZeptoView software version 3.1 was utilized to calculate the relative fluorescence intensity. All samples were normalized to the background values reported in the negative control containing secondary antibody only. Duplicates were aver-aged, samples normalised by global rank invariant method and the data log2-normalised; log2 fold changes compared to the parental MEL-XY3 cell line were presented. The RPPA heat map was generated with Plotly software (https://chart-studio.plot.ly/create/#/).
Western blotting
Cell pellets were lysed with RIPA buffer (150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0) plus protease inhibitor (cat. no. P8340) and phosphatase inhibitor (cat. no. P5726; both Sigma-Aldrich; Merck KGaA) at recommended rates. Western blotting was conducted as described previously (20). Total protein was determined using a BCA assay (Bio-Rad Laboratories, Inc.). Blocking of membranes was performed at room temperature for 1 h; primary antibody incubations were performed overnight at 4°C, with secondary antibody incubations performed at room temperature for 1 h. Labelled bands were detected by Clarity ECL kit (Bio-Rad Laboratories, Inc.) and images were captured by the ChemiDoc MP Bio-Rad image system. Antibodies directed against HDAC6 (1:1,000; cat. no. 7558), DNMT3a (1:1,000; cat. no. 3598), DNMT3b (1:1,000; cat. no. 67259) and enhancer of zeste homolog 2 (1:2,000; EZH2; cat. no. 5246) were purchased from Cell Signaling Technology, Inc. Anti-β-actin antibody (1:3,000; cat. no. AC-74) was acquired from Sigma-Aldrich (Merck KGaA). The secondary antibodies used were horseradish peroxidase-conjugated goat anti-mouse IgG (H+L) and goat-anti-rabbit IgG (H+L) antibodies (1:3,000; cat. nos. 1706516 and 1706515; Bio-Rad Laboratories, Inc.).
Transmission electron microscopy (TEM)
MEL-XY3 cells, MEL-XY3 SUR cells, and MEL-XY3 SUR cells at 2 and 7 days after PLX4032 removal were analyzed by TEM. Cells were harvested, fixed for 4 h at 4°C in 2.5% glutaraldehyde, post-fixed in 1% (v/v) osmium tetroxide. Subsequently, samples were embedded in Durcupan Epoxy resin (Sigma-Aldrich; Merck KGaA), and ultrathin sections (70-90 nm) were mounted on copper grids and stained with uranyl acetate. Stained samples on grids were visualized using a Zeiss EM 109T TEM (Carl Zeiss AG), and digital micrographs were captured with a Gatan ES1000W digital camera (Gatan, Inc.) in the LANAIS-MIE facility (Buenos Aires, Argentina).
Cell viability assay
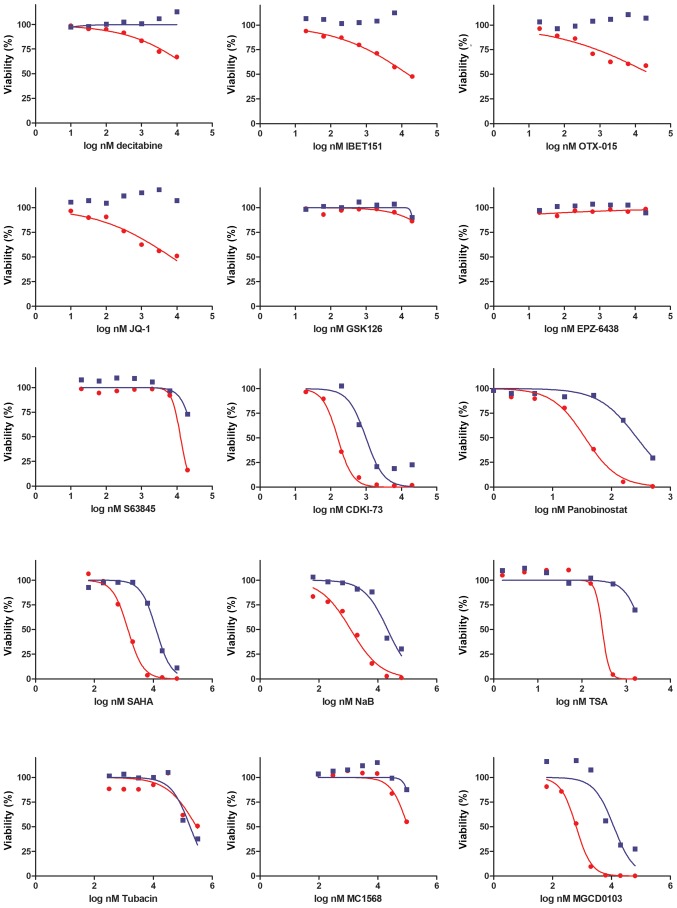
Cells were seeded at a density of 3,000 cells/well into 96-well plates in complete medium. After 24 h, different inhibitors were added, and cells were maintained at 37°C in a 5% CO2 humidified atmosphere. For each inhibitor, 7 different concentrations were assayed, starting from 10 µM (decitabine, JQ-1), 20 µM (IBET-151, OTX-015, S63845, CDKI-73, EPZ-6438 and GSK126), 500 nM (pano-binostat), 1.581 µM (TSA), 63.245 µM (SAHA, MGCD0103, sodium butyrate), 94.868 µM (MC1568) and 316.22 µM (tubacin), with subsequent 3.16-fold serial dilutions. After 72 h, cell viability was assessed using CellTiter-Glo® reagent (Promega Corporation). Luminescence was recorded with a POLARstar Omega microplate reader (BMG Labtech). Data generated using this assay are presented in Fig. 2.
Figure 2.
Sensitivity of MEL-XY3 and MEL-XY3 SUR cells to epigenetic inhibitors. Viability was determined with a CellTiter-Glo assay after 72 h of drug treatment. DNA methyltransferase (decitabine), bromodomain and extra-terminal motif inhibitors (IBET151, OTX-015, JQ-1), enhancer of zeste homolog 2 (GSK126, EPZ-6438), myeloid chronic leukaemia-1 (S63845), cyclin-dependent kinase 9 (CDKI-73) and histone deacetylase inhibitors (panobinostat, SAHA, NaB, TSA, tubacin, MGCD0103, MC1568) were assayed. Representative inhibition curves of 2 independent experiments are shown. Lines represent the nonlinear regression curve fit. Red, parental cells; blue, SUR cells. NaB, sodium butyrate; SUR, surviving cells following long-term PLX4032 treatment; TSA, trichostatin A.
Alternatively, after 72 h, cells were stained with 2 µg/ml Hoechst for 15 min at room temperature to visualize the nuclei and plates were scanned (six randomly selected fields/well) using an automated In Cell Analyzer 2200 microscope (GE Healthcare Life Sciences). Quantitative analysis of the acquired images was performed using In Cell Analyzer 1000 Workstation 3.7.3 (GE Healthcare Life Sciences). Data generated using this assay are presented in Fig. 4. Non-linear regression analysis and IC50 determination were performed using GraphPad Prism 5 software.
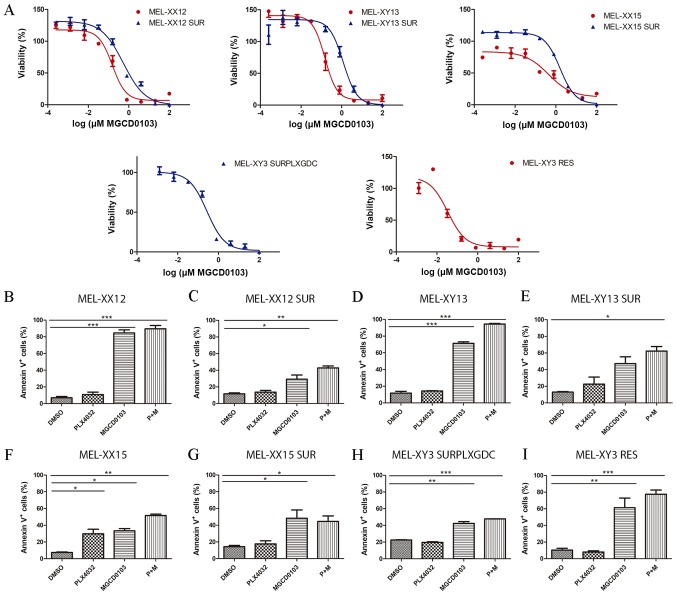
Figure 4.
Sensitivity of SUR melanoma cell lines to MGCD0103. (A) Sensitivity to MGCD0103 of parental (red) and SUR (blue) cells obtained from BRAF-mutated melanoma cell lines. Viability was determined after 72 h of drug treatment. Lines represent the nonlinear regression curve fit. Percentage of Annexin V+ apoptotic cells after 72 h treatment with 10 µM PLX4032, 2 µM MGCD0103 or combined treatment is shown for (B) MEL-XX12, (C) MEL-XX12 SUR, (D) MEL-XY13, (E) MEL-XY13 SUR, (F) MEL-XX15, (G) MEL-XX15 SUR, (H) MEL-XY3 SURPLXGDC and (I) MEL-XY3 RES cells. *P<0.05, **P<0.01, ***P<0.001. RES, resistant to PLX4032; SUR, surviving cells following long-term PLX4032 treatment; SURPLXGDC, cells that survive long-term PLX4032 and GDC-0973 treatment.
In order to study the effect of inhibitors for longer periods of time, 50,000 cells were seeded in MW24 plates in complete medium. After 24 h, DMSO, 10 µM PLX4032, 2 µM MGCD0103 or 10 µM PLX4032 + 2 µM MGCD0103 were added, and cells were maintained at 37°C for 10 days, changing media every 3 days. At day 10, cells were fixed with 4% paraformaldehyde at room temperature for 15 min and stained with 0.05% cristal violet solution at room temperature for 20 min.
Analysis of apoptosis and cell cycle
Cell cycle and apoptosis were analysed using BD FACS Canto II or BD FACScalibur flow cytometers (both BD Biosciences). Cells were seeded in a MW12 plate (50,000 cells/well) and 24 h later, the tested drugs were added to the media. Apoptotic cells were quantified using Annexin V/propidium iodide (PI) staining according to the manufacturer's protocol (BD Biosciences) incubating cells for 15 min at room temperature. For cell cycle analysis, cells were stained with PI staining solution (50 µg/ml PI, RNAse 100 µg/ml, 0.1% Triton-X, 1 mM Na3Citrate) for 30 min on ice. The cell cycle was analysed using PI-stained cells, and the cell cycle was fitted to viable cells using FlowJo v10.2 software (FlowJo LLC) to exclude dead cells from analysis. Experiments were performed in triplicate and 20,000 events were acquired for each point.
NRAS status of MEL-XY3 BRAF inhibitor resistant (RES) cells
MEL-XY3 RES cells were obtained from resistant colonies generated in one experiment when MEL-XY3 were treated with PLX4032 for several weeks to obtain SUR cells. The NRAS status of MEL-XY3 RES cells was determined after DNA extraction with DNAzol™ (Invitrogen; Thermo Fisher Scientific, Inc.), PCR amplification and Sanger sequencing (performed by Macrogen, Inc.). For PCR amplification, GoTaq polymerase (Promega coproration) was used and the thermocycling conditions used were: Initial denaturalization for 2 min at 95°C, then 35 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec, with a final extension at 72°C for 5 min. The primers used were: Forward, 5′-CCC CTT ACC CTC CAC ACC-3′ and reverse, 5′-CAC AAA GAT CAT CCT TTC AGA GAA-3′.
Three-dimensional (3D) spheroid cultures
A total of 3,000 cells were seeded in 96-well ultra-low attachment plates to induce anchorage-independent spheroid formation. After 72 h, multiple spheroids were harvested and embedded in 0.5 ml of collagen solution (Cultrex bovine collagen 1; Trevigen, Inc.) in a 24-well plate, and left to solidify at 37°C. After 15 min, 1 ml of media was added on top of the collagen layer in order to obtain a final concentration of 10 µM PLX4032, 2 µM SAHA, 2 µM MGCD0103 and 2 µM CDKI-73. Spheroids were main-tained at 37°C in a 5% CO2 humidified atmosphere. Following 5 days of treatment, spheroids were photographed using a Nikon Ti inverted microscope (magnification, 40×) and Image J software v3.5.0 (National Institutes of Health) was used to establish the spheroid diameter.
To assess the viability of cells grown in a 3D culture, spheroids were prepared. After 72 h, spheroids were grown for an additional 72 h in the presence of drugs without collagen. For each condition, 24 spheroids were disaggregated with trypsin-EDTA, stained with Annexin V/PI and analysed by flow cytometry as aforementioned. Three independent experiments were performed. Percentage of viability was determined normalizing the percentage of viable cells (Annexin V−/PI−) to DMSO treated cells.
Statistical analysis
Experiments were performed with at least two independent repeats; representative experiments were selected for inclusion in figures. GraphPad Prism version 5.0 software was used to graph data and perform statistical analysis. Data are presented as the mean ± SD. The significance of differences between experimental data was determined via ANOVA with Tukey's multiple comparison tests. The significance level was set as P<0.05.
Results
Molecular characterization of SUR cells
Preliminary characterization of SUR cells obtained after 5 weeks of PLX4032 treatment has been previously described (20). In order to obtain a more complete picture of the changes that accompany the transition from the actively dividing MEL-XY3 tumour cells to the resting SUR cells, TEM was performed to characterize SUR cell morphology and intracellular structure. Parental MEL-XY3 cells presented round nuclei and smooth plasma membranes (Fig. S1A). In contrast, SUR cells presented irregular nuclei, a large degree of membrane blebbing and an increased number of probably fragmented mitochondria (Fig. S1B). After drug removal, SUR cells started to resemble parental cells, with less distorted nuclei and a smoothed plasma membrane, even though a large number of mitochondria were still observed (Fig. S1C and D).
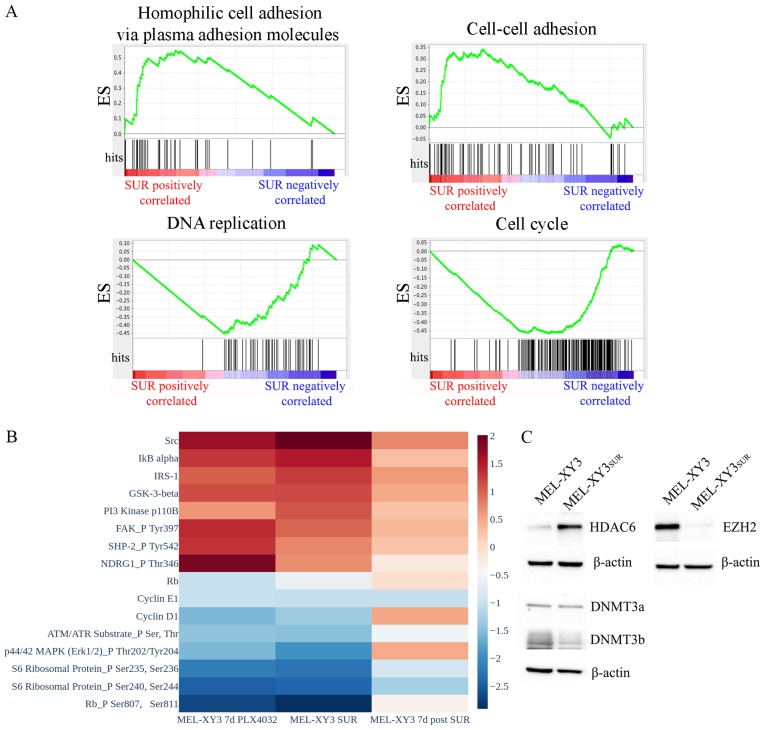
Next, SUR cells were subjected to RNA-seq analysis and their expression profiles were compared to the parental population (Fig. 1A). Considerable differences in gene expression were observed as 1,415 genes had significantly modified expression levels; 784 genes were downregulated and 631 genes were upregulated in SUR cells (P<0.05). GSEA revealed that SUR cells exhibited significantly enriched GO molecular signatures associated with cell adhesion (cell-cell adhesion, homophilic cell adhesion via plasma membrane adhesion molecules, cell-cell adhesion via plasma membrane adhesion molecules and calcium-dependent cell-cell adhesion via plasma membrane cell-adhesion molecules). In contrast, gene sets associated with proliferation and the cell cycle were downregulated (cell cycle, cell division and DNA replication; FDR <0.25, P<0.01), as predicted based on their phenotype.
Figure 1.
Characterization of SUR cells. (A) Gene set enrichment analysis of RNA sequencing data. The Gene Ontology molecular signatures 'Homophilic cell adhesion via plasma membrane adhesion molecules' and 'Cell-cell adhesion' were positively associated in SUR cells, while 'DNA replication' and 'Cell cycle' were negatively associated (false discovery rate <0.25; P<0.01). Red indicates positive association with SUR cells, whereas blue indicates negative association with SUR cells. (B) Reverse phase protein array heatmap representing the levels of proteins in MEL-XY3 cells treated for 7 days with PLX4032, SUR cells and SUR cells at 7 days after drug removal, normalized to MEL-XY3 parental cell expression levels. Proteins with log2 fold change >1 or <1 were shown. (C) Western blot analysis of epigenetic modifiers. DNMT3, DNA methyltransferase; ES, enrichment score; EZH2, enhancer of zeste homolog 2; HDAC, histone deacetylase; SUR, surviving cells following long-term PLX4032 treatment.
RPPA experiments validated the downregulation of proteins related to cell cycle progression (cyclin D1, cyclin E1, retinoblastoma protein total and phosphorylation levels, S6 ribosomal protein phosphorylation) in SUR cells. At 7 days after drug removal, the levels of cell cycle proteins started to return towards those of the parental MEL-XY3 cells (Figs. 1B and S2), confirming the reversibility of the SUR phenotype. Phosphorylated ERK1 levels decreased in SUR cells and returned to higher levels once the BRAF inhibitor was removed from culture media, indicative of continued but reversible suppression of MAPK signalling in SUR cells. Other proteins were upregulated in SUR cells, including proteins associated with proliferation pathways and drug resistance (Fig. 1B). These included Src and phosphorylated NDRG1, which have been linked to resistance to MAPK inhibitors and chemotherapy, respectively (31,32). PI3K protein was increased in SUR cells, possibly indicating a survival mechanism compensating for the loss of MAPK signalling. Even though there was an increase in the expression levels of these proteins or phosphorylated proteins, SUR cells remained in a quiescent state, suggesting that these changes were insufficient to promote proliferation and complete drug resistance. Once the drug was removed from culture media, the expression levels of these upregulated proteins decreased. Expression levels were similar in cells maintained with PLX4032 for 7 days and SUR cells, but a more pronounced change was observed in SUR cells.
In order to look for epigenetic changes in SUR cells with respect to the parental line XY3, the expression of different epigenetic enzymes were evaluated via western blotting. SUR cells presented a strong decrease in the expression of EZH2 (Fig. 1C). DNMT3b expression was slightly decreased in SUR cells, whereas DNMT3a expression remained stable. In contrast, HDAC6 was markedly increased in SUR cells compared with parental cells. Consistently, RNA-seq data from GSE50509 from patients with metastatic melanoma resistant to BRAF inhibitors revealed significant upregulation of several HDACs after BRAF inhibitor treatment, including HDAC6 (P<0.05; Fig. S3). These results demonstrated that SUR cells presented altered expression of epigenetic modifiers and, together with the apparently reversible changes observed in gene and protein expression, these data suggested that the SUR phenotype emerging after MAPK inhibition could be associated with epigenetic regulation.
Sensitivity of SUR cells to epigenetic inhibitors
To investigate whether SUR cells could be eliminated, the sensitivity of MEL-XY3 parental and SUR cells to different epigenetic inhibitors were analysed in vitro. Both cell types shared similar sensitivity to the assayed drugs. SUR cells were resistant to EZH2 histone methylase inhibitors (EPZ-6438 and GSK126), BET inhibitors (IBET151, JQ-1 and OTX-015), DNMT inhibi-tors (decitabine), HDAC class II inhibitors (MC1568) and HDAC6 inhibitors (tubacin; Fig. 2). Moreover, SUR cells were resistant to inhibition of the antiapoptotic protein member of Bcl-2 family myeloid cell leukaemia-1 (S63845).
However, parental and SUR MEL-XY3 cells were sensitive to certain pan-HDAC inhibitors, including SAHA (IC50: MEL-XY3 1.3 µM, SUR 6.98 µM), panobinostat (IC50: MEL-XY3 35 nM, SUR 269 nM) and sodium butyrate (IC50: MEL-XY3 1.2 µM, SUR 21 µM), whilst not exhibiting sensitivity to others, such as TSA. In addition, they were sensitive to MGCD0103 (IC50: MEL-XY3 0.63 µM, SUR 1.20 µM), an HDAC inhibitor which preferentially inhibits class I enzymes, with the exception of HDAC8 (33). Further, both parental and SUR cells were sensitive to the CDK9 inhibitor CDKI-73 (IC50: MEL-XY3 0.15 µM, SUR 0.99 µM; Fig. 2).
Even though PLX4032 did not induce apoptosis in MEL-XY3 parental cells or SUR cells, treatment with HDAC inhibitors and CDK9 inhibitors induced apoptosis both in parental and (to a lesser degree) SUR cells (Figs. 3A and B, and S4A). No significant differences were observed in the proportion of necrotic (Annexin V+/PI+) and apoptotic (Annexin V+/PI-) cells between the different treatments (data not shown).
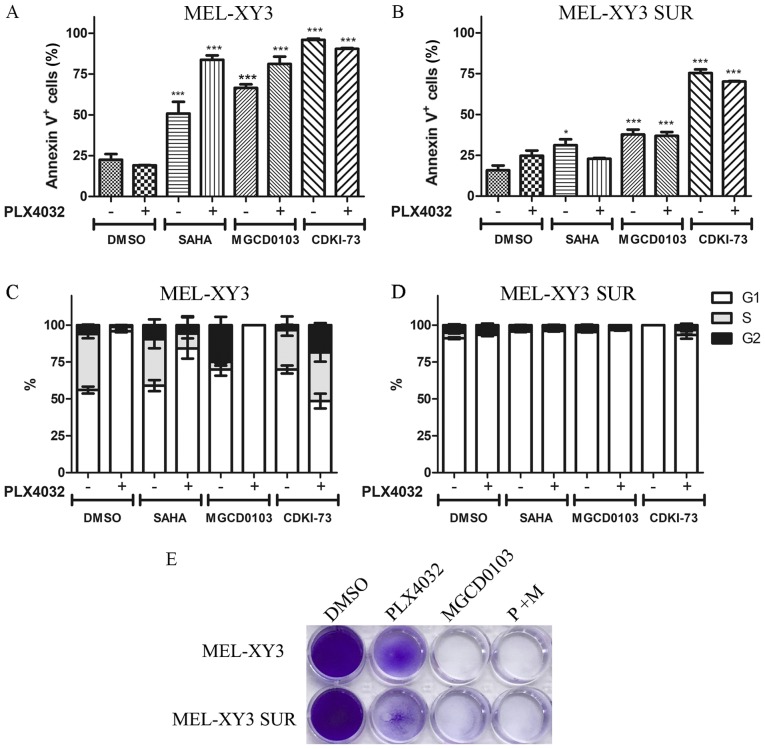
Figure 3.
Apoptosis and cell cycle distribution after treatment with histone deacetylase and cyclin-dependent kinase 9 inhibitors. Cells were treated for 72 h with 2 µM SAHA, 2 µM MGCD0103 or 2 µM CDKI-73, alone or in combination with PLX4032. Percentages of Annexin V+ apoptotic (A) MEL-XY3 cells and (B) MEL-XY3 SUR cells are shown. *P<0.05, ***P<0.001 vs. DMSO alone. Cell cycle distribution of (C) MEL-XY3 cells and (D) MEL-XY3 SUR cells. (E) MEL-XY3 parental and SUR cells treated with DMSO, PLX4720 (10 µM), MGCD0103 (2 µM) or the two inhibitors in combination for 10 days. After treatment, cells were fixed and stained with crystal violet. Photographs are representative of three independent experiments. SUR, surviving cells following long-term PLX4032 treatment.
PLX4032 induced G1 cell cycle arrest in MEL-XY3 and SUR cells maintained in the presence of the different inhibitors tested (Figs. 3D and S4B). MGCD0103 alone markedly increased the proportion of G2 cells, whereas SAHA and CDKI-73 alone did not notably alter cell cycle distribution (Fig. 3C). Although some melanoma cells were still viable after 72 h of treatment, after 10 days of treatment with MGCD0103, both parental MEL-XY3 cells and SUR cells were completely eliminated (Fig. 3E). Additionally, the presence of a sub-G0/G1 population was observed in cell cycle analysis, indicating the presence of dead cells, consistent with Annexin V/PI staining and the results obtained after 10 days of treatment (data not shown).
To determine whether the findings from the MEL-XY3 and MEL-XY3 SUR cells could be extended to other V600BRAF-mutated cell lines, the effects of MGCD0103 against three other BRAFV600E melanoma cells, and their corresponding SUR populations obtained after treatment with PLX4032 for 5 weeks, were evaluated. SUR cells obtained from MEL-XX12, MEL-XY13 and MEL-XX15 cells were sensitive to MGCD0103 (IC50: MEL-XX12 SUR 0.25 mM, MEL-XY13 SUR 0.82 mM and Mel-XX15 SUR 1.04 mM; Fig. 4A) as well as their parental cells. Apoptotic cell death was induced both in parental and SUR cells obtained from the different cell lines (Fig. 4B-G) by MGCD0103, alone or in combination with PLX4032. Additionally, MEL-XY3 SUR cells obtained after combined BRAF and MEK inhibitor treatment were sensitive to MGCD0103 (IC50: 0.27 mM; Fig. 4A), with apoptosis induced (Fig. 4H).
Although MEL-XY3 cells predominantly turned into SUR cells following PLX4032 treatment, a permanent BRAF inhibitor-resistant MEL-XY3 RES cell population capable of proliferating in the presence of PLX4032 was obtained in one of the experiments, when MEL-XY3 cells were cultured for several weeks in the presence of PLX4032. These cells presented a Q61K mutation in NRAS, a mutation already reported to confer resistance to BRAF inhibitors (4). These cells underwent apoptosis when treated with MGCD0103 (Figs. 4A and I, and S4C).
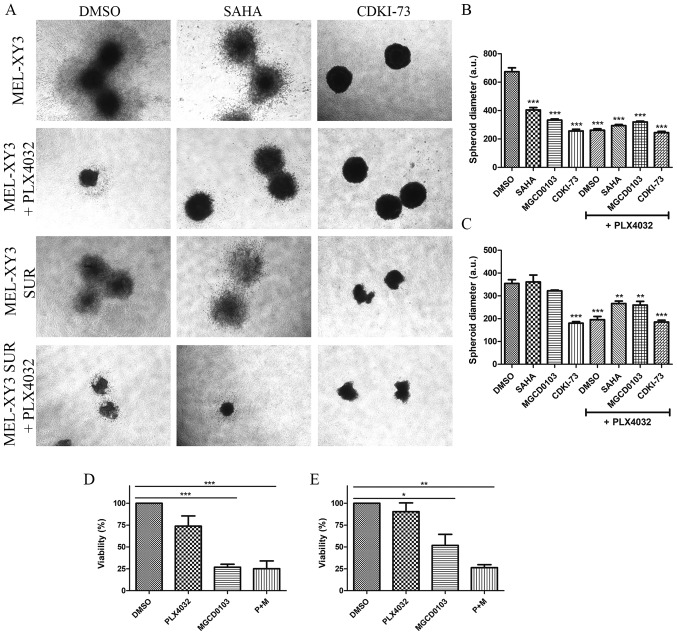
To determine the efficacy of the epigenetic inhibitors in a 3D growth model, the effects of HDAC and CDK9 inhibitors in spheroids that mimic tumour architecture were evaluated. Spheroids were incubated in a collagen matrix with the different drugs in order to analyse their impact on cell proliferation and migration. Treatment with PLX4032 significantly reduced spheroid diameter both in parental and SUR cells, consistent with cell cycle arrest observed after PLX4032 treatment (Fig. 5A-C). Moreover, the results suggested that BRAF inhibitor treatment also inhibited the migration of cells into the surrounding collagen matrix, as the spheroids appeared more compact. HDAC and CDK9 inhibitors both significantly reduced the diameter of MEL-XY3 spheroids (Fig. 5A and B); combined treatment with PLX4032 did not alter the effects observed with monotherapy. SUR cell spheroid diameter (Fig. 5A and C) was decreased by CDK9 inhibitor treatment; however, HDAC inhibitors did not significantly alter SUR cell spheroid diameter, except when they were combined with PLX4032. SUR cell spheroids treated with HDAC inhibitors may exhibit invasive behaviour, with morphology suggestive of an invasive phenotype. It has been previously demonstrated that several HDAC inhibitors, such as SAHA, TSA, valproic acid and sodium butyrate, promote melanoma cell invasion in vitro via the upregulation of N-cadherin and inhibition of Ras homolog A activity (34). Using flow cytometry, it was found that parental and SUR cell spheroids treated with MGCD0103 alone or in combination with PLX4032 for 72 h had significantly reduced viability, whereas PLX4032 treatment alone did not lead to significant loss of viability (Fig S4D; Fig. 5D and E;).
Figure 5.
Effect of treatment with epigenetic inhibitors on melanoma cell spheroids. Collagen-embedded spheroids obtained from MEL-XY3 and MEL-XY3 SUR cells were treated for 5 days with DMSO, 2 µM SAHA, 2 µM MGCD0103 or 2 µM CDKI-73 in the presence or absence of 10 µM PLX4032. (A) Phase contrast images of collagen-embedded spheroids after 5 days of treatment. Magnification, ×40. Diameters of (B) MEL-XY3 and (C) MEL-XY3 SUR spheroids. Data are presented as the mean ± SD. **P<0.01, ***P<0.001 vs. DMSO. Percentage of viable cells obtained from (D) MEL-XY3 and (E) MEL-XY3 SUR spheroids after 72 h of treatment relative to DMSO-treated cells. Following treatment, spheroids within each treatment were pooled, and the viability of cells was determined by Annexin V/PI staining and flow cytometry. *P<0.05, **P<0.01, ***P<0.001. a.u., arbitrary units; SUR, surviving cells following long-term PLX4032 treatment.
Discussion
Our previous study described SUR cells, a population of BRAF-mutated melanoma cells that persists after MAPK inhibitor treatment, and which represent a reversible state characterized by changes in gene expression, cell cycle arrest and SA β-galactosidase activity (20). To expand upon this previous study, the present study involved a molecular characterization of SUR cells by RNA-seq and RPPA experiments. RNA-seq showed that SUR cells exhibited decreased expression of proliferation-associated genes and increased expression of cell adhesion molecules. This is consistent with their phenotype; SUR cells do not proliferate and are strongly attached to culture dishes. At 7 days after removal of MAPK inhibitor, SUR cells revert to a proliferative state (20). RPPA data showed that the expression levels of proteins that are up- or downregulated in SUR cells compared with parental cells return to their basal expression levels after drug removal, suggesting phenotypic plasticity (21,35). Previous studies showed that aberrant expression of epigenetic modifiers such as EZH2, SET domain bifurcated histone lysine methyltransferase 1, G9a histone methyltransferase and HDACs led to adaptive resistance in cancer cells, including melanoma (21,35,36). In the present study, SUR cells presented changes in the protein expression levels of different epigenetic enzymes, such as EZH2 and HDAC6. These findings suggested that the SUR phenotype could be determined by epigenetic changes. It has been suggested that changes in nuclear morphology and structure could cause global epigenetic changes (37). The alterations observed by TEM in the nuclear morphology of SUR cells may be consistent with this hypothesis.
Previously, our study showed that SUR cells present SA characteristics, such as increased SA β-galactosidase activity and changes in the expression levels of several proteins associated with senescence, such as p16ink4 and p21 (20). Induction of senescence by BRAF inhibition with PLX4032 has also been reported by other groups (38,39). The acquisition of a senescent phenotype could contribute to resistance to apoptosis; cells that are resistant to apoptosis often exhibit an increase in senescence features (40). The induction of a senescent state could explain the limited proapoptotic effect of BRAF inhibition, which was observed in the different SUR cell lines. Furthermore, it has been shown that melanoma cells treated with PLX4032 present an SA secretory phenotype, which leads to the secretion of factors that stimulate naïve melanoma cells and fibroblasts, thereby limiting the antitumorigenic effect of BRAF inhibition (41). On the other hand, epigenetic control is a key factor in the induction of senescence. Recently, it has been proposed that oncogene-induced senescent melanocytes can bypass the senescent state through the activation of epigenetic modifying agents (including histone demethylases such as lysine-specific histone demethylase 1 and Jumonji domain-containing protein 2C), re-entering into the cell cycle as malignant melanoma cells (42). Moreover, the inhibition of these enzymes in melanoma cells restored senescence.
In the present study, in vitro SUR cell sensitivity to different epigenetic inhibitors was analysed, including DNMT, BET, HDAC, EZH2 and CDK9 inhibitors. It was found that both HDAC inhibitors (SAHA and MGCD0103) and a CDK9 inhibitor (CDKI-73) were able to induce apoptosis in SUR cells. Although parental cells were more sensitive than SUR cells to HDAC and CDK9 inhibitors, following 10 days of treatment with MGCD0103, complete elimination of SUR cells was observed in vitro. It should be stressed that parental non-treated cells are actively proliferating, whereas SUR cells are in a resting state; therefore the observed differences in the sensitivity to the assayed drugs may be based on these conditions. The main aim was to find drugs or combinations that resulted in complete elimination of SUR cells. The fact that the same inhibitors also effectively eliminated the parental cells is a therapeutic advantage that may increase the efficacy of first-line treatment. Regarding the spheroid growth assay results, PLX4032 efficiently reduced spheroid diameter in parental and SUR cells, and this effect was not reverted by the presence of the other inhibitors.
Different types of epigenetic inhibitors are currently being applied for cancer treatment. Decitabine inhibits DNMTs, producing DNA hypomethylation (43). It is approved for treatment of haematological malignancies, such as myelo-dysplastic syndrome, chronic myelomonocitic leukaemia and acute myeloid leukaemia. An EZH2 inhibitor, tazemetostat, has exhibited promising results in a phase I clinical trial, both in lymphoma and solid tumours such as sarcoma (44). Different BET inhibitors have been tested for the treatment of haematological malignancies and solid tumours, presenting limited efficacy as single agents; however, recent studies have proposed combination treatments to obtain improved clinical results (45,46).
To date, monotherapy with HDAC inhibitors has had limited success in the treatment of solid tumours; however, combination therapy has shown more encouraging results (47). It has been previously shown in different tumour models that drug-tolerant persister cells can be eliminated by HDAC inhibitors such as TSA (36,48). SAHA, also known as vorinostat, is a pan-HDAC inhibitor. Even though treatment of patients with advanced melanoma with SAHA as a monotherapy did not produce encouraging results (49), combination or sequential treatment with BRAF inhibitors could be a promising alternative. Wang et al (16) demonstrated that BRAF inhibitor-resistant melanoma cells present increased reactive oxygen species (ROS) levels, and that this characteristic produces vulnerability to HDAC inhibitors, as HDAC inhibitors yield a further increase in ROS levels, thereby leading to DNA damage and apoptosis. They proposed a sequential treatment (BRAF + MEK inhibitors followed by SAHA) to reach lethal ROS levels, with the benefit of avoiding toxicity due to combined treatment. A phase I clinical trial analysing SAHA in resistant BRAF-mutated advanced melanoma is currently ongoing (clinical trial no. NCT02836548). In the present study, it was observed that SAHA alone induced greater apoptosis than the combination of SAHA + PLX4032 in SUR cells, consistent with Wang et al (16); however, this effect was not significant.
Epigenetic alterations have been described in the pathogenesis of autoimmunity (50). Evidence suggests that HDAC inhibitors, including SAHA, may also be applicable to the treatment of autoimmune diseases (51). HDAC inhibitors have been shown to inhibit inflammatory mediator production (including cytokines, chemokines and nitric oxide) and lead to M2 polarization of macrophages (52). Moreover, experiments conducted in murine models suggest that DNMT inhibitors could be deployed in the treatment of autoimmune diseases (53,54).
MGCD0103, also known as mocetinostat, is a class I and IV (isoforms 1, 2, 3 and 11) HDAC-selective inhibitor (33). Preclinical studies have shown potent antitumor activity of MGCD0103 against various human cancer cell lines and human tumour xenografts in mice (33). A clinical trial demonstrated MGCD0103 clinical activity and acceptable safety profiles in patients with haematological malignancies (55). MGCD0103 increases antigen presentation and decreases immune-suppressive cell types (56). Currently, mocetinostat is being tested in combination with durvalumab, an anti-programmed cell death (PD)-ligand 1 antibody, in patients with non-small cell lung cancer (clinical trial no. NCT02805660). Further, a phase 1b clinical study of MGCD0103 in combination with ipilimumab (anti-cytotoxic T-lymphocyte-associated protein 4) and nivolumab (anti-PD-1) in patients with stage IV unresectable melanoma is being conducted (cat. no. NCT03565406). The present results showed that SUR cells present a stronger response to MGCD0103 than SAHA, with the advantage that MGCD0103 is a more selective inhibitor. MGCD0103 induced cytotoxicity in populations of SUR cells obtained from different melanoma cell lines, as well as in a PLX4032-resistant cell line harbouring an acquired NRAS mutation.
Although it was observed that HDAC6 was upregulated in SUR cells, these cells were not highly sensitive to HDAC6 inhibition. HDAC6 may not be the primary driver of cell survival in the SUR cells. Other alterations were observed, including EZH2 downregulation. SAHA and MGCD0103 are pan and class I/IV HDAC inhibitors, respectively, and they were effective in the elimination of SUR cells. These drugs can not only directly inhibit HDAC, but could have additional effects on HDAC-associated molecules.
CDK9, the catalytic subunit of positive transcription elongation factor b, is a key regulatory kinase of RNA polymerase II (57). Recently, it was reported that CDK9 is essential for maintaining gene silencing at heterochromatic loci, validating its role as an epigenetic target. CDK9 targeting reactivates tumour-suppressor genes and induces a cellular immune response that may sensitize to checkpoint inhibitors (57). CDKI-73 is a very potent CDK9 inhibitor that showed high cytotoxicity against different human cancer cell lines (58), and the present findings indicated that it effectively eliminates SUR cells.
The main limitation of the present study is the lack of in vivo experiments. It would be of major interest to perform in vivo experiments to validate the observed in vitro results. One potential experiment would be to grow xenograft tumours derived from human melanoma cells in the skin of immuno-compromised NSG mice. After the tumours have established, the mice should be treated with the BRAF inhibitor PLX4032 to reduce the size of the tumours and allow generation of SUR cells in vivo. Afterwards, mice should be treated with DMSO, PLX4032, MGCD0103 or PLX4032 + MGCD0103. It is predicted that tumours from mice treated with either MGCD0103 alone or in combination with BRAF inhibitor will not regrow, as these treatments should effectively kill the SUR cell population based on the present in vitro results.
In conclusion, in the present study a molecular characterization of SUR cells was performed, and evidence indicating that the SUR phenotype may be determined by epigenetic changes was found. Subsequently, it was investigated as to whether SUR cells were sensitive to epigenetic inhibitors, with the results suggesting that co-targeting of BRAF and HDAC/CDK9 can efficiently eliminate BRAF-mutated melanoma cells, whilst also eliminating the SUR population that remains viable after prolonged MAPK inhibitor treatment. The present study is limited to in vitro and 3D spheroid models; future preclinical studies will aid in designing therapeutic modalities that may provide a solid base for clinical trials aiming to overcome the persistence of melanoma cells after MAPK inhibition.
Supplementary Data
Acknowledgments
CDKI-73 was kindly provided by Dr Shudong Wang (University of South Australia). We acknowledge Dr Patricio Yankilevich (IBioBA-CONICET) for RNA-seq data analysis, and Professor Kaylene J. Simpson, Ms. Arthi Macpherson and Ms. Lucy Liu (Victorian Centre for Functional Genomics RPPA platform) for assistance with the RPPA experiments in this study.
Funding
This work was supported by grants from CONICET, Agencia Nacional de Promoción Científica y Tecnológica (grant nos. PICT 2014-1932 and PICT 2017-1141) and Fundación Sales, as well as by an Australian National Health and Medical Research Council program (grant no. 633004). The Victorian Centre for Functional Genomics RPPA platform is funded by the Australian Cancer Research Foundation, the Australian Phenomics Network through funding from the Australian Government's National Collaborative Research Infrastructure Strategy program, the Peter MacCallum Cancer Foundation and the University of Melbourne Collaborative Research Infrastructure Program.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126960).
Author contributions
FPMR participated in the conception and design of the study, collection and assembly of data, data analysis and interpre-tation, and manuscript writing. AB and EMVE participated in sample collection, and the assembly, analysis and interpretation of data. SJG, AAE and PH provided reagents, and contributed to the analysis and interpretation of data. MMB and JM conceived and designed the study, and contributed to data analysis and interpretation, and manuscript writing. All authors contributed to manuscript revision, read and approved the submitted version.
Ethics approval and consent to participate
Ethical approval was granted prior to the establishment of the MEL-XX12 and MEL-XX15 cell lines by the Comite de Etica en Investigacion of the Instituto Alexander Fleming, and informed consent was obtained from patients prior to the collection of samples.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Villanueva J, Vultur A, Herlyn M. Resistance to BRAF inhibitors: Unraveling mechanisms and future treatment options. Cancer Res. 2011;71:7137–7140. doi: 10.1158/0008-5472.CAN-11-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, Ng C, Chodon T, Scolyer RA, Dahlman KB, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arozarena I, Wellbrock C. Overcoming resistance to BRAF inhibitors. Ann Transl Med. 2017;5:387. doi: 10.21037/atm.2017.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levesque MP, Cheng PF, Raaijmakers MI, Saltari A, Dummer R. Metastatic melanoma moves on: Translational science in the era of personalized medicine. Cancer Metastasis Rev. 2017;36:7–21. doi: 10.1007/s10555-017-9658-0. [DOI] [PubMed] [Google Scholar]
- 9.Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, Karras P, Brown D, Chang YH, Debiec-Rychter M, et al. Toward minimal residual disease-directed therapy in melanoma. Cell. 2018;174:843–855 e819. doi: 10.1016/j.cell.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Strauss J, Figg WD. Epigenetic approaches to overcoming chemotherapy resistance. Lancet Oncol. 2015;16:1013–1015. doi: 10.1016/S1470-2045(15)00231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeller C, Dai W, Steele NL, Siddiq A, Walley AJ, Wilhelm-Benartzi CS, Rizzo S, van der Zee A, Plumb JA, Brown R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene. 2012;31:4567–4576. doi: 10.1038/onc.2011.611. [DOI] [PubMed] [Google Scholar]
- 12.Giessrigl B, Schmidt WM, Kalipciyan M, Jeitler M, Bilban M, Gollinger M, Krieger S, Jäger W, Mader RM, Krupitza G. Fulvestrant induces resistance by modulating GPER and CDK6 expression: Implication of methyltransferases, deacetylases and the hSWI/SNF chromatin remodelling complex. Br J Cancer. 2013;109:2751–2762. doi: 10.1038/bjc.2013.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Y, Tong M, Liang Q, Guo Y, Sun HQ, Zheng W, Ao L, Guo Z, She F. Epigenomics alternations and dynamic transcriptional changes in responses to 5-fluorouracil stimulation reveal mechanisms of acquired drug resistance of colorectal cancer cells. Pharmacogenomics J. 2018;18:23–28. doi: 10.1038/tpj.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song C, Piva M, Sun L, Hong A, Moriceau G, Kong X, Zhang H, Lomeli S, Qian J, Yu CC, et al. Recurrent tumor Cell-intrinsic and -extrinsic alterations during MAPKi-induced melanoma regression and early adaptation. Cancer Discov. 2017;7:1248–1265. doi: 10.1158/2159-8290.CD-17-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strub T, Ghiraldini FG, Carcamo S, Li M, Wroblewska A, Singh R, Goldberg MS, Hasson D, Wang Z, Gallagher SJ, et al. SIRT6 haploinsufficiency induces BRAFV600E melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat Commun. 2018;9:3440. doi: 10.1038/s41467-018-05966-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Leite de Oliveira R, Huijberts S, Bosdriesz E, Pencheva N, Brunen D, Bosma A, Song JY, Zevenhoven J, et al. An acquired vulnerability of Drug-resistant melanoma with therapeutic potential. Cell. 2018;173:1413–1425.e14. doi: 10.1016/j.cell.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Gallagher SJ, Gunatilake D, Beaumont KA, Sharp DM, Tiffen JC, Heinemann A, Weninger W, Haass NK, Wilmott JS, Madore J, et al. HDAC inhibitors restore BRAF-inhibitor sensitivity by altering PI3K and survival signalling in a subset of melanoma. Int J Cancer. 2018;142:1926–1937. doi: 10.1002/ijc.31199. [DOI] [PubMed] [Google Scholar]
- 18.Zhao B, Cheng X, Zhou X. The BET-bromodomain inhibitor JQ1 mitigates vemurafenib drug resistance in melanoma. Melanoma Res. 2018;28:521–526. doi: 10.1097/CMR.0000000000000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakharia Y, Monga V, Swami U, Bossler AD, Freesmeier M, Frees M, Khan M, Frydenlund N, Srikantha R, Vanneste M, et al. Targeting epigenetics for treatment of BRAF mutated metastatic melanoma with decitabine in combination with vemurafenib: A phase lb study. Oncotarget. 2017;8:89182–89193. doi: 10.18632/oncotarget.21269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madorsky Rowdo FP, Barón A, von Euw EM, Mordoh J. In vitro long-term treatment with MAPK inhibitors induces melanoma cells with resistance plasticity to inhibitors while retaining sensitivity to CD8 T cells. Oncol Rep. 2017;37:1367–1378. doi: 10.3892/or.2017.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravindran Menon D, Das S, Krepler C, Vultur A, Rinner B, Schauer S, Kashofer K, Wagner K, Zhang G, Bonyadi Rad E, et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene. 2015;34:4448–4459. doi: 10.1038/onc.2014.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher SJ, Tiffen JC, Hersey P. Histone modifications, modifiers and readers in melanoma resistance to targeted and immune therapy. Cancers (Basel) 2015;7:1959–1982. doi: 10.3390/cancers7040870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long GV, Hauschild A, Santinami M, Atkinson V, Mandalà M, Chiarion-Sileni V, Larkin J, Nyakas M, Dutriaux C, Haydon A, et al. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N Engl J Med. 2017;377:1813–1823. doi: 10.1056/NEJMoa1708539. [DOI] [PubMed] [Google Scholar]
- 24.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2015. https://www.R-project.org/ [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar R, Domrachev M, Lash EM. Gene expression omnibus: NCBI Gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, Haydu LE, Mijatov B, Becker TM, Boyd SC, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin Cancer Res. 2014;20:1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 31.Girotti MR, Lopes F, Preece N, Niculescu-Duvaz D, Zambon A, Davies L, Whittaker S, Saturno G, Viros A, Pedersen M, et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell. 2015;27:85–96. doi: 10.1016/j.ccell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiler M, Blaes J, Pusch S, Sahm F, Czabanka M, Luger S, Bunse L, Solecki G, Eichwald V, Jugold M, et al. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc Natl Acad Sci USA. 2014;111:409–414. doi: 10.1073/pnas.1314469111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fournel M, Bonfils C, Hou Y, Yan PT, Trachy-Bourget MC, Kalita A, Liu J, Lu AH, Zhou NZ, Robert MF, et al. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol Cancer Ther. 2008;7:759–768. doi: 10.1158/1535-7163.MCT-07-2026. [DOI] [PubMed] [Google Scholar]
- 34.Diaz-Nunez M, Diez-Torre A, De Wever O, Andrade R, Arluzea J, Silió M, Aréchaga J. Histone deacetylase inhibitors induce invasion of human melanoma cells in vitro via differential regulation of N-cadherin expression and RhoA activity. BMC Cancer. 2016;16:667. doi: 10.1186/s12885-016-2693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emran Al A, Marzese DM, Menon DR, Stark MS, Torrano J, Hammerlindl H, Zhang G, Brafford P, Salomon MP, Nelson N, et al. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget. 2017;9:8206–8222. doi: 10.18632/oncotarget.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guler GD, Tindell CA, Pitti R, Wilson C, Nichols K, KaiWai Cheung T, Kim HJ, Wongchenko M, Yan Y, Haley B, et al. Repression of Stress-induced LINE-1 expression protects cancer cell subpopulations from lethal drug exposure. Cancer Cell. 2017;32:221–237 e213. doi: 10.1016/j.ccell.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Easwaran HP, Baylin SB. Role of nuclear architecture in epigenetic alterations in cancer. Cold Spring Harb Symp Quant Biol. 2010;75:507–515. doi: 10.1101/sqb.2010.75.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haferkamp S, Borst A, Adam C, Becker TM, Motschenbacher S, Windhövel S, Hufnagel AL, Houben R, Meierjohann S. Vemurafenib induces senescence features in melanoma cells. J Invest Dermatol. 2013;133:1601–1609. doi: 10.1038/jid.2013.6. [DOI] [PubMed] [Google Scholar]
- 39.Krayem M, Najem A, Journe F, Morandini R, Sales F, Awada A, Ghanem GE. Acquired resistance to BRAFi reverses senescence-like phenotype in mutant BRAF melanoma. Oncotarget. 2018;9:31888–31903. doi: 10.18632/oncotarget.25879. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Childs BG, Baker DJ, Kirkland JL, Campisi J, van Deursen JM. Senescence and apoptosis: Dueling or complementary cell fates? EMBO Rep. 2014;15:1139–1153. doi: 10.15252/embr.201439245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grimm J, Hufnagel A, Wobser M, Borst A, Haferkamp S, Houben R, Meierjohann S. BRAF inhibition causes resilience of melanoma cell lines by inducing the secretion of FGF1. Oncogenesis. 2018;7:71. doi: 10.1038/s41389-018-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Y, Schleich K, Yue B, Ji S, Lohneis P, Kemper K, Silvis MR, Qutob N, van Rooijen E, Werner-Klein M, et al. Targeting the Senescence-overriding cooperative activity of structurally unrelated H3K9 demethylases in melanoma. Cancer Cell. 2018;33:322–336.e8. doi: 10.1016/j.ccell.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duchmann M, Itzykson R. Clinical update on hypomethylating agents. Int J Hematol. 2019;110:161–169. doi: 10.1007/s12185-019-02651-9. [DOI] [PubMed] [Google Scholar]
- 44.Italiano A, Soria JC, Toulmonde M, Michot JM, Lucchesi C, Varga A, Coindre JM, Blakemore SJ, Clawson A, Suttle B, et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: A first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649–659. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- 45.Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: A novel epigenetic approach. Ann Oncol. 2017;28:1776–1787. doi: 10.1093/annonc/mdx157. [DOI] [PubMed] [Google Scholar]
- 46.Pawar A, Gollavilli PN, Wang S, Asangani IA. Resistance to BET inhibitor leads to alternative therapeutic vulnerabilities in castration-resistant prostate cancer. Cell Rep. 2019;29:1397. doi: 10.1016/j.celrep.2019.10.037. [DOI] [PubMed] [Google Scholar]
- 47.Suraweera A, O'Byrne KJ, Richard DJ. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: Achieving the full therapeutic potential of HDACi. Front Oncol. 2018;8:92. doi: 10.3389/fonc.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas NB, Quirt I, Hotte S, McWhirter E, Polintan R, Litwin S, Adams PD, McBryan T, Wang L, Martin LP, et al. Phase II trial of vorinostat in advanced melanoma. Invest New Drugs. 2014;32:526–534. doi: 10.1007/s10637-014-0066-9. [DOI] [PubMed] [Google Scholar]
- 50.Jeffries MA, Sawalha AH. Autoimmune disease in the epigenetic era: How has epigenetics changed our understanding of disease and how can we expect the field to evolve? Expert Rev Clin Immunol. 2015;11:45–58. doi: 10.1586/1744666X.2015.994507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohammadi A, Sharifi A, Pourpaknia R, Mohammadian S, Sahebkar A. Manipulating macrophage polarization and function using classical HDAC inhibitors: Implications for autoimmunity and inflammation. Crit Rev Oncol Hematol. 2018;128:1–18. doi: 10.1016/j.critrevonc.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 52.Patel U, Rajasingh S, Samanta S, Cao T, Dawn B, Rajasingh J. Macrophage polarization in response to epigenetic modifiers during infection and inflammation. Drug Discov Today. 2017;22:186–193. doi: 10.1016/j.drudis.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mangano K, Fagone P, Bendtzen K, Meroni PL, Quattrocchi C, Mammana S, Di Rosa M, Malaguarnera L, Coco M, Magro G. Hypomethylating agent 5-aza-2′-deoxycytidine (DAC) ameliorates multiple sclerosis in mouse models. J Cell Physiol. 2014;229:1918–1925. doi: 10.1002/jcp.24641. [DOI] [PubMed] [Google Scholar]
- 54.Fagone P, Mazzon E, Chikovani T, Saraceno A, Mammana S, Colletti G, Mangano K, Bramanti P, Nicoletti F. Decitabine induces regulatory T cells, inhibits the production of IFN-gamma and IL-17 and exerts preventive and therapeutic efficacy in rodent experimental autoimmune neuritis. J Neuroimmunol. 2018;321:41–48. doi: 10.1016/j.jneuroim.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 55.Younes A, Oki Y, Bociek RG, Kuruvilla J, Fanale M, Neelapu S, Copeland A, Buglio D, Galal A, Besterman J, et al. Mocetinostat for relapsed classical Hodgkin's lymphoma: An open-label, single-arm, phase 2 trial. Lancet Oncol. 2011;12:1222–1228. doi: 10.1016/S1470-2045(11)70265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Briere D, Sudhakar N, Woods DM, Hallin J, Engstrom LD, Aranda R, Chiang H, Sodré AL, Olson P, Weber JS, Christensen JG. The class I/IV HDAC inhibitor mocetinostat increases tumor antigen presentation, decreases immune suppressive cell types and augments checkpoint inhibitor therapy. Cancer Immunol Immunother. 2018;67:381–392. doi: 10.1007/s00262-017-2091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Pandey S, Travers M, Sun H, Morton G, Madzo J, Chung W, Khowsathit J, Perez-Leal O, Barrero CA, et al. Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell. 2018;175:1244–1258.e1226. doi: 10.1016/j.cell.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lam F, Abbas AY, Shao H, Teo T, Adams J, Li P, Bradshaw TD, Fischer PM, Walsby E, Pepper C, et al. Targeting RNA transcription and translation in ovarian cancer cells with phar-macological inhibitor CDKI-73. Oncotarget. 2014;5:7691–7704. doi: 10.18632/oncotarget.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126960).