Abstract
The aim of this study was to determine the use of the lipid profile of patients with sarcoidosis and compare it with healthy subjects. We assume that lipid profile of serum in sarcoidosis differs from the lipid profile of control subjects. Serum was collected from 14 patients with II stage of sarcoidosis and 14 control subjects (healthy volunteers). Proton NMR spectroscopy combined with discriminant analyses, OPLS-DA (orthogonal partial least squares projections to latent structures discriminant analysis), was used. Thirty four NMR signals of lipid compounds were selected. OPLS-DA model consisted of three components and very good explain the data and also predict the data. Discriminant analysis correctly classified patients according to their groups for 92.9% of sarcoidose and 100% of control. From multivariate discriminant analysis we obtain a list of potentialbiomarkers which are statistically significant and which separate one class from another. These biomarkers are statistically significant, but not necessarily biochemically significant. They may have biochemical significance and they may be the biomarkers we are interested in, however, this must be established through extensive testing. Presented method allows distinguishing between healthy subject and sarcoidosis patients. (Sarcoidosis Vasc Diffuse Lung Dis 2018; 35: 150-153)
Keywords: lipidomics, sarcoidosis, diagnostics
Introduction
Sarcoidosis is a systemic granulomatous inflammatory disease that primarily affects the lungs and the lymphatic system (1). This disease is often accompanied by alterations in the lipoprotein profile (2). Decreased HDL-cholesterol level (HDL-c) without any significant changes in total cholesterol (TC), LDL-cholesterol (LDL-c) and triglycerides levels has been observed in untreated sarcoidosis (3). Lipidomics, an emerging tool for basic and clinical sciences, is based on high-content and usually high-throughput analysis of all lipids present in a biological system, collectively known as the lipidome. The cellular lipidome consists of subdivides into species subsets that help assess various cellular processes, including energy storage, membrane integrity and cell signaling processes involved in cell proliferation, metabolism and apoptosis induction.
Recently, in patients with chronic pulmonary diseases, Telenga et al. reported on lipid expression with liquid chromatography and high-resolution quadrupole time-of-flight mass spectrometry in induced sputum, in which over 1,500 lipid compounds were identified. In the context of tobacco smoking in COPD patients, the smokers’ sputum contained significantly higher levels of lipid species, both at the single-compound level with 168 sphingolipids, 36 phosphatidylethanolamine lipids, and 5 tobacco-related compounds, together with changes in the whole lipid-subsets levels, with the class of sphingolipids higher expressed in smokers than in non-smokers (4). These data demonstrate a high analytical potential of lipidomics in identifying lipid components involved in disorders of the human respiratory system.
So far little is known about the role and the impact of the lipidome and lipid signaling in sarcoidosis, therefore we applied the lipidomics approach to systemically define the inscrutable role of lipid species in this devastating disease.
Material and methods
Study subjects
All patients gave their informed consent following the guidelines of the 2008 revision of the Declaration of Helsinki and the local Ethics Committee of the Medical University od Silesia, Poland. An approval of the lipidomics analysis to be performed on clinical samples was obtained from the local Ethics Committee of the Medical University of Silesia, Poland (KNW/0022/KB1/123/15).
Fourteen non-smoking, newly diagnosed sarcoidosis patients, aged 34-63 (mean 46.0±9.6), referred to the Dpt. of Lung Diseases and Tuberculosis between January 2011 and December 2015, were included in this study. Patients had been diagnosed based on consistent clinical features, the bronchoalveolar lavage fluid analysis, and/or biopsy- proven noncaseating epithelioid cell granulomas, according to the WASOG guidelines (1). No one had received prednisone or any other immunosuppressive agent prior to the study. Selected patients were within normal body-mass index, BMI (18.5-24.9), without any serious co-morbidities, like metabolic or diabetic disorders.
A healthy control group (n=14) was recruited from hospital employees, matching in terms of age and gender. The data were used as a reference for the serum analysis.
Study design
To obtain the lipid profile and phospholipids concentrations, proton- and phosphorus NMR spectra were used.
The multivariate projection method for data exploration - partial least squares (PLS) (5-8) was used to interpret the systematic changes existing between multiple samples, characterized by the relative concentrations of many metabolites. The variable-importance in the projection (VIP) value of each variable for both models was calculated to indicate its contribution to the classification. We validated the model by applying the test based on CV-Anova (Analysis of Variance testing of Cross-Validated predictive residuals).
The multivariate analysis (OPLS-DA) was performed using the SIMCA-P software package (Version 12, Umetrics AB, Sweden) (9).
Methods
Sample preparation and spectra acquisition
All serum samples were kept frozen at -80°C until the NMR analysis was performed. For lipid extraction 500μl of serum was used. Lipids from serum samples were extracted using Blight and Dyer method (10), modified by the addition of HCl to the extraction mixture (0.05:1:2 HCl:methanol:chloroform).
All NMR spectra were acquired at 20°C using a Varian Inova 400 spectrometer (Varian Inc., USA). Proton NMR spectra were collected using standard one-pulse sequence (5s delay time, 90° pulse and 128 repetitions). Zero-filling to 32k data points, line broadening of 0.5, baseline and phase correction were applied to each spectrum using commercial software of the spectrometer.
Analysis
Data analysis
Quantities of metabolites were expressed in terms of relative intensity (based on the magnitude of spectral peaks, and relative to the rest signal of chloroform). The signal magnitudes measured corresponded to the compounds concentration.
Spectra were normalized to the rest signal of chloroform prior to statistical analysis using custom software. We selected 25 signals of the NMR spectrum for the statistical analysis of the lipid extracts samples. Signals assignments were made based upon our own database and literature data (11, 12).
In discriminant analysis (OPLS-DA), mean centering, log transformation and Pareto scaling were applied.
Results
The OPLS-DA analysis of the lipid profile of sarcoidosis patients and control samples indicated multiple significant differences. For the purpose of model, building 25 variables and 26 observations were used (two control samples were outliers). The built model consisted of one perpendicular and one orthogonal component and described good data fit and good data prediction (R2Xcum - 0.81, R2Ycum - 0.718, Q2cum - 0.469). The misclassification table indicated that upon data from lipid profile samples, 96.43% of subjects were classified correctly to their groups, with an accuracy of 100% for controls, and 92.86% for sarcoidosis patients (Fishers probability – 3.7e-7). The analysis indicated significant differences between the lipid profiles (Fig. 1). The validation test by CV-ANOVA calculated p value as less than 0.01.
Fig. 1.
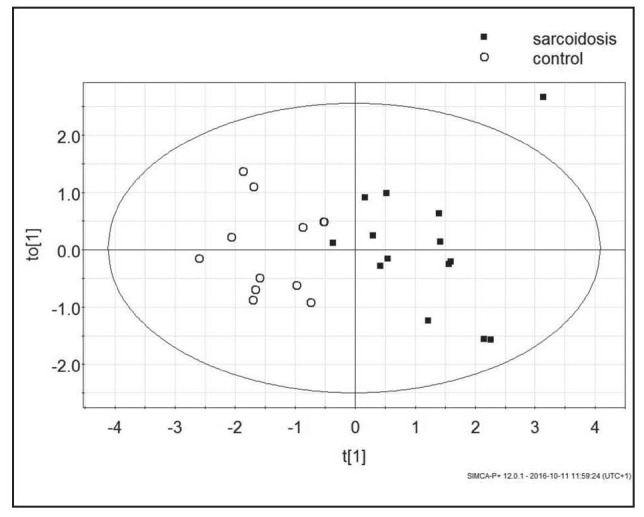
The scores plot of the two-component OPLS-DA model; to[1] represents within class variation in the first orthogonal component, whereas t[1] represents between class variation in the first predictive component. Ellipse represents Hotelling T2 with 95% confidence in score plots
The VIP analysis showed that in order to compare sarcoidosis to the control group, the elements that were most important for group differentiations were NMR signal magnitudes from Phosphatidylocholine/Sphigomyelin (PC/SM), Total Cholesterol, Triglycerides, Free Cholesterol (FC), Sphingomyelin, Phosphatidylcholine, Fatty Acids and an unassigned signal at 3.50 ppm. FC (0.86 ppm) and PC/SM (3.65 ppm) had high statistical reliability as discriminating variables (putative biomarkers).
Discussion
Our report is the first systematic and lipidome-wide systematic analysis on profiles of serum lipids that aimed to compare sarcoidosis patients with control subjects, in which we examined the serum-lipids profile based on the NMR spectroscopy.
Our studies provided a set of putative biomarkers that differed the most in sarcoidosis patients in comparison to control subjects. The most outstanding were: free cholesterol and, measured collectively, phosphatidylcholine and sphingomyeline. Phospholipids compounds bind cholesterol in LDL or HDL. Free cholesterol could be extracted from either the LDL core or outer layer (13, 14).
The so-called omics field is a term that was first used to define the studies of genomes (genomics) and gene expression (transcriptomics) of cells, tissues, organs and organisms and has subsequently been adopted for studies of proteins (proteomics), lipids (lipidomics) and metabolites (metabolomics) (15). Lipidomics is a large-scale study of acquisition of lipid compositions in biological systems where lipids are broadly defined as a hydrophobic or amphipathic small molecules that may originate entirely or partially from carbanion-based condensations of thioesters and/or from carbocation-based condensations of isoprene units (15). The relatively poor ability to identify and quantify individual lipid species remains the key obstacle in most lipidomics studies. However, it is expected, that in the future technical advances will significantly increase our ability to quantify such complex lipidomics profiles.
Conclusion
We observed significant differences in the serum lipids profile between control samples and sarcoidosis patients. Therefore the lipidome analysis may become an important research tool that can lead to a possible new diagnostic method.
Acknowledgments
This study was supported by grant KNW-1-093/K/6/0 from the Medical University of Silesia, Poland.
References
- 1.Hunninghake GW, Costabel U, Ando M, Baughman R, Cordier JF, du Bois R, Eklund A, Kitaichi M, Lynch J, Rizzato G, Rose C, Selroos O, Semenzato G, Sharma OP. ATS/ERS/WASOG statement on sarcoidosis. American Thoracic Society/European Respiratory Society/World Association of Sarcoidosis and other Granulomatous Disorders. Sarcoidosis, Vasculitis, and Diffuse Lung Diseases. 1999;16(2):149–173. [PubMed] [Google Scholar]
- 2.Salazar A, Mañá J, Pintó X, Argimón JM, Hurtado I, Pujol R. Corticosteroid therapy increases HDL-cholesterol concentrations in patients with active sarcoidosis and hypoalphalipoproteinemia. Clin Chim Acta. 2002;320(5):9–64. doi: 10.1016/s0009-8981(02)00046-3. [DOI] [PubMed] [Google Scholar]
- 3.Salazar A, Pintó X, Mañá J. Serum amyloid A and high-density lipoprotein cholesterol: serum markers of inflammation in sarcoidosis and other systemic disorders. Eur J Clin Invest. 2001;31:1070–7. doi: 10.1046/j.1365-2362.2001.00913.x. [DOI] [PubMed] [Google Scholar]
- 4.Telenga ED1, Hoffmann RF, Ruben t’Kindt, Hoonhorst SJ, Willemse BW, van Oosterhout AJ, Heijink IH, van den Berge M, Jorge L, Sandra P, Postma DS, Sandra K, ten Hacken NH. Untargeted lipidomic analysis in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;190(2):155–164. doi: 10.1164/rccm.201312-2210OC. [DOI] [PubMed] [Google Scholar]
- 5.Toczylowska B, Piotrowski M, Chalimoniuk M. P-31 High Resolution NMR Spectroscopy in Analysis of Phosphate-containing Compounds of Bile. Biocybernetics and Biomedical Engineering. 2011;31(1):63–71. [Google Scholar]
- 6.Motta A, Paris D, D’Amato M, Melck D, Calabrese C, Vitale C, Stanziola AA, Corso G, Sofia M, Maniscalco M. NMR metabolomic analysis of exhaled breath condensate of asthmatic patients at two different temperatures. J Proteome Res. 2014;13(12):6107–6120. doi: 10.1021/pr5010407. [DOI] [PubMed] [Google Scholar]
- 7.Weckwerth W, Morgenthal K. Metabolomics: from pattern recognition to biological interpretation. Drug Discov Today. 2005;10(22):1551–1558. doi: 10.1016/S1359-6446(05)03609-3. [DOI] [PubMed] [Google Scholar]
- 8.Niu QY, Li ZY, Du GH, Qin XM. (1)H NMR based metabolomic profiling revealed doxorubicin-induced systematic alterations in a rat model. J Pharm Biomed Anal. 2016;118:338–348. doi: 10.1016/j.jpba.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Ellis DI, Dunn WB, Griffin JL, Allwood JW, Goodacre R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics. 2007;8(9):1243–1266. doi: 10.2217/14622416.8.9.1243. [DOI] [PubMed] [Google Scholar]
- 10.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 11.Naughton DP, Haywood R, Blake DR, Edmonds S, Hawkes GE, Grootveld M. A comparative evaluation of the metabolic profiles of normal and inflammatory knee-joint synovial fluids by high resolution proton NMR spectroscopy. FEBS Lett. 1993;332(3):221–225. doi: 10.1016/0014-5793(93)80636-9. [DOI] [PubMed] [Google Scholar]
- 12.Blundell CD, Reed MAC, Almond A. Complete assignment of hyaluronan oligosaccharides up to hexasaccharides. Carbohydrate Research. 2006;341(17):2803–2815. doi: 10.1016/j.carres.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Sommer A, Prenner E, Gorges R, Stutz H, Grillhofer H, Kostner GM, et al. Organization of phosphatidylcholine and sphingomyelin in the surface monolayer of low density lipoprotein and lipoprotein(a) as determined by time-resolved fluorometry. J Biol Chem. 1992;267(34):24217–24222. [PubMed] [Google Scholar]
- 14.Hevonoja T, Pentikainen MO, Hyvonen MT, Kovanen PT, la-Korpela M. Structure of low density lipoprotein (LDL) particles: basis for understanding molecular changes in modified LDL. Biochim Biophys Acta. 2000;1488(3):189–210. doi: 10.1016/s1388-1981(00)00123-2. [DOI] [PubMed] [Google Scholar]
- 15.Wheelock CE1, Goss VM, Balgoma D, Nicholas B, Brandsma J, Skipp PJ, Snowden S, Burg D, D‘Amico A, Horvath I, Chaiboonchoe A, Ahmed H, Ballereau S, Rossios C, Chung KF, Montuschi P, Fowler SJ, Adcock IM, Postle AD, Dahlén SE, Rowe A, Sterk PJ, Auffray C, Djukanovic R. U-BIOPRED Study Group: Application of ’omics technologies to biomarker discovery in inflammatory lung diseases. Eur Respir J. 2013;42(3):802–25. doi: 10.1183/09031936.00078812. [DOI] [PubMed] [Google Scholar]


